Abstract
A core set of 190 rice landraces were used to decipher the genetic structure and to discover the chromosomal regions containing QTLs, affecting the grain micro-nutrients, fatty acids, and yield-related traits by using 148 molecular markers in this study. Landraces were categorized into three sub-groups based on population stratification study and followed by neighbor-joining tree and principal component analysis. Analysis of variance revealed abundant variations among the landraces for studied traits with less influence of environmental factors. Genome Wide Association Studies (GWAS) revealed 22 significant and consistent QTLs through marker trait association (MTAs) for 12 traits based on 2 years and pooled analysis. Out of 22 QTLs, three have been reported earlier while 19 QTLs are novel. Interestingly, 13 QTLs out of 22 were explained more than 10% phenotypic variance. Association of RM1148 and RM205 with Days to 50% flowering was comparable with flowering control genes Ghd8/qDTH8 and qDTH9, respectively. Similarly, Zn content was associated with RM44, which is situated within the QTL qZn8-1. Moreover, significant association of RM25 with oleic acid content was closely positioned with QTL qOle8. Association of RM7434 with grain yield/plant; RM184 with spikelet fertility %; R3M10, R9M42 with hundred seed weight; RM536, RM17467, RM484, RM26063 with Fe content; RM44, RM6839 with Zn content are the major outcomes of this study. In addition, association of R11M23 with days to 50% flowering, panicle length and total spikelets per panicle are explained the possible occurrence of pleiotropism among these traits. Prominent rice landraces viz., Anjani (early maturity); Sihar (extra dwarf); Gangabaru (highest grain yield/plant); Karhani (highest iron content); Byalo-2 (highest zinc content) and Kadamphool (highest oleic acid) were identified through this study. The present study will open many avenues towards utilization of these QTLs and superior landraces in rice breeding for developing nutrition-rich high yielding varieties.
Electronic supplementary material
The online version of this article (10.1007/s13205-020-02467-z) contains supplementary material, which is available to authorized users.
Keywords: Association mapping, Fatty acids, Micronutrients, Qtls, Rice landraces
Introduction
Rice (Oryza sativa L.) has been considered as an important cereal crop for the food security of over half of the world’s population (~ 3.5 billion), with Asia accounting for 90% of global rice consumption. In lower and middle-income countries, rice serves as a means of livelihood for millions of rural households and source of dietary energy and nourishment for more than 70% people (FAOSTAT 2020). However, rice is a rich source of carbohydrate but poor in some essential micro-nutrients, proteins, fats and vitamins therefore, heavy dependency on it increases the possibilities of hidden hunger among the rice consuming children and people (Descalsota et al. 2018; Zhang et al. 2014; Garcia-Oliveira et al. 2009). According to the latest estimates, about 2 billion people (26.4%) of the world are suffering from moderate to severe levels of food insecurity and hidden hunger (FAOSTAT 2020). Constant increment in the world population with limiting land resource, gives intimation to us for improvising the yield potential and nutritional quality of rice consequently. Therefore, efforts to combat the food deficiency and malnutrition at global level is one of the prime goals for the rice breeders. Elevating the nutritional quality of food crops with maintaining yield potential through traditional breeding, modern biotechnology and advanced genomic approaches, is one of the vital and balanced approach to overcome the problems (Descalsota et al. 2018; Norton et al. 2018; Juan et al. 2018). As rice being the most consumed food crop throughout the world, attentiveness for enhancement in their nutritive value is as much essential as yield (Descalsota et al. 2018; Huang et al. 2015). Some nutritionally enriched and high yielding rice varieties viz., CR Dhan-311, DRR Dhan-45, Chhattishgarh Zinc Rice-1, Chhattisgarh Zinc Rice-2, Zinco Rice-MS, Protezein, CG Madhuraj-55, ARC10063 were recently developed in India through traditional breeding strategies (Sarawgi et al. 2019; Pradhan et al. 2019, 2020; Sanjeeva Rao et al. 2020).
Availability of abundant genetic variability and diversity in the gene pool is the fundamental need for genetic enhancement of rice. Traditional rice landraces best adapted in their inhabitant environment possess vast genetic diversity, variability and huge number of valuable genes for nutritional, agronomic traits along with resistance to biotic and abiotic stresses (Agrama et al. 2007; Sahu et al. 2017). Several rice landraces viz., Laicha, Gathuvan, Maharaji, Bhejri, Danwar, Bora, Karangi, Chapti Gurmatiya, Red Kavuni, Kaivara Samba, Kuruvi Kar, Poongar, Kattu Yanam, Koliyal, Maappillai Samba etc. have been reported and are very popular for their high nutritional and medicinal properties in India (Rathna Priya et al. 2019; Rahman et al. 2006). The indigenous rice landraces have long been served as a source of several valuable genes, often used in genetic improvement of rice. They have also played a significant role in maintaining the food security in the light of changing climate (Del Cruz and Khush 2000).
Rice, being a model plant with fully sequenced genome provides an opportunity to use genomic approaches for studying the genetic control of traits, exploring new genes, its domestication, adaptive selection and the genetic basis of adaptation across the varying environments (Zhang et al. 2014; Agrama et al. 2007). Advancement in molecular technologies, availability of genome sequences, accessibility of genome-wide molecular markers, economical genotyping facilities, and advances in statistical analysis have made it possible to unravel the genetic basis and linkages of complex traits. In rice, mapping of grain nutritional and yield-related traits have been done with the help of linkage based mapping method (Zhang et al. 2019; Zhao et al. 2011). However, linkage mapping approach does not sample the larger pool of genetic variation that may contribute to phenotypic variation within a species. In addition to the bi-parental mapping strategies used in QTL mapping of important agronomic and nutritional traits, GWAS is widely used in the recent times (Suman et al. 2020; Haritha et al. 2020; Donde et al. 2020; Pradhan et al. 2019, 2020; Zhang et al. 2014, 2019; Norton et al. 2018; Kadam et al. 2018; Juan et al. 2018; Prasanth et al. 2017; Swamy et al. 2017; Xu et al. 2016; Lu et al. 2015; Zhao et al. 2011; Borba et al. 2010; Agrama et al. 2007). GWAS maximizes the utilization of genetic variation of unrelated individuals in the natural population for large and more representative set of loci without the additional requirement of mapping population development (Kadam et al. 2018; Swamy et al. 2017). It has been considered as a feasible approach to decipher and map the multiple loci for many traits simultaneously with high accuracy and fine resolution in crop plants. Identifying the QTLs in multi-environments may possibly provide an accurate information for gene cloning and molecular breeding (Norton et al. 2018; Pradhan et al. 2020).
In this study, we employed GWAS in a large sample of cultivated rice landraces (190 accessions) with following major objectives (1) To dissect the genetic structure and diversity of traditional rice landraces, (2) to identify the chromosomal regions containing QTLs affecting the grain micro-nutrients (Fe content and Zn content), fatty acids (palmitic acid, stearic acid, oleic acid, linoleic acid and linolenic) and yield-related traits, (3) to select the landraces with high nutritional value and yield potential for further breeding use. Our results revealed a number of important donor landraces and novel QTLs for grain nutritional and yield-related traits that can serve as foci of future studies to characterize the molecular basis of the observed variations and for developing nutritional rich, high yielding rice varieties.
Materials and methods
Experimental plant materials
The experimental materials comprised of 188 traditional rice landraces and two improved check varieties viz., Rajeshwari (IGKV R-1) and Mahamaya (Supplementary Table 1) of Chhattisgarh, India. Rice landraces were procured from R.H. Richharia Rice Gene Bank, Department of Genetics and Plant Breeding, Indira Gandhi Krishi Vishwavidyalaya (IGKV), Raipur-492012 (Chhattisgarh), India for the study.
Field layout and experimental design
Rice genotype were evaluated in field for two consecutive years (2015 and 2016) during wet season (June–September) at research farm, college of agriculture, IGKV, Raipur to acquire the precise morphological data. Plant materials were transplanted in field by following the Augmented Block Design (ABD) (Federer 1956) during both the years. Two improved cultivars viz., Rajeshwari (IGKV R-1) and Mahamaya were used as repeated checks in each block and randomized within the blocks. Field was divided into 13 blocks and each block had 17 genotypes (15 test genotypes and two check varieties) except for block 13 which had total 10 genotypes (eight test genotypes + two checks). Each genotype was transplanted in two rows of two meter length at spacing of 0.2 m between rows and 0.15 m between plants. Distance between entries was kept 0.3 m and block to block was maintained at 0.5 m. Each block had 9.50 m in length and 2 m width (Supplementary Fig. S1). Standard agronomic practices were adopted for normal crop growth and proper expression of genotypes throughout the crop season during both years.
Phenotyping for yield-related traits and grain nutritional traits
Eleven morphological traits viz., days to 50% flowering (DFF), plant height (PH) (cm), total tillers/plant (TTP), productive tillers/plant (PTP), panicle length (PL), fertile spikelets/panicle (FSP), sterile spikelets/panicle (SSP), total spikelets/panicle (TSP), spikelet fertility percent (SF%), 100 seed weight (g) (HSW) and grain yield (g/plant) (GY) were recorded by following the Standard Evaluation System of International Rice Research Institute (IRRI), Philipines (SES IRRI 2013). The observations for grain yield and its contributing traits were recorded on ten random plants in each genotype at specific stage.
Seven nutritional traits (micronutrients and fatty acids) were also recorded in this study. Micronutrient content viz., iron (Fe), zinc (Zn) were estimated from hulled rice (brown rice) by energy dispersive x-ray fluorescence (ED-XRF) (OXFORD Instruments X-Supreme 8000, Abingdon, UK) at Indian Institute of Rice Research (IIRR), Hyderabad, India (Babu et al. 2014; Rao et al. 2014). Rough rice samples were hulled by a single pass through standard rubber roll huller (Satake Engineering Co. Ltd. Tokyo, Japan) to eliminate the mineral contamination from huller machine. Similarly, milling of hulled rice was done in a standard miller to extract the bran fractions (Satake Engineering Co. Ltd. Tokyo, Japan). Components of five fatty acids viz., palmitic acid (PA), stearic acid (SA), oleic acid (OA), linoleic acid (LA) and linolenic acid (LnA) were estimated from bran fraction by base-catalyzed trans-esterification method (Mondal et al. 2018) in Gas Chromatography (Shimadzu, Kyoto, Japan) at Nuclear Agriculture and Biotechnology Division (NA&BTD), Bhabha Atomic Research Centre (BARC), Mumbai, India. Bran of each sample was collected after milling of hulled rice in a standard miller (Satake Engineering Co. Ltd. Tokyo, Japan). Rice bran of each genotype was packed into zipper polythene bag, properly labelled and kept immediately at 40C to avoid the harmful activities of lipase enzyme. The machine was cleaned after every sample run to avoid sample contamination.
Analysis of statistical parameters and variation components
Analysis of Augmented Randomized Block Design was done with the help of SAS v9.4 software by following GLM procedure (SAS, Inc., Cary, NC, USA). Shapiro–Wilk test for normal distribution of observations, descriptive statistics and heritability estimates were analyzed by software WINDOSTAT v9.3. Phenotypic correlations among studied traits were estimated with the help of PAST v3.14 software (Hammer et al. 2001). Mean data (adjusted mean) of all the traits for both the years along with the pooled data were used in bio-statistical analysis and association mapping study.
Genomic DNA extraction and marker locus selection
The total genomic DNA was isolated from young leaf tissue using GenElute™ Plant Genomic DNA miniprep kit (Sigma-Aldrich, Missouri, USA) as following the manufacturer's protocol. Young leaf tissues from 10 plants of each genotype were used for DNA extraction to capture the landrace heterogeneity. The quality and quantity of DNA were estimated with a NanoDrop system (ND-1000, Thermo Scientific, USA). Total 148 molecular markers including 107 SSR markers and 41 InDel markers were used for genotyping of 190 rice landraces (Table 1). Total 106 SSR markers were adopted from ‘Gramene: a genomics and genetics resource for rice marker database’ (https://www.gramene.org/markers/microsat/) in which, the panel of 50 standard SSR markers of Generation Challenge Program for rice diversity analysis was included (Table 1) and one SSR marker was manually designed for this study (Supplementary Table 2). InDel markers were taken from the published data (Shen et al. 2004).
Table 1.
List of molecular markers (SSRs and InDels) used in the study along with their map positions and genetic diversity parameters
| S. no | Marker name | Chromosome No | Map position (bp)b | Major allele frequency | Allele No | Gene diversity (He) | Landraces (proportion) showing > 1 alleles | PICa | Marker typec | Availabilityd |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | RM495 | 1 | 2,15,956 | 0.56 | 8 | 0.64 | 0.01 | 0.6 | SSR | GDB |
| 2 | RM1 | 1 | 46,35,793 | 0.18 | 17 | 0.9 | 0 | 0.89 | SSR | GDB |
| 3 | RM283 | 1 | 48,85,915 | 0.43 | 9 | 0.68 | 0 | 0.63 | SSR | GDB |
| 4 | RM259 | 1 | 74,45,627 | 0.22 | 10 | 0.84 | 0 | 0.82 | SSR | GDB |
| 5 | RM312 | 1 | 91,06,659 | 0.42 | 5 | 0.67 | 0 | 0.62 | SSR | GDB |
| 6 | RM23 | 1 | 91,06,659 | 0.32 | 6 | 0.77 | 0 | 0.73 | SSR | GDB |
| 7 | R1M7 | 1 | 1,05,18,002 | 0.98 | 2 | 0.05 | 0.01 | 0.05 | InDel | PBS |
| 8 | RM229 | 1 | 1,84,07,879 | 0.41 | 8 | 0.72 | 0 | 0.67 | SSR | GDB |
| 9 | RM5 | 1 | 2,39,71,321 | 0.28 | 10 | 0.8 | 0 | 0.77 | SSR | GDB |
| 10 | R1M30 | 1 | 2,46,60,853 | 0.7 | 2 | 0.42 | 0.05 | 0.33 | InDel | PBS |
| 11 | RM237 | 1 | 2,83,39,373 | 0.37 | 9 | 0.76 | 0 | 0.73 | SSR | GDB |
| 12 | RM212 | 1 | 3,30,53,493 | 0.64 | 6 | 0.53 | 0 | 0.49 | SSR | GDB |
| 13 | RM1003 | 1 | 3,34,75,908 | 0.52 | 6 | 0.61 | 0 | 0.54 | SSR | GDB |
| 14 | RM3403 | 1 | 3,49,95,620 | 0.3 | 22 | 0.85 | 0 | 0.84 | SSR | GDB |
| 15 | R1M47 | 1 | 3,68,28,634 | 0.89 | 2 | 0.2 | 0.01 | 0.18 | InDel | PBS |
| 16 | RM431 | 1 | 3,88,93,890 | 0.3 | 8 | 0.79 | 0.01 | 0.76 | SSR | GDB |
| 17 | RM154 | 2 | 10,83,820 | 0.18 | 13 | 0.88 | 0.01 | 0.87 | SSR | GDB |
| 18 | RM279 | 2 | 28,82,052 | 0.16 | 15 | 0.91 | 0.03 | 0.9 | SSR | GDB |
| 19 | R2M10 | 2 | 71,07,574 | 0.99 | 2 | 0.01 | 0 | 0.01 | InDel | PBS |
| 20 | RM53 | 2 | 77,06,705 | 0.28 | 15 | 0.84 | 0 | 0.82 | SSR | GDB |
| 21 | RM452 | 2 | 95,63,257 | 0.37 | 7 | 0.73 | 0.01 | 0.69 | SSR | GDB |
| 22 | RM521 | 2 | 1,08,04,792 | 0.49 | 12 | 0.72 | 0 | 0.7 | SSR | GDB |
| 23 | R2M24 | 2 | 1,12,09,429 | 0.89 | 2 | 0.2 | 0.01 | 0.18 | InDel | PBS |
| 24 | RM300 | 2 | 1,31,91,380 | 0.37 | 12 | 0.75 | 0.01 | 0.72 | SSR | GDB |
| 25 | R2M26 | 2 | 1,51,04,418 | 0.84 | 2 | 0.27 | 0.01 | 0.23 | InDel | PBS |
| 26 | RM29 | 2 | 1,74,84,665 | 0.5 | 9 | 0.67 | 0 | 0.62 | SSR | GDB |
| 27 | R2M37 | 2 | 2,35,63,428 | 0.51 | 2 | 0.5 | 0.05 | 0.37 | InDel | PBS |
| 28 | RM263 | 2 | 2,58,65,334 | 0.4 | 13 | 0.8 | 0 | 0.78 | SSR | GDB |
| 29 | RM221 | 2 | 2,76,09,877 | 0.23 | 11 | 0.86 | 0 | 0.84 | SSR | GDB |
| 30 | R2M50 | 2 | 3,04,06,330 | 0.88 | 2 | 0.21 | 0.01 | 0.19 | InDel | PBS |
| 31 | RM60 | 3 | 1,05,852 | 0.37 | 11 | 0.73 | 0 | 0.69 | SSR | GDB |
| 32 | RM81 | 3 | 19,45,837 | 0.29 | 9 | 0.78 | 0 | 0.75 | SSR | GDB |
| 33 | RM489 | 3 | 43,33,680 | 0.48 | 14 | 0.73 | 0 | 0.71 | SSR | GDB |
| 34 | R3M10 | 3 | 60,44,614 | 0.99 | 2 | 0.02 | 0 | 0.02 | InDel | PBS |
| 35 | RM7 | 3 | 1,24,07,382 | 0.39 | 8 | 0.72 | 0 | 0.68 | SSR | GDB |
| 36 | RM338 | 3 | 1,32,21,482 | 0.68 | 7 | 0.5 | 0 | 0.46 | SSR | GDB |
| 37 | R3M23 | 3 | 1,56,78,406 | 0.89 | 2 | 0.2 | 0.01 | 0.18 | InDel | PBS |
| 38 | R3M30 | 3 | 1,98,88,471 | 0.96 | 2 | 0.07 | 0 | 0.07 | InDel | PBS |
| 39 | RM6832 | 3 | 2,24,00,917 | 0.66 | 11 | 0.53 | 0 | 0.5 | SSR | GDB |
| 40 | R3M37 | 3 | 2,64,74,984 | 0.63 | 3 | 0.51 | 0.03 | 0.44 | InDel | PBS |
| 41 | RM55 | 3 | 2,82,31,692 | 0.39 | 9 | 0.75 | 0 | 0.72 | SSR | GDB |
| 42 | R3M53 | 3 | 3,24,85,490 | 0.64 | 2 | 0.46 | 0.02 | 0.36 | InDel | PBS |
| 43 | RM514 | 3 | 3,52,81,232 | 0.27 | 12 | 0.84 | 0 | 0.83 | SSR | GDB |
| 44 | R4M13 | 4 | 81,15,751 | 0.89 | 2 | 0.2 | 0 | 0.18 | InDel | PBS |
| 45 | RM307 | 4 | 86,10,617 | 0.25 | 12 | 0.84 | 0 | 0.82 | SSR | GDB |
| 46 | R4M17 | 4 | 1,16,36,507 | 0.54 | 2 | 0.5 | 0.04 | 0.37 | InDel | PBS |
| 47 | RM16623 | 4 | 1,24,31,839 | 0.09 | 30 | 0.95 | 0 | 0.95 | SSR | GDB |
| 48 | R4M30 | 4 | 1,81,72,382 | 0.56 | 2 | 0.49 | 0.01 | 0.37 | InDel | PBS |
| 49 | RM471 | 4 | 1,88,24,746 | 0.28 | 10 | 0.79 | 0 | 0.75 | SSR | GDB |
| 50 | R4M43 | 4 | 2,45,94,268 | 0.87 | 2 | 0.23 | 0 | 0.2 | InDel | PBS |
| 51 | RM1388 | 4 | 2,50,35,173 | 0.09 | 28 | 0.95 | 0 | 0.95 | SSR | GDB |
| 52 | RM241 | 4 | 2,68,57,374 | 0.16 | 19 | 0.92 | 0.01 | 0.91 | SSR | GDB |
| 53 | R4M50 | 4 | 2,89,89,250 | 0.74 | 2 | 0.39 | 0.02 | 0.31 | InDel | PBS |
| 54 | RM17467 | 4 | 3,06,74,329 | 0.39 | 24 | 0.8 | 0.04 | 0.79 | SSR | GDB |
| 55 | RM124 | 4 | 3,47,39,636 | 0.49 | 5 | 0.64 | 0.01 | 0.57 | SSR | GDB |
| 56 | RM507 | 5 | 1,02,742 | 0.39 | 6 | 0.73 | 0 | 0.69 | SSR | GDB |
| 57 | RM413 | 5 | 22,12,736 | 0.28 | 13 | 0.86 | 0 | 0.84 | SSR | GDB |
| 58 | RM3322 | 5 | 42,62,731 | 0.27 | 14 | 0.83 | 0 | 0.81 | SSR | GDB |
| 59 | R5M13 | 5 | 59,50,163 | 0.87 | 2 | 0.22 | 0 | 0.2 | InDel | PBS |
| 60 | R5M30 | 5 | 2,14,47,905 | 0.64 | 2 | 0.46 | 0.01 | 0.35 | InDel | PBS |
| 61 | RM178 | 5 | 2,51,01,829 | 0.55 | 6 | 0.64 | 0 | 0.6 | SSR | GDB |
| 62 | RM87 | 5 | 2,68,48,154 | 0.59 | 7 | 0.61 | 0 | 0.58 | SSR | GDB |
| 63 | RM161 | 5 | 2,68,48,154 | 0.5 | 8 | 0.68 | 0.01 | 0.64 | SSR | GDB |
| 64 | RM334 | 5 | 2,73,42,022 | 0.15 | 13 | 0.89 | 0 | 0.88 | SSR | GDB |
| 65 | RM133 | 6 | 4,41,616 | 0.75 | 4 | 0.41 | 0 | 0.38 | SSR | GDB |
| 66 | RM510 | 6 | 28,31,443 | 0.61 | 5 | 0.56 | 0 | 0.5 | SSR | GDB |
| 67 | RM585 | 6 | 37,22,966 | 0.43 | 9 | 0.76 | 0.01 | 0.74 | SSR | GDB |
| 68 | RM217 | 6 | 42,12,289 | 0.14 | 21 | 0.92 | 0.01 | 0.91 | SSR | GDB |
| 69 | RM253 | 6 | 54,25,408 | 0.23 | 15 | 0.88 | 0 | 0.87 | SSR | GDB |
| 70 | RM2126 | 6 | 59,05,238 | 0.28 | 15 | 0.8 | 0 | 0.77 | SSR | GDB |
| 71 | RM7488 | 6 | 69,98,987 | 0.27 | 19 | 0.84 | 0.01 | 0.82 | SSR | GDB |
| 72 | R6M14 | 6 | 74,36,963 | 0.68 | 2 | 0.43 | 0.01 | 0.34 | InDel | PBS |
| 73 | RM3330 | 6 | 1,10,64,125 | 0.22 | 10 | 0.87 | 0 | 0.86 | SSR | GDB |
| 74 | RM7434 | 6 | 2,39,34,663 | 0.37 | 7 | 0.71 | 0 | 0.66 | SSR | GDB |
| 75 | RM7434 | 6 | 2,39,34,663 | 0.25 | 8 | 0.79 | 0.01 | 0.76 | SSR | GDB |
| 76 | RM162 | 6 | 2,40,35,491 | 0.41 | 8 | 0.71 | 0.01 | 0.66 | SSR | GDB |
| 77 | R6M44 | 6 | 2,57,35,724 | 0.89 | 2 | 0.2 | 0.01 | 0.18 | InDel | PBS |
| 78 | RM454 | 6 | 2,85,32,453 | 0.44 | 5 | 0.65 | 0 | 0.58 | SSR | GDB |
| 79 | RM481 | 7 | 57,35,196 | 0.26 | 16 | 0.88 | 0.04 | 0.87 | SSR | GDB |
| 80 | R7M7 | 7 | 65,81,196 | 0.76 | 2 | 0.36 | 0 | 0.3 | InDel | PBS |
| 81 | RM501 | 7 | 67,79,215 | 0.57 | 14 | 0.61 | 0 | 0.56 | SSR | GDB |
| 82 | RM125 | 7 | 67,79,215 | 0.46 | 7 | 0.68 | 0 | 0.64 | SSR | GDB |
| 83 | R7M20 | 7 | 1,12,79,013 | 0.5 | 2 | 0.5 | 0.01 | 0.38 | InDel | PBS |
| 84 | RM418 | 7 | 1,81,32,231 | 0.37 | 14 | 0.79 | 0 | 0.77 | SSR | GDB |
| 85 | RM11 | 7 | 1,92,56,914 | 0.38 | 9 | 0.75 | 0 | 0.71 | SSR | GDB |
| 86 | RM455 | 7 | 2,23,50,593 | 0.65 | 3 | 0.48 | 0 | 0.39 | SSR | GDB |
| 87 | R7M37 | 7 | 2,36,24,859 | 0.99 | 2 | 0.03 | 0.01 | 0.03 | InDel | PBS |
| 88 | RM505 | 7 | 2,51,01,758 | 0.42 | 4 | 0.65 | 0 | 0.58 | SSR | GDB |
| 89 | RM234 | 7 | 2,54,72,688 | 0.52 | 10 | 0.66 | 0 | 0.62 | SSR | GDB |
| 90 | RM118 | 7 | 2,93,39,845 | 0.36 | 7 | 0.73 | 0 | 0.69 | SSR | GDB |
| 91 | RM408 | 8 | 1,25,275 | 0.42 | 6 | 0.67 | 0 | 0.61 | SSR | GDB |
| 92 | RM407 | 8 | 5,22,394 | 0.45 | 12 | 0.74 | 0 | 0.71 | SSR | GDB |
| 93 | RM152 | 8 | 6,82,963 | 0.36 | 8 | 0.78 | 0 | 0.75 | SSR | GDB |
| 94 | RM22254 | 8 | 6,91,385 | 0.44 | 22 | 0.69 | 0 | 0.64 | SSR | GDB |
| 95 | RM44 | 8 | 21,14,843 | 0.45 | 9 | 0.67 | 0 | 0.62 | SSR | GDB |
| 96 | RM25 | 8 | 21,14,843 | 0.38 | 9 | 0.78 | 0 | 0.75 | SSR | GDB |
| 97 | RM1148 | 8 | 37,39,008 | 0.39 | 8 | 0.74 | 0 | 0.7 | SSR | GDB |
| 98 | RM6999 | 8 | 39,84,260 | 0.14 | 19 | 0.91 | 0 | 0.91 | SSR | GDB |
| 99 | RM3481 | 8 | 91,35,139 | 0.22 | 12 | 0.88 | 0 | 0.86 | SSR | GDB |
| 100 | RM3395 | 8 | 1,02,93,664 | 0.58 | 6 | 0.59 | 0 | 0.54 | SSR | GDB |
| 101 | R8M23 | 8 | 1,34,82,749 | 0.95 | 2 | 0.2 | 0 | 0.01 | InDel | PBS |
| 102 | R8M33 | 8 | 2,07,97,429 | 0.65 | 2 | 0.45 | 0.01 | 0.35 | InDel | PBS |
| 103 | RM284 | 8 | 2,11,42,348 | 0.62 | 5 | 0.57 | 0 | 0.53 | SSR | GDB |
| 104 | RM433 | 8 | 2,55,90,576 | 0.29 | 11 | 0.8 | 0 | 0.77 | SSR | GDB |
| 105 | RM447 | 8 | 2,65,46,992 | 0.41 | 10 | 0.66 | 0 | 0.6 | SSR | GDB |
| 106 | RM316 | 9 | 12,71,123 | 0.32 | 11 | 0.82 | 0 | 0.8 | SSR | GDB |
| 107 | R9M10 | 9 | 45,15,585 | 0.74 | 2 | 0.38 | 0 | 0.31 | InDel | PBS |
| 108 | R9M20 | 9 | 93,78,728 | 0.89 | 2 | 0.2 | 0.01 | 0.18 | InDel | PBS |
| 109 | RM105 | 9 | 1,18,07,446 | 0.58 | 5 | 0.58 | 0 | 0.53 | SSR | GDB |
| 110 | RM6839 | 9 | 1,45,66,026 | 0.7 | 5 | 0.46 | 0 | 0.42 | SSR | GDB |
| 111 | R9M30 | 9 | 1,46,47,925 | 0.84 | 2 | 0.27 | 0 | 0.23 | InDel | PBS |
| 112 | RM257 | 9 | 1,77,19,660 | 0.21 | 12 | 0.85 | 0.01 | 0.83 | SSR | GDB |
| 113 | R9M42 | 9 | 1,91,15,701 | 0.99 | 2 | 0.02 | 0 | 0.02 | InDel | PBS |
| 114 | RM215 | 9 | 2,11,89,110 | 0.35 | 7 | 0.75 | 0 | 0.71 | SSR | GDB |
| 115 | RM205 | 9 | 2,27,20,624 | 0.39 | 18 | 0.77 | 0.01 | 0.74 | SSR | GDB |
| 116 | RM474 | 10 | 18,18,800 | 0.42 | 19 | 0.78 | 0 | 0.77 | SSR | GDB |
| 117 | R10M10 | 10 | 52,07,502 | 0.52 | 2 | 0.5 | 0.06 | 0.37 | InDel | PBS |
| 118 | RM271 | 10 | 53,52,766 | 0.32 | 10 | 0.81 | 0 | 0.79 | SSR | GDB |
| 119 | R10M17 | 10 | 90,04,292 | 0.97 | 2 | 0.06 | 0 | 0.06 | InDel | PBS |
| 120 | RM184 | 10 | 1,63,58,895 | 0.32 | 7 | 0.75 | 0 | 0.7 | SSR | GDB |
| 121 | R10M30 | 10 | 1,71,91,366 | 0.54 | 2 | 0.5 | 0.04 | 0.37 | InDel | PBS |
| 122 | RM171 | 10 | 1,90,48,795 | 0.3 | 12 | 0.81 | 0 | 0.78 | SSR | GDB |
| 123 | R10M40 | 10 | 1,96,17,284 | 0.15 | 2 | 0.1 | 0 | 0.01 | InDel | PBS |
| 124 | RM484 | 10 | 2,10,66,729 | 0.46 | 5 | 0.61 | 0 | 0.53 | SSR | GDB |
| 125 | RM228 | 10 | 2,22,43,157 | 0.33 | 21 | 0.85 | 0.01 | 0.84 | SSR | GDB |
| 126 | RM591 | 10 | 2,28,99,617 | 0.17 | 27 | 0.91 | 0 | 0.9 | SSR | GDB |
| 127 | RM590 | 10 | 2,30,43,156 | 0.33 | 7 | 0.78 | 0 | 0.75 | SSR | GDB |
| 128 | RM26063 | 11 | 22,56,991 | 0.18 | 20 | 0.9 | 0.01 | 0.89 | SSR | GDB |
| 129 | RM552 | 11 | 48,43,013 | 0.12 | 17 | 0.91 | 0 | 0.91 | SSR | GDB |
| 130 | RM536 | 11 | 89,85,132 | 0.27 | 9 | 0.82 | 0 | 0.8 | SSR | GDB |
| 131 | RM287 | 11 | 1,67,67,319 | 0.29 | 9 | 0.82 | 0 | 0.8 | SSR | GDB |
| 132 | RM26771 | 11 | 1,73,59,392 | 0.41 | 18 | 0.75 | 0.01 | 0.72 | SSR | GDB |
| 133 | RM21 | 11 | 1,83,18,826 | 0.2 | 16 | 0.88 | 0 | 0.87 | SSR | GDB |
| 134 | R11M23 | 11 | 2,01,48,709 | 0.89 | 2 | 0.2 | 0 | 0.18 | InDel | PBS |
| 135 | RM26981 | 11 | 2,16,89,935 | 0.09 | 38 | 0.96 | 0 | 0.96 | SSR | GDB |
| 136 | R11M40 | 11 | 2,48,11,386 | 0.71 | 2 | 0.41 | 0.04 | 0.33 | InDel | PBS |
| 137 | RM144 | 11 | 2,82,81,693 | 0.31 | 13 | 0.84 | 0 | 0.83 | SSR | GDB |
| 138 | RiceSSR PODIP | 12 | 6,23,664 | 0.26 | 6 | 0.81 | 0 | 0.78 | SSR | MD |
| 139 | RM19 | 12 | 31,85,384 | 0.23 | 11 | 0.86 | 0 | 0.85 | SSR | GDB |
| 140 | RM27601 | 12 | 35,53,566 | 0.48 | 16 | 0.71 | 0 | 0.68 | SSR | GDB |
| 141 | R12M10 | 12 | 42,89,075 | 0.99 | 2 | 0.02 | 0.02 | 0.02 | InDel | PBS |
| 142 | RM277 | 12 | 88,26,555 | 0.67 | 6 | 0.5 | 0 | 0.45 | SSR | GDB |
| 143 | RM7102 | 12 | 1,32,11,325 | 0.38 | 8 | 0.72 | 0.01 | 0.68 | SSR | GDB |
| 144 | R12M27 | 12 | 1,73,79,856 | 0.77 | 2 | 0.35 | 0 | 0.29 | InDel | PBS |
| 145 | RM2972 | 12 | 1,91,38,384 | 0.45 | 7 | 0.71 | 0 | 0.67 | SSR | GDB |
| 146 | RM519 | 12 | 1,99,03,791 | 0.36 | 7 | 0.78 | 0 | 0.75 | SSR | GDB |
| 147 | R12M33 | 12 | 2,00,06,587 | 0.55 | 2 | 0.5 | 0 | 0.37 | InDel | PBS |
| 148 | R12M43 | 12 | 2,61,02,497 | 0.89 | 2 | 0.19 | 0.01 | 0.17 | InDel | PBS |
| Mean | 0.49 | 8.75 | 0.62 | 0.01 | 0.58 |
aPolymorphic Information Content
bPhysical locations of SSR markers were taken from Gramene: a genomics and genetics resource for rice marker database’ (https://www.gramene.org/markers/microsat/) whereas physical locations of InDel markers were accessed from MSU Rice Genome Annotation Project (https://rice.plantbiology.msu.edu/analyses_search_blast.shtml) based on the BAC accession ID given in the published study of Shen et al. (2004)
cSSR: Simple Sequence Repeats, InDel: Insertion Deletion,
dGDB: adopted from ‘Gramene: a genomics and genetics resource for rice marker database’ (https://www.gramene.org/markers/microsat/), PBS: adopted from the published study of Shen et al. (2004), MD: manually designed for this study
PCR amplification and electrophoresis analysis
PCR reaction was performed in a 10 µl reaction mixture containing 4 µl (2.5 ɳg/µl) template DNA, 2 µl of 5 × assay buffer, 2 mM MgCl2 (Promega, Madison, USA), 0.2 µM of each forward and reverse SSR/InDel primer, 200 µM dNTPs (Roche, Indianapolis, USA) and 1.5 U of Taq DNA polymerase (BRIT, Mumbai, India). PCR reactions were performed in a thermal-cycler (Eppendorf, Hamburg, Germany). The amplification profile consisted of initial denaturation for 5 min at 95 °C; 10 cycles of 40 s denaturation at 95 °C, 30 s annealing at 60 °C followed by a decrement of temperature @ − 0.5 °C per cycle (for 10 cycles) and 40 s extension at 72 °C. Remaining 25 cycles were used to amplify DNA with denaturation at 94 °C for the 40 s, 40 s annealing at 55 °C and amplification for 40 s at 72 °C. After that, the final extension was carried out at 72 °C for 7 min. PCR products of SSR markers were resolved on a capillary gel electrophoresis system (Qiagen, Hamburg, Germany). Whereas PCR products of InDel markers were resolved on 2.5% agarose gel (containing 1.75% Methaphor® agarose + 0.75% normal agarose) (Sigma-Aldrich, Missouri, USA). Initially, allele size of all the InDel primer pairs was checked on capillary gel electrophoresis (Qiagen, Hamburg, Germany).
Genetic diversity parameters and informativeness of the molecular markers
Marker utility information and genetic diversity parameters such as the number of alleles per locus, Landraces (proportion) showing > 1 alleles, major allele frequency, gene diversity and the polymorphic information content (PIC) for each marker were estimated by using software POWERMARKER v3.25 (Liu and Muse 2005).
Analysis of population structure and genetic differentiation among the rice landraces
Population structure of the 190 rice landraces based on 148 molecular markers was analyzed by following the Bayesian statistical model in the program STRUCTURE v2.3.4 (Pritchard et al. 2000). The run-length was specified as 100,000 burning period followed by 100,000 MCMC replication with an admixture model and correlated allele frequencies for inferring the number (K) of subpopulations, ranging from K = 1 to 10. The analysis was performed using five replicates per K value. The result generated from structure analysis was used to identify optimum K value based on LnP(D) and Evanno’s ΔK method in STRUCTURE HARVESTER (Earl 2012). The genotypes with the probability of more than ≥ 0.80 scores (inferred ancestry) were considered as pure and less than 0.80 scores (inferred ancestry) as an admixture (Supplementary Table 1).
Genetic differentiation among subpopulations was estimated by analysis of molecular variance (AMOVA) using software GenAlEx 6.503 (Smouse et al. 2015). Unrooted neighbor joining (NJ) tree based on Jaccard similarity coefficient and UPGMA algorithm was constructed by SAS v9.4 software (SAS, Inc., Cary, NC, USA). A principal component analysis (PCA) for assessing genetic diversity among landraces was conducted based on variance–covariance matrix using PAST v3.15 software (Hammer et al. 2001). The principal component axes were drawn according to eigenvalues. The coefficients of kinship between pairs of accessions were determined using the data from above, 148 markers in TASSEL v3.0 software (Bradbury et al. 2007).
Genome-wide association studies (GWAS) for grain nutritional and yield-related traits
Genome-wide association mapping was performed using the TASSEL v3.0 software (Bradbury et al. 2007) by following the mixed linear model (MLM) using 50,000-time permutations for the correction of multiple testing (Pritchard et al. 2000). MLM analysis was performed with molecular marker data, phenotypic data, population inferred ancestry (Q matrix) and kinship (K) matrix (Yu et al. 2006). Significant marker-trait associations were identified by considering the Bonferroni threshold and it was calculate by standard method in MSExcel (Moran, 2003). Bonferroni threshold level for this study was 1/148 = 0.0067, where 148 is the number of markers used for association tests for each trait in this study. Therefore, QTLs with the probability p value 0.0067 or less than 0.0067 were considered as significant.
Results
Phenotypic variation in grain yield and nutritional traits among the rice landraces
The factors imparting variance in the population for studied nutritional and yield-related traits laid out in augmented block design were duly analyzed under unidirectional elimination of heterogeneity separately for blocks and treatments respectively during both years (Supplementary Table 3a, b). The critical differences (CD) per se briefed the response of the augmented treatments for all the studied traits. The CD of controls confirms the uniform contrast expression of traits across the blocks. CD of augmented treatment between the blocks, being higher with respect to within the blocks for all the traits during both years, implicitly inferred that environmental heterogeneity was insignificant within the blocks for all the traits. Comparable CD for the augmented treatments within block and between control and augmented treatment for all the traits during both years also infer lesser micro-environment effect within the blocks.
Under block adjustment, variance due to augmentation, control versus augmentation and block per se were found to be significant for most of the traits during both years. It implies that considerable environmental interaction effect contributes to the overall variation for studied traits. Under heterogeneity elimination for the treatments and control + control versus augmentation showed significant variance for most of the traits during both the years. The controls were found to be significant and equal mean sum of squares (MSS) under both cases heterogeneity elimination (adjustment) and non-adjustment for most of the traits during both years. It implied that some fixed effect was making the controls to stabilize the performance. The MSS of the treatments was found to be significant and comparable both under adjustment of blocks and treatment per se for most of the traits during both years, which was an intriguing result. Significant variation in the population for most of the studied traits due to genotypic effect (treatment) under both the cases of heterogeneity elimination and equivalent MSS of the treatments indicated towards the sound genetic component of the respective trait (Supplementary Table 3a, b). The residual MSS was observed fewer for all the traits during both years, indicated the minor difference in phenotypic variance and genotypic variance of the traits. It implied the less environmental contribution in the total variance. Lower value of the residual MSS under both systems of heterogeneity elimination during both the years confirmed the precision of the experimental set-up, methodology and research findings.
Testing of normal distribution by Shapiro–Wilk test of pooled data revealed that panicle length (W = 0.995, p = 0.804), grain yield/plant (W = 0.988, p = 0.117), stearic acid (W = 0.986, p = 0.075) and linolenic acid (W = 0.991, p = 0.327) were following the normal distribution pattern (Supplementary Fig. 2). This indicated the quantitative nature of traits which are controlled by many genes. Mean, range and standard deviation of all rice landraces for all the traits revealed a wide range of phenotypic variation. More than 10% coefficient of variation among the landraces for all the traits were recorded except for palmitic acid, oleic acid, and linoleic acid. Heritability in the broad sense was recorded very high (> 70%) for all the traits except the oleic acid and linoleic acid which showed moderate heritability (Table 2).
Table 2.
Summary statistics and genetic parameters of variation for grain nutritional and yield attributing traits in rice landraces
| Traits | Year | Mean | Range | Standard deviation | Coefficient of variation (%) | Heritability (bs) | |
|---|---|---|---|---|---|---|---|
| Minimum | Maximum | ||||||
| Days to 50% Flowering (DFF) | 2015–2016 | 112.41 | 75.45 | 131.25 | 13.56 | 12.06 | 0.97 |
| 2016–2017 | 111.94 | 72.00 | 135.00 | 12.52 | 11.18 | 0.98 | |
| Pooled | 112.18 | 73.73 | 133.63 | 12.81 | 11.42 | 0.98 | |
| Plant height (cm) (PH) | 2015–2016 | 148.66 | 82.20 | 175.46 | 21.91 | 14.74 | 0.96 |
| 2016–2017 | 143.84 | 86.00 | 172.00 | 19.76 | 13.74 | 0.94 | |
| Pooled | 146.25 | 85.10 | 170.26 | 20.42 | 13.96 | 0.95 | |
| Total Tiller/plant (TTP) | 2015–2016 | 9.87 | 5.60 | 19.80 | 2.81 | 28.47 | 0.79 |
| 2016–2017 | 10.06 | 5.00 | 19.00 | 2.56 | 25.45 | 0.74 | |
| Pooled | 9.97 | 6.20 | 18.50 | 2.46 | 24.67 | 0.77 | |
| Prod Tiller/plant (PTP) | 2015–2016 | 9.52 | 4.96 | 17.86 | 2.52 | 26.47 | 0.75 |
| 2016–2017 | 9.33 | 4.00 | 18.00 | 2.60 | 27.87 | 0.75 | |
| Pooled | 9.43 | 5.38 | 16.83 | 2.29 | 24.28 | 0.75 | |
| Panicle length (cm) (PL) | 2015–2016 | 25.60 | 15.59 | 35.35 | 3.06 | 11.95 | 0.92 |
| 2016–2017 | 25.01 | 17.00 | 34.05 | 3.09 | 12.36 | 0.91 | |
| Pooled | 25.31 | 16.97 | 34.70 | 2.92 | 11.54 | 0.92 | |
| Fertile spikelets/plant (FSP) | 2015–2016 | 133.18 | 28.31 | 353.72 | 53.64 | 40.28 | 0.99 |
| 2016–2017 | 132.98 | 23.31 | 359.72 | 54.47 | 40.96 | 0.99 | |
| Pooled | 133.08 | 25.81 | 356.72 | 53.90 | 40.50 | 0.99 | |
| Sterile spikelets/plant (SSP) | 2015–2016 | 44.25 | 5.91 | 114.65 | 21.90 | 49.49 | 0.98 |
| 2016–2017 | 45.83 | 4.58 | 120.65 | 23.53 | 51.34 | 0.99 | |
| Pooled | 45.04 | 6.58 | 117.65 | 22.49 | 49.93 | 0.99 | |
| Total spikelets/plant (TSP) | 2015–2016 | 177.43 | 48.82 | 468.37 | 65.15 | 36.72 | 0.99 |
| 2016–2017 | 178.81 | 39.82 | 480.37 | 66.79 | 37.35 | 0.98 | |
| Pooled | 178.12 | 44.32 | 474.37 | 65.71 | 36.89 | 0.99 | |
| Spikelet fertility % (SF%) | 2015–2016 | 74.68 | 40.65 | 92.30 | 9.69 | 12.98 | 0.96 |
| 2016–17 | 74.03 | 35.49 | 94.68 | 10.59 | 14.31 | 0.95 | |
| Pooled | 74.36 | 39.16 | 92.19 | 9.99 | 13.43 | 0.96 | |
| 100 seed weight (g) (HSP) | 2015–2016 | 2.49 | 1.25 | 3.67 | 0.58 | 23.29 | 0.98 |
| 2016–2017 | 2.50 | 1.03 | 3.67 | 0.58 | 23.20 | 0.98 | |
| Pooled | 2.50 | 1.14 | 3.59 | 0.57 | 22.80 | 0.98 | |
| Grain Yield (g/plant)(GY) | 2015–2016 | 30.01 | 10.63 | 43.83 | 6.08 | 20.26 | 0.95 |
| 2016–2017 | 29.84 | 10.78 | 43.46 | 6.19 | 20.74 | 0.96 | |
| Pooled | 29.92 | 12.24 | 41.83 | 5.85 | 19.55 | 0.96 | |
| Iron content (ppm) (Fe) | 2015–2016 | 11.31 | 7.25 | 22.30 | 2.43 | 21.49 | 0.95 |
| 2016–2017 | 11.26 | 7.00 | 23.21 | 2.44 | 21.67 | 0.95 | |
| Pooled | 11.28 | 7.13 | 22.76 | 2.43 | 21.54 | 0.95 | |
| Zinc content (ppm)(Zn) | 2015–2016 | 25.19 | 14.80 | 44.05 | 4.34 | 17.23 | 0.94 |
| 2016–2017 | 25.14 | 14.70 | 44.30 | 4.36 | 17.34 | 0.95 | |
| Pooled | 25.16 | 14.75 | 44.18 | 4.35 | 17.29 | 0.95 | |
| Palmitic acid (%) (PA) | 2015–2016 | 16.59 | 12.49 | 20.44 | 1.29 | 7.78 | 0.71 |
| 2016–2017 | 18.46 | 11.14 | 23.25 | 2.32 | 12.57 | 0.93 | |
| Pooled | 17.53 | 12.59 | 20.25 | 1.34 | 7.64 | 0.82 | |
| Stearic acid (%) (SA) | 2015–2016 | 1.68 | 0.56 | 2.81 | 0.36 | 21.43 | 0.88 |
| 2016–2017 | 1.91 | 1.05 | 2.09 | 0.43 | 22.51 | 0.93 | |
| Pooled | 1.80 | 1.01 | 2.90 | 0.29 | 16.11 | 0.91 | |
| Oleic acid (%) (OA) | 2015–2016 | 44.33 | 40.29 | 50.25 | 1.62 | 3.65 | 0.35 |
| 2016–2017 | 43.39 | 38.22 | 52.66 | 2.86 | 6.59 | 0.67 | |
| Pooled | 43.86 | 37.60 | 49.17 | 1.83 | 4.17 | 0.51 | |
| Linoleic acid (%) (LA) | 2015–2016 | 36.11 | 32.33 | 41.28 | 1.33 | 3.68 | 0.31 |
| 2016–2017 | 35.12 | 28.00 | 43.47 | 3.46 | 9.85 | 0.69 | |
| Pooled | 35.61 | 31.55 | 44.67 | 1.88 | 5.28 | 0.50 | |
| Linolenic acid (%) (LnA) | 2015–2016 | 1.28 | 0.49 | 2.07 | 0.21 | 16.41 | 0.98 |
| 2016–2017 | 1.07 | 0.42 | 1.70 | 0.25 | 23.36 | 0.98 | |
| Pooled | 1.18 | 0.55 | 1.83 | 0.17 | 14.41 | 0.98 | |
Phenotypic correlation analysis
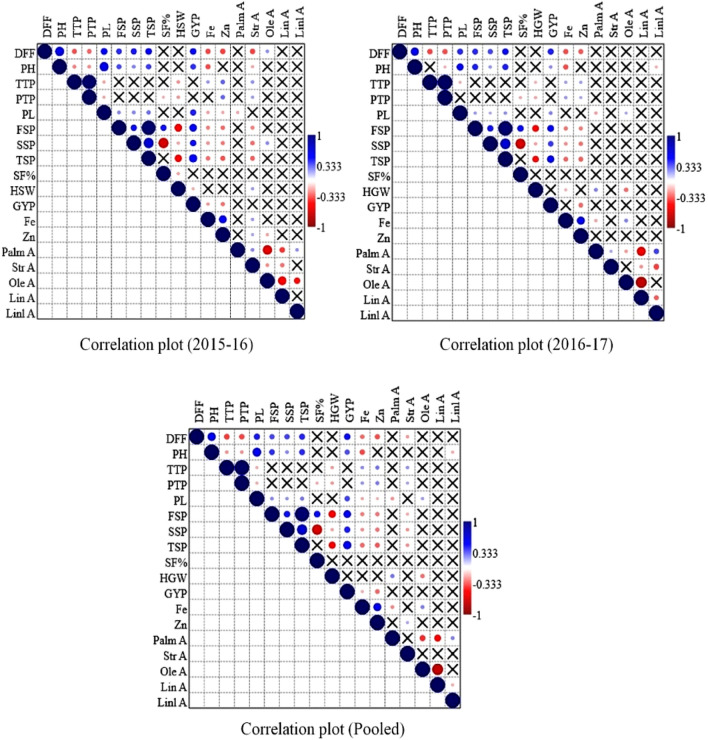
Present study revealed significant (p < 0.05) positive correlations of DFF with PH, PL, FSP, SSP, TSP and GY; PH with PL, FSP, SSP, TSP and GY; TTP with Fe and Zn; PL with FSP, SSP, TSP and GY; TSP with GY; Fe content with Zn content during both years and pooled analysis. Furthermore, significant (P < 0.05) negative correlations were found between DFF with TTP, PTP, Fe, Zn and SA; PH with Fe content; PL with PA; FSP with HSP, Fe and Zn content; SSP with SF%, Fe and Zn content; GY with Fe and Zn content; PA with OA and LA; OA with LA (Fig. 1).
Fig. 1.
Correlations among the grain nutritional and yield related traits in rice landraces evaluated in two years along with pooled analysis. The blue colored circle shows the positive association, red colored circle indicates the negative association and the intensity of the color indicates degree of association (as explained in the right side of the each diagram). Cross sign shows no correlation between traits. The test of significance was calculated based on 0.05 probability level. DFF days to 50% flowering, PH plant height, TTP total tillers/plant, PTP productive tillers/plant, PL panicle length, FSP fertile spikelets/panicle, SSP sterile spikelets/panicle, TSP total spikelets/panicle, SF% spikelet fertility percent, HSW-100 seed weight, GY grain yield, Fe iron content, Zn zinc content, PA palmitic acid, SA stearic acid, OA oleic acid, LA linoleic acid and LnA linolenic acid
Genetic diversity parameters and informativeness of the molecular markers
Total 190 rice landraces were used for genotyping with 148 molecular markers (107 SSRs and 41 InDels markers). A total of 1294 bands (alleles) were amplified among tested landraces. The number of alleles per locus varied from 2 to 38 (RM26981) with an average of 8.7 alleles per locus. Polymorphic Information Content (PIC) value ranged from 0.01 (R2M10, R8M23, R10M40) to 0.96 (RM26981) with an average of 0.58. The percentage of landraces that were found to exhibit more than one allele ranged from 0 to 0.06 (R10M10) with an average of 0.01 which showed that several loci did not detect any level of heterozygosity. Expected heterozygosity or gene diversity (He) computed according to Nei (1973) varied from 0 to 0.96 (RM26981) with an average of 0.62. Genotype number ranged from 1 to 41 (RM26981) with an average of 9.77. The major allele frequency ranged from 0.09 (RM1388) to 1 (R10M40) with an average of 0.49 (Table 1).
Population structure and genetic diversity analysis of rice landraces
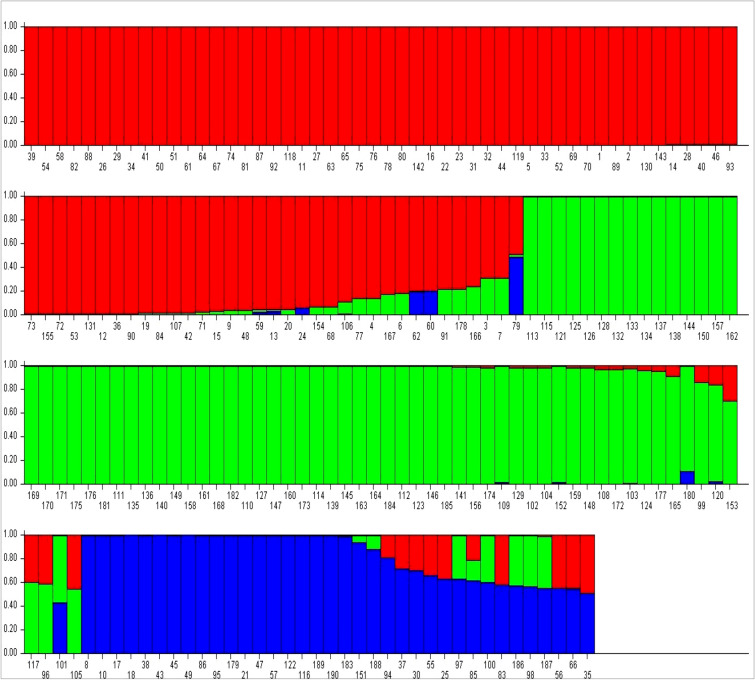
Based on both LnP(D) and Evanno’s ΔK values, optimum K value was found as three (Supplementary Fig. 3). This indicated that 190 rice landraces had a genetic structure of three sub-populations viz., SG-1, SG-2 and SG-3. The SG1, SG2 and SG3 had total 77 (40.52%), 64 (33.68%) and 22 (11.57%) pure genotypes whereas had eight, five and 14 admixture genotypes (14.21%), respectively (Supplementary Table 1 and Fig. 2).
Fig. 2.
The bar plot presentation the genetic structure of 190 rice landraces based on population stratification at three sub-population level (K = 3). Red colored bar shows the genotypes of Sub Group-1, green colored bar shows the genotypes of Sub Group-2 and Blue colored bar shows the genotypes of Sub Group-3. Bars with mixed colored represent the mixed ancestry of genotypes (admixtures). X axis shows the serial number of genotypes and Y axis shows the membership proportion of genotype on respective sub-group
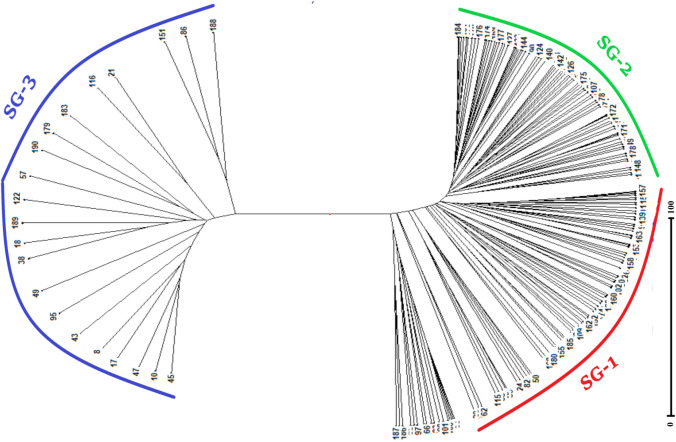
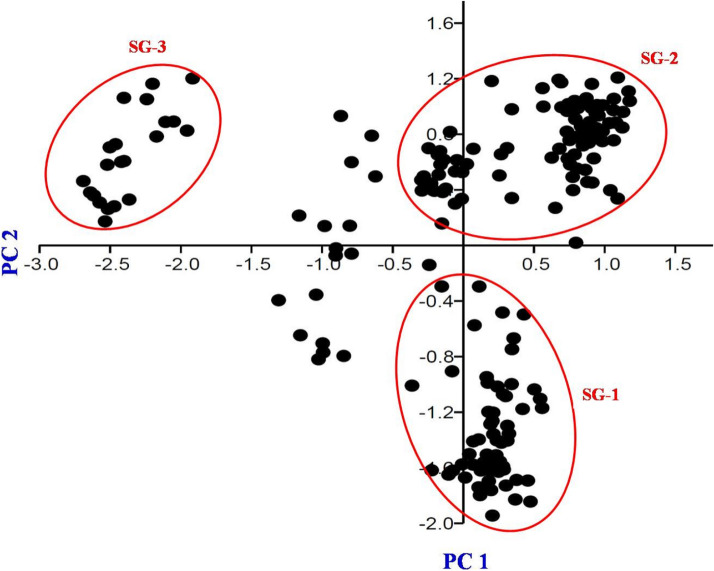
Population-specific mean Fst value of the three subgroups SG-1, SG-2 and SG-3 was recorded 0.10, 0.22 and 0.17, respectively, with an average of 0.165. The mean alpha value of the population was 0.043, indicating the less number of admixture type genotypes. The allele frequencies (divergence among sub-populations based on net nucleotide distance) were 0.09 between SG-1 and SG-2; 0.16 between SG-1 and SG-3 and 0.20 between SG-2 and SG-3. Further, analysis of molecular variance (AMOVA) detected a significant genetic differentiation (ɸPT = 0.162, p = 0.000) between three sub-populations. Total molecular variance was partitioned into two, of which, 16% explained variation among populations and the remaining 84% explained variation within the populations (Supplementary Table 4). Furthermore, the results of the unrooted neighbor-joining tree (NJ tree) and principal component analysis (PCA) were coinciding with the result of model based population structure analysis. NJ tree (Fig. 3) and PC scatter diagram (Fig. 4) grouped the rice landraces into three main clusters/groups. Genotypes of different clusters/groups are well matched with the genotypes of different sub groups of population structure analysis.
Fig. 3.
Neighbor joining (NJ) tree based on UPGMA algorithm and Jaccard’s similarity coefficient showing the pattern of grouping of 190 rice landraces. Rice landraces were grouped into three clusters as marked in NJ tree. SG-1, SG-2 and SG-3 represent the genotypes of cluster-1, cluster-2 and cluster-3, respectively. Genotypes of cluster-3 (SG-3) are clearly distinguished from cluster-1 (SG-1) and cluster-2 (SG-2). The serial number against each spoke of the figure represents genotype as per the supplementary Table 1
Fig. 4.
Principal Component (PC) scatter diagram showing the grouping pattern of 190 rice landraces based on the PC1 and PC2. X axis represents the PC1 and Y axis represents the PC2. Rice landraces were grouped into three sub-groups as marked with red coloured circle. SG-1, SG-2 and SG-3 represent the genotypes of subgroup-1, subgroup-2 and subgroup-3, respectively. Genotypes of subgroup-3 (SG-3) are clearly distinguished from subgroup-1 (SG-1) and subgroup-2 (SG-2)
Effect of Mixed Linear Model (MLM) for controlling type-1 error
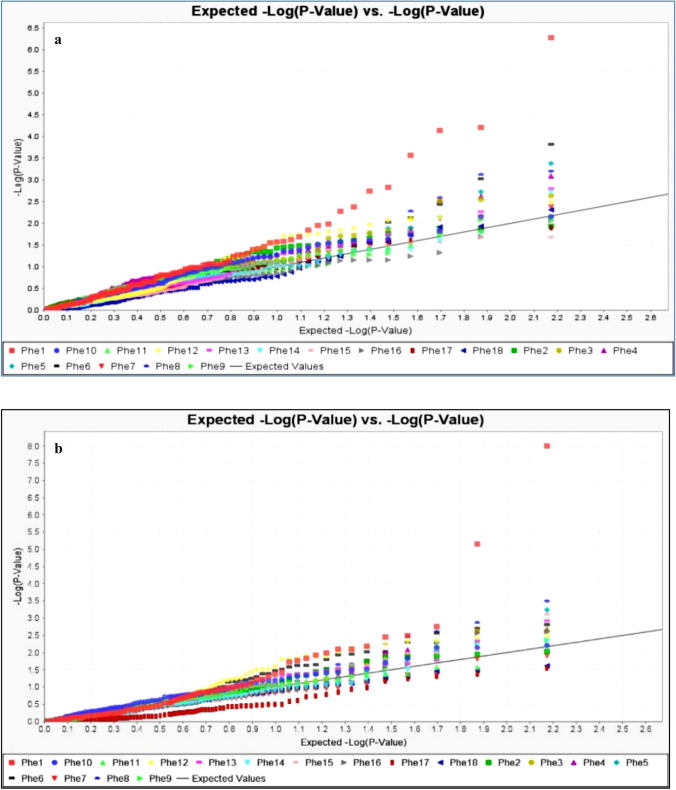
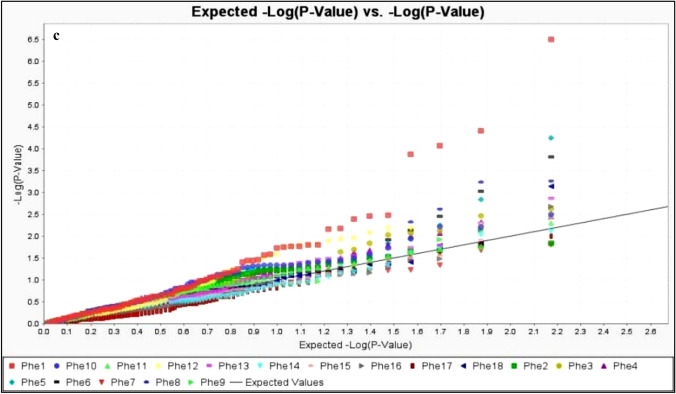
Observed versus expected p values for each marker-trait association were plotted in the quantile–quantile plot (Q–Q plot) to assess the control of type-I errors. Uniform distributions of the observed and expected p values for all traits were observed during both the years. The false positives were well controlled by MLM model which indicates the robustness of results obtained through statistical analysis (Fig. 5a–c).
Fig. 5.
a Quantile–Quantile (QQ) plot of observed versus expected p value for all the marker-trait association (MTA) identified by GWAS from the experimental year 2015. Y axis shows the observed − log10(p values) and X axis shows the expected − log10(p values) for all the identified MTAs. Each trait has assigned a colored shape (different for each trait). b Quantile–Quantile (QQ) plot of observed versus expected p value for all the marker-trait association (MTA) identified by GWAS from the experimental year 2016. Y axis shows the observed − log10(p values) and X axis shows the expected − log10(p values) for all the identified MTAs. Each trait has assigned a colored shape (different for each trait). c Quantile–Quantile (QQ) plot of observed versus expected p value for all the marker-trait association (MTA) identified by GWAS from the pooled analysis. Y axis shows the observed − log10(p values) and X axis shows the expected − log10(p values) for all the identified MTAs. Each trait has assigned a coloured shape (different for each trait). Phe1 days to 50% flowering, Phe2 plant height, Phe3 total tillers/plant, Phe4 productive tillers/plant, Phe5 panicle length, Phe6 fertile spikelets/panicle, Phe7 sterile spikelets/panicle, Phe8 total spikelets/panicle, Phe9 spikelet fertility percent, Phe10 hundred seed weight, Phe11 grain yield, Phe12 iron content, Phe13 zinc content, Phe14 palmitic acid, Phe15 stearic acid, Phe16 oleic acid, Phe17 linoleic acid, Phe18 linolenic acid
Identification and mapping of QTLs for grain yield and nutritional traits through genome-wide association studies
Genome-wide association studies (GWAS) revealed total 22 consistent and significant QTLs for 12 traits (out of 18 traits investigated) based on 2 years and pooled analysis (Table 3). Of which, total 14 significant and consistent QTLs were detected for grain yield and attributing traits during both years and pooled analysis which are further explained here. DFF was significantly associated with RM1148 (chr-8), RM205 (chr-9), and R11M23(chr-11) in all three conditions. Association of RM118 (chr-7) with TTP was significant and consistent. Two InDel markers R11M23 (chr-11) and R12M43 (chr-12) were significantly linked with PL. TSP, an important yield contributing traits was significantly associated with three markers, RM234 (chr-7), R11M23 (chr-11) and R1M47 (chr-1). Furthermore, it was found that SSR markers RM234 (chr-7) significantly associated with fertile spikelets/panicle during both years and pooled analysis. Another marker RM184(chr-10) was also found to have association with spikelet fertility%. HSW was significantly associated with two markers R3M10 (chr-3) and R9M42 (chr-9) in all the three tested conditions. It was found that grain yield per plant was significantly associated with RM7434 (chr-6) with more than 10% explained phenotypic variance (R2-value). No significant and consistent associations were detected for PH and PTP. Interestingly, the InDel marker R11M23 was significantly associated with three traits viz., DFF, PL and TSP in this study.
Table 3.
List of Significant Marker Trait Associations (MTAs) identified under the study during both the years and pooled analysis along with reports of previous findings
| Traits | Year | Marker name | Chromosome number | p_marker value | R2 _ Marker | Reports of QTLs for same or similar traits |
|---|---|---|---|---|---|---|
| Days to fifty % flowering (DFF) | 2015–2016 | RM1148 | 8 | 5.264E–07 | 0.260 | – |
| RM205 | 9 | 6.15E–05 | 0.318 | – | ||
| R11M23 | 11 | 7.23E–05 | 0.087 | – | ||
| 2016–2017 | RM1148 | 8 | 1.01E–08 | 0.323 | – | |
| RM205 | 9 | 6.88E–06 | 0.367 | – | ||
| R11M23 | 11 | 0.0066 | 0.040 | – | ||
| Pooled | RM1148 | 8 | 3.16E–07 | 0.267 | – | |
| R11M23 | 11 | 3.91E–05 | 0.094 | – | ||
| RM205 | 9 | 8.63E–05 | 0.311 | Suh et al. (2005) (qDTH9) | ||
| Total tillers/plant (TTP) | 2015–2016 | RM118 | 7 | 0.00148 | 0.085 | – |
| 2016–17 | RM118 | 7 | 0.0061 | 0.097 | – | |
| Pooled | RM118 | 7 | 0.0024 | 0.112 | – | |
| Panicle length (PL) | 2015–2016 | R12M43 | 12 | 4.22E–04 | 0.107 | – |
| R11M23 | 11 | 0.00190 | 0.053 | – | ||
| 2016–2017 | R12M43 | 12 | 5.73E–04 | 0.103 | – | |
| R11M23 | 11 | 0.0060 | 0.131 | – | ||
| Pooled | R12M43 | 12 | 5.75E–05 | 0.110 | – | |
| R11M23 | 11 | 0.0015 | 0.056 | – | ||
| Fertile spikelets/Plant (FSP) | 2015–2016 | RM234 | 7 | 1.50E–04 | 0.197 | Hua et al. (2002) (gp7b) |
| 2016–2017 | RM234 | 7 | 0.0015 | 0.160 | ||
| Pooled | RM234 | 7 | 1.54E–04 | 0.196 | ||
| Total spikelets/Plant (TSP) | 2015–2016 | R11M23 | 11 | 6.28E–04 | 0.064 | – |
| RM234 | 7 | 7.62E–04 | 0.172 | – | ||
| R1M47 | 1 | 0.00261 | 0.065 | – | ||
| 2016–2017 | RM234 | 7 | 3.14E–04 | 0.187 | – | |
| R11M23 | 11 | 0.0013 | 0.057 | – | ||
| R1M47 | 1 | 0.0056 | 0.057 | – | ||
| Pooled | R11M23 | 11 | 5.53E–04 | 0.066 | – | |
| RM234 | 7 | 5.84E–04 | 0.176 |
Xing et al. (2001) (sp7b & sp7c) Feng et al. (2015) (qSP-7c) |
||
| R1M47 | 1 | 0.0024 | 0.066 | – | ||
| Spikelet fertility % (SF%) | 2015–2016 | RM184 | 10 | 0.00643 | 0.108 | – |
| 2016–2017 | RM184 | 10 | 0.0043 | 0.105 | – | |
| Pooled | RM184 | 10 | 0.0038 | 0.107 | – | |
| Hundred seed weight (HSW) | 2015–2016 | R3M10 | 3 | 0.00610 | 0.040 | – |
| R9M42 | 9 | 0.00610 | 0.040 | – | ||
| 2016–17 | R3M10 | 3 | 0.0061 | 0.140 | – | |
| R9M42 | 9 | 0.0061 | 0.140 | – | ||
| Pooled | R3M10 | 3 | 0.0061 | 0.041 | – | |
| R9M42 | 9 | 0.0061 | 0.041 | – | ||
| Grain yield/Plant (GY) | 2015–2016 | RM7434 | 6 | 0.00179 | 0.102 | – |
| 2016–17 | RM7434 | 6 | 0.0058 | 0.108 | – | |
| Pooled | RM7434 | 6 | 0.0050 | 0.110 | – | |
| Iron content (Fe) | 2015–2016 | RM536 | 11 | 0.00327 | 0.138 | – |
| RM17467 | 4 | 0.00601 | 0.304 | – | ||
| RM484 | 10 | 0.00639 | 0.076 | – | ||
| RM26063 | 11 | 0.00637 | 0.244 | – | ||
| 2016–2017 | RM536 | 11 | 0.0033 | 0.138 | – | |
| RM17467 | 4 | 0.0045 | 0.316 | – | ||
| RM26063 | 11 | 0.0047 | 0.258 | – | ||
| RM484 | 10 | 0.0057 | 0.079 | – | ||
| Pooled | RM536 | 11 | 0.0033 | 0.138 | – | |
| RM17467 | 4 | 0.0056 | 0.310 | – | ||
| RM26063 | 11 | 0.0062 | 0.251 | – | ||
| RM484 | 10 | 0.0065 | 0.077 | – | ||
| Zinc content (Zn) | 2015–2016 | RM44 | 8 | 0.00158 | 0.148 | – |
| RM6839 | 9 | 0.00541 | 0.090 | – | ||
| 2016–2017 | RM44 | 8 | 0.0012 | 0.153 | – | |
| RM6839 | 9 | 0.0047 | 0.092 | – | ||
| Pooled | RM44 | 8 | 0.0014 | 0.150 | – | |
| RM6839 | 9 | 0.0051 | 0.091 | – | ||
| Oleic acid (OA) | 2015–2016 | RM25 | 8 | 0.00595 | 0.134 | – |
| 2016–2017 | RM25 | 8 | 0.0024 | 0.129 | – | |
| Pooled | RM25 | 8 | 0.0021 | 0.131 | – | |
| Linolenic acid (LnA) | 2015–2016 | RM495 | 1 | 0.00503 | 0.122 | – |
| 2016–2017 | RM495 | 1 | 0.0015 | 0.104 | – | |
| Pooled | RM495 | 1 | 7.43E–04 | 0.151 | – |
Similarly, totally eight significant and consistent QTLs were detected for studied grain nutritional traits. GWAS based marker-trait association analysis revealed four common markers (RM536 at chr-11, RM17467 at chr-4, RM484 at chr-10 and RM26063 at chr-11) for grain Fe content; two (RM44 at chr-8 and RM6839 at chr-9) for grain Zn content; one (RM25 at chr-8) for oleic acid content and one (RM495 at chr-1) for linolenic acid content based on 2 years and pooled analysis. Two markers RM536 and RM26063 linked with Fe content on chromosome-11 were closely linked and were 6.73 Mb apart from each other. Similarly, Zn content was associated with RM44, which is closely spaced with QTL qZn8-1. Moreover, significant QTL for oleic acid content was found around RM25 that was closely located with QTL qOle8 in chromosome 8. None of the markers were significantly and consistently associated with linoleic acid, palmitic acid and stearic acid.
Interestingly, 13 QTLs out of 22, explained more than 10% phenotypic variation (R2 value) for respective traits. Of which, three (RM536, RM1746, RM484) for Fe content; two (RM1148 and RM205) for DFF and one for PL(R12M43), FSP(RM234), TSP (RM234), SF% (RM184), GY (RM7434), Zn(RM44), OL(RM25) and LnL (RM495) were found major QTLs in the present study (Table 3). Of the 22 identified associated QTLs, 19 are novel and reported first time in this study.
Discussion
Since rice is the staple food crop of the developing world, a lot of efforts are being made to develop nutritionally enriched high yielding genotypes. The first pre-requisite for such breeding program is to explore the genetic variation and diversity in available germplasm, which can be used later in hybridization, genetic studies, marker development, and marker-trait association studies. Natural population with high phenotypic variation is crucial for the success of GWAS study. Sufficient genetic variation among the augmented landraces for all the traits were recorded through ANOVA, making of population appropriate for this study. Minor differences in phenotypic variance and genotypic variance for all the traits during both the years indicated the less influence of environment. CD values for controls over the blocks, CD of augmented treatment between the blocks, CD for augmented treatment within the blocks and CD for control and augmented treatment for all the studied traits during both years revealed the insignificant environmental heterogeneity and lesser micro-environment effect (Supplementary Table 3a, b). These all indicated the precision of the experimental methodology and research finding. Mondal et al. (2017) also implied augmented block design in 300 Recombinant Inbred Lines (RILs) for QTL mapping for early ground cover in wheat (Triticum aestivum L.) under drought stress condition and reported significant and reliable informations.
Presence of abundant variability for studied traits might be due to diverse landraces possessing several unknown natural recombinations and mutations accumulated over many generations. Coefficient of variation was also higher for most of the agronomic (Agrama et al. 2007; Swamy et al. 2017; Zhang et al. 2019) and nutritional traits (Norton et al. 2014; Descalsota et al. 2018; Huang et al. 2015).This provides a good opportunity to select the superior genotypes for grain yield and nutritional traits to be utilized in breeding programs. Most of the traits showed high heritability, indicates the less influence of environmental factors. Direct selections in breeding program would be rewarding for improvement of these traits. Present study revealed significant positive correlation of GY with most of the yield attributing traits. However, GY showed negative or no correlation with nutritional traits. This negative or no correlation may be even more likely due to the increased starch content in seed that dilute concentration of nutritive compounds (Rabson et al. 1978). The higher negative correlation between oleic acid and linoleic acid exist due to possible natural mutation in oleyl phosphatidyl choline desaturase gene which converts oleic acid to linoleic acid (Mondal et al. 2018).
Some prominent rice landraces have been identified for important traits through this study. Based on the observations for two years, landraces, Anjani and Maran Dhan (early maturity); Sihar and Kanakbhog (extra dwarf); Korma, Manmohan and Raja Banga (longest panicles); Sichar and Chhindmauri (highest spikelet fertility%); Gangabaru and Bhusu (highest grain yield per plant); Karhani and Bathrash (highest Fe content); Byalo-2 and Petgadi (highest Zn content); Kadamphool and Rudra Dhan (highest oleic acid content) and Gangachur and Antarved (highest linoleic acid content) were recognized in this study (Supplementary Table 5). Landraces identified for various traits may play an important role in rice breeding program for nurturing desirable genotypes.
SSR and InDel markers used in this study were distributed in entire 12 chromosomes, with an average of 12 markers per chromosome (ranging from 10 to 15 markers per chromosome). Average allele per marker or locus was high due to diverse landraces and usage of capillary gel electrophoresis to resolve the SSR alleles. Heterozygosity was found to be very low due to the autogamous nature of rice. Total 66 highly polymorphic, useful and informative markers were identified by considering the parameters of PIC value (> 0.70), gene diversity (> 0.7) and the number of polymorphic alleles (> 0.6) (Table 1). Amount of molecular diversity existing within the current population panel is comparable with the earlier reports in rice germplasm (Agrama et al. 2007; Swamy et al. 2017; Sahu et al. 2017; Descalsota et al. 2018; Norton et al. 2018; Zhang et al. 2019). The informative markers identified in this study may be further effectively used for various genetical studies in rice genotypes.
It is essential to determine the genetic structure of the population to reduce the possibilities of spurious associations in GWAS (Pritchard et al, 2000). Population stratification revealed three sub-populations (SG-1, SG-2, and SG-3) with few admixtures among the 190 landraces. Total 86% (163) genotypes separated into any one of the three subgroups, while 14% (27) genotypes showed allelic reshuffling or allelic sharing with genotypes of different subgroups. Few admixtures detected due to accumulation of spontaneous mutations in the genotypes from different geographical areas (Sahu et al. 2017; Swamy et al. 2017; Agrama et al. 2007). Based on the results of our previous experiment (Sahu et al. 2017), genotypes appeared in SG-3 contain some japonica oriented alleles while genotype of SG-1 and SG-2, carried indica-oriented alleles (Sahu et al. 2017). Genotypes of SG-3 were differentiated as ‘close to japonica’ type whereas genotypes of SG-1 and SG-2 were categorized under ‘indica type’ and ‘close to indica type’ category based on ‘InDel Molecular Index’ (Sahu et al. 2017). Richharia et al. (1960) also reported the allelic reshuffling between indica and japonica genotypes due to previous hybridization and migration phenomena in the landraces of Chhattisgarh state, which is very well depicted in the findings of present study. However, genotypes of SG-1 and SG-2 were not clearly distinguishable based on plant or grain morphology. Therefore, it is assumed that they may fall in two subgroups due to their different evolutionary pattern and adaptation behavior in the environment. Moderate to high genetic variation (based on Fst statistics) among sub-populations indicated the possibilities of generating super rice hybrids with better adaptability while crossing the genotypes of SG-3 with genotypes of SG-1 and SG-2. Earlier, several scientists also reported two to five sub-groups in rice population panel and assumed the evolutionary pattern and adaptation behavior to environment as main reason for differentiation (Agrama et al. 2007; Zhao et al. 2011; Swamy et al. 2017; Sahu et al. 2017; Juan et al. 2018; Kadam et al. 2018; Zhang et al. 2019). Interestingly, neighbor joining tree and principal component based scatter diagram showed the similar distribution of landraces in three sub-groups as explained in population structure analysis, indicating the robustness of population stratification.
Association mapping has shown great promise and power of mapping the complex quantitative traits in plants as compared to traditional bi-parental mapping. Several scientists employed GWAS in the rice population panel for various traits and reported novel QTLs (Suman et al. 2020; Haritha et al. 2020; Donde et al. 2020; Pradhan et al. 2019, 2020; Zhang et al. 2014, 2019; Norton et al. 2018; Kadam et al. 2018; Juan et al. 2018; Prasanth et al. 2017; Swamy et al. 2017; Xu et al. 2016; Lu et al. 2015; Zhao et al. 2011; Borba et al. 2010; Agrama et al. 2007). But such study in diverse rice landrace is very meager. Present study reported total 22 consistent and significant QTLs for 12 different grain yield and nutritional trait in those studied landraces. Out of 22 QTLs, association of three markers (RM205 with DFF; RM234 with FSP and RM234 with TSP) were previously reported in rice. While rest 19 QTLs are novel and reported first time in this study. Marker RM205 was significantly associated with QTL qDTH9 (days to heading) at position 173.5–177.7 cM in chromosome-9 (Suh et al. 2005). Similarly, Hua et al. (2002) reported the significant association of RM234 with QTL gp7b (grain/panicle) at 129 cM genomic positions of chromosome-7 which is comparable to present reports. Xing et al. (2001) and Feng et al. (2015) also reported the association of RM234 with QTLs qsp7b and qsp7c (total spikelets/panicle) and qSP-7c (spikelets/plant), respectively on chromosome-7. These four QTLs for three above traits have been validated co-incidentally through the present investigation.
Apart from these, some noteworthy results were also found in this study. SSR marker RM1148 (physical position: 3.739 Mb) consistently associated with DFF, is closely positioned with RM22475 (physical position: 4.288 Mb) in chromosome 8. RM22475 is linked to the Ghd8/DTH8 (days to heading) gene in rice (Wei et al. 2010). This study revealed an association of RM44 (chr-8) with Zn content in rice. Moreover, Xiang-Dong et al. (2016) reported the close linkage between markers RM44 and RM152 (chr-8) during the QTL mapping for cold tolerance in Oryza rufipogon Griff. at early seedling stage. Interestingly, marker RM152 was also reported to be associated with QTL qZn8-1 (Zn content) in another QTL analysis (Garcia-Oliveira et al. 2009). Until now, no attempts have been made to map QTLs for fatty acid components in rice by GWAS. Ying et al. (2012) identified a QTL qOle8 for oleic acid in the proximity of RM1148 on chromosome-8 by analyzing an F2 population. Later close linkage (4.4 cM) of RM25 and RM1148 was established by Xing et al. (2015) during the tagging of a new rice blast resistance gene in a RIL population. Thus, identification of RM25 as a significantly associated marker for oleic acid content in these rice landrace has immense opportunity to use this marker for the MAS program in the future breeding for high oleic acid content in rice bran. In addition, association of R11M23 with DFF, PL and TSP explained the possible occurrence of pleiotropism among these traits. Moreover, significant and positive correlations among these traits were also observed in the study (Fig. 1).
In GWAS, importance of QTLs depends on the explained phenotypic variance (R2 value) by the marker. Wide range (3.4–36.7%) of R2 value was obtained by the associated markers/QTLs. Of 22, 13 QTLs explained more than 10% phenotypic variation and considered as major QTLs. The wider range of phenotypic variance explained for most of the traits entail genome-wide significance and high statistical power of the current panel of 190 rice landraces.
Conclusion
Landraces, a reservoir of unique and important genes, are the most imperative component in the crop improvement program. Therefore, landraces identified for grain yield and nutritional traits can be utilized as donors in cross breeding, construction of mapping population or can be used to breed new cultivars through conventional breeding methods. Genetic dissection of rice landraces revealed abundant genetic variation and diversity for the grain micronutrients, fatty acids and yield-related traits with less influence of environment on their inheritance. Population stratification based grouping of rice panel offers a way to breed the super hybrids by crossing the divergent genotypes among and within different sub-groups (Fig. 2). This study reported 19 novel QTLs for 12 grain yield-related traits, micronutrients and fatty acid components while three earlier reported QTLs were also validated. We do hope that the present research work will open many avenues towards utilization of these QTLs and superior landraces in rice breeding for developing nutrition-rich high yielding varieties.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
Department of Science and Technology (DST), Ministry of Science and Technology, Government of India is sincerely acknowledged for providing financial support as INSPIRE fellowship to PKS. Indian Institute of Rice Research, Hyderabad is duly acknowledged for technical and scientific cooperation. Authors are also thankful to Head, NA&BTD; Associate Director, BSG, BARC, Mumbai and Head, Department of Genetics and Plant Breeding, IGKV, Raipur for technical support and cooperation during the experiment.
Author contributions
Conceptualization of work: DS, SM, BKD; Data curation: PKS, SM, GV, RS, VK; Data analysis: PKS, SM; Funding acquisition: DS, BKD; Investigation: PKS, SM; Methodology: PKS, SM, GV, VK; Resources: DS, SM, BKD, VK; Supervision: DS, BKD; Writing ± original draft: PKS, SM, RS; Review and editing: SM, DS, BKD, VK, GV. All authors read and approved the final manuscript.
Funding
Department of Science and Technology, Ministry of Science and Technology, Grant number [IF150523].
Compliance with ethical standards
Conflict of interest statement
The authors declare that they have no conflict of interest in the publication.
Contributor Information
Parmeshwar K. Sahu, Email: parmeshwarsahu1210@gmail.com, https://scholar.google.co.in/citations?user=FkLaQKkAAAAJ&hl=en
Suvendu Mondal, Email: suvenduhere@yahoo.co.in, https://scholar.google.com/citations?user=_IlDZdAAAAAJ&hl=en.
Richa Sao, Email: richasao.agro@gmail.com.
Gautam Vishwakarma, Email: gtmvish@barc.gov.in.
Vikash Kumar, Email: vikash007barc@gmail.com, https://www.researchgate.net/profile/Vikash_Kumar21.
B. K. Das, Email: bkdas@barc.gov.in
Deepak Sharma, Email: deepakigkv@gmail.com, https://scholar.google.com/scholar?hl=en&as_sdt=0%2C5&q=deepakigkv%40gmail.com&btnG.
References
- Agrama H, Eizenga G, Yan W. Association mapping of yield and its components in rice cultivars. Mol Breed. 2007;19(4):341–356. doi: 10.1007/s11032-006-9066-6. [DOI] [Google Scholar]
- Babu RV, Neeraja CN, Sanjeeva Rao D, Sundaram RM, Longvah T, Usharani G, Padmavathi G, Balachandran SM, Nirmala Devi G, Bhadana VP, Suneetha K, Rao KV, Surekha K, Sarla N, Brajendra P, Raghuveer Rao P, Girish C, Shashidhar HE, Bijan A. and Viraktamath BC (2014) Bioforotification in Rice. DRR Technical Bulletin No. 81/2014. Hyderabad: Directorate of Rice Research. p 86
- Borba C, Oliveria D, Brondani RPV, Breseghello F, Coelho ASG, Mendonca JA, Rangel PHN, Brondani C. Association mapping for yield and grain quality traits in rice (Oryza sativa L.) Genet Mol Biol. 2010;33:515–524. doi: 10.1590/S1415-47572010005000065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradbury PJ, Zhang Z, Kroon DE, Casstevens TM, Ramdoss Y, Buckler ES. TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics. 2007;23(19):2633–2635. doi: 10.1093/bioinformatics/btm308. [DOI] [PubMed] [Google Scholar]
- DelCruz N, Khush GS. Rice grain quality evaluation procedures. In: Singh RK, Singh US, Khush GS, editors. Aromatic rices. New Delhi: Oxford & IBH Publishing Co Pvt Ltd; 2000. pp. 16–28. [Google Scholar]
- Descalsota GIL, Swamy BPM, Zaw H, Asilo MAI, Amparado A, Mauleon R, Mohanty PC, Arocena EC, Raghavan C, Leung H, Hernandez JE, Lalusin AB, Mendioro MS, Diaz MGQ, Reinke R. Genome-wide association mapping in rice MAGIC plus population detects QTLs and genes useful for biofortification. Front Plant Sci. 2018;9(1347):1–20. doi: 10.3389/fpls.2018.01347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donde R, Mohapatra S, Baksh SKY, Padhy B, Mukherjee M, Roy S, Chattopadhyay K, Anandan A, Swain P, Sahoo KK, Singh ON, Behera L, Dash SK. Identification of QTLs for high grain yield and component traits in new plant types of rice. PLoS ONE. 2020;15(7):e0227785. doi: 10.1371/journal.pone.0227785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earl DA. STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour. 2012;4(2):359–361. doi: 10.1007/s12686-011-9548-7. [DOI] [Google Scholar]
- FAOSTAT (2020) https://www.fao.org/state-of-food-security-nutrition/en/. Accessed 12 May 2020
- Federer WT. Augmented (or Hoonuiaku) Designs. Hawaii Plr Rec. 1956;55:191–208. [Google Scholar]
- Feng Y, Zhai RR, Li ZC, Cao LY, Wei XH, Cheng SH. Quantitative trait locus analysis for rice yield traits under two nitrogen levels. Rice Sci. 2015;22(3):108–115. doi: 10.1016/j.rsci.2015.05.014. [DOI] [Google Scholar]
- Garcia-Oliveira AL, Tan L, Fu Y, Sun C. Genetic identification of quantitative trait loci for contents of mineral nutrients in rice grain. J Integr Plant Biol. 2009;51(1):84–92. doi: 10.1111/j.1744-7909.2008.00730.x. [DOI] [PubMed] [Google Scholar]
- Hammer Q, Harper DAT, Ryan PD. PAST: Paleontological statistics software package for education and data analysis. Palaeontol Electron. 2001;4(1):9. [Google Scholar]
- Haritha B, Yadav AK, Vinod KK, Gopala Krishnan S, Bhowmick PK, Nagarajan M, Neeraja CN, Ellur RK, Singh AK. Genome-wide association study reveals novel marker-trait associations (MTAs) governing the localization of Fe and Zn in the rice grain. Front Genet. 2020;11:213. doi: 10.3389/fgene.2020.00213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua JP, Xing YZ, Xu CG, Sun XL, Yu SB, Zhang Q. Genetic dissection of an elite rice hybrid revealed that heterozygotes are not always advantageous for performance. Genet. 2002;162(4):1885–1895. doi: 10.1093/genetics/162.4.1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Sun C, Min J, Chen Y, Tong C, Bao J. Association mapping of quantitative trait loci for mineral element contents in whole grain rice (Oryza sativa L.) J Agric Food Chem. 2015;63(50):10885–10892. doi: 10.1021/acs.jafc.5b04932. [DOI] [PubMed] [Google Scholar]
- Juan LR, Marques L, Talon M, Domingo C. Genome-wide association study of agronomic traits in rice cultivated in temperate regions. BMC Plant Biol. 2018;19:706. doi: 10.1186/s12864-018-5086-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadam NN, Struik PC, Rebolledo CM, Yin X, Jagadish SVK. Genome-wide association reveals novel genomic loci controlling rice grain yield and its component traits under water-deficit stress during the reproductive stage. J Exp Bot. 2018;69(16):4017–4032. doi: 10.1093/jxb/ery186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K, Muse SV. Power Marker: an integrated analysis environment for genetic marker analysis. Bioinformatics. 2005;21(9):2128–2129. doi: 10.1093/bioinformatics/bti282. [DOI] [PubMed] [Google Scholar]
- Lu Q, Zhang M, Niu X, Wang S, Xu Q, Feng Y, Wang C, Deng H, Yuan X, Yuan H, Wang Y, Wei X. Genetic variation and association mapping for 12 agronomic traits in indica rice. BMC Genom. 2015;16:1067. doi: 10.1186/s12864-015-2245-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondal BJ, Singh A, Yadav A, Tomar RSS, SinghPrakash VGSKV. QTL mapping for early ground cover in wheat (Triticum aestivum L.) under drought stress. Current Sci. 2017;112(6):1266–1271. doi: 10.18520/cs/v112/i06/1266-1271. [DOI] [Google Scholar]
- Mondal S, Nazareth J, Bhad PG, Badigannavar AM. Isolation of high oleate recombinants in peanut by near infra-red spectroscopy and confirmation with allele specific polymerase chain reaction marker. J American Oil Chem Soc. 2018;95:113–121. doi: 10.1002/aocs.12012. [DOI] [Google Scholar]
- Moran MD. Arguments for rejecting the sequential Bonferroni in ecological studies. Oikos. 2003;100:403–405. doi: 10.1034/j.1600-0706.2003.12010.x. [DOI] [Google Scholar]
- Norton GJ, Douglas A, Lahner B, Yakubova E, Guerinot ML, Pinson SRM, Tarpley L, Eizenga GC, McGrath SP, Zhao FJ, Rafiqul Islam M, Islam S, Duan G, Zhu Y, Salt DE, Meharg AA, Price AH. Genome wide association mapping of grain arsenic, copper, molybdenum and zinc in rice (Oryza sativa L.) grown at Four International Field Sites. PLoS ONE. 2014;9(2):e89685. doi: 10.1371/journal.pone.0089685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norton GJ, Travis AJ, Douglas A, Fairley S, De Paiva AE, Ruang-areerate P, Naredo MEB, McNally KL, Hossain M, Islam MR, Price AH. Genome wide association mapping of grain and straw biomass traits in the rice Bengal and Assam Aus Panel (BAAP) Grown Under Alternate Wetting and Drying and Permanently Flooded Irrigation. Front Plant Sci. 2018;9:1223. doi: 10.3389/fpls.2018.01223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pradhan SK, Pandit E, Pawar S, Bharati B, Chatopadhyay K, Singh S, Dash P, Reddy JN. Association mapping reveals multiple QTLs for grain protein content in rice useful for biofortification. Mol Geneti Genom MGG. 2019;294(4):963–983. doi: 10.1007/s00438-019-01556-w. [DOI] [PubMed] [Google Scholar]
- Pradhan SK, Pandit E, Pawar S, Naveenkumar R, Barik SR, Mohanty SP, Nayak DK, Ghritlahre SK, Sanjiba Rao D, Reddy JN, Patnaik SSC. Linkage disequilibrium mapping for grain Fe and Zn enhancing QTLs useful for nutrient dense rice breeding. BMC Plant Biol. 2020;20:57. doi: 10.1186/s12870-020-2262-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasanth VV, Babu MS, Basava RK, Tripura Venkata VGN, Mangrauthia SK, Voleti SR, Neelamraju S. Trait and marker associations in Oryza nivara and O. rufipogon derived rice lines under two different heat stress conditions. Front Plant Sci. 2017;26:1819. doi: 10.3389/fpls.2017.01819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genet. 2000;155(2):945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabson R, Bhatia C, Mitra RK. Crop productivity, grain protein and energy. In: Joint FAO/IAEA, editor. Seed protein improvement by nuclear techniques. (Proc. Meet. Bader, 1977) Vienna, Austria: lnternational Atomic Energy Agency; 1978. pp. 3–20. [Google Scholar]
- Rahman S, Sharma MP, Sahai S. Nutritional and medicinal values of some indigenous rice varieties. Indian J Tradit Knowl. 2006;5:454–458. [Google Scholar]
- Rao SD, Babu PM, Swarnalatha P, Kota S, Bhadana VP, Surekha K, Neerja CN, Babu RV. Assessment of grain zinc and iron variability in rice germplasm using energy dispersive X-ray fluorescence spectrophotometer (ED-XRF) J Rice Res. 2014;7:45–52. [Google Scholar]
- Rathna Priya TS, Nelson ARLE, Ravichandran K, Antony U. Nutritional and functional properties of coloured rice varieties of South India: a review. J Ethn Foods. 2019;6:1–11. doi: 10.1186/s42779-019-0002-x. [DOI] [Google Scholar]
- Richharia RH, Misro B, Butany WT, Seetharaman R. Linkage studies in rice (Oryza sativa L.) Euphytica. 1960;9(1):122–126. [Google Scholar]
- Sahu PK, Mondal S, Sharma D, Vishwakarma G, Kumar V, Das BK. InDel marker based genetic differentiation and genetic diversity in traditional rice (Oryza sativa L.) landraces of Chhattisgarh, India. PLoS ONE. 2017;12(11):e0188864. doi: 10.1371/journal.pone.0188864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjeeva Rao D, Neeraja CN, Madhu Babu P, Nirmala B, Suman K, Rao L, Surekha K, Raghu P, Longvah T, Surendra P, Kumar R, Babu VR, Voleti SR. Zinc biofortified rice varieties: challenges, possibilities, and progress in India. Front Nutr. 2020;7:26. doi: 10.3389/fnut.2020.00026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarawgi AK, Gauraha D, Nair SK, Sao A, Bhandarkar S, Verma R, Sharma B, Chandel G, Sharma D, Kar S. Development of improved varieties of rice- 1987 to 2019 (Dhan ki unnat kishmo ka vikas-1987 to 2019) Raipur, India: IGKV publication\2019; 2019. pp. 1–32. [Google Scholar]
- SES, IRRI . Standard evaluation system for rice. Manila: International Rice Research Institute; 2013. [Google Scholar]
- Shen YJ, Jiang H, Jin JP, Zhang ZB, Xi B, He YY, Wang G, Wang C, Qian L, Li X, Yu QB, Liu HJ, Chen DH, Gao JH, Huang H, Shi TL, Yang ZN. Development of genome-wide DNA polymorphism database for mapbased cloning of rice genes. Plant Physiol. 2004;135:1198–1205. doi: 10.1104/pp.103.038463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smouse PE, Whitehead MR, Peakall R. An informational diversity framework, illustrated with sexually deceptive orchids in early stages of speciation. Mol Ecol Resour. 2015;15(6):1375–1386. doi: 10.1111/1755-0998.12422. [DOI] [PubMed] [Google Scholar]
- Suh JP, Ahn SN, Cho YC, Kang KH, Choi IS, Kim YG, Suh HS, Hong HC. Mapping of QTLs for yield traits using an advanced backcross population from a cross between Oryza sativa and O. glaberrima. Korean J Breed. 2005;37(4):214–220. [Google Scholar]
- Suman K, Madhubabu P, Rathod R, Sanjeeva Rao D, Rojarani A, Prashant S, Subbarao LV, Ravindrababu V, Neeraja CN. Variation of grain quality characters and marker-trait association in rice (Oryza sativa L.) J Genet. 2020;99:5. doi: 10.1007/s12041-019-1164-4. [DOI] [PubMed] [Google Scholar]
- Swamy BPM, Shamsudin NAA, Rahman SNA, Mauleon R, Ratnam W, Cruz MTS, Kumar A. Association mapping of yield and yield related traits under reproductive stage drought stress in rice (Oryza sativa L.) Rice. 2017;10(21):1–13. doi: 10.1186/s12284-017-0161-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X, Xu J, Guo H, Jiang L, Chen S, Yu C, Zhou J, Hu P, Zhai H, Wan J. DTH8 suppresses flowering in rice, influencing plant height and yield potential simultaneously. Plant Physiol. 2010;153:1747–1758. doi: 10.1104/pp.110.156943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang-dong L, Jun Z, Liang-fang D, Fan-tao Z, Yi Z, Yong W, Jina-kun X. Linkage map construction and QTL mapping for cold tolerance in Oryza rufipogon Griff. at early seedling stage. J Integr Agric. 2016;15:60345–60347. [Google Scholar]
- Xing YZ, Xu CG, Hua JP, Tan YF. Analysis of QTL x environment interaction for rice panicle characteristics. Acta Genet Sin. 2001;28:439–446. [PubMed] [Google Scholar]
- Xing J, Jia MH, Correll JC, Yuan LP, Deng H, Jia Y. Confirming and identifying new loci for rice blast disease resistance using Magnaporthe oryzae field isolates in the US. Crop Sci. 2015;55:2620–2627. doi: 10.2135/cropsci2015.02.0077. [DOI] [Google Scholar]
- Xu F, Jin L, Huang Y, Tong C, Chen YL, Bao JS. Association mapping of quantitative trait loci for yield-related agronomic traits in rice (Oryza sativa L.) J Integr Agric. 2016;15(10):2192–2202. doi: 10.1016/S2095-3119(15)61244-8. [DOI] [Google Scholar]
- Ying JZ, Shan JX, Gao JP, Zhu MZ, Shi M, Lin HX. Identification of quantitative trait loci for lipid metabolism in rice seeds. Mol Plant. 2012;5(4):865–875. doi: 10.1093/mp/ssr100. [DOI] [PubMed] [Google Scholar]
- Yu J, Pressoir G, Briggs WH, Bi IV, Yamasaki M, Doebley JF, McMullen MD, Gaut BS, Nielsen DM, Holland JB. A unified mixed model method for association mapping that accounts for multiple levels of relatedness. Nat Genet. 2006;38(2):203–208. doi: 10.1038/ng1702. [DOI] [PubMed] [Google Scholar]
- Zhang M, Pinson SRM, Tarpley L, Huang XY, Lahner B, Yakubova E, et al. Mapping and validation of quantitative trait loci associated with concentrations of 16 elements in unmilled rice grain. Theor Appl Genet. 2014;127:137–165. doi: 10.1007/s00122-013-2207-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, Zhong K, Zhong Z, Tong H. Genome-wide association study of important agronomic traits within a core collection of rice (Oryza sativa L.) BMC Plant Biol. 2019;19:259. doi: 10.1186/s12870-019-1842-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao K, Tung CW, Eizenga GC, Wright MH, Ali ML, Price AH, Norton GJ, Islam MR, Rynolds A, Mezey J, McClung AM, Bustamante CD, McCouch SR. Genome-wide association mapping reveals a rich genetic architecture of complex traits in Oryza sativa L. Nat Commun. 2011;2(467):1–10. doi: 10.1038/ncomms1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.