Abstract
Apicomplexan parasites include the causative agents of malaria and toxoplasmosis. Cell-based screens in Toxoplasma previously identified a chemical modulator of calcium signaling (ENH1) that blocked parasite egress from host cells and exhibited potent antiparasitic activity. To identify the targets of ENH1, we adapted thermal proteome profiling to Toxoplasma, which revealed calcium-dependent protein kinase 1 (CDPK1) as a target. We confirmed the inhibition of CDPK1 by ENH1 in vitro and in parasites by comparing alleles sensitive or resistant to ENH1. CDPK1 inhibition explained the block in egress; however, the effects of ENH1 on calcium homeostasis and parasite viability were CDPK1-independent, implicating additional targets. Thermal proteome profiling of lysates from parasites expressing the resistant allele of CDPK1 identified additional candidates associated with the mitochondrion and the parasite pellicle—compartments that potentially function in calcium release and homeostasis. Our findings illustrate the promise of thermal profiling to identify druggable targets that modulate calcium signaling in apicomplexan parasites.
INTRODUCTION
Apicomplexans are early-branching eukaryotic intracellular parasites that cause important human diseases including malaria (Plasmodium spp.), cryptosporidiosis (Cryptosporidium spp.), and toxoplasmosis (Toxoplasma gondii). Calcium regulates key stages of the parasite infection cycle.1 Parasites move through tissues and invade host cells with a special form of movement called gliding motility, which depends on calcium-induced release of adhesins followed by their rear movement by actomyosin motors.2 Following several rounds of intracellular replication within the host cell, parasite movement may be triggered again to effectuate egress and lysis of the host. Parasite intracellular calcium levels rise prior to invasion and egress, and chemicals that block parasite calcium signaling, through chelation or inhibition of store release3,4, disrupt the parasite lytic cycle.
Calcium signaling and homeostasis in apicomplexan parasites diverge from mammalian systems, presenting both opportunities and challenges for therapeutics. For example, apicomplexans encode an expanded repertoire of calcium-mobilized proteins, including calcium-dependent protein kinases (CDPKs)5, which are absent from animals and consequently make appealing targets for pharmacological inhibition.6,7 On the other hand, due to the evolutionary distance separating apicomplexans and better studied organisms, the mechanisms regulating calcium signaling and homeostasis remain poorly characterized1, precluding design of inhibitors against specific proteins.
To address this gap, we previously used parasites expressing genetically encoded calcium indicators as a platform for phenotypic screens of chemical libraries. We identified several small molecules that perturb calcium homeostasis in the model apicomplexan, T. gondii.8 Phenotypic screens have the advantage of identifying promising compounds even when the complexity of pathogenesis is incompletely understood, but suffer from the need for eventual target deconvolution to efficiently and rationally optimize a chemical scaffold and elucidate the mechanism of action.9
In parasites, target deconvolution is predominantly achieved by directed evolution, which requires culturing under compound exposure for several months followed by whole-genome sequencing to identify loci associated with resistance.10 In recent years, several label-free mass spectrometry approaches such as thermal proteome profiling11–13 have been developed to identify drug-target interactions, often through ligand-induced thermal stabilization of the target.14 Such unbiased methods are especially useful for divergent organisms with limited genome annotation and have been implemented successfully for other eukaryotic parasites including Leishmania15 and Plasmodium.12,16
In our previous study, we characterized a chemical modulator, ENH1, that exhibited potent antiparasitic properties against T. gondii and the malaria parasite, Plasmodium falciparum. ENH1 inhibited various stages of the lytic cycle and induced asynchronous calcium oscillations in T. gondii; however, the target(s) of this compound eluded identification.8 Here, we employed thermal proteome profiling to identify the targets of ENH1 in T. gondii. This work illustrates the potential of thermal proteome profiling to de-orphan antiparasitic agents and identify new components of calcium signaling cascades in apicomplexan parasites.
RESULTS & DISCUSSION
Thermal proteome profiling identifies CDPK1 as a cellular target of ENH1.
Enhancer 1 (ENH1; Figure 1a) is an ATP analog isolated from a GlaxoSmithKline protein kinase inhibitor set (PKIS: GSK260205A) as an inducer of calcium oscillations in T. gondii with antiparasitic activity.8 To determine the molecular target of ENH1, we performed thermal proteome profiling, which is proteome-wide implementation of the cellular thermal shift assay (CETSA).11,13 In principle, ligand binding induces a shift in the thermal denaturation curve of a protein target. Such shifts can be identified if the thermal denaturation profiles of thousands of proteins are determined by mass spectrometry (Figure 1b).
Figure 1. Thermal profiling identifies putative targets of ENH1.
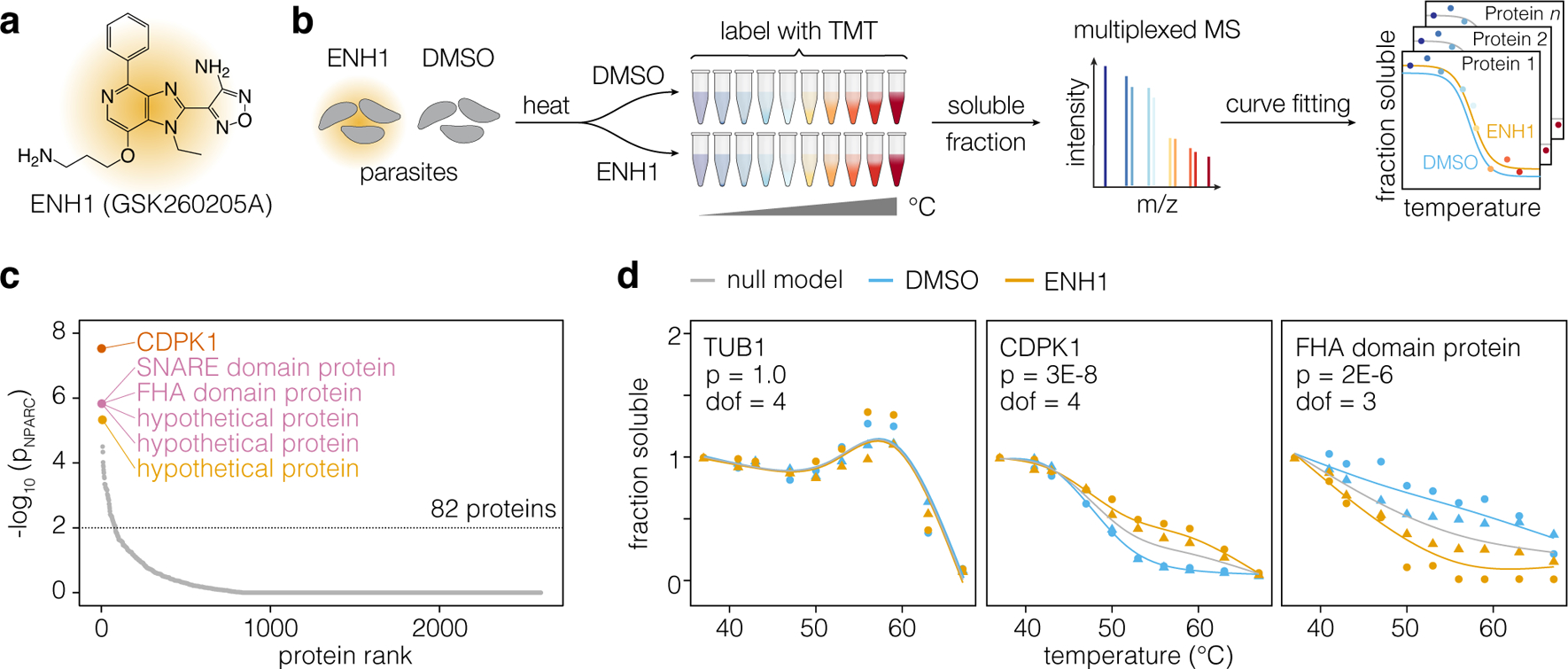
(a) Structure of ENH1. (b) Thermal profiling strategy. Parasites (gray) treated with ENH1 or vehicle (DMSO) were heated, and soluble protein was extracted following lysis. Peptide abundance at each temperature was quantified by mass spectrometry to generate protein stability curves. (c) Proteins ranked by p-value, based on the responsiveness of their thermal profiles to ENH1. (d) Representative stability curves of proteins unaffected (TUB1: TGGT1_316400B), stabilized (CDPK1: TGGT1_301440), or destabilized (FHA domain-containing protein: TGGT1_267600) by ENH1. The p-value (p) corresponds to an F-statistic based nonparametric model comparing the shapes of protein stability curves corrected for multiple hypothesis testing; the degrees of freedom (dof) required for the curves is indicated. Circles and triangles denote replicates.
We treated extracellular parasites with vehicle (DMSO) or ENH1 for 15 minutes at 37°C, under conditions previously determined to induce calcium fluxes in T. gondii.8 Parasites were then heated at 10 different temperatures to induce thermal denaturation. Following lysis and high-speed centrifugation, soluble protein was isolated and analyzed by mass spectrometry to generate thermal stability profiles from each condition. Across two biological replicates (Figure S1), we detected 2740 proteins, 2147 of which had sufficient data points to generate separate DMSO- and ENH1-treated thermal stability profiles (Figure 1c). Hits from thermal proteome profiling experiments can be evaluated by several metrics, including a single-parameter melting point shift (ΔTm) or via a comparison of curves by nonparametric curve fitting, which has a lower false-negative rate.13,17 Using the nonparametric analysis, we identified 82 proteins exhibiting a significant (p < 0.01) ENH1-dependent change in thermal stability (Figure 1c). The top hit was the calcium-dependent protein kinase CDPK1 (TGGT1_301440), followed by a SNARE domain-containing protein (TGGT1_204060), FHA domain-containing protein (TGGT1_267600), and three hypothetical proteins (TGGT1_289150, TGGT1_293360 and TGGT1_246982; Table S1). Figure 1d shows representative stability curves of individual proteins unaffected, stabilized, and destabilized by ENH1, respectively.
A chemical genetic strategy confirms ENH1 binds to and inhibits CDPK1.
To validate CDPK1 as a target of ENH1, we confirmed the results of the thermal profiling experiment by measuring CDPK1 thermal stability using immunoblots of soluble fractions. Under the same conditions used for the global mass spectrometry–based experiment, CDPK1 was strongly stabilized by ENH1 (CDPK1G, Figure 2a). As a negative control, α-tubulin (TUB1) did not exhibit ENH1-induced changes in its stability (Figure 2b). Moreover, the magnitude of the thermal stabilization of CDPK1 by ENH1 was similar to that seen in other target-based CDPK1 CETSA studies.18
Figure 2. A chemical-genetic strategy confirms CDPK1 as a target of ENH1.
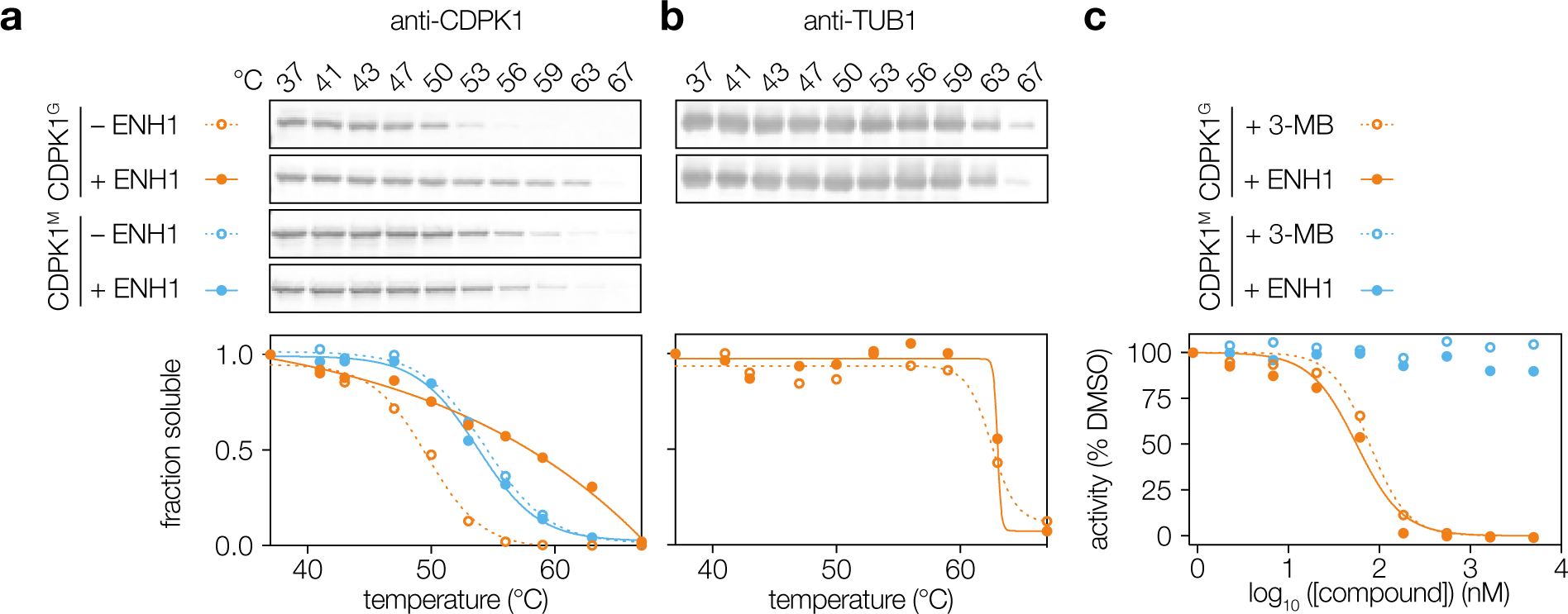
(a) Addition of ENH1 to wild-type CDPK1G and CDPK1M parasites followed by Western blot detection of protein thermal stability. (b) TUB1 thermal stability in the presence of ENH1. (c) Kinase assays of recombinantly expressed and purified CDPK1G or CDPK1M in the presence of ENH1 or the known gatekeeper-specific competitive inhibitor 3-MB.
Next, we determined the effect of ENH1 binding on CDPK1 activity. CDPK1 undergoes calcium-induced conformational changes19,20, which could indirectly arise from ENH1-induced calcium fluxes in live parasites. To disentangle these multiple effects, we performed kinase assays with recombinantly expressed and purified enzymes. In the presence of excess calcium, ENH1 potently inhibited kinase activity with an IC50 of 56 nM—comparable to the activity of the known CDPK1 inhibitor 3-MB, also known as 3-MB-PP121, which inhibited kinase activity with an IC50 of 79 nM (Figure 2c). Together, these results suggest that the ENH1-induced stability change of CDPK1 is due to direct binding of ENH1, leading to CDPK1 inhibition.
We used chemical genetics to determine the nature of ENH1 binding to CDPK1. In T. gondii, CDPK1 has a glycine at the kinase gatekeeper position (designated CDPK1G), resulting in an atypically expanded ATP-binding pocket.6,7,19,21 This feature enables selective binding and inhibition of CDPK1 by bulky pyrazolo [3,4-d] pyrimidine (PP) derivatives like 3-MB. Mutation of the gatekeeper to a methionine renders CDPK1 insensitive to 3-MB while maintaining kinase activity, and we have previously generated parasite strains expressing CDPK1G128M (herein abbreviated as CDPK1M).21 To determine whether ENH1 binds the active site of the kinase in a gatekeeper-dependent manner, we probed CDPK1M from parasites treated with ENH1 under the same conditions used for the previous assays. CDPK1M did not exhibit thermal stabilization (CDPK1M, Figure 2a), suggesting that ENH1 binding depends on the expanded ATP-binding pocket of CDPK1. Concordantly, neither 3-MB nor ENH1 inhibited CDPK1M kinase activity in vitro (Figure 2c). Collectively, our results suggest that ENH1 functions similarly to 3-MB, despite its distinct chemical scaffold.
ENH1 inhibits parasite growth through CDPK1-dependent and -independent pathways.
Next, we determined whether the antiparasitic effect of ENH1 was entirely dependent on CDPK1 inhibition. Chemical inhibition of CDPK1 blocks parasite invasion and egress but has not been associated with fluctuations in parasite calcium.21,22 Because ENH1 was previously reported to inhibit parasite egress and growth8, we used parasites with the sensitive and insensitive alleles of CDPK1 to determine whether these effects were due to its inhibition.
We investigated the inhibitory properties of ENH1 on parasite egress using a high-content imaging assay.23 Briefly, the assay monitors entry of the cell-impermeant dye DAPI into lysed host cells following stimulated parasite egress. ENH1 inhibited parasite egress in a dose-dependent manner (Figure 3a). Over the same concentration range, CDPK1M parasite egress was not affected. Overall, these results suggest that the ability of ENH1 to inhibit T. gondii egress arises from inhibition of CDPK1, and not due solely to the depletion of parasite intracellular calcium stores, as proposed.8
Figure 3. ENH1 inhibits parasite growth through CDPK1-dependent and -independent pathways.
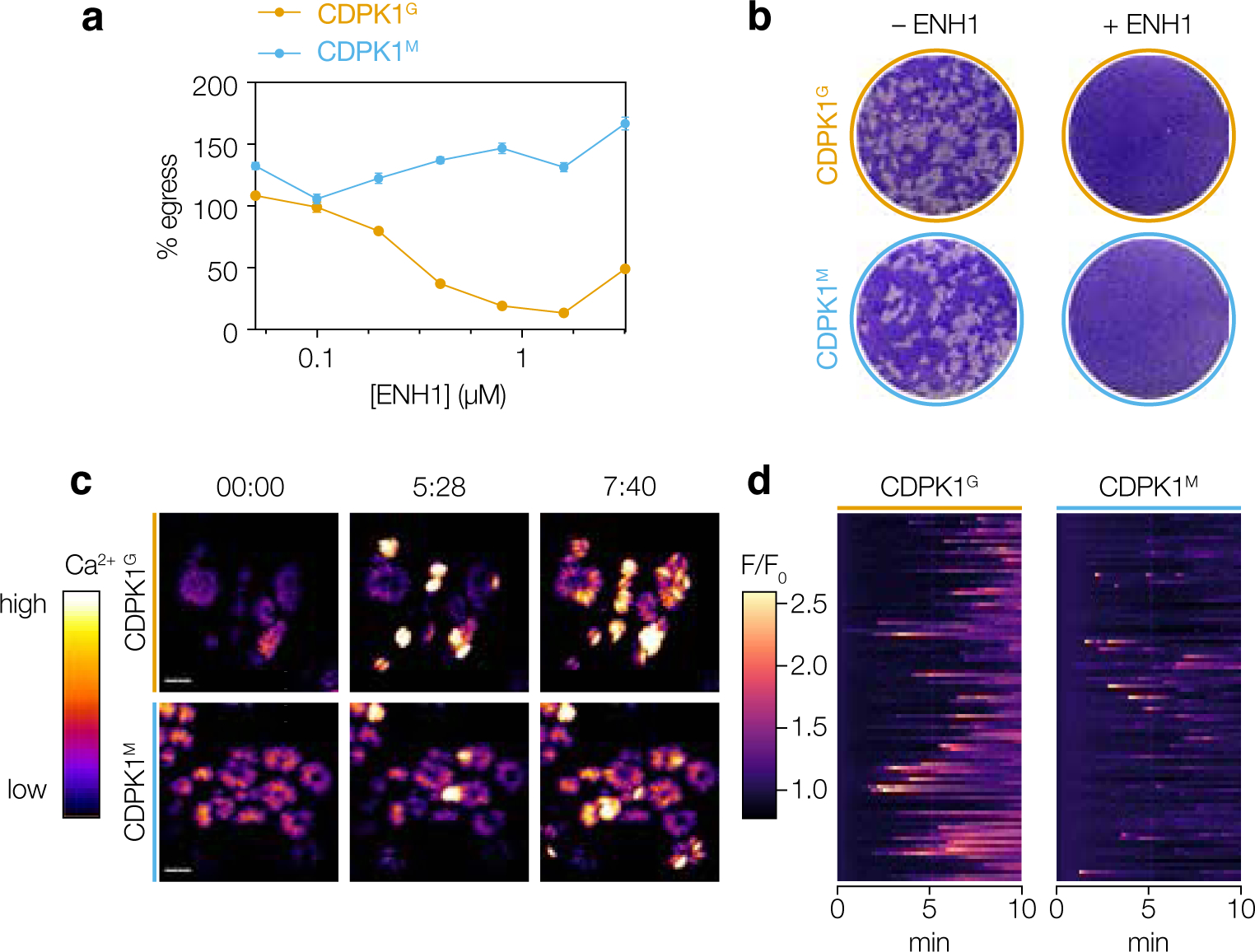
(a) Egress assays of wild-type CDPK1G and CDPK1M parasites treated with ENH1 followed by the egress agonist zaprinast. (b) Plaque assays of CDPK1G and CDPK1M parasites treated with ENH1. (c) Selected frames of time-lapse microscopy of CDPK1G and CDPK1M parasites expressing the genetically encoded calcium indicator GCaMP6f with ENH1. Scale bar is 10 μm. Time is indicated as minutes:seconds. (d) Kymographs showing the median fluorescence intensities relative to the initial intensity of 99 parasite vacuoles (rows) 10 minutes following treatment with ENH1.
Previously, we reported that ENH1 exhibited potent antiparasitic properties against both T. gondii and the related apicomplexan parasite, P. falciparum. Since the P. falciparum ortholog of TgCDPK1, PfCDPK4, is dispensable during blood stage infection5, we investigated the possibility that CDPK1-independent mechanisms contribute to the antiparasitic effects of ENH1.
We therefore examined the effect of ENH1 on repeated cycles of parasite invasion, replication, and egress, by performing plaque assays on host-cell monolayers. As observed earlier, ENH1 potently inhibited T. gondii plaque formation (Figure 3b). This inhibition occurred independently of the CDPK1 gatekeeper, as CDPK1M plaque formation was also inhibited, despite the inability of ENH1 to block parasite egress in this background. Overall, these results show that ENH1 kills T. gondii through both CDPK1-dependent and -independent pathways.
Calcium fluxes caused by ENH1 occur independently of CDPK1 inhibition.
We next sought to determine how ENH1 perturbs T. gondii physiology. Since ENH1 was previously identified for its ability to induce asynchronous calcium fluxes in parasites, we assessed whether this phenotype arose from inhibition of CDPK1. To monitor intracellular calcium release, we integrated the genetically encoded calcium indicator GCaMP6f24 into a defined, neutral locus of CDPK1G and CDPK1M parasites using CRISPR/Cas9.8,25
Intracellular parasites treated with ENH1 exhibited asynchronous calcium fluxes across the population (Figure 3c and Movie S1), as previously described.8 Calcium spikes were observed even in CDPK1M parasites, although their duration was decreased (Figure 3d), consistent with the proposed function of CDPK1 in dampening cytosolic calcium during stimulated egress.22 This observation indicates that CDPK1 inhibition does not trigger calcium spikes and suggests alternative targets of ENH1 are responsible for this effect.
Thermal proteome profiling in parasite lysates identifies other potential targets of ENH1.
Signaling effectors act dynamically by altering protein states, including triggering conformational changes, post-translational modifications, re-localization, and low-affinity protein-protein interactions.26–28 In principle, all of these processes may be captured by changes in a protein’s thermal stability profile. Our initial thermal profiling experiment may therefore have identified proteins exhibiting ENH1-dependent stability changes downstream of the increase in intracellular calcium or CDPK1 function. To circumvent these additional effects, we performed thermal profiling with the lysates of parasites expressing CDPK1M, which is not inhibited by ENH1, and in excess calcium (Figure 4a). Across two biological replicates, we detected 1825 proteins, 1246 of which had sufficient data points for a comparison of DMSO- and ENH1-treated thermal stability profiles (Table S2). Validating our approach, CDPK1M did not exhibit an ENH1-induced thermal shift (Figure 4b). To identify potential targets of ENH1, we compared the ENH1-dependent thermal stability changes in both the CDPK1M lysate and CDPK1G parasite experiments (Figure 4c); proteins exhibiting significant ENH1-dependent stability changes in both experiments are candidates for mediating the ENH1 effects on parasite calcium, and are summarized in Figure 4d.
Figure 4. Thermal profiling in CDPK1M parasite lysates reveals other potential targets of ENH1.
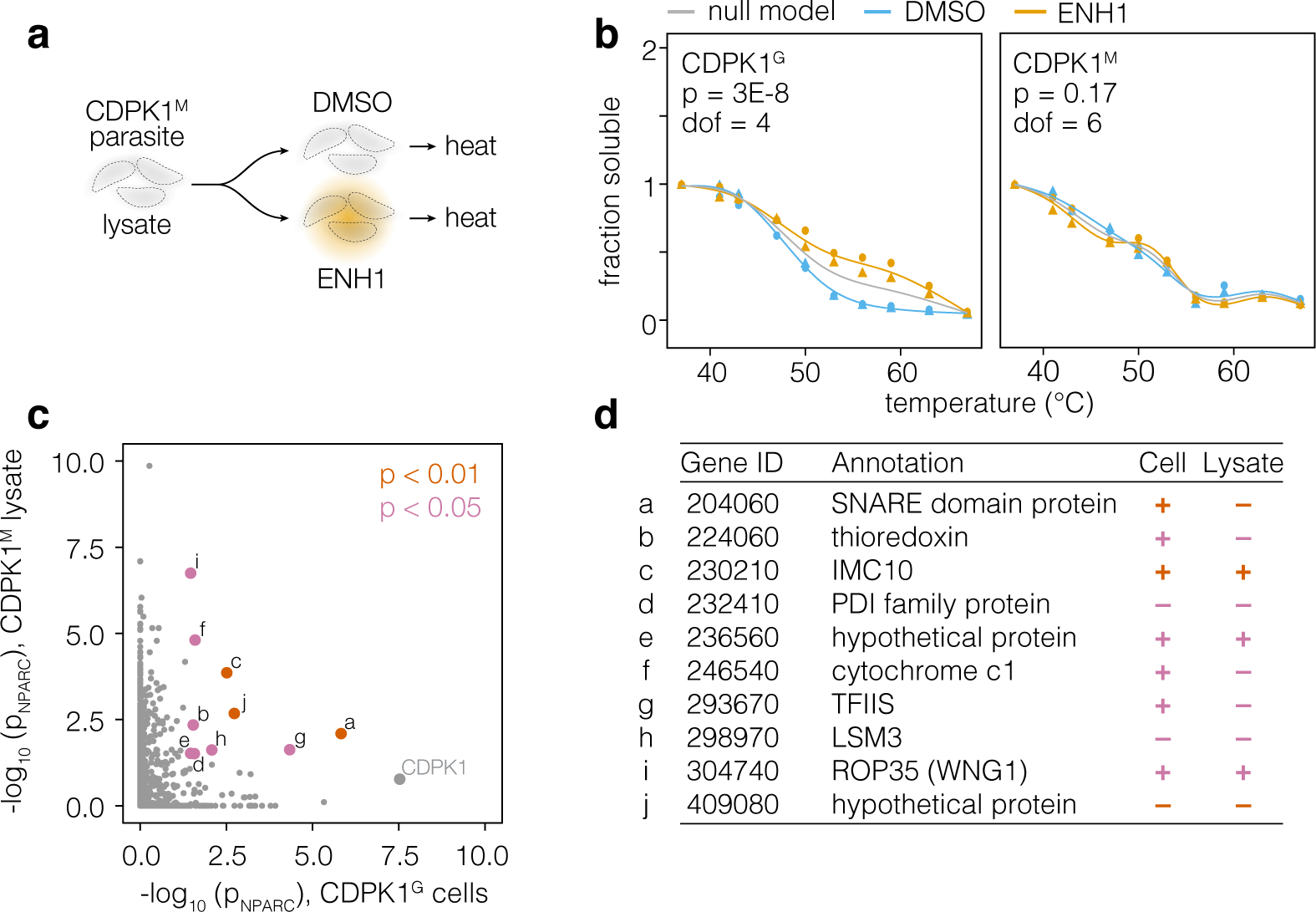
(a) Thermal profiling strategy in CDPK1M parasite lysates with 2 mM calcium. Detergent-solubilized parasite lysates were combined with ENH1 or vehicle, heated, and processed analogously to the intact-cell profiling experiment. (b) A side-by-side comparison of the CDPK1 thermal stability profiles obtained from the in-cell (CDPK1G) and lysate (CDPK1M) experiment. Circles and triangles denote replicates. (c) A comparison of thermal stability shifts measured in the in-cell (CDPK1G) and lysate (CDPK1M) experiment. Proteins exhibiting significant shifts in both experiments are highlighted. (d) A table of proteins exhibiting significant ENH1-dependent changes in thermal stabilization. The direction of alteration to the thermal profile in each experiment is indicated by + (stabilization) or – (destabilization).
A few of the candidates localized to compartments associated with calcium release and homeostasis, such as the mitochondrion and the parasite pellicle, known as the inner membrane complex (IMC). Cytochrome c1 is a component of Complex III in the mitochondrial electron transport chain. Many small molecules have been shown to target Complex III, including the antimalarial compound atovaquone29, and ENH1 binding to the complex could result in a thermal-stability shift for cytochrome c1. Since mitochondria buffer cytosolic calcium30, depolarization of the inner membrane could impact calcium homeostasis and affect parasite survival; however, this hypothesis will await further testing. In alveolates such as Paramecium, the pellicle operates as an additional calcium store.31,32 A similar role has been posited for the cognate IMC in apicomplexans but has not been confirmed.33 IMC10 (PfIMC1j in P. falciparum), a component of the IMC, was also affected by ENH1 in a CDPK1-independent manner; however, at this time, it is unclear how it would bind to the compound or what its role in calcium homeostasis might be.
Several of these candidates may bind directly to ENH1. For example, the thioredoxin and the PDI-domain–containing protein, which has a thioredoxin-like domain, may bind ENH1 in place of NADPH. ROP35, recently renamed WNG1, is a secreted effector kinase that phosphorylates T. gondii proteins in the parasitophorous vacuole, the membrane-enclosed compartment in which parasites reside in the host.34 WNG kinases lack the canonical Gly-loop and therefore have an open binding pocket that may accompany bulky ATP analogs such as ENH1. However, given the predicted function of these candidates, none is likely to be responsible for the ENH1-induced calcium fluxes.
CONCLUSION
We have applied unbiased thermal proteome profiling to the widespread parasite T. gondii. In contrast to other target identification approaches such as affinity pulldown, one attractive feature of thermal proteome profiling is that it does not require any chemical modification of the compound employed. Using mass spectrometry in combination with molecular, genetic, and phenotypic-level studies, we showed that CDPK1 is a target of the antiparasitic compound ENH1. However, other as-yet unverified targets were responsible for ENH1-induced calcium fluxes. The polypharmacology of ENH1 perturbs calcium homeostasis and other signaling pathways beyond the inhibition of any single molecular target, which may explain its potent toxicity to multiple apicomplexan parasites, including P. falciparum. Because apicomplexan calcium signaling pathways are conserved in the parasite phylum but divergent from those of their mammalian hosts, perturbagens targeting parasite calcium networks may hold untapped therapeutic potential.
In the course of our studies, we confronted the challenges of disentangling the causes of ENH1-dependent stability changes, which may arise from direct effects, i.e. binding, or indirect effects downstream of ENH1 target engagement, i.e. calcium release and signaling. We adapted our approach by performing thermal profiling in CDPK1M lysates, which are expected to have fewer of these indirect effects, and identified a list of candidates exhibiting ENH1-dependent stability changes in all conditions. At the same time, our proof-of-principle study highlights the potential to use thermal proteome profiling to identify calcium-responsive proteins in parasites, independent of genomic sequence or annotation. Ongoing efforts in our group will define the cryptic calcium-responsive proteome of T. gondii and related apicomplexan parasites.
Supplementary Material
ACKNOWLEDGEMENTS
We thank E. Shortt for technical assistance. This research was supported by funds from a National Institutes of Health grant (1R01AI144369) to SL, National Science Foundation Graduate Research Fellowship (174530) to ALH, the Boehringer Ingelheim Fonds PhD fellowship to BMM, and the Structural Genomics Consortium (DHD). The Structural Genomics Consortium is a registered charity (number 1097737) that receives funds from AbbVie, Bayer Pharma AG, Boehringer Ingelheim, Canada Foundation for Innovation, Eshelman Institute for Innovation, Genome Canada, Genentech, Innovative Medicines Initiative (EU/EFPIA) [ULTRA-DD grant no. 115766], Janssen, Merck KGaA Darmstadt Germany, MSD, Novartis Pharma AG, Ontario Ministry of Economic Development and Innovation, Pfizer, São Paulo Research Foundation-FAPESP [2013/50724-5, 2014/5087-0 and 2016/17469-0], Takeda, and Wellcome [106169/ZZ14/Z].
Footnotes
Supporting Information
This information is available free of charge through the journal’s website, including experimental methods, supplemental figures and tables, and associated references.
The authors declare no competing financial interests.
REFERENCES
- (1).Lourido S; Moreno SNJ The Calcium Signaling Toolkit of the Apicomplexan Parasites Toxoplasma Gondii and Plasmodium Spp. Cell Calcium 2015, 57 (3), 186–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Frénal K; Dubremetz JF; Lebrun M; Soldati-Favre D Gliding Motility Powers Invasion and Egress in Apicomplexa. Nat. Rev. Microbiol 2017, 15 (11), 645–660. [DOI] [PubMed] [Google Scholar]
- (3).Carruthers VB; Giddings OK; Sibley LD Secretion of Micronemal Proteins Is Associated with Toxoplasma Invasion of Host Cells. Cell. Microbiol 1999, 1 (3), 225–235. [DOI] [PubMed] [Google Scholar]
- (4).Wiersma HI; Galuska SE; Tomley FM; Sibley LD; Liberator PA; Donald RGK A Role for Coccidian cGMP-Dependent Protein Kinase in Motility and Invasion. Int. J. Parasitol 2004, 34, 369–380. [DOI] [PubMed] [Google Scholar]
- (5).Billker O; Lourido S; Sibley LD Calcium-Dependent Signaling and Kinases in Apicomplexan Parasites. Cell Host Microbe 2009, 5 (6), 612–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Johnson SM; Murphy RC; Geiger JA; Derocher AE; Zhang Z; Ojo KK; Larson ET; Perera BGK; Dale EJ; He P; Reid MC; Fox AMW; Mueller NR; Merritt EA; Fan E; Parsons M; Van Voorhis WC; Maly DJ Development of Toxoplasma Gondii Calcium-Dependent Protein Kinase 1 (Tg CDPK1) Inhibitors with Potent Anti-Toxoplasma Activity. J. Med. Chem 2012, 55 (5), 2416–2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Lourido S; Zhang C; Lopez MS; Tang K; Barks J; Wang Q; Wildman SA; Shokat KM; Sibley LD Optimizing Small Molecule Inhibitors of Calcium-Dependent Protein Kinase 1 to Prevent Infection by Toxoplasma Gondii. J. Med. Chem 2013, 56 (7), 3068–3077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Sidik SM; Triana MAH; Paul AS; El Bakkouri M; Hackett CG; Tran F; Westwood NJ; Hui R; Zuercher WJ; Duraisingh MT; Moreno SNJ; Lourido S Using a Genetically Encoded Sensor to Identify Inhibitors of Toxoplasma Gondii Ca2+signaling. J. Biol. Chem 2016, 291 (18), 9566–9580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Schürmann M; Janning P; Ziegler S; Waldmann H Small-Molecule Target Engagement in Cells. Cell Chem Biol 2016, 23 (4), 435–441. [DOI] [PubMed] [Google Scholar]
- (10).Cowell AN; Istvan ES; Lukens AK; Gomez-Lorenzo MG; Vanaerschot M; Sakata-Kato T; Flannery EL; Magistrado P; Owen E; Abraham M; LaMonte G; Painter HJ; Williams RM; Franco V; Linares M; Arriaga I; Bopp S; Corey VC; Gnädig NF; Coburn-Flynn O; Reimer C; Gupta P; Murithi JM; Moura PA; Fuchs O; Sasaki E; Kim SW; Teng CH; Wang LT; Akidil A; Adjalley S; Willis PA; Siegel D; Tanaseichuk O; Zhong Y; Zhou Y; Llinás M; Ottilie S; Gamo F-J; Lee MCS; Goldberg DE; Fidock DA; Wirth DF; Winzeler EA Mapping the Malaria Parasite Druggable Genome by Using in Vitro Evolution and Chemogenomics. Science 2018, 359 (6372), 191–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Savitski MM; Reinhard FBM; Franken H; Werner T; Savitski MF; Eberhard D; Molina DM; Jafari R; Dovega RB; Klaeger S; Kuster B; Nordlund P; Bantscheff M; Drewes G Tracking Cancer Drugs in Living Cells by Thermal Profiling of the Proteome. Science 2014, 346 (6205). 10.1126/science.1255784. [DOI] [PubMed] [Google Scholar]
- (12).Dziekan JM; Yu H; Chen D; Dai L; Wirjanata G; Larsson A; Prabhu N; Sobota RM; Bozdech Z; Nordlund P Identifying Purine Nucleoside Phosphorylase as the Target of Quinine Using Cellular Thermal Shift Assay. Sci. Transl. Med 2019, eaau3174 (January). [DOI] [PubMed] [Google Scholar]
- (13).Franken H; Mathieson T; Childs D; Sweetman GMA; Werner T; Tögel I; Doce C; Gade S; Bantscheff M; Drewes G; Reinhard FBM; Huber W; Savitski MM Thermal Proteome Profiling for Unbiased Identification of Direct and Indirect Drug Targets Using Multiplexed Quantitative Mass Spectrometry. Nat. Protoc 2015, 10 (10), 1567–1593. [DOI] [PubMed] [Google Scholar]
- (14).Kaur U; Meng H; Lui F; Ma R; Ogburn RN; Johnson JHR; Fitzgerald MC; Jones LM Proteome-Wide Structural Biology: An Emerging Field for the Structural Analysis of Proteins on the Proteomic Scale. J. Proteome Res 2018, 17 (11), 3614–3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Corpas-Lopez V; Moniz S; Thomas M; Wall RJ; Torrie LS; Zander-Dinse D; Tinti M; Brand S; Stojanovski L; Manthri S; Hallyburton I; Zuccotto F; Wyatt PG; De Rycker M; Horn D; Ferguson MAJ; Clos J; Read KD; Fairlamb AH; Gilbert IH; Wyllie S Pharmacological Validation of N-Myristoyltransferase as a Drug Target in Leishmania Donovani. ACS Infectious Diseases 2019, 5 (1), 111–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Lu K-Y; Quan B; Sylvester K; Srivastava T; Fitzgerald MC Plasmodium Chaperonin TRiC / CCT Identified as a Target of the Antihistamine Clemastine Using Parallel Chemoproteomic Strategy. Proc. Natl. Acad. Sci. U. S. A 2020, 117 (11), 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Childs D; Bach K; Franken H; Anders S; Kurzawa N; Bantscheff M; Savitski M; Huber W Non-Parametric Analysis of Thermal Proteome Profiles Reveals Novel Drug-Binding Proteins. 10.1101/373845. [DOI] [PMC free article] [PubMed]
- (18).Scheele S; Geiger JA; DeRocher AE; Choi R; Smith TR; Hulverson MA; Vidadala RSR; Barrett LK; Maly DJ; Merritt EA; Ojo KK; Van Voorhis WC; Parsonsa M Toxoplasma Calcium-Dependent Protein Kinase 1 Inhibitors: Probing Activity and Resistance Using Cellular Thermal Shift Assays. Antimicrob. Agents Chemother 2018, 62 (6), e00051–e00118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Wernimont AK; Artz JD; Finerty P; Lin Y-H; Amani M; Allali-Hassani A; Senisterra G; Vedadi M; Tempel W; Mackenzie F; Chau I; Lourido S; Sibley LD; Hui R Structures of Apicomplexan Calcium-Dependent Protein Kinases Reveal Mechanism of Activation by Calcium. Nat. Struct. Mol. Biol 2010, 17 (5), 596–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Ingram JR; Knockenhauer KE; Markus BM; Mandelbaum J; Ramek A; Shan Y; Shaw DE; Schwartz TU; Ploegh HL; Lourido S Allosteric Activation of Apicomplexan Calcium-Dependent Protein Kinases. Proceedings of the National Academy of Sciences 2015, 112 (36), E4975–E4984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Lourido S; Shuman J; Zhang C; Shokat KM; Hui R; Sibley LD Calcium-Dependent Protein Kinase 1 Is an Essential Regulator of Exocytosis in Toxoplasma. Nature 2010, 465 (7296), 359–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Stewart RJ; Lessene G; Sleebs BE; Mcconville MJ; Rogers KL; Tonkin CJ Analysis of Ca 2 + Mediated Signaling Regulating Toxoplasma Infectivity Reveals Complex Relationships between Key Molecules. Cell. Microbiol 2016, 19 (e12685). 10.1111/cmi.12685. [DOI] [PubMed] [Google Scholar]
- (23).Shortt E; Lourido S Plate-Based Quantification of Stimulated Toxoplasma Egress. Methods Mol. Biol 2020, 2071, 171–186. [DOI] [PubMed] [Google Scholar]
- (24).Chen T-W; Wardill TJ; Sun Y; Pulver SR; Renninger SL; Baohan A; Schreiter ER; Kerr RA; Orger MB; Jayaraman V; Looger LL; Svoboda K; Kim DS Ultrasensitive Fluorescent Proteins for Imaging Neuronal Activity. Nature 2013, 499 (7458), 295–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Markus BM; Bell GW; Lorenzi HA; Lourido S Optimizing Systems for Cas9 Expression in Toxoplasma Gondii. mSphere 2019, 4 (3), 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Mateus A; Bobonis J; Kurzawa N; Stein F; Helm D; Hevler J; Typas A; Savitski MM Thermal Proteome Profiling in Bacteria: Probing Protein State in Vivo. Mol. Syst. Biol 2018, 14 (7), e8242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Becher I; Andrés-Pons A; Romanov N; Stein F; Schramm M; Baudin F; Helm D; Kurzawa N; Mateus A; Mackmull MT; Typas A; Müller CW; Bork P; Beck M; Savitski MM Pervasive Protein Thermal Stability Variation during the Cell Cycle. Cell 2018, 173 (6), 1495–1507.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Dai L; Zhao T; Bisteau X; Sun W; Prabhu N; Lim YT; Sobota RM; Kaldis P; Nordlund P Modulation of Protein-Interaction States through the Cell Cycle. Cell 2018, 173 (6), 1481–1494.e13. [DOI] [PubMed] [Google Scholar]
- (29).Korsinczky M; Chen N; Kotecka B; Saul A; Rieckmann K; Cheng Q Mutations in Plasmodium Falciparum Cytochrome B That Are Associated with Atovaquone Resistance Are Located at a Putative Drug-Binding Site. Antimicrob. Agents Chemother 2000, 44 (8), 2100–2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Wacquier B; Combettes L; Dupont G Cytoplasmic and Mitochondrial Calcium Signaling: A Two-Way Relationship. Cold Spring Harb. Perspect. Biol 2019, 11 (10). 10.1101/cshperspect.a035139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Stelly N; Mauger JP; Claret M; Adoutte A Cortical Alveoli of Paramecium: A Vast Submembranous Calcium Storage Compartment. J. Cell Biol 1991, 113 (1), 103–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Plattner H; Habermann A; Kissmehl R; Klauke N; Majoul I; Söling HD Differential Distribution of Calcium Stores in Paramecium Cells. Occurrence of a Subplasmalemmal Store with a Calsequestrin-like Protein. Eur. J. Cell Biol 1997, 72 (4), 297–306. [PubMed] [Google Scholar]
- (33).Holder AA; Mohd Ridzuan MA; Green JL Calcium Dependent Protein Kinase 1 and Calcium Fluxes in the Malaria Parasite. Microbes Infect. 2012, 14 (10), 825–830. [DOI] [PubMed] [Google Scholar]
- (34).Beraki T; Hu X; Broncel M; Young JC; Shaughnessy WJO; Borek D Divergent Kinase Regulates Membrane Ultrastructure of the Toxoplasma Parasitophorous Vacuole. Proceedings of the National Academy of Sciences 2019, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


