Abstract
Sex/gender disparity has been shown in the incidence and prognosis of many types of diseases, probably due to differences in genes, physiological conditions such as hormones, and lifestyle between the sexes. The mortality and survival rates of many cancers, especially liver cancer, differ between men and women. Due to the pronounced sex/gender disparity, considering sex/gender may be necessary for the diagnosis and treatment of liver cancer. By analyzing research articles through a PubMed literature search, the present review identified 12 genes which showed practical relevance to cancer and sex disparities. Among the 12 sex-specific genes, 7 genes (BAP1, CTNNB1, FOXA1, GSTO1, GSTP1, IL6, and SRPK1) showed sex-biased function in liver cancer. Here we summarized previous findings of cancer molecular signature including our own analysis, and showed that sex-biased molecular signature CTNNB1High, IL6High, RHOAHigh and GLIPR1Low may serve as a female-specific index for prediction and evaluation of OS in liver cancer patients. This review suggests a potential implication of sex-biased molecular signature in liver cancer, providing a useful information on diagnosis and prediction of disease progression based on gender.
Keywords: Liver cancer, Sex/gender, Overall survival, Gene expression, Molecular signature
INTRODUCTION
Accumulating data demonstrate that the incidence and prognosis of many types of diseases differ depending on patients’ sex/gender. The sex/gender disparity may be due to genetic differences on the X chromosome, differences in physiological conditions such as hormone levels, and other factors (Spatz et al., 2004; Bottarelli et al., 2007; Scosyrev et al., 2009; Klinge, 2012; Gabriele et al., 2016). It should be noted that ‘sex’ and ‘gender’ are not mutually exclusive terms (Clayton and Tannenbaum, 2016; Heidari et al., 2016). In general, ‘sex’ is used for biological expression, such as gene expression or hormone-related symptoms, while ‘gender’ is used when lifestyle, behavior, and environment are reflected (Clayton and Tannenbaum, 2016; Heidari et al., 2016; Pelletier et al., 2016). Sex/gender-biased diseases include autoimmune diseases, neurodegenerative diseases, and cardiovascular diseases (Skavdahl et al., 2005; Cantuti-Castelvetri et al., 2007; Gleicher and Barad, 2007; Haley et al., 2010). Diseases showing sex/gender disparities also include several types of cancers, including liver cancer, melanoma, and thyroid cancer (Bray et al., 2018). Liver cancer and melanoma have lower incidence rates and better prognosis in females than in males (Bray et al., 2018; Smalley, 2018). In contrast, the incidence rate of thyroid cancer is three times higher in females than in males, even though there is not much difference in mortality rate (Bray et al., 2018).
Liver cancer, the fourth leading cause of cancer death worldwide (Bray et al., 2018), shows a prominent sex/gender disparity. For liver cancer, the incidence is 3-5 times higher in males, while the mortality rates is twice as high in males than in females (Bray et al., 2018; Jung et al., 2019). The reasons for this sex/gender disparity are complex. Sex hormones, metabolic factors, and behavioral factors have been suggested as risk factors for liver cancer (Lin et al., 2013; Welzel et al., 2013; Kohi, 2016). Due to the pronounced sex/gender disparity, considering sex/gender may be necessary for the diagnosis and treatment of liver cancer (Guy and Peters, 2013).
Cancer is a complex disease and cancer genes do not act separately and deregulation of various genes from different pathways can lead to cancer initiation or progression (Hanahan and Weinberg, 2011). Gene signature or gene expression signature is a single or combined group of genes in a cell with different pattern of gene expression that occurs as a result of pathogenic condition. Many studies have been made to predict gene signatures related to cancer (van de Vijver et al., 2002; Allahyar and de Ridder, 2015) and produce informative genes or sub-networks by considering a predefined biological network (Babaei et al., 2013). Gene signature that can be applied for a broad range of cancers could be highly useful in research and clinical settings as a biomarker. The discovery of molecular signatures is proving to be a powerful tool for disease diagnosis and drug discovery.
The predictive effect of single gene biomarkers is not sufficiently specific (Zhao et al., 2019). When a cell becomes cancerous, the changes in the gene or protein expression patterns that causes the biological characteristics as a cancer cell occur. The cluster of genes or proteins representing these changes is called a cancer molecular signature (Nilsson et al., 2009; Sung et al., 2012). Significant clinical phenotypes resulting from these changes can be predicted by cancer molecular signature. They include the progression of diseases and the consequent increase in risk rates (Mehrabian et al., 2007; Hur et al., 2011), the response to drugs used to treat diseases and the resulting toxicity evaluation (Hines et al., 2010; Cohen et al., 2011; Xie et al., 2012), and the prediction of recurrence or death of diseases (Pittman et al., 2004; Bøvelstad et al., 2007).
In this review, we summarized cancer molecular signature which reported previously and cancer biomarker candidates reflecting sex disparity. In addition, we described how we identified 12 sex-biased genes and their expression patterns in liver cancer patient. More importantly, we discussed that sex-biased molecular signature CTNNB1High, IL6High, RHOAHigh and GLIPR1Low was correlated with overall survival (OS) in liver cancer patients with sex-dependency.
CANCER MOLECULAR SIGNATURE
Recently, many significant cancer molecular signatures have been published (Tang et al., 2017; Erstad et al., 2018; So et al., 2020). This is due to the development of various omics capable of analyzing a large number of samples at the same time and bioinformatics capable of analyzing a database reflecting the research results and clinical information accumulated over a long period of time.
All 1207 differentially expressed genes (DEGs) between recurrent and non-recurring samples in colon cancer patients were analyzed (Xu et al., 2017). Through support vector machine (SVM) analysis and verification of gene expression profiling, molecular signatures including 15 genes (HES5, ZNF417, GLRA2, OR8D2, HOXA7, FABP6, MUSK, HTR6, GRIP2, KLRK1, VEGFA, AKAP12, RHEB, NCRNA00152, PMEPA1) were identified as indicators that inform the prognosis and recurrence risk in colon cancer patients (Xu et al., 2017).
Wang et al. (2019) analyzed 332 DEGs between normal ovarian tissue and ovarian cancer tissue and observed the associated prognosis. Sixty-four of them were significantly correlated with the OS of ovarian cancer patients, and five genes, IGF2, PEG3, DCN, LYPD1, and RARRES1, were selected and screened to construct a 5-gene signature (Wang et al., 2019). As a result of clinical analysis, patients with low expressed 5-gene signature had significantly better OS compared to patients with the high expression (p=0.0004).
CD44-high and CD24-low cells not only express cancer stem cell-related genes, but also represent epithelial-mesenchymal transition (EMT) properties. When the relative expression of CD44 and CD24 was observed in the clinical samples of oral squamous cell carcinoma patients, CD44 expression was high in tumor tissues, but CD24 was significantly low. Therefore, CD44high and CD24low have high potential to be used as a molecular signature of cancer stem-like cells in oral squamous cell carcinoma (Ghuwalewala et al., 2016).
After neoadjuvant chemotherapy for operable gastroesophageal cancer, lymph node metastasis is known as the only proven variable that can predict the prognosis (Smyth et al., 2016). In the high- and low-risk groups of OS in The Medical Research Council Adjuvant Gastric Infusional Chemotherapy trial samples, 7-gene signature (CDH1, ELOVL5, EGFR, PIP5K1B, FGF1, CD44v8.10, TBCEL) could independently predict the patient’s prognosis (Smyth et al., 2018). These results suggest that stratification of patients using this 7-gene signature may help in postoperative chemotherapy selection.
In hepatocellular carcinoma (HCC), there have been reported on several molecular signatures related to cancer growth and malignancy. Molecular signature including five genes, ANGPT2, NETO2, ESM1, NR4A1 and DLL4, can be showed HCC growth, invasion into blood vessels, recurrence of cancer, and degree of intrahepatic metastasis (Villa et al., 2016). MicroRNA signature related to intravenous invasion and metastasis can be used to predict disease free survival and OS: highly upregulation of miR-219, miR-207 and miR-338, and extremely downregulation of miR-34, miR-30, and miR-148 (Budhu et al., 2008). The gene signature reflecting abnormal DNA methylation in HCC, SCAND3, SGIP1 and PI3, can be used to determine the risk of recurrence in patients with resected early-stage HCC (Qiu et al., 2017). Molecular signatures in HCC can contribute to the development of targeted treatment regimens. In addition, more accurate prognosis can be predicted after treatment, selective and intensive monitoring of patients with poor prognosis can be performed, and clinical trial design can be improved, such as subdividing diseases with similar advanced stages (Erstad et al., 2018).
SEX-BIASED CANCER BIOMARKER CANDIDATES
Men and women show distinct sex/gender-biased differences in various areas, including growth rate and lifespan, metabolism and immune mechanisms, which are affected by carcinogenesis and cancer progression, treatment mechanisms, and survival. This is caused by sexual dimorphism that occurs due to sex-biased differences including genetic and epigenetic mechanisms as well as sex hormones that circulate in the body and induce sex disparities. Sex disparity have also been demonstrated in the expression of biomarkers that predict prognosis and diagnosis of cancer and the rate and pattern of cancer metastasis, and the response to various trials of treatment in different cancer types (Pal and Hurria, 2010; Mervic, 2012).
There are sex disparities in the epigenetic mechanisms of autosomal and sex chromosome genes (El-Maarri et al., 2007; Tobi et al., 2009; Reviewed in Yuasa, 2010). And impairment of epigenetic regulation is known as an important mechanism for the incidence and progression of cancer (Tobi et al., 2009). DNA methylation is the most extensively applied epigenetic marker that represented by sex-biased expression of gene such as DNA methyltransferases (DNMTs) (Nugent et al., 2015; Mosley et al., 2017). Reizel’s group reported for DNA methylation in the liver, where males are hypomethylated compared to females due to testosterone exposure, and demonstrated that it was regulated by DNA methylation with sex disparity (Reizel et al., 2015).
One of the most striking differences of epigenetics in male and female is the inactivation of additional X-chromosomes in female cells (Rubin et al., 2020). Some long non-coding RNA (lncRNA), such as five prime to Xist (FTX), and a lot of epigenetic modifiers located on X chromosome, such as lysine demethylases KDM6A and KDM5C, are concerned with inactivation of X chromosome and has been known as presumed tumor suppressors in HCC (Wijchers and Festenstein, 2011; Liu et al., 2016). They are highly expressed in female HCC patients and suppress the proliferation and invasion of HCC cells (Xu et al., 2008; Liu et al., 2016; Snell and Turner, 2018). Their expressions correlate positively with cancer survival and decrease the risk of liver cancer in females.
Sex-biased metabolic pathways also influence cancer progression and treatment mechanisms. In the overall metabolic mechanisms, the expression of molecules involved in carbohydrate and amino acid metabolism is increased in men, and the expression of molecules involved in fatty acid metabolism is increased in women (Mittelstrass et al., 2011; Garcia-Herreros et al., 2012; Krumsiek et al., 2015; Ippolito et al., 2017). In spite of the need for higher mitochondrial activity, female mitochondria produce less reactive oxygen species (ROS), one of the causes of cell damage (Borras et al., 2003; Harish et al., 2013). Fundamental sex disparities in metabolic pathways, such as the use of nutrients in the body and mitochondrial function, can lead to sex-biased differences in the incidence and progression of cancer, and further, the mechanism of application of chemotherapy.
In general, females have a stronger and more adaptable immune response system than males (Cook et al., 2009; Klein and Flanagan, 2016). These immune responses with sex disparity may contribute to differences in cancer progression and mortality according to sex. Females have a greater number of neutrophils and macrophages and more active phagocytosis than males due to the inhibited secretion of inflammatory cytokines by estrogen (Scotland et al., 2011; Laffont et al., 2017). These researches have suggested that estrogen decrease the risk of cancer in females. The X chromosome contains the largest number of immune-related genes in the entire human genome such as FOXP3 and CD40L (Fish, 2008; Libert et al., 2010; Pinheiro et al., 2011; Bianchi et al., 2012). Given the differences of expression of sex chromosome genes in male and female, X-linked immunoregulatory genes and sex hormones are expected to play an important role in mediating sex-biased immune response.
Sex hormones additionally affect sex disparity in the angiogenesis process in cancer via different expression of circulating angiogenic factors by intrinsically different endothelial cells (ECs) (Addis. et al., 2014). As shown in Evanson’s study, platelet-rich plasma of adult females has more pro-angiogenic factors, including vascular endothelial growth factor (VEGF), epidermal growth factor (EGF), platelet-derived growth factors (PDGFs), and so on (Evanson et al., 2014). On the other hand, several angiogenic growth factors, such as basic fibroblast growth factor (bFGF), transforming growth factor-beta 1 (TGF-β1) and tumor necrosis factor-alpha (TNF-α), were higher expressed in male plasma (Xiong et al., 2018). However, there is no clear elucidation of how certain growth factors and cytokines have sex-biased expression, and how this sex disparity translates into differential angiogenesis signals and functions (Rubin et al., 2020). These findings suggest the need for analysis of correlation between pro- or anti-angiogenic factors and sex.
The sex-biased molecular differences induced by the various causes of sex disparity mentioned earlier were identified via systematic evaluation of omics and big data analysis. These identified molecules applied as sex-biased potential biomarker candidates (Li et al., 2018; Shin et al., 2019).
Efforts have been made to elucidate sex-biased gene expression and functions in liver cancer. Wu et al. (2019) suggested CDK1, CCNB1, CYP3A4 and SERPINA4 as sex-biased cancer molecular signature which have been identified as DEGs with sex dimorphism in HCC via gene expression profiling, the most frequent type of liver cancer (Wu et al., 2019). Phosphoglucomutase-like protein 5, encoded by the PGM5 gene, was shown to have potential as a male-specific prognostic biomarker reflecting overall survival (OS) probability in liver cancer (Jiao et al., 2019). However, there is no known marker indicating the risk of liver cancer in female patients.
Although sex-biased differences in HCC development risk are well recognized (Setiawan et al., 2016; Wu et al., 2018; Yu et al., 2019; Rich et al., 2020), the prognosis between sexes remains unclear. Sex hormones are expected to play an important role in the sex disparity of malignancy. Androgen/androgen receptor signaling is known to be involved in tumor promotion as well as estrogen/estrogen receptor signaling is involved in tumor protection in mouse models (Li et al., 2012). Function of sex hormones in HCC has been suggested as restrain of interleukin-6 and STAT3 inactivation (Naugler et al., 2007; Hou et al., 2013). Therefore, in order to understand why male and female patients show difference in HCC development and prognosis, sex-biased molecular signature would be required to predict and evaluate the prognosis of liver cancer.
We conducted a PubMed search (https://www.ncbi.nlm.nih.gov/pubmed/) for papers describing sex-biased genes known to be related to cancer. Using five keywords, ‘cancer,’ ‘malignancy,’ ‘sex,’ ‘gender,’ and ‘gene,’ the PubMed search resulted in 598 related papers. After carefully checking these papers, 12 cancer-related genes that showed sex-biased differences were selected for further analysis (Table 1). These genes code for proteins with critical roles in the cell cycle, cellular differentiation, regulation of cell death and growth, or cancer development processes such as angiogenesis and metastasis.
Table 1.
List of previously reported cancer biomarker candidates with sex disparity identified by a PubMed search
| Symbol | Gene name | Sex-biased function in cancer | References |
|---|---|---|---|
| BAP1 | BRCA1 Associated Protein 1 | - Regulation of cell cycle, cellular differentiation, and DNA damage | Li et al., 2018; Masoomian et al., 2018 |
| - More frequent mutation in female-derived HCC | |||
| BRUCE (BIRC6) | BIR Repeat-Containing Ubiquitin-Conjugating Enzyme | - Regulation of tumor cell death | Salehi et al., 2017 |
| - Higher levels of expression specific in female | |||
| CTNNB1 | Catenin Beta 1 | - Regulation of cell growth and adhesion between cells | Xia et al., 2006; Li et al., 2018 |
| - More frequent mutation in male liver cancer patients | |||
| FOXA1 | Forkhead Box Protein A1 | - Regulation of apoptosis and cell cycle | Li et al., 2012, 2017 |
| - Significantly expressed higher in female HCC | |||
| GLIPR1 | Glioma pathogenesis-related protein 1 | - Regulation of cell growth and chemokine secretion | Li et al., 2011; Zhang et al., 2015 |
| - Lower expressed specific in male thyroid cancer patients | |||
| GSTO1 | Glutathione S-transferase omega-1 | - Protection of normal cells against damage induced by carcinogens in HCC | Niu et al., 2009; Qu et al., 2015 |
| GSTP1 | Glutathione S-transferase pi-1 | - Significantly associated with overall survival in HCC patients | |
| IL6 | Interleukin 6 | - Strong correlation between inflammation and cancer | Naugler et al., 2007; Liu and Liu, 2014; Kumari, et al., 2016 |
| - Low expression reduced risk of cancer in female liver cancer patients | |||
| KISS1R | Kisspeptin receptor 1 | - Suppression of cancer metastasis | Shirasaki et al., 2001; Yaron et al., 2015 |
| - Significantly expressed highly in female pituitary tumors patients | |||
| PER1 | Period 1 | - Regulation of cell cycle and promotion of DNA repair | Wang et al., 2015 |
| - Higher expression in female colon cancer patients | |||
| RHOA | Ras Homolog Family Member A | - Promotion of tumor cell proliferation and metastasis | De Rienzo et al., 2016 |
| - Significant reduction in survival of the entire cohort and across gender subgroups | |||
| SRPK1 | Serine-arginine protein kinase 1 | - Regulation of mRNA splicing | Graveley, 2000; Zhang et al., 2016 |
| - Higher expression significantly correlated with sex specific to male HCC patients |
SEX/GENDER-BIASED OVERALL SURVIVAL PROBABILITY OF LIVER CANCER PATIENTS WITH SELECTED CANCER-RELATED GENES
To alleviate the aggressive progression of liver cancer in women, timely and appropriate diagnosis that reflects individual differences, such as sex/gender disparities, is essential. Identified 12 genes which showed practical relevance to cancer and sex disparities via a PubMed literature search were investigated the correlation between expression of these genes and cancer malignancy by Kaplan-Meier (KM) plot analysis using clinical data from liver cancer patients in The Cancer Genome Atlas (TCGA) database. Among the 12 sex-biased genes, 7 genes (BAP1, CTNNB1, FOXA1, GSTO1, GSTP1, IL6, and SRPK1) showed sex-biased function in liver cancer (Table 1), suggesting that cancer biomarker candidates with sex disparity may be reliable in liver cancer.
The repositories in TCGA datasets of male and female liver cancer patients with available survival data were analyzed for the 12 genes. In order to obtain survival data for male and female liver cancer patients, the transcriptomic dataset in TCGA (version 2016_01_28; https://cancergenome.nih.gov/) was analyzed and plotted to KM plots using The Kaplan Meier Plotter (http://kmplot.com). Male (N=246) and female (N=118) liver cancer patients were divided using the auto-select best cutoff criteria. The cutoff values for high and low expression for each gene were as follows: catenin beta 1 (CTNNB1, female, 7932; male, 10396), IL6 (female, 3; male, 2), glioma pathogenesis related 1 (GLIPR1, female, 153; male, 399), and Ras Homolog Family Member A (RHOA, female, 13302; male, 13613). A detailed description of the patient populations is given in Table 2. Based on TCGA data, KM plots were generated to check the correlation between expression of the 12 genes and OS probability in male and female liver cancer patients (N=246 and 118, respectively). The OS hazard ratio (HR) of the 12 genes and the statistical significance (logrank P) in the KM plots are listed in Table 3 and depicted in Fig. 1.
Table 2.
The detailed description of sample populations of male and female liver cancer patients from TCGA
| Cohort | RNA-seq | ||
|---|---|---|---|
| Platform | Illumina HiSeq 2000 | ||
| Patients | Total N | 364 | |
| Sex | Male | 246 | |
| Female | 118 | ||
| Race | White/Caucasian | 184 | |
| Black or African-American | 17 | ||
| Asian | 158 | ||
| Pathology | Stage | I | 171 |
| II | 86 | ||
| III | 85 | ||
| IV | 5 | ||
| Vascular Invasion | None | 205 | |
| Micro | 93 | ||
| Macro | 16 | ||
Table 3.
Values of hazard ratio (HR) and specificity (logrank P) of 12 genes in male and female liver cancer patients by the KM plot analysis
| Symbol | Gene name | Male | Female | |||
|---|---|---|---|---|---|---|
| HR (95% CI) | Logrank p | HR (95% CI) | Logrank p | |||
| BAP1 | BRCA1 Associated Protein 1 | 0.77 (0.49-1.2) | 0.25 | 1.47 (0.85-2.54) | 0.17 | |
| BRUCE (BIRC6) | BIR Repeat-Containing Ubiquitin-Conjugating Enzyme | 0.75 (0.47-1.19) | 0.22 | 0.83 (0.48-1.45) | 0.52 | |
| CTNNB1* | Catenin Beta 1 | 0.67 (0.39-1.14) | 0.14 | 1.91 (1.07-3.4) | 0.026 | |
| FOXA1 | Forkhead Box Protein A1 | 0.71 (0.44-1.15) | 0.16 | 0.68 (0.35-1.34) | 0.26 | |
| GLIPR1* | Glioma pathogenesis-related protein 1 | 1.51 (0.97-2.36) | 0.069 | 0.54 (0.3-0.96) | 0.034 | |
| GSTO1 | Glutathione S-transferase omega-1 | 0.67 (0.38-1.16) | 0.15 | 0.76 (0.4-1.45) | 0.41 | |
| GSTP1 | Glutathione S-transferase pi-1 | 0.83 (0.53-1.29) | 0.41 | 0.59 (0.32-1.1) | 0.091 | |
| IL6* | Interleukin 6 | 0.61 (0.38-0.96) | 0.029 | 2.34 (1.13-4.83) | 0.018 | |
| KISS1R | Kisspeptin receptor 1 | 1.38 (0.86-2.21) | 0.17 | 1.68 (0.96-2.93) | 0.066 | |
| PER1 | Period 1 | 0.39 (0.25-0.62) | 3.6e-05 | 0.51 (0.28-0.93) | 0.025 | |
| RHOA# | Ras Homolog Family Member A | 1.65 (1.06-2.56) | 0.026 | 1.86 (1.06-3.27) | 0.029 | |
| SRPK1 | Serine-arginine protein kinase 1 | 1.74 (1.1-2.75) | 0.017 | 2.15 (1.22-3.8) | 0.0066 | |
*Gene with a sex disparity in the correlation between expression and overall survival in liver cancer patients.
#Gene with a high HR value in both male and female liver cancer patients, but prominent sex differences with longer survival periods.
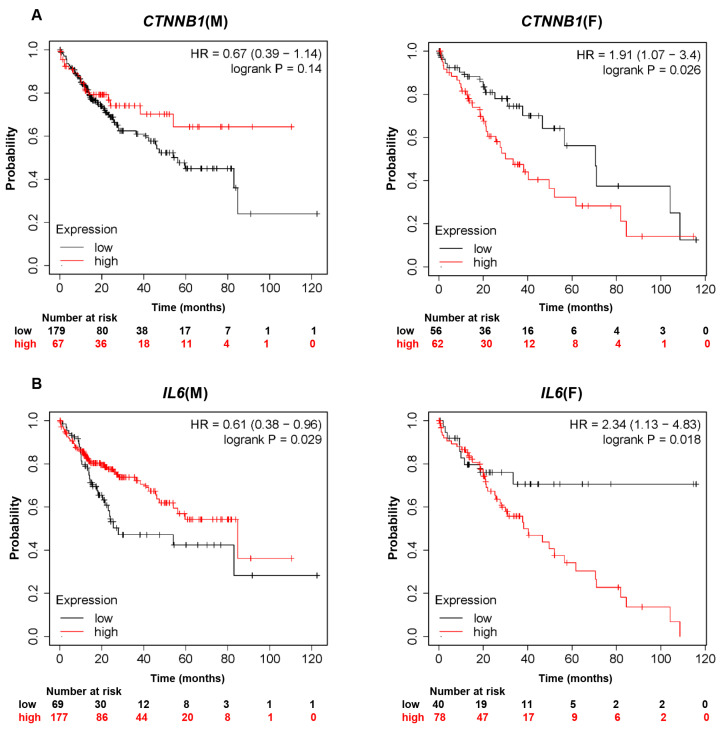
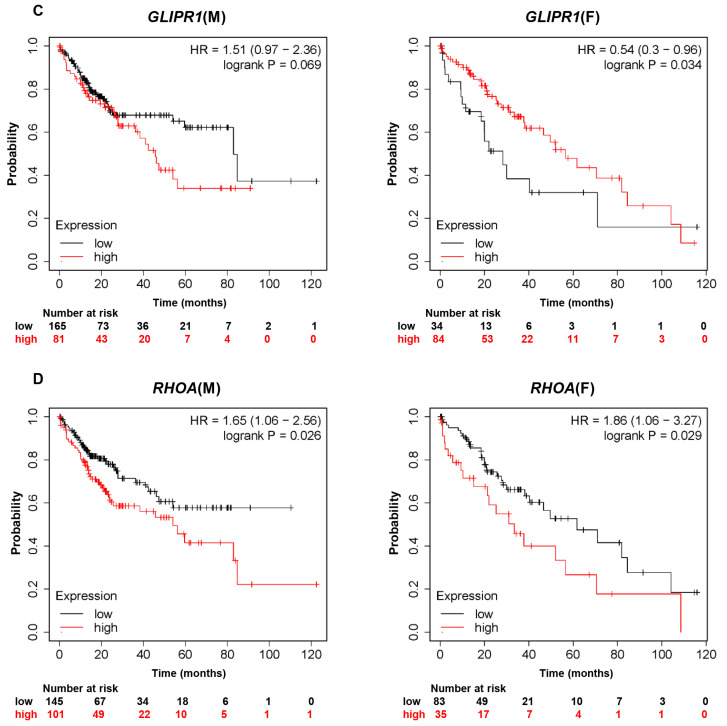
Fig. 1.
Kaplan-Meier analysis assessing overall survival time according to gene expression in male and female liver cancer patients using TCGA databases. Kaplan-Meier survival curves comparing overall survival probability with expression levels of (A) CTNNB1, (B) IL6, (C) GLIPR1, or (D) RHOA in male and female liver cancer patients. CTNNB1, catenin beta 1; IL6, interleukin 6; GLIPR1, glioma pathogenesis-related protein 1; RHOA, Ras homolog family member A; HR, hazard ratio.
Three out of the 12 genes, CTNNB1, IL6 and GLIPR1, showed sex disparity in OS probability in liver cancer patients (Fig. 1A-1C). Male liver cancer patients with relatively high expression of CTNNB1 showed a higher OS probability than did male liver cancer patients with low expression of CTNNB1 (HR=0.67). In contrast, female liver cancer patients with relatively high expression of CTNNB1 showed a lower OS probability and higher risk than did female liver cancer patients with low expression of CTNNB1 (HR=1.91, logrank p=0.026). These data indicate that there is a positive correlation between CTNNB1 expression and OS probability in male liver cancer patients, but there is a negative correlation in female liver cancer patients.
As shown in Fig. 1B, male liver cancer patients with relatively high expression of IL6 showed a significantly higher OS probability than did male liver cancer patients with low expression of IL6 (HR=0.61, logrank p=0.029). In females, liver cancer patients with relatively high expression of IL6 showed a significantly lower OS probability than did patients with low expression of IL6 (HR=2.34, logrank p=0.018). These results suggest a positive correlation between IL6 expression and OS probability in male liver cancer patients, but a negative correlation in female liver cancer patients, as in the case for CTNNB1. These data show that female liver cancer patients with higher expression of IL6 had poorer prognoses than those with lower levels of IL6.
In the analysis of GLIPR1 (Fig. 1C), male liver cancer patients with relatively high expression of GLIPR1 showed a lower OS probability than did male liver cancer patients with low expression of GLIPR1 (HR=1.51). Female liver cancer patients with relatively high expression of GLIPR1 showed a significantly higher OS probability than did female liver cancer patients with low expression of GLIPR1 (HR=0.54, logrank p=0.034). Unlike CTNNB1 and IL6, there is a negative correlation between GLIPR1 expression and OS probability in male liver cancer patients, whereas a positive correlation exists in female liver cancer patients.
There was no apparent sex difference between males and females in HR for RHOA (HR=1.65 in males and HR=1.86 in females). Of note, the difference in OS probability between high RHOA expression and low RHOA expression was more drastic in male liver cancer patients than in female liver cancer patients (Fig. 1D). Taken together, our analysis demonstrates a sex-dependent correlation between OS probability in liver cancer patients and expression levels of CTNNB1, IL6, GLIPR1, and RHOA.
CTNNB1 codes for β-catenin, a known key molecule in canonical WNT signaling (Behari, 2010). The WNT/β-catenin pathway is a critical regulator in cancers including liver cancer, and β-catenin plays an important role in liver regeneration (Barker, 2008; Behari, 2010; Li et al., 2018). CTNNB1 mutations, which are observed more frequently in male liver cancer patients (Xia et al., 2006; Li et al., 2018), induce high β-catenin activity leading to liver cancer (Rebouissou et al., 2016).
IL6 is overexpressed in almost all types of cancer; its increased expression indicates a strong correlation between inflammation and cancer (Kumari et al., 2016). IL6 expression is inhibited by estrogen in Kupffer cells in the liver, which may explain the lower incidence of liver cancer in women than men (Naugler et al., 2007; Liu and Liu, 2014). In the diethylnitrosamine-induced animal model of HCC, higher production of IL6 was reported in males than in females (Naugler et al., 2007). Our results showed that the higher expression levels of CTNNB1 and IL6, the lower the OS probability, and the difference in female liver cancer patients were all significant (Fig. 1A, 1B). It would be worthwhile to examine whether female liver cancer patients with high IL6 levels have low estrogen levels.
The Rho family is involved in cellular proliferation and metastasis in cancer via reorganization of the actin cytoskeleton and regulation of related signaling (Heasman and Ridley, 2008; De Rienzo et al., 2016). In male malignant pleural mesothelioma patients, expression of RHOA was significantly higher in non-epithelioid tumors, and was associated with a significant reduction in survival (De Rienzo et al., 2016). In this study, high RHOA expression was associated with low OS probability in both male and female liver cancer patients. When the survival period was extended to 80 months or longer, the difference in OS probability between the RHOA-high and RHOA-low expression groups was markedly greater in male than in female liver cancer patients, indicating a sex-biased effect of this gene (Fig. 1D).
The protein GLIPR1 has been shown to act as a tumor suppressor in thyroid and prostate cancers. Thyroid cancer occurs more frequently in women, but is more aggressive in men (Rahbari et al., 2010). In thyroid cancer, testosterone promotes cancer progression by reducing GLIPR1 expression and cancer immune mechanisms (Zhang et al., 2015). In prostate cancer, GLIPR1 inhibits cancer development by destructing cytosolic β-catenin and c-Myc (Li et al., 2011). It would be worthwhile to further investigate the interaction between GLIPR1 and β-catenin. In glioma, GLIPR1 functions as an oncoprotein (Murphy et al., 1995; Ren et al., 2004; Awasthi et al., 2013). The role of GLIPR1 in liver cancer has not yet been determined. This study shows a sex-biased correlation between GLIPR1 expression and OS in liver cancer patients: a negative correlation in males and a positive correlation in females (Fig. 1C).
In this study, correlations between sex-biased OS probability and the levels of expression of the four genes develop into a meaningful result. It can be expected that this is due to the influence of sex hormones, especially estrogen. Because the action of the female sex hormone estrogen is the main reason why liver cancer is superior to male (Bray et al., 2018; Jung et al., 2019). Of the four sex-biased genes from liver cancer through literature search and database analysis, CTNNB1 and IL6 have been reported to have estrogen correlation.
Crosstalk between Wnt signaling pathway, the representative signaling mechanisms related to CTNNB1, and estrogen signaling pathway increase cell growth by inducing the transcription of cyclin D1 gene, CCND1, and stabilization of cyclin D1 protein (Kouzmenko et al., 2004; Mulligan et al., 2017). As mentioned earlier in the introduction, the previous studies showed that the risk of liver cancer is lower in female by suppressing IL6 secretion from Kupffer cells due to the estrogen signal (Naugler et al., 2007; Liu and Liu, 2014). There are also some study showing that estrogen decreases the activity of the transcription factors nuclear factor κB (NF-κB) and CCAAT/Enhancer Binding Protein (C/EBPβ), thereby reducing the promoter activity of IL6 (Stein and Yang, 1995).
There are few studies of the association between estrogen and RHOA or GLIPR1 in liver cancer. According to Sailland’s study, inhibition of estrogen-related receptor α (ERRα) in breast cancer increases the stability and activation of RhoA protein, which affects cell migration (Sailland et al., 2014). In another study, RHOA and sex hormones were found to have no significant relevance in the primary breast cancer (Bellizzi et al., 2008). There are no studies on the relationship between GLIPR1 and estrogen. In this study, the relationship between RHOA or GLIPR1 expression and sex-biased OS in liver cancer patients shows the need for further study.
SEX-BIASED MOLECULAR SIGNATURE FOR OVERALL SURVIVAL OF LIVER CANCER PATIENTS
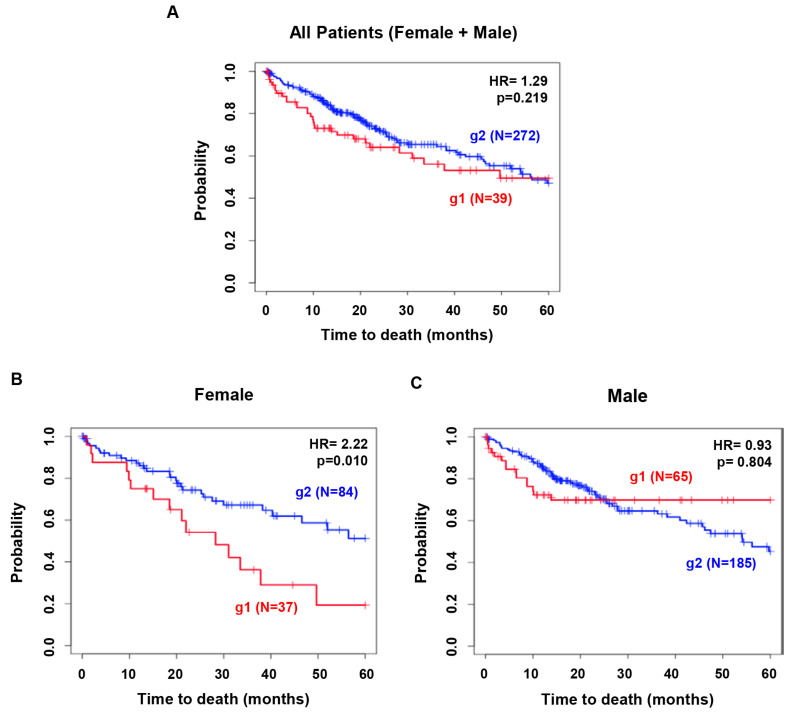
In order to predict clinical outcomes of liver cancer in a sex-biased manner, four genes in this study were investigated further the possibility as a sex-biased molecular signature. To this end, these four genes grouped together and analyzed the TCGA database for overall survival of male and female liver cancer patients. The patients were stratified into two groups based on expression of the genes: group1 (g1) had the profile CTNNB1High, IL6High, RHOAHigh and GLIPR1Low, whereas group 2 (g2) included the other patients. For each gene, high and low expression were determined by comparison with the average expression level for each gene.
As shown in Fig. 2A, KM plot analysis exhibited no significant difference in OS probability between g1 and g2 when male and female data were analyzed together. However, when data from males and females were analyzed individually, different results were obtained. In female patients (N=121), g1 exhibited worse prognostic outcomes compared to g2 (HR=2.224, p=0.010, Fig. 2B). In contrast, in male patients (N=250), there was no significant difference in OS between the two groups (HR=0.931, p=0.805, Fig. 2C). Taken together, these results suggest that high expression of CTNNB1, IL6, and RHOA and low expression of GLIPR1 may serve as a prognostic biomarker for female liver cancer patients. The sex-biased molecular signature CTNNB1High, IL6High, RHOAHigh, and GLIPR1Low proposed in this study may serve as an index for prediction and evaluation of OS in liver cancer, specifically in female liver cancer patients.
Fig. 2.
Sex difference of the marker genes in predicting the clinical outcomes of liver cancer patients. Group 1 (g1), liver cancer patients with high expression of CTNNB1, IL6, and RHOA and low expression of GLIPR1; Group 2 (g2), the other patients. (A) In all liver cancer patients; (B) In female and (C) male liver cancer patients.
CONCLUSION
Sex/gender is an important biological variable that should be considered in all cancer research that aims to improve targeted therapies (Gabriele et al., 2016). Diseases cover both sex and gender concepts. In the present study, we demonstrated that the expression pattern of a set of genes (CTNNB1High, IL6High, RHOAHigh, and GLIPR1Low) was associated with poor OS probability in female liver cancer patients. The sex-biased molecular signature proposed in this study can be used to predict and evaluate the prognosis of liver cancer specifically in female patients. Further studies are needed to elucidate the molecular mechanisms underlying our findings.
ACKNOWLEDGMENTS
The present study was supported by the Bio & Medical Technology Development Program (no. 2015M3A9B6074045) of the NRF funded by the Korean Government.
Footnotes
CONFLICT OF INTEREST
The authors declare no conflict of interest.
REFERENCES
- Addis R., Campesi I., Fois M., Capobianco G., Dessole S., Fenu G., Montella A., Cattaneo M. G., Vicentini L. M., Franconi F. Human umbilical endothelial cells (HUVECs) have a sex: characterisation of the phenotype of male and female cells. Biol. Sex Differ. 2014;5:18. doi: 10.1186/s13293-014-0018-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allahyar A., de Ridder J. FERAL: network-based classifier with application to breast cancer outcome prediction. Bioinformatics. 2015;31:i311–i319. doi: 10.1093/bioinformatics/btv255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awasthi A., Woolley A. G., Lecomte F. J., Hung N., Baguley B. C., Wilbanks S. M., Jeffs A. R., Tyndall J. D. Variable expression of GLIPR1 correlates with invasive potential in melanoma cells. Front. Oncol. 2013;3:225. doi: 10.3389/fonc.2013.00225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babaei S., Hulsman M., Reinders M., de Ridder J. Detecting recurrent gene mutation in interaction network context using multi-scale graph diffusion. BMC Bioinformatics. 2013;14:29. doi: 10.1186/1471-2105-14-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker N. The canonical Wnt/beta-catenin signalling pathway. Methods Mol. Biol. 2008;468:5–15. doi: 10.1007/978-1-59745-249-6_1. [DOI] [PubMed] [Google Scholar]
- Behari J. The Wnt/β-catenin signaling pathway in liver biology and disease. Expert Rev. Gastroenterol. Hepatol. 2010;4:745–756. doi: 10.1586/egh.10.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellizzi A., Mangia A., Chiriatti A., Petroni S., Quaranta M., Schittulli F., Malfettone A., Cardone R. A., Paradiso A., Reshkin S. J. RhoA protein expression in primary breast cancers and matched lymphocytes is associated with progression of the disease. Int. J. Mol. Med. 2008;22:25–31. doi: 10.3892/ijmm.22.1.25. [DOI] [PubMed] [Google Scholar]
- Bianchi I., Lleo A., Gershwin M. E., Invernizzi P. The X chromosome and immune associated genes. J. Autoimmun. 2012;38:J187–J192. doi: 10.1016/j.jaut.2011.11.012. [DOI] [PubMed] [Google Scholar]
- Borrás C., Sastre J., García-Sala D., Lloret A., Pallardó F. V., Viña J. Mitochondria from females exhibit higher antioxidant gene expression and lower oxidative damage than males. Free Radic. Biol. Med. 2003;34:546–552. doi: 10.1016/S0891-5849(02)01356-4. [DOI] [PubMed] [Google Scholar]
- Bottarelli L., Azzoni C., Necchi F., Lagrasta C., Tamburini E., D'Adda T., Pizzi S., Sarli L., Rindi G., Bordi C. Sex chromosome alterations associate with tumor progression in sporadic colorectal carcinomas. Clin. Cancer Res. 2007;13:4365–4370. doi: 10.1158/1078-0432.CCR-06-2736. [DOI] [PubMed] [Google Scholar]
- Bøvelstad H. M., Nygård S., Størvold H. L., Aldrin M., Borgan Ø., Frigessi A., Lingjaerde O. C. Predicting survival from microarray data-a comparative study. Bioinformatics. 2007;23:2080–2087. doi: 10.1093/bioinformatics/btm305. [DOI] [PubMed] [Google Scholar]
- Bray F., Ferlay J., Soerjomataram I., Siegel R. L., Torre L. A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- Budhu A., Jia H. L., Forgues M., Liu C., Goldstein D., Lam A., Zanetti K. A., Ye Q., Qin L., Croce C. M., Tang Z., Wang X. W. Identification of metastasis-related microRNAs in hepatocellular carcinoma. Hepatology. 2008;47:897–907. doi: 10.1002/hep.22160. [DOI] [PubMed] [Google Scholar]
- Cantuti-Castelvetri I., Keller-McGandy C., Bouzou B., Asteris G., Clark T. W., Frosch M. P., Standaert D. G. Effects of gender on nigral gene expression and parkinson disease. Neurobiol. Dis. 2007;26:606–614. doi: 10.1016/j.nbd.2007.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton J. A., Tannenbaum C. Reporting sex, gender, or both in clinical research? JAMA. 2016;316:1863–1864. doi: 10.1001/jama.2016.16405. [DOI] [PubMed] [Google Scholar]
- Cohen A. L., Soldi R., Zhang H., Gustafson A. M., Wilcox R., Welm B. E., Chang J. T., Johnson E., Spira A., Jeffrey S. S., Bild A. H. A pharmacogenomic method for individualized prediction of drug sensitivity. Mol. Syst. Biol. 2011;7:513. doi: 10.1038/msb.2011.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook M. B., Dawsey S. M., Freedman N. D., Inskip P. D., Wichner S. M., Quraishi S. M., Devesa S. S., McGlynn K. A. Sex disparities in cancer incidence by period and age. Cancer Epidemiol. Biomarkers Prev. 2009;18:1174–1182. doi: 10.1158/1055-9965.EPI-08-1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rienzo A., Archer M. A., Yeap B. Y., Dao N., Sciaranghella D., Sideris A. C., Zheng Y., Holman A. G., Wang Y. E., Dal Cin P. S., Fletcher J. A., Rubio R., Croft L., Quackenbush J., Sugarbaker P. E., Munir K. J., Battilana J. R., Gustafson C. E., Chirieac L. R., Ching S. M., Wong J., Tay L. C., Rudd S., Hercus R., Sugarbaker D. J., Richards W. J., Bueno R. Gender-specific molecular and clinical features underlie malignant pleural mesothelioma. Cancer Res. 2016;76:319–328. doi: 10.1158/0008-5472.CAN-15-0751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Maarri O., Becker T., Junen J., Manzoor S. S., Diaz-Lacava A., Schwaab R., Wienker T., Oldenburg J. Gender specific differences in levels of DNA methylation at selected loci from human total blood: a tendency toward higher methylation levels in males. Hum. Genet. 2007;122:505–514. doi: 10.1007/s00439-007-0430-3. [DOI] [PubMed] [Google Scholar]
- Erstad D. J., Fuchs B. C., Tanabe K. K. Molecular signatures in hepatocellular carcinoma: a step toward rationally designed cancer therapy. Cancer. 2018;124:3084–3104. doi: 10.1002/cncr.31257. [DOI] [PubMed] [Google Scholar]
- Evanson J. R., Guyton M. K., Oliver D. L., Hire J. M., Topolski R. L., Zumbrun S. D., McPherson J. C., Bojescul J. A. Gender and age differences in growth factor concentrations from platelet-rich plasma in adults. Mil. Med. 2014;179:799–805. doi: 10.7205/MILMED-D-13-00336. [DOI] [PubMed] [Google Scholar]
- Fish E. N. The X-files in immunity: sex-based differences predispose immune responses. Nat. Rev. Immunol. 2008;8:737–744. doi: 10.1038/nri2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriele L., Buoncervello M., Ascione B., Bellenghi M., Matarrese P., Carè A. The gender perspective in cancer research and therapy: novel insights and on-going hypotheses. Ann. Ist. Super. Sanita. 2016;52:213–222. doi: 10.4415/ANN_16_02_13. [DOI] [PubMed] [Google Scholar]
- Garcia-Herreros M., Aparicio I. M., Rath D., Fair T., Lonergan P. Differential glycolytic and glycogenogenic transduction pathways in male and female bovine embryos produced in vitro. Reprod. Fertil. Dev. 2012;24:344–352. doi: 10.1071/RD11080. [DOI] [PubMed] [Google Scholar]
- Ghuwalewala S., Ghatak D., Das P., Dey S., Sarkar S., Alam N., Panda C. K., Roychoudhury S. CD44(high)CD24(low) molecular signature determines the Cancer Stem Cell and EMT phenotype in Oral Squamous Cell Carcinoma. Stem Cell Res. 2016;16:405–417. doi: 10.1016/j.scr.2016.02.028. [DOI] [PubMed] [Google Scholar]
- Gleicher N., Barad D. H. Gender as risk factor for autoimmune diseases. J. Autoimmun. 2007;28:1–6. doi: 10.1016/j.jaut.2006.12.004. [DOI] [PubMed] [Google Scholar]
- Graveley B. R. Sorting out the complexity of SR protein functions. RNA. 2000;6:1197–1211. doi: 10.1017/S1355838200000960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guy J., Peters M. G. Liver disease in women: the influence of gender on epidemiology, natural history, and patient outcomes. Gastroenterol. Hepatol. 2013;10:633–639. [PMC free article] [PubMed] [Google Scholar]
- Haley W. E., Roth D. L., Howard G., Safford M. M. Caregiving strain and estimated risk for stroke and coronary heart disease among spouse caregivers: differential effects by race and sex. Stroke. 2010;41:331–336. doi: 10.1161/STROKEAHA.109.568279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D., Weinberg R. A. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Harish G., Venkateshappa C., Mahadevan A., Pruthi N., Bharath M. M., Shankar S. K. Mitochondrial function in human brains is affected by pre- and post mortem factors. Neuropathol. Appl. Neurobiol. 2013;39:298–315. doi: 10.1111/j.1365-2990.2012.01285.x. [DOI] [PubMed] [Google Scholar]
- Heasman S. J., Ridley A. J. Mammalian Rho GTPases: new insights into their functions from in vivo studies. Nat. Rev. Mol. Cell Biol. 2008;9:690–701. doi: 10.1038/nrm2476. [DOI] [PubMed] [Google Scholar]
- Heidari S., Babor T. F., De Castro P., Tort S., Curno M. Sex and gender equity in research: rationale for the SAGER guidelines and recommended use. Res. Integr. Peer Rev. 2016;1:2. doi: 10.1186/s41073-016-0016-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hines A., Staff F. J., Widdows J., Compton R. M., Falciani F., Viant M. R. Discovery of metabolic signatures for predicting whole organism toxicology. Toxicol. Sci. 2010;115:369–378. doi: 10.1093/toxsci/kfq004. [DOI] [PubMed] [Google Scholar]
- Hou J., Xu J., Jiang R., Wang Y., Chen C., Deng L., Huang X., Wang X., Sun B. Estrogen-sensitive PTPRO expression represses hepatocellular carcinoma progression by control of STAT3. Hepatology. 2013;57:678–688. doi: 10.1002/hep.25980. [DOI] [PubMed] [Google Scholar]
- Hur J., Sullivan K. A., Pande M., Hong Y., Sima A. A. F., Jagadish H. V., Kretzler M., Feldman E. L. The identification of gene expression profiles associated with progression of human diabetic neuropathy. Brain. 2011;134:3222–3235. doi: 10.1093/brain/awr228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ippolito J. E., Yim A. K., Luo J., Chinnaiyan P., Rubin J. B. Sexual dimorphism in glioma glycolysis underlies sex differences in survival. JCI Insight. 2017;2:e92142. doi: 10.1172/jci.insight.92142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y., Li Y., Jiang P., Han W., Liu Y. PGM5: a novel diagnostic and prognostic biomarker for liver cancer. PeerJ. 2019;7:e7070. doi: 10.7717/peerj.7070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung K. W., Won Y. J., Kong H. J., Lee E. S. Cancer statistics in Korea: incidence, mortality, survival, and prevalence in 2016. Cancer Res. Treat. 2019;51:417–430. doi: 10.4143/crt.2019.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein S. L., Flanagan K. L. Sex differences in immune responses. Nat. Rev. Immunol. 2016;16:626–638. doi: 10.1038/nri.2016.90. [DOI] [PubMed] [Google Scholar]
- Klinge C. M. miRNAs and estrogen action. Trends Endocrinol. Metab. 2012;23:223–233. doi: 10.1016/j.tem.2012.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohi M. P. Gender-related differences in hepatocellular carcinoma: does sex matter? J. Vasc. Interv. Radiol. 2016;27:1338–1341. doi: 10.1016/j.jvir.2016.06.035. [DOI] [PubMed] [Google Scholar]
- Kouzmenko A. P., Takeyama K., Ito S., Furutani T., Sawatsubashi S., Maki A., Suzuki E., Kawasaki Y., Akiyama T., Tabata T., Kato T. Wnt/beta-catenin and estrogen signaling converge in vivo. J. Biol. Chem. 2004;279:40255–40258. doi: 10.1074/jbc.C400331200. [DOI] [PubMed] [Google Scholar]
- Krumsiek J., Mittelstrass K., Do K. T., Stückler F., Ried J., Adamski J., Peters A., Illig T., Kronenberg F., Friedrich N., Nauck M., Pietzner M., Mook-Kanamori D. O., Suhre K., Gieger C., Grallert H., Theis F. J., Kastenmüller G. Gender-specific pathway differences in the human serum metabolome. Metabolomics. 2015;11:1815–1833. doi: 10.1007/s11306-015-0829-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumari N., Dwarakanath B. S., Das A., Bhatt A. N. Role of interleukin-6 in cancer progression and therapeutic resistance. Tumour Biol. 2016;37:11553–11572. doi: 10.1007/s13277-016-5098-7. [DOI] [PubMed] [Google Scholar]
- Laffont S., Blanquart E., Guéry J. C. Sex differences in asthma: a key role of androgen-signaling in group 2 innate lymphoid cells. Front. Immunol. 2017;8:1069. doi: 10.3389/fimmu.2017.01069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C. H., Haider S., Shiah Y. J., Thai K., Boutros P. C. Sex differences in cancer driver genes and biomarkers. Cancer Res. 2018;78:5527–5537. doi: 10.1158/0008-5472.CAN-18-0362. [DOI] [PubMed] [Google Scholar]
- Li L., Ren C., Yang G., Fattah E. A., Goltsov A. A., Kim S. M., Lee J. S., Park S., Demayo F. J., Ittmann M. M., Troncoso P., Thompson T. C. GLIPR1 suppresses prostate cancer development through targeted oncoprotein destruction. Cancer Res. 2011;71:7694–7704. doi: 10.1158/0008-5472.CAN-11-1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Kang K., Krahn J. M., Croutwater N., Lee K., Umbach D. M., Li L. A comprehensive genomic pan-cancer classification using The Cancer Genome Atlas gene expression data. BMC Genomics. 2017;18:508. doi: 10.1186/s12864-017-3906-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Tuteja G., Schug J., Kaestner K. H. Foxa1 and Foxa2 are essential for sexual dimorphism in liver cancer. Cell. 2012;148:72–83. doi: 10.1016/j.cell.2011.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libert C., Dejager L., Pinheiro I. The X chromosome in immune functions: when a chromosome makes the difference. Nat. Rev. Immunol. 2010;10:594–604. doi: 10.1038/nri2815. [DOI] [PubMed] [Google Scholar]
- Lin C. W., Lin C. C., Mo L. R., Chang C. Y., Perng D. S., Hsu C. C., Lo G. H., Chen Y. S., Yen Y. C., Hu J. T., Yu M. L., Lee P. H., Lin J. T., Yang S. S. Heavy alcohol consumption increases the incidence of hepatocellular carcinoma in hepatitis B virus-related cirrhosis. J. Hepatol. 2013;58:730–735. doi: 10.1016/j.jhep.2012.11.045. [DOI] [PubMed] [Google Scholar]
- Liu F., Yuan J. H., Huang J. F., Yang F., Wang T. T., Ma J. Z., Zhang L., Zhou C. C., Wang F., Yu J., Zhou W. P., Sun S. H. Long noncoding RNA FTX inhibits hepatocellular carcinoma proliferation and metastasis by binding MCM2 and miR-374a. Oncogene. 2016;35:5422–5434. doi: 10.1038/onc.2016.80. [DOI] [PubMed] [Google Scholar]
- Liu W. C., Liu Q. Y. Molecular mechanisms of gender disparity in hepatitis B virus-associated hepatocellular carcinoma. World J. Gastroenterol. 2014;20:6252–6261. doi: 10.3748/wjg.v20.i20.6252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masoomian B., Shields J. A., Shields C. L. Overview of BAP1 cancer predisposition syndrome and the relationship to uveal melanoma. J. Curr. Ophthalmol. 2018;30:102–109. doi: 10.1016/j.joco.2018.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehrabian M., Allayee H., Stockton J., Lum P. Y., Drake T. A., Castellani L. W., Suh M., Armour C., Edwards S., Lamb J., Lusis A. J., Schadt E. E. Integrating genotypic and expression data in a segregating mouse population to identify 5-lipoxygenase as a susceptibility gene for obesity and bone traits. Nat. Genet. 2007;37:1224–1233. doi: 10.1038/ng1619. [DOI] [PubMed] [Google Scholar]
- Mervic L. Time course and pattern of metastasis of cutaneous melanoma differ between men and women. PLoS ONE. 2012;7:e32955. doi: 10.1371/journal.pone.0032955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittelstrass K., Ried J. S., Yu Z., Krumsiek J., Gieger C., Prehn C., Roemisch-Margl W., Polonikov A., Peters A., Theis F. J., Meitinger T., Kronenberg F., Weidinger S., Wichmann H. E., Suhre K., Wang-Sattler R., Adamski J., Illig T. Discovery of sexual dimorphisms in metabolic and genetic biomarkers. PLoS Genet. 2011;7:e1002215. doi: 10.1371/journal.pgen.1002215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosley M., Weathington J., Cortes L. R., Bruggeman E., Castillo-Ruiz A., Xue B., Forger N. G. Neonatal inhibition of DNA methylation alters cell phenotype in sexually dimorphic regions of the mouse brain. Endocrinology. 2017;158:1838–1848. doi: 10.1210/en.2017-00205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulligan M. A., Wegner K. A., Keil K. P., Mehta V., Taketo M. M., Vezina C. M. Beta-catenin and estrogen signaling collaborate to drive cyclin D1 expression in developing mouse prostate. Differentiation. 2017;93:66–71. doi: 10.1016/j.diff.2016.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy E. V., Zhang Y., Zhu W., Biggs J. The human glioma pathogenesis-related protein is structurally related to plant pathogenesis-related proteins and its gene is expressed specifically in brain tumors. Gene. 1995;159:131–135. doi: 10.1016/0378-1119(95)00061-A. [DOI] [PubMed] [Google Scholar]
- Naugler W. E., Sakurai T., Kim S., Maeda S., Kim K., Elsharkawy A. M., Karin M. Gender disparity in liver cancer due to sex differences in MyD88-dependent IL-6 production. Science. 2007;317:121–124. doi: 10.1126/science.1140485. [DOI] [PubMed] [Google Scholar]
- Niu D., Zhang J., Ren Y., Feng H., Chen W. N. HBx genotype D represses GSTP1 expression and increases the oxidative level and apoptosis in HepG2 cells. Mol. Oncol. 2009;3:67–76. doi: 10.1016/j.molonc.2008.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson R., Björkegren J., Tegnér J. On reliable discovery of molecular signatures. BMC Bioinformatics. 2009;10:38. doi: 10.1186/1471-2105-10-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nugent B. M., Wright C. L., Shetty A. C., Hodes G. E., Lenz K. M., Mahurkar A., Russo S. J., Devine S. E., McCarthy M. M. Brain feminization requires active repression of masculinization via DNA methylation. Nat. Neurosci. 2015;18:690–697. doi: 10.1038/nn.3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal S. K., Hurria A. Impact of age, sex, and comorbidity on cancer therapy and disease progression. J. Clin. Oncol. 2010;28:4086–4093. doi: 10.1200/JCO.2009.27.0579. [DOI] [PubMed] [Google Scholar]
- Pelletier R., Khan N. A., Cox J., Daskalopoulou S. S., Eisenberg M. J., Bacon S. L., Lavoie K. L., Daskupta K., Rabi D., Humphries K. H., Norris C. M., Thanassoulis G., Behlouli H., Pilote L. GENESIS-PRAXY Investigators, . Sex versus gender-related characteristics: which predicts outcome after acute coronary syndrome in the young? J. Am. Coll. Cardiol. 2016;67:127–135. doi: 10.1016/j.jacc.2015.10.067. [DOI] [PubMed] [Google Scholar]
- Pinheiro I., Dejager L., Libert C. X-chromosome-located microRNAs in immunity: might they explain male/female differences? The X chromosome-genomic context may affect X-located miRNAs and downstream signaling, thereby contributing to the enhanced immune response of females. Bioessays. 2011;33:791–802. doi: 10.1002/bies.201100047. [DOI] [PubMed] [Google Scholar]
- Pittman J., Huang E., Dressman H., Horng C. F., Cheng S. H., Tsou M., Chen C., Bild A., Iversen E. S., Huang A. T., Nevins J. R., West M. Integrated modeling of clinical and gene expression information for personalized prediction of disease outcomes. Proc. Natl. Acad. Sci. U.S.A. 2004;101:8431–8436. doi: 10.1073/pnas.0401736101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu J., Peng B., Tang Y., Qian Y., Guo P., Li M., Luo J., Chen B., Tang H., Lu C., Cai M., Ke Z., He W., Zheng Y., Xie D., Li B., Yuan Y. CpG methylation signature predicts recurrence in early-stage hepatocellular carcinoma: results from a multicenter study. J. Clin. Oncol. 2017;35:734–742. doi: 10.1200/JCO.2016.68.2153. [DOI] [PubMed] [Google Scholar]
- Qu K., Liu S. S., Wang Z. X., Huang Z. C., Liu S. N., Chang H. L., Xu X. S., Lin T., Dong Y. F., Liu C. Polymorphisms of glutathione S-transferase genes and survival of resected hepatocellular carcinoma patients. World J. Gastroenterol. 2015;21:4310–4322. doi: 10.3748/wjg.v21.i14.4310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahbari R., Zhang L., Kebebew E. Thyroid cancer gender disparity. Future Oncol. 2010;6:1771–1779. doi: 10.2217/fon.10.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebouissou S., Franconi A., Calderaro J., Letouzé E., Imbeaud S., Pilati C., Nault J. C., Couchy G., Laurent A., Balabaud C., Bioulac-Sage P., Zucman-Rossi J. Genotype-phenotype correlation of CTNNB1 mutations reveals different ß-catenin activity associated with liver tumor progression. Hepatology. 2016;64:2047–2061. doi: 10.1002/hep.28638. [DOI] [PubMed] [Google Scholar]
- Reizel Y., Spiro A., Sabag O., Skversky Y., Hecht M., Keshet I., Berman B. P., Cedar H. Gender-specific postnatal demethylation and establishment of epigenetic memory. Genes Dev. 2015;29:923–933. doi: 10.1101/gad.259309.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren C., Li L., Yang G., Timme T. L., Goltsov A., Ren C., Ji X., Addai J., Luo H., Ittmann M. M., Thompson T. C. RTVP-1, a tumor suppressor inactivated by methylation in prostate cancer. Cancer Res. 2004;64:969–976. doi: 10.1158/0008-5472.CAN-03-2592. [DOI] [PubMed] [Google Scholar]
- Rich N. E., Murphy C. C., Yopp A. C., Tiro J., Marrero J. A., Singal A. G. Sex disparities in presentation and prognosis of 1110 patients with hepatocellular carcinoma. Aliment. Pharmacol. Ther. 2020;52:701–709. doi: 10.1111/apt.15917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin J. B., Lagas J. S., Broestl L., Sponagel J., Rockwell N., Rhee G., Rosen S. F., Chen S., Klein R. S., Imoukhuede P., Luo J. Sex differences in cancer mechanisms. Biol. Sex Differ. 2020;11:17. doi: 10.1186/s13293-020-00291-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sailland J., Tribollet V., Forcet C., Billon C., Barenton B., Carnesecchi J., Bachmann A., Gauthier K. C., Yu S., Giguère V., Chan F. L., Vanacker J. Estrogen-related receptor α decreases RHOA stability to induce orientated cell migration. Proc. Natl. Acad. Sci. U.S.A. 2014;111:15108–15113. doi: 10.1073/pnas.1402094111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salehi S., Jafarian A. H., Montazer M., Moghbeli M., Forghanifard M. M. BRUCE protein, new marker for targeted therapy of gastric carcinoma. J. Gastrointest. Cancer. 2017;48:151–155. doi: 10.1007/s12029-016-9874-9. [DOI] [PubMed] [Google Scholar]
- Scosyrev E., Noyes K., Feng C., Messing E. Sex and racial differences in bladder cancer presentation and mortality in the US. Cancer. 2009;115:68–74. doi: 10.1002/cncr.23986. [DOI] [PubMed] [Google Scholar]
- Scotland R. S., Stables M. J., Madalli S., Watson P., Gilroy D. W. Sex differences in resident immune cell phenotype underlie more efficient acute inflammatory responses in female mice. Blood. 2011;118:5918–5927. doi: 10.1182/blood-2011-03-340281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setiawan V. W., Lim U., Lipworth L., Lu S. C., Shepherd J., Ernst T., Wilkens L. R., Henderson B. E., Le Marchand L. Sex and ethnic differences in the association of obesity with risk of hepatocellular carcinoma. Clin. Gastroenterol. Hepatol. 2016;14:309–316. doi: 10.1016/j.cgh.2015.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J. Y., Jung H. J., Moon A. Molecular markers in sex differences in cancer. Toxicol. Res. 2019;35:331–341. doi: 10.5487/TR.2019.35.4.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirasaki F., Takata M., Hatta N., Takehara K. Loss of expression of the metastasis suppressor gene KiSS1 during melanoma progression and its association with LOH of chromosome 6q16.3-q23. Cancer Res. 2001;61:7422–7425. [PubMed] [Google Scholar]
- Skavdahl M., Steenbergen C., Clark J., Myers P., Demianenko T., Mao L., Rockman H. A., Korach K. S., Murphy E. Estrogen receptor-beta mediates male-female differences in the development of pressure overload hypertrophy. Am. J. Physiol. Heart Circ. Physiol. 2005;288:469–476. doi: 10.1152/ajpheart.00723.2004. [DOI] [PubMed] [Google Scholar]
- Smalley K. S. Why do women with melanoma do better than men? Elife. 2018;7:e33511. doi: 10.7554/eLife.33511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth E. C., Fassan M., Cunningham D., Allum W. H., Okines A. F. C., Lampis A., Hahne J. C., Rugge M., Peckitt C., Nankivell M., Langley R., Ghidini M., Braconi C., Wotherspoon A., Grabsch H. I., Valeri N. Effect of pathologic tumor response and nodal status on survival in the medical research council adjuvant gastric infusional chemotherapy trial. J. Clin. Oncol. 2016;34:2721–2727. doi: 10.1200/JCO.2015.65.7692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth E. C., Nyamundanda G., Cunningham D., Fontana E., Ragulan C., Tan I. B., Lin S. J., Wotherspoon A., Nankivell M., Fassan M., Lampis A., Hahne J. C., Davies A. R., Lagergren J., Gossage J. A., Maisey N., Green M., Zylstra J. L., Allum W. H., Langley R. E., Tan P., Valeri N., Sadanandam A. A seven-Gene Signature assay improves prognostic risk stratification of perioperative chemotherapy treated gastroesophageal cancer patients from the MAGIC trial. Ann. Oncol. 2018;29:2356–2362. doi: 10.1093/annonc/mdy407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snell D. M., Turner J. M. A. Sex chromosome effects on male-female differences in mammals. Curr. Biol. 2018;28:R1313–R1324. doi: 10.1016/j.cub.2018.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- So A. R., Si J. M., Lopez D., Pellegrini M. Molecular signatures for inflammation vary across cancer types and correlate significantly with tumor stage, sex and vital status of patients. PLoS ONE. 2020;15:e0221545. doi: 10.1371/journal.pone.0221545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spatz A., Borg C., Feunteun J. X-chromosome genetics and human cancer. Nat. Rev. Cancer. 2004;4:617–629. doi: 10.1038/nrc1413. [DOI] [PubMed] [Google Scholar]
- Stein B., Yang M. X. Repression of the interleukin-6 promoter by estrogen receptor is mediated by NF-kappa B and C/EBP beta. Mol. Cell. Biol. 1995;15:4971–4979. doi: 10.1128/MCB.15.9.4971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung J., Wang Y., Chandrasekaran S., Witten D. M., Price N. D. Molecular signatures from omics data: from chaos to consensus. Biotechnol. J. 2012;7:946–957. doi: 10.1002/biot.201100305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H., Wang S., Xiao G., Schiller J., Papadimitrakopoulou V., Minna J., Wistuba I., Xie Y. Comprehensive evaluation of published gene expression prognostic signatures for biomarker-based lung cancer clinical studies. Ann. Oncol. 2017;28:733–740. doi: 10.1093/annonc/mdw683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobi E. W., Lumey L. H., Talens R. P., Kremer D., Putter H., Stein A. D., Slagboom P. E., Heijmans B. T. DNA methylation differences after exposure to prenatal famine are common and timing- and sex-specific. Hum. Mol. Genet. 2009;18:4046–4053. doi: 10.1093/hmg/ddp353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Vijver M. J., He Y. D., van't Veer L. J., Dai H., Hart A. A., Voskuil D. W., Schreiber G. J., Peterse J. L., Roberts C., Marton M. J., Parrish M., Atsma D., Witteveen A., Glas A., Delahaye L., van der Velde T., Bartelink H., Rodenhuis S., Rutgers E. T., Friend S. H., Bernards R. A gene-expression signature as a predictor of survival in breast cancer. N. Engl. J. Med. 2002;347:1999–2009. doi: 10.1056/NEJMoa021967. [DOI] [PubMed] [Google Scholar]
- Villa E., Critelli R., Lei B., Marzocchi G., Cammà C., Giannelli G., Pontisso P., Cabibbo G., Enea M., Colopi S., Caporali C., Pollicino T., Milosa F., Karampatou A., Todesca P., Bertolini E., Maccio L., Martinez-Chantar M. L., Turola E., Del Buono M., De Maria N., Ballestri S., Schepis F., Loria P., Gerunda G. E., Losi L., Cillo U. Neoangiogenesis-related genes are hallmarks of fast-growing hepatocellular carcinomas and worst survival. Results from a prospective study. Gut. 2016;65:861–869. doi: 10.1136/gutjnl-2014-308483. [DOI] [PubMed] [Google Scholar]
- Wang R., Ye X. H., Zhao X. L., Liu J. L., Zhang C. Y. Development of a five-gene signature as a novel prognostic marker in ovarian cancer. Neoplasma. 2019;66:343–349. doi: 10.4149/neo_2018_180705N447. [DOI] [PubMed] [Google Scholar]
- Wang Y., Xing T., Huang L., Song G., Sun X., Zhong L., Fan J., Yan D., Zhou C., Cui F., Yu F., Chen J., Yu Y., Li C., Tang H., Peng Z., Wang X. Period 1 and estrogen receptor-beta are downregulated in Chinese colon cancers. Int. J. Clin. Exp. Pathol. 2015;8:8178–8188. [PMC free article] [PubMed] [Google Scholar]
- Welzel T. M., Graubard B. I., Quraishi S., Zeuzem S., Davila J. A., El-Serag H. B., McGlynn K. A. Population-attributable fractions of risk factors for hepatocellular carcinoma in the United States. Am. J. Gastroenterol. 2013;108:1314–1321. doi: 10.1038/ajg.2013.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijchers P. J., Festenstein R. J. Epigenetic regulation of autosomal gene expression by sex chromosomes. Trends Genet. 2011;27:132–140. doi: 10.1016/j.tig.2011.01.004. [DOI] [PubMed] [Google Scholar]
- Wu E. M., Wong L. L., Hernandez B. Y., Ji J. F., Jia W., Kwee S. A., Kalathil S. Gender differences in hepatocellular cancer: disparities in nonalcoholic fatty liver disease/steatohepatitis and liver transplantation. Hepatoma Res. 2018;4:66. doi: 10.20517/2394-5079.2018.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Yao N., Feng Y., Tian Z., Yang Y., Zhao Y. Identification and characterization of sexual dimorphism-linked gene expression profile in hepatocellular carcinoma. Oncol. Rep. 2019;42:937–952. doi: 10.3892/or.2019.7217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia J., Urabe K., Moroi Y., Koga T., Duan H., Li Y., Furue M. beta-Catenin mutation and its nuclear localization are confirmed to be frequent causes of Wnt signaling pathway activation in pilomatricomas. J. Dermatol. Sci. 2006;41:67–75. doi: 10.1016/j.jdermsci.2005.09.005. [DOI] [PubMed] [Google Scholar]
- Xie L., Xie L., Kinnings S. L., Bourne P. E. Novel computational approaches to polypharmacology as a means to define responses to individual drugs. Annu. Rev. Pharmacol. Toxicol. 2012;52:361–379. doi: 10.1146/annurev-pharmtox-010611-134630. [DOI] [PubMed] [Google Scholar]
- Xiong G., Lingampalli N., Koltsov J. C. B., Leung L. L., Bhutani N., Robinson W. H., Chu C. R. Men and women differ in the biochemical composition of platelet-rich plasma. Am. J. Sports Med. 2018;46:409–419. doi: 10.1177/0363546517740845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G., Zhang M., Zhu H., Xu J. A 15-gene signature for prediction of colon cancer recurrence and prognosis based on SVM. Gene. 2017;604:33–40. doi: 10.1016/j.gene.2016.12.016. [DOI] [PubMed] [Google Scholar]
- Xu J., Deng X., Watkins R., Disteche C. M. Sex-specific differences in expression of histone demethylases Utx and Uty in mouse brain and neurons. J. Neurosci. 2008;28:4521–4527. doi: 10.1523/JNEUROSCI.5382-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaron M., Renner U., Gilad S., Stalla G. K., Stern N., Greenman Y. KISS1 receptor is preferentially expressed in clinically non-functioning pituitary tumors. Pituitary. 2015;18:306–311. doi: 10.1007/s11102-014-0572-y. [DOI] [PubMed] [Google Scholar]
- Yu L., Liu X., Wang X., Dang Z., Jiang Y., Wang X., Yang Z. Impact of gender as a prognostic factor in HBV-related hepatocellular carcinoma: the survival strength of female patients in BCLC stage 0-B. J. Cancer. 2019;10:4237–4244. doi: 10.7150/jca.33430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuasa Y. Epigenetics in molecular epidemiology of cancer a new scope. Adv. Genet. 2010;71:211–235. doi: 10.1016/B978-0-12-380864-6.00007-9. [DOI] [PubMed] [Google Scholar]
- Zhang J., Jiang H., Xia W., Jiang Y., Tan X., Liu P., Jia H., Yang X., Shen G. Serine-arginine protein kinase 1 is associated with hepatocellular carcinoma progression and poor patient survival. Tumour Biol. 2016;37:283–290. doi: 10.1007/s13277-015-3771-x. [DOI] [PubMed] [Google Scholar]
- Zhang L. J., Xiong Y., Nilubol N., He M., Bommareddi S., Zhu X., Jia L., Xiao Z., Park J. W., Xu X., Patel D., Willingham M. C., Cheng S. Y., Kebebew E. Testosterone regulates thyroid cancer progression by modifying tumor suppressor genes and tumor immunity. Carcinogenesis. 2015;36:420–428. doi: 10.1093/carcin/bgv001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L., Jiang L., He L., Wei Q., Bi J., Wang Y., Yu L., He M., Zhao L., Wei M. Identification of a novel cell cycle-related gene signature predicting survival in patients with gastric cancer. J. Cell. Physiol. 2019;234:6350–6360. doi: 10.1002/jcp.27365. [DOI] [PubMed] [Google Scholar]