Abstract
We examined the anticancer effects of a novel sirtuin inhibitor, MHY2256, on HCT116 human colorectal cancer cells to investigate its underlying molecular mechanisms. MHY2256 significantly suppressed the activity of sirtuin 1 and expression levels of sirtuin 1/2 and stimulated acetylation of forkhead box O1, which is a target protein of sirtuin 1. Treatment with MHY2256 inhibited the growth of the HCT116 (TP53 wild-type), HT-29 (TP53 mutant), and DLD-1 (TP53 mutant) human colorectal cancer cell lines. In addition, MHY2256 induced G0/G1 phase arrest of the cell cycle progression, which was accompanied by the reduction of cyclin D1 and cyclin E and the decrease of cyclin-dependent kinase 2, cyclin-dependent kinase 4, cyclin-dependent kinase 6, phosphorylated retinoblastoma protein, and E2F transcription factor 1. Apoptosis induction was shown by DNA fragmentation and increase in late apoptosis, which were detected using flow cytometric analysis. MHY2256 downregulated expression levels of procaspase-8, -9, and -3 and led to subsequent poly(ADP-ribose) polymerase cleavage. MHY2256-induced apoptosis was involved in the activation of caspase-8, -9, and -3 and was prevented by pretreatment with Z-VAD-FMK, a pan-caspase inhibitor. Furthermore, the autophagic effects of MHY2256 were observed as cytoplasmic vacuolation, green fluorescent protein-light-chain 3 punctate dots, accumulation of acidic vesicular organelles, and upregulated expression level of light-chain 3-II. Taken together, these results suggest that MHY2256 could be a potential novel sirtuin inhibitor for the chemoprevention or treatment of colorectal cancer or both.
Keywords: SIRT inhibitor, MHY2256, Colorectal cancer cells, Cell cycle arrest, Apoptosis, Autophagy
INTRODUCTION
Colorectal cancer (CRC), which occurs primarily occurs in the colon and rectum, is the third most common type of cancer worldwide, accounting for 10.2% of all cases (Bray et al., 2018). Currently, effective methods of treating CRC include surgery, chemotherapy, radiation, and a combination of chemotherapy and radiation therapy, but many patients are still dying from CRC (Bray et al., 2018). Therefore, there is a continuing need to investigate and develop new targeted therapy or treatment options for CRC.
Recently, histone deacetylase (HDAC) inhibitors have been extensively studied for the development as therapeutic agents against CRC (Bultman, 2017). HDACs play an important role in upregulating tumor suppressor genes in CRC cells (Bultman, 2017). HDAC inhibitors have been tested in several clinical trials for different types of human cancers (Jiang et al., 2017; Zhou et al., 2018; Wang et al., 2019). We hypothesize that more specific targeted inhibitors for class III HDACs [sirtuins (SIRTs)] will facilitate personalized treatment plans for CRC.
A novel SIRT inhibitor, MHY2256, was synthesized and tested against several types of human cancer cells. Park and his colleagues reported that MHY2256 shows anticancer effects against human MCF-7 breast cancer cells via regulation of MDM-2 p53 binding (Park et al., 2016). In addition, De and her colleagues also reported that MHY2256 induces apoptosis and autophagic cell death of human Ishikawa endometrial cancer cells (De et al., 2018). Therefore, we investigated its anticancer activity against the following three human CRC cell lines: HCT116 (TP53 wild-type), HT-29 (TP53 mutant), and DLD-1 (TP53 mutant). To evaluate the ability of MHY2256 to inhibit SIRT, cell viability, cell cycle regulation, and levels of apoptosis- and autophagy-related protein molecules were measured.
MATERIALS AND METHODS
Chemicals
5-(3,5-Di-tert-butyl-4-hydroxybenzylidene)-2-thioxodihydropyrimidine-4,6(1H,5H)-dione (MHY2256, Fig. 1A) was synthesized as previously reported (De et al., 2018) and kindly provided by Prof. Hyung Ryong Moon (Division of Pharmacy, College of Pharmacy, Pusan National University, Busan, Korea). The compound was solubilized in sterile dimethyl sulfoxide (DMSO) to prepare a 10-mM stock solution, which was stored at –20°C until use. Subsequent dilutions were made in Roswell Park Memorial Institute (RPMI)-1640 medium (GE Healthcare Life Sciences, Logan, UT, USA), and the maximum concentration of DMSO did not exceed 0.1% (v/v), which is the concentration below which there is no effect on cell viability. DMSO and 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) were purchased from Amresco LLC (Solon, OH, USA). Antibodies specific for SIRT1 (sc-15404), SIRT2 (sc-20966), Ac-FoxO1 (sc-49437), FoxO1 (sc-67140), procaspase-3 (sc-7272), procaspase-8 (sc-7890), and procaspase-9 (sc-7885), PARP (sc-7150), cyclin D1 (sc-246), cyclin E (sc-481), cyclin-dependent kinase 2 (Cdk2, sc-163), Cdk4 (sc-260), Cdk6 (sc-177), retinoblastoma (Rb, sc-50), E2F transcription factor 1 (E2F1, sc-193), glycogen synthase kinase-3β (GSK3β, sc-9166), and Z-VAD-FMK were obtained from Santa Cruz Biotechnology (Dallas, TX, USA). The polyclonal antibodies against phosphorylated Rb (p-RB, #9308), p-GSK3β (#5558), γ-H2A histone family member X (H2AX, #9718), and light chain 3 (LC3B, #3868) were purchased from Cell Signaling Technology (Danvers, MA, USA). Lithium chloride, propidium iodide (PI), acridine orange, and a monoclonal antibody against β-actin were obtained from Sigma-Aldrich (St. Louis, MO, USA).
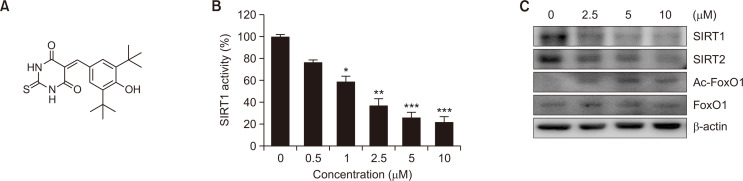
Fig. 1.
Identification of MHY2256 as a novel sirtuin (SIRT) inhibitor. (A) Chemical structure of MHY2256. (B) HCT116 cells were treated with various concentrations of MHY2256 and in vitro SIRT1 deacetylase activity was measured using Fluor-de-Lys fluorescent assays. Data are shown as the mean ± SD of three independent experiments. *p<0.05, **p<0.01 and ***p<0.001 vs. vehicle-treated cells. (C) HCT116 cells were treated with MHY2256 for 24 h, and cell lysates were immunoblotted with SIRT1, SIRT2, Ac-forkhead box O1 (FoxO1), or FoxO1 antibodies. Representative results from three independent experiments are shown.
Cell culture and viability assay
The human HCT116, HT-29, and DLD-1 cell lines were cultured in RPMI-1640 supplemented with 10% fetal bovine serum (FBS) (Thermo Fisher Scientific, Waltham, MA, USA), 100 units/mL of penicillin and 100 μg/mL of streptomycin (GE Healthcare Life Sciences) at 37°C in a humidified 5% CO2 as previously described (Heo et al., 2019). The non-transformed rat IEC-18 intestinal epithelial cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM, GE Healthcare Life Sciences) supplemented with 10% FBS. Cell viability was determined using the MTT assay as previously described (Hwangbo et al., 2020). The cells were seeded in 24-well culture plates, incubated for 24 h in the growth medium, and then treated with or without MHY2256 for the indicated concentrations. The cells were then incubated in the dark with 0.5 mg/mL MTT at 37°C for 2 h. The formed formazan granules were subsequently dissolved in DMSO, and absorbance of the final solution was measured at 540 nm wavelength using a microplate reader (Thermo Fisher Scientific).
SIRT1 deacetylase activity assay
SIRT1 activity was determined using a SIRT1 fluorometric drug discovery kit (BML-AK555; Enzo Life Sciences, Farmingdale, NY, USA) according to the manufacturer’s protocol. Briefly, total protein (10 μg) was dissolved in the assay buffer and incubated with the acetylated substrate (Fluor de Lys-SIRT1 substrate, 25 μM) and NAD+ (100 μM) at 37°C for 45 min. The deacetylated substrates were measured after the developer was added and then the fluorescence intensity was detected using a fluorescence plate reader (GENios, TECAN Instrument, Salzburg, Austria) with excitation at 369 nm and emission at 460 nm.
Cell cycle analysis
To determine the effect of MHY2256 on the cell cycle regulation, cell cycle analysis was performed using flow cytometry as previously described (Jang et al., 2018). Briefly, the cells were treated for 24 h at the appropriate conditions, trypsinized, washed once with cold phosphate-buffered saline (PBS), and then stored in 70% ethanol overnight at –20°C. The fixed cells were then stained with cold PI solution (50 μg/mL in PBS) for 30 min at room temperature in the dark. The stained DNA contents of these cells were examined using an Accuri C6 flow cytometer (BD Biosciences, Franklin Lakes, NJ, USA).
Annexin V-FITC/PI double staining
To determine the induction of apoptosis, Annexin V-FICT/PI double staining was performed using flow cytometry as previously described (Jang et al., 2018). Briefly, the cells were treated for 24 h at the appropriate conditions with or without MHY2256, trypsinized, harvested, washed one time with cold PBS, and then suspended in binding buffer (BD Biosciences). The cells were stained in PI and Annexin V-fluorescein isothiocyanate (FITC) solution (BD Pharmingen FITC Annexin V apoptosis detection kit, BD Biosciences) at room temperature for 15 min in the dark. The stained cells were analyzed within 1 h using an Accuri C6 flow cytometer (BD Biosciences).
DNA fragmentation assay
To detect DNA breakages in apoptotic cells, DNA fragmentation assay was performed as previously described (Jang et al., 2018). Briefly, the cells treated with or without MHY2256 were lysed in buffer containing 5 mM Tris-HCl (pH 7.5), 5 mM ethylenediaminetetraacetic acid (EDTA), and 0.5% Triton X-100 on ice for 30 min. The fragmented DNA was treated with RNase, followed by proteinase K digestion, extraction using a phenol/chloroform/isoamyl alcohol mixture (25:24:1, v/v/v), and then isopropanol precipitation. The DNA was separated using a 1.6% agarose gel, stained with 0.1 μg/mL ethidium bromide, and visualized using an ultraviolet source.
Western blot analysis
Western blot analysis was performed as previously described (Jang et al., 2018). The cells were lysed in lysis buffer [25 mM Tris (pH 7.5), 250 mM sodium chloride (NaCl), 5 mM EDTA, 1% Nonidet P-40 (NP-40), 100 μg/mL phenylmethylsulfonyl fluoride, and protease inhibitor cocktail (Sigma-Aldrich)]. The same amount of protein was denatured by boiling at 100°C for 5 min in the sample buffer (Bio-Rad Laboratories, Hercules, CA, USA). Total proteins were separated using 8-12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred onto polyvinylidene difluoride (PVDF) membranes. The membranes were probed with primary antibodies overnight, incubated with horseradish peroxidase-conjugated secondary antibodies (Santa Cruz Biotechnology), and then visualized using an enhanced chemiluminescence (ECL) detection system (GE Healthcare Life Sciences).
Caspase activity assay
The activities of caspases in CRC cells were detected as previously described (Choi et al., 2015). The cells were incubated with the lysis buffer (R&D Systems, Minneapolis, MN, USA) for 10 min on ice. Total protein (100 μg) samples were incubated with 2x reaction buffer and substrates of colorimetric tetrapeptides, Z-DEVD, Z-IETD, and Ac-LEHD for caspase-3, -8, and -9, respectively. The reaction mixtures were incubated at 37°C for 2 h, and then the enzyme-catalyzed release of p-nitroaniline was quantified at 405 nm using a multi-well reader (Thermo Fisher Scientific).
Detection of acidic vesicular organelles
The formation of acidic vesicular organelles (AVOs) is known as one of the main features of autophagy. We detected the formation of AVOs using a previously published method (Lee et al., 2014; Choi et al., 2015). Briefly, the cells were treated with or without MHY2256 under the appropriate conditions for 24 h, stained with acridine orange (Sigma-Aldrich, 1 μg/mL) for 15 min, trypsinized and then washed with PBS. The stained cells were then analyzed with an Accuri C6 flow cytometer (BD Biosciences).
Green fluorescent protein-LC3 assay
The formation of punctate LC3 was detected using green fluorescent protein-LC3 assay as previously described (Jang et al., 2018). Cells were transfected with the LC3-green fluorescent protein (GFP) plasmid using Lipofectamine 2000 reagent (Invitrogen Corporation, Grand Island, NY, USA) according to the manufacturer’s protocol. The cells were seeded in Lab-Tek II chamber slides (Thermo Fisher Scientific), transfected for 24 h, treated with or without MHY2256 for 24 h, washed with PBS twice, and then fixed with 4% paraformaldehyde for 20 min. The formation of punctate LC3-positive structures was analyzed by imaging with a FV10i FLUOVIEW confocal microscope (Olympus Corporation, Tokyo, Japan).
Statistical analysis
Data are presented as means ± standard deviation (SD). For three separate experiments and were statistically analyzed using the Student’s t-tests. The mean was considered significantly different if *p<0.05, **p<0.01, and ***p<0.001.
RESULTS
MHY2256 inhibits SIRT1 activity and induces acetylation of FoxO1 in HCT116 cells
To investigate the SIRT1 inhibitory activity of MHY2256, we first examined the in vitro effects using a fluorogenic substrate (BML-AK555, Enzo Life Sciences). Treatment with MHY2256 inhibited SIRT1 deacetylase activity in a concentration-dependent manner (Fig. 1B). The half-maximal inhibitory activity (IC50) of MHY2256 against the SIRT1 enzyme activity was 1.02 μM. Next, the effect of MHY2256 treatment on SIRT protein expression was determined using western blot analysis. MHY2256 significantly decreased the expression levels of SIRT1 and SIRT2 proteins in HCT116 cells (Fig. 1C). Previous studies have established that SIRT1 interacts with FoxO1 and regulates transcriptional activity by deacetylation (Park et al., 2016). Therefore, we assessed whether MHY2256 affects FoxO1 acetylation and found that HCT116 cells exposed to MHY2256 showed increased levels of acetylated FoxO1 (Fig. 1C). These findings indicate that MHY2256 might induce FoxO1 acetylation by inhibiting SIRT1 activity in HCT116 cells.
MHY2256 suppresses proliferation of CRC cells
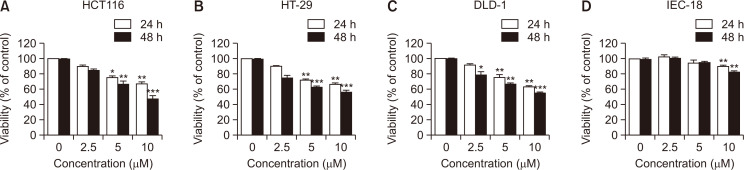
To determine whether MHY2256 inhibited the cell viability of HCT116 (TP53 wild type), HT-29 (TP53 mutant) and DLD-1 (TP53 mutant) human colorectal cancer cells, they were treated with increasing concentrations of MHY2256 for 24 h or 48 h. As shown in Fig. 2A-2C, MHY2256 treatment reduced the proliferation of CRC cells in a concentration- and time-dependent manner. The HCT116 cell line was the most sensitive to the effects of MHY2256 treatment in comparison to the HT-29 and DLD-1 cell lines. Therefore, we used the HCT116 cell line in subsequent experiments. The effect of MHY2256 on the normal IEC-18 cell line were also analyzed (Fig. 2D). MHY2256 (10 μM at 48 h) inhibited the cell growth in HCT116, HT-29, and DLD-1 cells by >45%, whereas little growth inhibition was observed even with MHY2256 treatment in the non-transformed rat IEC-18 intestinal epithelial cells.
Fig. 2.
Effect of MHY2256 on the viability of human colorectal cancer (CRC) cell lines. (A-C) Human CRC cells were incubated with increasing concentrations of MHY2256 for 24 h and 48 h. (D) IEC-18 rat intestinal epithelial cells were treated with MHY2256, and percentage of cell survival was determined using the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) assay. Results were expressed as a percentage of vehicle treated control ± SD of three separate experiments. The significance was determined using the Student’s t-test (*p<0.05, **p<0.01 and ***p<0.001 compared with vehicle-treated cells).
MHY2256 changes cell cycle progression in HCT116 cells
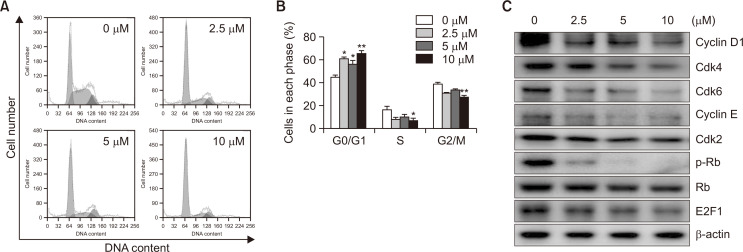
To determine whether MHY2256 treatment modulates cell cycle distribution, HCT116 cells were treated with various concentrations of MHY2256 for 24 h, and then the distribution of cell cycle was studied with flow cytometry. As shown in Fig. 3A, 3B, cells treated with MHY2256 showed G0/G1 phase cell cycle arrest. A total of 64.47% of the cells cultured with 10 µM MHY2256 were in the G0/G1 phase compared to 43.13% of the control cells. Next, we determined whether MHY2256 treatment changed the expression of G0/G1 cell cycle regulator proteins. Cells were treated with various concentrations of MHY2256 for 24 h, and then the levels of G0/G1 cell cycle regulatory proteins were examined using western blot analysis. MHY2256 markedly reduced not only the expression levels of the early G1 phase cyclin and Cdk proteins such as cyclin D1, Cdk4, and Cdk6 but also the late G1 phase cyclin and Cdk protein such as cyclin E and Cdk2 (Fig. 3C). In addition, MHY2256 significantly downregulated p-Rb and, subsequently, the E2F1 transcription factor (Fig. 3C).
Fig. 3.
Effects of MHY2256 on cell cycle and cell cycle regulatory proteins in HCT116 cells. (A) Cells were treated for 24 h with the indicated concentrations of MHY2256, stained with propidium iodide (PI), and then flow cytometry was used by analyze their distribution in each phase of the cell cycle. Representative results from three independent experiments are shown. (B) Data are shown as the mean ± SD of three independent experiments. Significance was determined using the Student’s t-test (*p<0.05, and **p<0.01 vs. vehicle-treated cells). (C) To detect the protein expression levels of cell cycle regulators, cells were incubated with MHY2256 for 24 h and then cellular proteins were separated and immunoblotted with indicated antibodies. Representative results from three independent experiments are shown. β-actin was used as an internal control.
GSK3β is required for MHY2256-induced downregulation of cyclin D1 and cyclin E in HCT116 cells
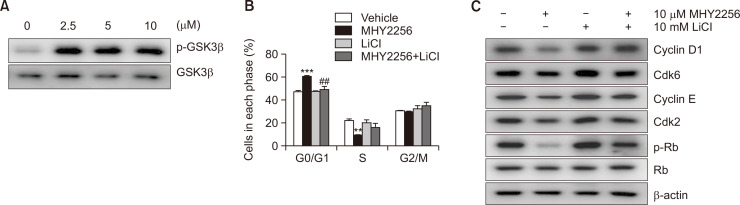
Degradation of cyclin D1 and cyclin E is activated by the phosphorylation by GSK3β and subsequent recognition by SCF E3 ligase family (Takahashi-Yanaga and Sasaguri, 2008; Liu et al., 2017). Therefore, it is necessary to elucidate whether the downregulation of cyclin D1, cyclin E, or both by MHY2256 is GSK3β-dependent. First, we investigated the expression level of GSK3β following MHY2256 treatment. HCT116 cells were incubated with different concentrations of MHY2256 for 12 h. As shown in Fig. 4A, the expression of total GSK3β was not perturbed, whereas that of p-GSK3β was upregulated in a concentration-dependent manner in HCT116 cells treated with MHY2256. To assess whether GSK3β is involved in the MHY2256-mediated cyclin D1 and cyclin E1 downregulation, we used the GSK3β inhibitor LiCl. HCT116 cells pretreated with 10 mM LiCl for 1 h and then co-incubated with 10 μM MHY2256 for 12 h, and we found that LiCl suppressed the MHY2256-induced G1 phase accumulation and induced the transition from G1 to S phase (Fig. 4B). In addition, LiCl suppressed the MHY2256-induced cyclin D1 and cyclin E downregulation (Fig. 4C). Moreover, the cell cycle regulators of the transition from G1 to S phase such as Cdk2, Cdk6, and p-Rb were upregulated. These data suggest that MHY2256-induced cyclin D1 and cyclin E downregulation and G0/G1 arrest are mediated by the activity of GSK3β.
Fig. 4.
Role of glycogen synthase kinase-3β (GSK3β) in MHY2256-induced G1 arrest of HCT116 cells. (A) Cells were treated with various concentration of MHY2256 for 24 h, and phosphorylated GSK3β (p-GSK3β) and GSK3β were analyzed using western blot analysis. (B) Cells were incubated with 10 µM MHY2256 for 24 h after pretreatment of 10 mM lithium chloride (LiCl) for 1 h and then cell cycle distribution was assessed using flow cytometry. The percentage of each cell population is shown as the mean ± SD of three independent experiments (**p<0.01, ***p<0.001, vs. vehicle-treated cells and ##p<0.01, vs. MHY2256-treated cells). (C) Total cell lysates were prepared and immunoblotted with G1 transition-related antibodies. β-actin was used as an internal control.
MHY2256 induces apoptosis in HCT116 cells
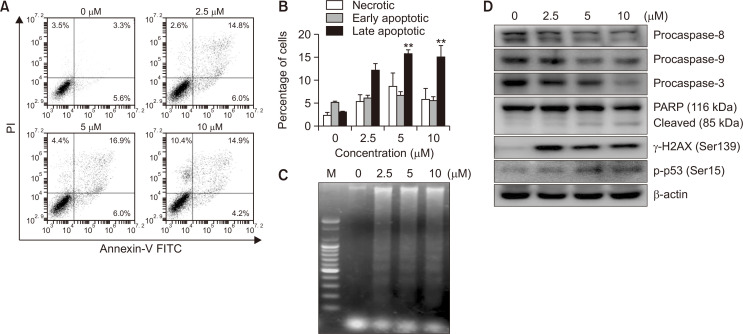
We investigated whether the MHY2256-dependent growth inhibition in HCT116 cells is mediated by apoptosis using flow cytometry using Annexin V and PI staining method. As shown in Fig. 5A, the early apoptotic proportion (shown in lower right quadrant) increased from 3.5% to 25.4%, and the late apoptotic proportion (shown in upper right quadrant) increased from 3.3% to 14.9% after 24 h treatment with 10 µM MHY2256. The flow cytometry results also indicated that MHY2256-induced apoptosis was concentration-dependent (Fig. 5B). Treatment of HCT116 cells with MHY2256 for 24 h resulted in a concentration-dependent internucleosomal DNA fragmentation (Fig. 5C). To examine the possible mechanism underlying these effects, we studied the influence of MHY2256 on apoptosis-related gene expression. In particular, we studied the potential of MHY2256 treatment to alter the expression levels of caspases in HCT116 cells. We found that MHY2256 treatment resulted in concentration-dependent decreases of procaspase-8, -9, and -3 levels and cleavage of their substrate PARP (Fig. 5D). As a result, due to the increase in the level of the active form caspases with the decrease of procaspases (Lee et al., 2014), the cleavage of their substrate PARP was also augmented in a concentration-dependent manner (Fig. 5D). In addition, we investigated the effects of MHY2256 on the DNA damage response by examining the expression of DNA damage response proteins such as γ-H2AX and p-p53 (Jang et al., 2018), which were increased after treatment with MHY2256 alone (Fig. 5D). These data suggest that MHY2256 induces apoptosis of HCT116 cells through activation of the caspase cascade and the DNA damage response pathway in HCT116 cells.
Fig. 5.
Effect of MHY2256 on the induction of apoptosis in HCT116 cells. (A, B) Cells were incubated with the indicated concentrations of MHY2256 for 24 h and stained with Annexin V-FITC//propidium iodide (PI) and analyzed using flow cytometry. Representative data from three separate experiments are shown (**p<0.01 vs. vehicle-treated cells). (C) Fragmented genomic DNA was extracted from the cells and monitored by 1.6% agarose gel electrophoresis. M, marker. (D) The expression levels of procaspase-8, -9, -3, PARP (116/85 kDa), and γ-H2AX protein were detected by western blot analysis. β-actin was used as a loading control. The experiments were performed three times and representative data are shown. PARP, poly(ADP-ribose) polymerase; γ-H2AX, γ-Histone H2AX (Ser139).
Caspases are involved in MHY2256-induced apoptosis in HCT116 cells
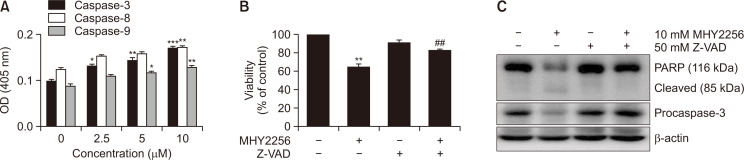
Procaspases are precursors of caspases, which are the executors of the apoptotic process (Kaufmann et al., 2008). However, a decrease in procaspase levels does not necessarily mean that caspases are activated. Therefore, we examine the effect of MHY2256 on caspase activation using specific substrates. The histogram in Fig. 6A shows that caspase-3 and caspase-8 activations were increased by almost 1.7-fold in HCT116 cells treated with 10 µM MHY2256, whereas caspase-9 was only increased by <1.5-fold (Fig. 6A). MHY2256-induced apoptosis of HCT116 cells cultured with or without the broad-spectrum caspase inhibitor Z-VAD-FMK was analyzed using flow cytometry and western blot analysis. As shown in Fig. 6B, pretreatment of cells with Z-VAD-FMK partially blocked the decrease of cell viability induced by MHY2256. To further demonstrate this result, PARP cleavage was analyzed using western blotting under the same experimental conditions. Consistent with the results of cell death measured using the MTT assay, the western blot analysis of PARP showed that pretreatment of cells with Z-VAD-FMK significantly inhibited the MHY2256-induced cleavage of PARP, an activated caspase-3 substrate protein (Fig. 6C). These results suggest that activation of the caspase cascade must precede the induction of apoptosis by MHY2256 treatment.
Fig. 6.
Effect of MHY2256 on caspase activation in HCT116 cells. (A) Cells were treated with increasing concentration of MHY2256 for 24 h, and in vitro caspase-3, -8, and -9 activity assay was assessed using Z-DEVD-pNA, Z-IETD-pNA, and Ac-LEHD-pNA substrates, respectively. Each point represents the mean ± standard deviation (SD) of three independent experiments (*p<0.05, **p<0.01 and ***p<0.001 vs. vehicle-treated cells). (B) HCT116 cells were incubated with 10 µM MHY2256 for 24 h after pretreatment of 50 µM Z-VAD-FMK for 30 min. The cell viability was measured using 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) assay. Data shown are representative of three independent experiments. Values represent mean ± SD (n=3), **p<0.01 vs. vehicle-treated cells and ##p<0.01 vs. MHY2256-treated cells. (C) Expression level of PARP and procaspase 3 were analyzed using western blotting after treatment with 50 µM Z-VAD-FMK, 10 µM MHY2256, or both for 24 h. Results represent three independent experiments. β-actin was used as a loading control.
MHY2256 induces autophagy in HCT116 cells
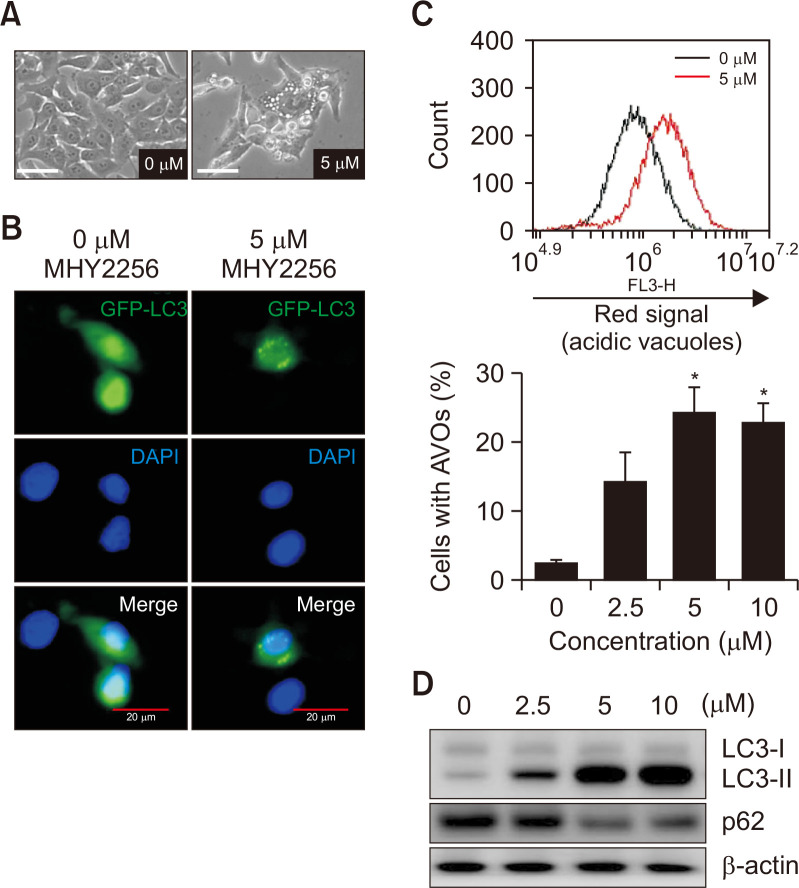
Recently, MHY2256 has been reported to induce autophagic cell death in human breast cancer cells (Park et al., 2016) and endometrial cancer cells (De et al., 2018). HCT116 cells were treated with or without MHY2256 for 24 h, and the formation of autophagic vacuoles was investigated using phase contrast microscopy (Zeiss Axiophot, Göttingen, Germany). As shown in Fig. 7A, MHY2256 (5 μM) induced the formation of autophagic vacuoles in HCT116 cells. Next, to further confirm that autophagy formation was induced by MHY2256, we studied the LC3 formation in MHY2256-treated HCT116 cells using fluorescence microscopy (FV10i FLUOVIEW, Olympus Corporation). As shown in Fig. 7B, GFP-tagged-LC3 (GFP-LC3) formed several cytoplasmic punctas in cells treated with MHY2256 (5 μM) but not in untreated control cells. Since the formation of AVOs in the cytoplasm is one of the typical features of autophagic cell death, to quantify AVO formation, cells were stained with acridine orange and flow cytometry analysis was performed. The number of AVOs in MHY2256-treated cells clearly increased in a concentration-dependent manner (Fig. 7C). In addition, western blot analysis showed that LC3-I (the soluble form) was converted to LC3-II (the lipidized form) in MHY2256-treated HCT116 cells, thus indicating the induction of autophagy formation (Fig. 7D). Downregulation of p62 by MHY2256 was also observed in HCT116 cells, as predicted by the results of LC3 conversion (Fig. 7D).
Fig. 7.
Effect of MHY2256 on the induction of autophagy in HCT116 cells. (A) Cells were treated with 5 µM MHY2256 for 24 h, and representative images were captured using phase contrast microscopy at a magnification of 400x. Scale bar, 20 µm. (B) HCT116 cells were transfected with the light-chain 3 (LC3)-green fluorescent protein (GFP) plasmid for 24 h, and transfected cells were treated with 5 µM MHY2256 for 24 h. The formation of GFP-LC3 puncta was examined using confocal microscopy. Representative data from three independent experiments are shown. Scale bar, 20 µm. (C) Cells were incubated with MHY2256 for 24 h, stained with acridine orange, and then analyzed using flow cytometry to quantify formation of acidic vesicular organelles (AVOs). The results are expressed as mean ± SD (n=3), *p<0.05 vs. vehicle-treated cells. (D) Cells were treated with the indicated concentrations of MHY2256 for 24 h, and expression levels of LC3 and p62 protein were monitored using western blot analysis.
DISCUSSION
Previous studies showed that MHY2256 exhibits anticancer effects against human MCF-7 breast cancer cells (Park et al., 2016) and human endometrial cancer Ishikawa cells (De et al., 2018). In the present study, we also discovered that synthetic MHY2256, an SIRT inhibitor, exerted antitumor effects on human HCT116 CRC cells. MHY2256 treatment not only inhibited SIRT1 activity and downregulated SIRT1 and SIRT2 protein levels but also induced G1 phase cell cycle arrest, apoptosis, and autophagy of HCT116 cells.
SIRTs (previously known as silent mating-type information regulator proteins) are a family of well-conserved proteins found in a variety of living organisms from bacteria to humans (O’Callaghan and Vassilopoulos, 2017). SIRTs are NAD+-dependent protein deacylases/deacetylases that remove acyl moieties from the lysine-modified α-amino group. In some cases, NAD+-dependent protein ADP-ribosylation is observed. There are seven known mammalian sirtuins (SIRT1-7) are segregated in different subcellular structures: the nuclei (SIRT1, -2, -6, and -7), cytoplasm (SIRT1 and SIRT2), and mitochondria (SIRT3, -4, and -5). SIRT1 protects against neurodegeneration in Alzheimer’s, Huntington’s and Parkinson’s diseases and acts as a redox sensor (O’Callaghan and Vassilopoulos, 2017). SIRTs have a profound effect on the maintenance of homeostasis and various types of cellular processes such as transcription, inflammation, apoptosis, stress management, aging, and energy metabolism as well as alertness in the event of nutrient deficiency (Dai et al., 2018). SIRTs can also control circadian rhythm and synthesis of mitochondria (oxidative metabolism) (Yanar et al., 2019).
Therefore, SIRT inhibitors are sought-after agents for use as therapeutics strategies for curing numerous human diseases. Potent inhibitors have been identified for SIRT1, 2, 3, and 5 (such as AGK2, cambinol, salermide, sirtinol, splitomicin, suramin, and tenovin) (Villalba and Alcaín, 2012; Hu et al., 2014; Jiang et al., 2017; Zhou et al., 2018; Wang et al., 2019). SIRT1 inhibition could be a potential option in the treatment of cancer, human immunodeficiency virus (HIV) infections, Fragile X syndrome, and some parasitic diseases (Villalba and Alcaín, 2012). SIRT2 inhibitors are implicated in the treatment of several types of cancer and neurodegenerative diseases (Zhang et al., 2020). Various SIRT1/2 inhibitors have been reported to date, but none have been approved as drugs (Zhang et al., 2020).
Several other studies have also reported the anticancer effects of SIRT inhibition (Heltweg et al., 2006; Lain et al., 2008; Peck et al., 2010; Mellini et al., 2012; Rotili et al., 2012; Mellini et al., 2013; Jiang et al., 2017; Zhou et al., 2018; Wang et al., 2019). However, because of the low efficacy and uncertain selectivity of SIRT inhibitors, there is admittedly some ambiguity in the exact correlation between the pharmacological effects in cancer cell lines and animal models observed in several studies and the specific types of SIRTs. However, one consensus is that dual SIRT1/2 inhibition appears to be necessary for cancer cells retaining the wild-type tumor suppressor protein p53, which could be because p53 is a common endogenous substrate for both SIRT1 and SIRT2 (Martínez-Redondo and Vaquero, 2013; Park et al., 2016; De et al., 2018). When we tested MHY2256 to three CRC cell lines, it showed higher cytotoxicity against HCT116 cells with TP53 wild-type than against the HT-29 and DLD-1 TP53 mutant cell lines (Fig. 2). Therefore, an effective SIRT inhibition strategy appears to be highly dependent on the gene profiling of cancer cell type.
In conclusion, when the treatment of MHY2256, as a novel SIRT inhibitor, inhibited the growth of HCT116 cells by inducing a DNA damage response, arrested the cell cycle at the G0/G1 phase, initiating apoptosis through activation of the caspase cascade, and inducing autophagy. Overall, these results suggest that MHY2256 may be a useful therapeutic agent for CRC.
ACKNOWLEDGMENTS
This work was supported by a 2-Year Research Grant of Pusan National University.
Footnotes
CONFLICT OF INTEREST
None.
REFERENCES
- Bray F., Ferlay J., Soerjomataram I., Siegel R. L., Torre L. A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- Bultman S. J. Interplay between diet, gut microbiota, epigenetic events, and colorectal cancer. Mol. Nutr. Food Res. 2017;61:1500902. doi: 10.1002/mnfr.201500902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi P. R., Kang Y. J., Sung B., Kim J. H., Moon H. R., Chung H. Y., Kim S. E., Park M. I., Park S. J., Kim N. D. MHY218-induced apoptotic cell death is enhanced by the inhibition of autophagy in AGS human gastric cancer cells. Int. J. Oncol. 2015;47:563–572. doi: 10.3892/ijo.2015.3031. [DOI] [PubMed] [Google Scholar]
- Dai H., Sinclair D. A., Ellis J. L., Steegborn C. Sirtuin activators and inhibitors: promises, achievements, and challenges. Pharmacol. Ther. 2018;188:140–154. doi: 10.1016/j.pharmthera.2018.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De U., Son J. Y., Sachan R., Park Y. J., Kang D., Yoon K., Lee B. M., Kim I. S., Moon H. R., Kim H. S. A new synthetic histone deacetylase inhibitor, MHY2256, induces apoptosis and autophagy cell death in endometrial cancer cells via p53 acetylation. Int. J. Mol. Sci. 2018;19:2743. doi: 10.3390/ijms19092743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heltweg B., Gatbonton T., Schuler A. D., Posakony J., Li H., Goehle S., Kollipara R., Depinho R. A., Gu Y., Simon J. A., Bedalov A. Antitumor activity of a small-molecule inhibitor of human silent information regulator 2 enzymes. Cancer Res. 2006;66:4368–4377. doi: 10.1158/0008-5472.CAN-05-3617. [DOI] [PubMed] [Google Scholar]
- Heo G., Kang D., Park C., Kim S. J., Choo J., Lee Y., Yoo J. W., Jung Y., Lee J., Kim N. D., Chung H. Y., Moon H. R., Im E. Pro-apoptotic effect of the novel benzylidene derivative MHY695 in human colon cancer cells. Oncol. Lett. 2019;18:3256–3264. doi: 10.3892/ol.2019.10664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J., Jing H., Lin H. Sirtuin inhibitors as anticancer agents. Future Med. Chem. 2014;6:945–966. doi: 10.4155/fmc.14.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwangbo H., Kim S. Y., Lee H., Park S. H., Hong S. H., Park C., Kim G. Y., Leem S. H., Hyun J. W., Cheong J., Choi Y. H. Auranofin enhances sulforaphane-mediated apoptosis in hepatocellular carcinoma Hep3B cells through inactivation of the PI3K/Akt signaling pathway. Biomol. Ther. (Seoul) 2020;28:443–455. doi: 10.4062/biomolther.2020.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang J. Y., Kang Y. J., Sung B., Kim M. J., Park C., Kang D., Moon H. R., Chung H. Y., Kim N. D. MHY440, a novel topoisomerase Ι inhibitor, induces cell cycle arrest and apoptosis via a ROS-dependent DNA damage signaling pathway in AGS human gastric cancer cells. Molecules. 2018;24:96. doi: 10.3390/molecules24010096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y., Liu J., Chen D., Yan L., Zheng W. Sirtuin inhibition: strategies, inhibitors, and therapeutic potential. Trends Pharmacol. Sci. 2017;38:459–472. doi: 10.1016/j.tips.2017.01.009. [DOI] [PubMed] [Google Scholar]
- Kaufmann S. H., Lee S. H., Meng X. W., Loegering D. A., Kottke T. J., Henzing A. J., Ruchaud S., Samejima K., Earnshaw W. C. Apoptosis-associated caspase activation assays. Methods. 2008;44:262–272. doi: 10.1016/j.ymeth.2007.11.005. [DOI] [PubMed] [Google Scholar]
- Lain S., Hollick J. J., Campbell J., Staples O. D., Higgins M., Aoubala M., McCarthy A., Appleyard V., Murray K. E., Baker L., Thompson A., Mathers J., Holland S. J., Stark M. J., Pass G., Woods J., Lane D. P., Westwood N. J. Discovery, in vivo activity, and mechanism of action of a small-molecule p53 activator. Cancer Cell. 2008;13:454–463. doi: 10.1016/j.ccr.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y., Sung B., Kang Y. J., Kim D. H., Jang J. Y., Hwang S. Y., Kim M., Lim H. S., Yoon J. H., Chung H. Y., Kim N. D. Apigenin-induced apoptosis is enhanced by inhibition of autophagy formation in HCT116 human colon cancer cells. Int. J. Oncol. 2014;44:1599–1606. doi: 10.3892/ijo.2014.2339. [DOI] [PubMed] [Google Scholar]
- Liu S. L., Liu Z., Zhang L. D., Zhu H. Q., Guo J. H., Zhao M., Wu Y. L., Liu F., Gao F. H. GSK3β-dependent cyclin D1 and cyclin E1 degradation is indispensable for NVP-BEZ235 induced G0/G1 arrest in neuroblastoma cells. Cell Cycle. 2017;16:2386–2395. doi: 10.1080/15384101.2017.1383577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Redondo P., Vaquero A. The diversity of histone versus nonhistone sirtuin substrates. Genes Cancer. 2013;4:148–163. doi: 10.1177/1947601913483767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellini P., Carafa V., Di Rienzo B., Rotili D., De Vita D., Cirilli R., Gallinella B., Provvisiero D. P., Di Maro S., Novellino E., Altucci L., Mai A. Carprofen analogues as sirtuin inhibitors: enzyme and cellular studies. ChemMedChem. 2012;7:1905–1908. doi: 10.1002/cmdc.201200318. [DOI] [PubMed] [Google Scholar]
- Mellini P., Kokkola T., Suuronen T., Salo H. S., Tolvanen L., Mai A., Lahtela-Kakkonen M., Jarho E. M. Screen of pseudopeptidic inhibitors of human sirtuins 1-3: two lead compounds with antiproliferative effects in cancer cells. J. Med. Chem. 2013;56:6681–6695. doi: 10.1021/jm400438k. [DOI] [PubMed] [Google Scholar]
- O'Callaghan C., Vassilopoulos A. Sirtuins at the crossroads of stemness, aging, and cancer. Aging Cell. 2017;16:1208–1218. doi: 10.1111/acel.12685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park E. Y., Woo Y., Kim S. J., Kim D. H., Lee E. K., De U., Kim K. S., Lee J., Jung J. H., Ha K. T., Choi W. S., Kim I. S., Lee B. M., Yoon S., Moon H. R., Kim H. S. Anticancer effects of a new SIRT inhibitor, MHY2256, against human breast cancer MCF-7 cells via regulation of MDM2-p53 binding. Int. J. Biol. Sci. 2016;12:1555–1567. doi: 10.7150/ijbs.13833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peck B., Chen C. Y., Ho K. K., Di Fruscia P., Myatt S. S., Coombes R. C., Fuchter M. J., Hsiao C. D., Lam E. W. SIRT inhibitors induce cell death and p53 acetylation through targeting both SIRT1 and SIRT2. Mol. Cancer Ther. 2010;9:844–855. doi: 10.1158/1535-7163.MCT-09-0971. [DOI] [PubMed] [Google Scholar]
- Rotili D., Tarantino D., Nebbioso A., Paolini C., Huidobro C., Lara E., Mellini P., Lenoci A., Pezzi R., Botta G., Lahtela-Kakkonen M., Poso A., Steinkühler C., Gallinari P., De Maria R., Fraga M., Esteller M., Altucci L., Mai A. Discovery of salermide-related sirtuin inhibitors: binding mode studies and antiproliferative effects in cancer cells including cancer stem cells. J. Med. Chem. 2012;55:10937–10947. doi: 10.1021/jm3011614. [DOI] [PubMed] [Google Scholar]
- Takahashi-Yanaga F., Sasaguri T. GSK-3beta regulates cyclin D1 expression: a new target for chemotherapy. Cell. Signal. 2008;20:581–589. doi: 10.1016/j.cellsig.2007.10.018. [DOI] [PubMed] [Google Scholar]
- Villalba J. M., Alcaín F. J. Sirtuin activators and inhibitors. Biofactors. 2012;38:349–359. doi: 10.1002/biof.1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T., Xu Z., Lu Y., Shi J., Liu W., Zhang C., Jiang Z., Qi B., Bai L. Recent progress on the discovery of Sirt2 inhibitors for the treatment of various cancers. Curr. Top. Med. Chem. 2019;19:1051–1058. doi: 10.2174/1568026619666190510103416. [DOI] [PubMed] [Google Scholar]
- Yanar K., Simsek B., Çakatay U. Integration of melatonin related redox homeostasis, aging, and circadian rhythm. Rejuvenation Res. 2019;22:409–419. doi: 10.1089/rej.2018.2159. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Anoopkumar-Dukie S., Arora D., Davey A. K. Review of the anti-inflammatory effect of SIRT1 and SIRT2 modulators on neurodegenerative diseases. Eur. J. Pharmacol. 2020;867:172847. doi: 10.1016/j.ejphar.2019.172847. [DOI] [PubMed] [Google Scholar]
- Zhou Z., Ma T., Zhu Q., Xu Y., Zha X. Recent advances in inhibitors of sirtuin1/2: an update and perspective. Future Med. Chem. 2018;10:907–934. doi: 10.4155/fmc-2017-0207. [DOI] [PubMed] [Google Scholar]