Abstract
Artificial intelligence is one of the most exciting methodological shifts in our era. It holds the potential to transform healthcare as we know it, to a system where humans and machines work together to provide better treatment for our patients. It is now clear that cutting edge artificial intelligence models in conjunction with high-quality clinical data will lead to improved prognostic and diagnostic models in neurological disease, facilitating expert-level clinical decision tools across healthcare settings. Despite the clinical promise of artificial intelligence, machine and deep-learning algorithms are not a one-size-fits-all solution for all types of clinical data and questions. In this article, we provide an overview of the core concepts of artificial intelligence, particularly contemporary deep-learning methods, to give clinician and neuroscience researchers an appreciation of how artificial intelligence can be harnessed to support clinical decisions. We clarify and emphasize the data quality and the human expertise needed to build robust clinical artificial intelligence models in neurology. As artificial intelligence is a rapidly evolving field, we take the opportunity to iterate important ethical principles to guide the field of medicine is it moves into an artificial intelligence enhanced future.
Keywords: artificial intelligence, neurology, augmented intelligence, deep learning, ethics
Artificial intelligence can transform healthcare as we know it, to a system where humans and machines work together to facilitating expert-level clinical decision tools across healthcare settings. We review contemporary deep-learning methods and ethical principles in artificial intelligence, as we are moving into the future of precision medicine.
Graphical Abstract
Graphical Abstract.
Background—AI emulates human intelligence, processed by computer programs
The history of AI stems back to the 1950s with the introduction of the perceptron model (Rosenblatt, 1958; Minsky et al., 2017); however, it was not until the 1990s that machine-learning techniques became more widely utilized (Crevier, 1993). The development of machine-learning tools including support vector machine and recurrent neural networks (Sarle, 1994; Cortes and Vapnik, 1995; Kohavi, 1995) allowed scientists to leverage the computational power available in this era to build statistical models robust to data variation, and to make new inferences about real-world problems (Obermeyer and Emanuel, 2016). However, arguably the biggest advances in AI to date have come in the last decade, as massive scale data and hardware suitable to process these data have become available, and sophisticated deep-learning methods—that aim to imitate the working of the human brain in processing data—became computationally feasible (Ngiam et al., 2011; LeCun et al., 2015; Schmidhuber, 2015; Goodfellow et al., 2016). Deep learning is now widely regarded as the foundation of contemporary AI (Sejnowski, 2020) (Fig. 1 and Box 1).
Figure 1.
Definitions of AI: AI encompasses both ‘traditional’ machine learning and ‘contemporary’ deep-learning concepts.
Box 1.
Definitions of AI quoted from the select committee on AI, Committee Office, House of Lords, London (see https://publications.parliament.uk/pa/ld201719/ldselect/ldai/100/10005.htm)
AI: Technologies with the ability to perform tasks that would otherwise require human intelligence, such as visual perception, speech recognition and language translation.
Machine learning: One particular form of AI, which gives computers the ability to learn from and improve with experience, without being explicitly programmed. When provided with sufficient data, a machine-learning algorithm can learn to make predictions or solve problems, such as identifying objects in pictures or winning at particular games, for example.
Neural network: Also known as an artificial neural network, this is a type of machine learning loosely inspired by the structure of the human brain. A neural network is composed of simple-processing nodes, or ‘artificial neurons’, which are connected to another layer. Each node will receive data from several nodes ‘above’ it and give data to several nodes ‘below’ it. Nodes attach a ‘weight’ to the data they receive and attribute a value to that data. If the data does not pass a certain threshold, it is not passed on to another node. The weights and thresholds of the nodes are adjusted when the algorithm is trained until similar data input results in stable outputs.
Deep learning: A more recent variation of neural networks, which uses many layers of artificial neurons to solve more difficult problems. Its popularity as a technique increased from the mid- 2000s onwards, as it is behind much of the wider interest in AI today. It is often used to classify images, text or sound.
In medicine, AI has been most successfully used for image classification and prediction including detecting lung cancer and stroke based on computed tomography scans (Zhou et al., 2002; Lee et al., 2017; Chilamkurthy et al., 2018; Zhu et al., 2018; Ardila et al., 2019), assessing the risk of sudden cardiac death and other severe heart diseases based on electrocardiograms and cardiac MRI (Rahhal et al., 2016; Zhang et al., 2017; Faust et al., 2018; Hannun et al., 2019) and classifying abnormal skin lesions based on dermatological images (Jafari et al., 2016; Premaladha and Ravichandran, 2016; Codella et al., 2017; Esteva et al., 2017).
There are preliminary examples of the value of AI in neurology, for example in detecting structural brain lesions on MRI (Brosch et al., 2014; Korfiatis et al., 2016; Akkus et al., 2017; Zaharchuk et al., 2018). A common limitation of clinical AI studies is the amount of available data with high-quality clinical outcome labels, rather the availability of robust AI algorithms and computational resources. AI and deep learning are a framework that can potentially answer many disease-related questions through application of existing complex and comprehensive model architectures, so long as training data of sufficient quantity and quality is available (Box 2).
Box 2.
The clinical potential of AI—a case in point
A recent study found that AI can transform a person’s brain waves recorded during speech production into real text. Makin et al. (2020) studied four people with epilepsy who underwent brain surgery and had implanted electrodes directly over the inferior frontal cortices where words and speech are produced. The four epilepsy patients read sentences aloud, and brain signals from intracranial electrodes recorded during speech production were the inputs into an encoder recurrent neural network often used for data containing temporal information such as spontaneous brain activity. The output of the network (after training and updating) generated written text from speech with 97% accuracy. This highlights the clinical potential of AI if we have the right type of data and models available to answer specific questions. This has therapeutical implications to expedite speech rehabilitation for disorders that affect people’s ability to communicate.
Deep learning to extract high-level information from large and complex data
There exist several deep neural network architectures including deep neural networks, deep belief networks, recurrent neural networks and convolutional neural networks (see Sainath et al., 2015). There are also methods such as Generative Adversarial Network approaches, which utilize a pair of generator and discriminator networks to improve performance (Xing et al., 2019). All of these networks can learn information from large and unstructured data such as images and words, including modelling non-linear and high-dimensional features. They circumvent several limitations that have hampered efforts to translate conventional machine-learning approaches into medical biomarker discovery tools over the last decades (Ngiam et al., 2011; LeCun et al., 2015; Schmidhuber, 2015; Goodfellow et al., 2016).
In short, deep learning deals with, and leverages, vast amounts of information whereas traditional machine-learning methods require human intervention to reduce the size of data using various feature reduction and feature selection techniques (Mwangi et al., 2014; Hestness et al., 2017). An intuitive way to appreciate how deep-learning works comes from understanding the firing patterns of a neuron in the brain (Savage, 2019). A neuron in the brain, as well as a node within a deep-learning network, receives inputs that they transform to an output according to a set of predefined rules that aids learning (Fig. 2; LeCun et al., 2015; Daubechies et al., 2019).
Figure 2.
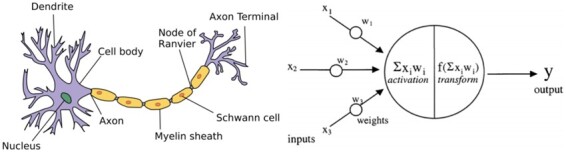
Biological and artificial neuron: on the left side of the figure is a biological neuron (reused under the terms of Creative Commons Attribution Licence—CC BY-SA 3.0—allowing for reproduction https://commons.wikimedia.org/wiki/ File: Neuron.svg), and on the right side of the figure is a model of an artificial neuron [reprinted from Agatonovic-Kustrin and Beresford (2000) with permission from Elsevier].
The similarity between neuronal function and AI is the reason why a deep-learning network is often called an artificial neural network (see Hassoun and Hassoun, 1995; Dreiseitl and Ohno-Machado, 2002 and Box 3). The sheer complexity of the brain, and deep-learning networks, arises from the interaction between multiple neurons in the brain, or multiple nodes in a deep-learning network, and how complex network interactions between multiple entities result in iterative learning. A deep-learning network learns by propagating information between multiple ‘hidden network layers’ (see Fig. 3, for a schematic overview). The hidden network layers comprise a non-linear transformation of the received input, and non-linearities make for very flexible transformations of the input data—i.e. a deep-learning neural network can ‘self-learn’ higher-order features from the input data.
Box 3.
Open question—biological relevance of artificial network back-propagation?
Although feed-forward propagation in deep neural networks mirrors the functioning of neurons in the brain, it is more difficult to reconcile how back-propagating errors updating in artificial networks is similar to the back-propagation of real neurons. Several theoretical accounts are attempting to outline the biological basis of the error updates that occur during back-propagation (e.g. Whittington and Bogacz, 2019; Lillicrap et al., 2020). For example, Lillicrap et al. (2020) proposed what they call backdrop-like learning with feedback network where neurons learn via feedback connections that convey errors transcoded with the changes in neuronal activity. Here, error updating of neurons is influenced by neuronal activations not directly involved in neural feed-forward propagation. While neurons in the brain and deep-learning networks appear to have different back-propagating mechanisms, this topic remains an active investigation in computational biology and AI.
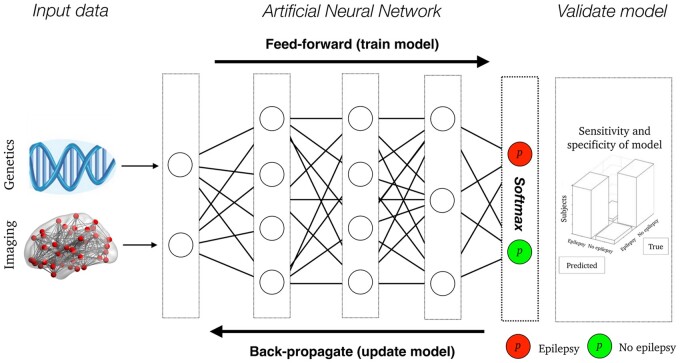
Figure 3.
An Artificial Neural Network example: here is a schematic overview of how high-dimensional genetics and brain imaging is used in a deep-learning model to make a probabilistic estimate (p) whether people are likely to develop epilepsy (red node) or not (green node). The lines between layers represent connections, each associated with a weight-adjusted during feed-forward training and updated during back-propagation until the optimal model performance.
To describe this process in more detail, the values of single nodes in a deep-learning model is the sum of all incoming nodes—analogous to dendrites of a neuron—multiplied by incoming edges—analogous to synaptic connections—with an added bias score—analogous a threshold for activity (action potential) as a neuron's resting membrane potential would be. This score is then entered into a non-linear activation function—analogous to a neuron’s membrane potential and the threshold required to generate an action potential. The most common activation function in contemporary AI is the rectified linear unit, a simple, fast and robust non-linear function, enabling learning within layers (Dahl et al., 2013). The reason why this function is similar to initiation of an action potential (or lack thereof) is that it turns negative input values into a score of zero—activation is not passed onto the subsequent layer—and for positive values, its gradient is always equal to its input—activation is passed onto the subsequent layer. Unlike the hidden layers, the output layer of a deep-learning network has a different activation function, usually Softmax (Gibbs, 2010). Softmax is popular as it provides a score across multiple output nodes with a sum of one. This means that a Softmax provides a probabilistic output that is ideal to use for prediction analysis between the deep-learning output and clinical labels of interest.
The performance of a deep-learning network is directed by a loss function that measures how accurate the output of the network is to the true clinical label value provided in the training data. There are various loss functions available including mean squared error loss, hinge loss and cross-entropy loss (Janocha and Czarnecki, 2017), all quantifying model performance in different ways, with the potential to up-weight or down-weight certain errors—allowing the trade-off between false positives and false negatives to be adjusted to the particular situation.
Once a loss function is chosen, the network learns how to perform the task by adjusting the weights between the neurons in the different layers to minimize the numerical value of the loss function over all the training examples. This is done using the back-propagation algorithm (Rojas, 1996), which determines the impact of each weight on the outcome and makes fine adjustments achieved by multiplying a pre-specified learning rate coefficient, usually a value in the range of 0.1–0.5, to the weights for each batch of training examples to improve the value of the loss function (Le et al., 2011). A low learning rate value provides a smooth gradient descent of the loss function across training examples and enables detection of robust local minima—the optimal point—of the loss function (Smith et al., 2018). Smith et al. raise a relevant point that researchers should not be tempted to increase learning rate in deep-learning model (i.e. >0.6). Higher learning rate provides faster but less reliable deep-learning prediction, as the local minimum is hard to find in a noisy gradient descent curve. A more reliable way to increase learning speed is to increase the batch size (the number of training examples utilized in one iteration of the deep-learning model).
Increase AI model prediction with multimodal data
There is evidence showing that including multiple data modalities into a single AI model can result in improved model performance and predictive accuracy [see Baltrušaitis et al. (2017) for a review]. The scientific proposition of combining several sources of data into a single AI model remains an active field of research due to the challenge of integrating data of varying dimensionality, time scales and scope, but progress is evident as ensemble methods that take advantage of collections of separately learned models have been shown to have consistently higher performance than a single monolithic model (D’Mello and Westlund, 2015).
An example where multimodal data are likely to be clinically effective is in epilepsy. High-dimensional brain imaging and genetics data are two types of data that have significantly enhanced our understanding of epilepsy over the last decades (Jackson, 1994; Kuzniecky et al., 1997; Scheffer and Berkovic, 1997; Marini et al., 2003; Dibbens et al., 2013; Pedersen et al., 2015; Jackson et al., 2017). Incorporating such multimodal data into a single classifier is likely to result in an improved predictive AI modelling of epilepsy than a classifier relying on only a single data type, as these data sources contain complementary information pertinent to the disease. Additional data sources, such as EEG (Hosseini et al., 2020; Reuben et al., 2020) and clinical documentation of patient characteristics (Cohen et al., 2016), may further enrich the modelling. These data are high-dimensional (Motsinger and Ritchie, 2006), so there is a lot of information that can be hard to interpret and compute with conventional statistical methods (Friston et al., 1994; Benjamini and Hochberg, 1995). By using deep learning, which is designed to deal with high-dimensional data, we can start asking questions pertinent to the diagnosis and treatment of epilepsy, questions that clinicians cannot answer with current tools (see Fig. 4).
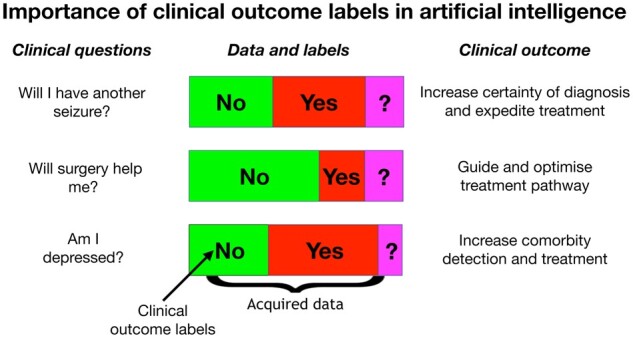
Figure 4.
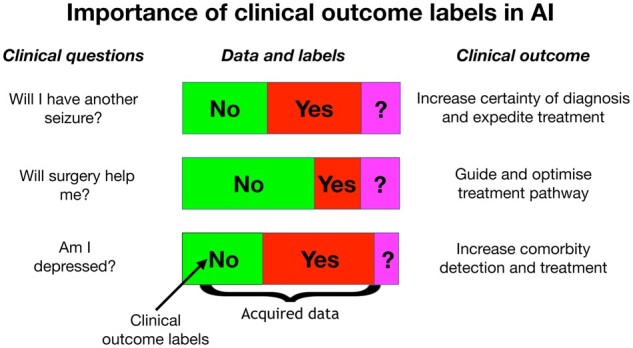
Importance of labels in AI: AI can answer difficult clinical questions in neurology.
Combining multimodal data in AI models is an active area of research (He et al., 2015; Badrinarayanan et al., 2017; Choi and Lee, 2019), where AI models learn inherent cross-relationships between data modalities [see also Duong et al. (2017) for an overview]. These approaches extract and join the most useful parts of each data modality, to improve AI model performance and prediction. For example, it is possible to perform an early fusion of data (Zeppelzauer and Schopfhauser, 2016). This requires a single deep-learning model where data modalities are correlated, and their intrinsic relationships are important contributors to the outcome. Here, the model is trained on the combined representations meaning that multiple data modalities are ‘fused’ throughout all layers of the model. Although early fusion allows for better joint feature learning, it is sensitive to missing data, which also reinforces that a focus on data quality and completeness is imperative in clinical AI. Another way of combining data modalities is a late fusion of data (Cui et al., 2010). This approach also requires one AI model but the assumption here is that data modalities are not significantly correlated, but their combined contribution is an important factor of the model outcome and accuracy. A newer model fusion technique is joint fusion (Duong et al., 2017) that incorporates data at different levels of the deep-learning model. This can work well for data of different sizes including text and images.
Validate AI models on previously unseen data by splitting data into train, test and validate sets
Any unimodal or multimodal dataset used for AI modelling needs to be divided into three different sub-categories, to ensure that we validate AI models based on unseen data (Kohavi, 1995). The data-splitting framework in AI consists of training data used to fit the AI model; testing data where the final accuracy and validity of the model is tested (Xu and Goodacre, 2018); and validation/development data separate from the training data instances enabling us to validate the model performance and tune parameters of the AI model (Ripley, 1996).
According to Liu and Cocea (2017), between 60–80% of the data is often employed to train an AI model and 20–40% of data used for testing. To fine-tune AI models and their hyper-parameters, it is important to avoid overlap contamination between training and testing data, to ensure that the AI model is tested with unseen and independent test data. It is advisable to withhold 10–30% of the training data as a validation/development dataset. The validation dataset is used to tune and optimize hyper-parameters of the AI model as this ensures that data leakage between training and test data does not occur, and therefore ensuring unbiased estimates of AI performance that are more likely to generalize to other datasets. The desired outcome of an AI model is to generate a good data-fit which is a model that resembles the underlying data. A well-fitted model also produces more accurate predictions about new data (Everitt and Skrondal, 2002; Goodfellow et al., 2016). There are fallacies in model fitting that are important to be aware of and to avoid in AI analyses. A model may fit the training data ‘too well’, leading to overfitting. This overfitting often occurs in homogenous datasets, and although resulting in a valid model, it is unlikely that such a model would be generalizable (Hawkins, 2004). A model that underfits the data has not learned the patterns in the data well enough; this is usually caused by insufficient sample size. An essential requirement to avoid problems with model fitting is to obtain sufficiently large, and diverse, datasets.
Transfer learning: previous AI models can be used as the starting point for new AI models
Transfer learning enables researchers to leverage the wealth of knowledge stored in the large and rich dataset to pre-train other AI models with (more limited) data, as this can solve other related problems or adapt to the characteristics of local data acquisition methods and demographics (Dai et al., 2009; Torrey and Shavlik, 2010; Weiss et al., 2016; Tan et al., 2018). Transfer learning may become an important part of AI-based neurology as we want to avoid re-developing models from scratch for all diagnostic and prognostic problems that clinicians face (Kouw and Loog, 2019). An effective transfer learning paradigm will support generalization of an AI model to different populations. Predictive AI models can be altered to the local context with a significantly smaller amount of data than that required to train a model from scratch.
A successful example of transfer learning comes from a study by Eitel et al. (2019) who wanted to develop a diagnostic deep-learning model based on structural MRI data from a small sample of 76 people with Multiple Sclerosis and 71 healthy control subjects. This number of subjects was insufficient to train a robust deep-learning model from scratch, so the authors deployed transfer learning to pre-train an AI model based on a previously acquired, and openly released, dataset that containing 921 subjects from the Alzheimer’s Disease Neuroimaging Initiative (Petersen et al., 2010). With ‘help’ from pre-trained Alzheimer’s disease data, Eitel and others were able to use transfer learning to classify people with Multiple Sclerosis from healthy control subjects with over 87% accuracy, providing a potential diagnostic test of Multiple Sclerosis based on their limited MRI data. This showcases how one can leverage large datasets and transfer learning for purposes well beyond the primary reason for acquiring the original data.
Domain adaptation also offers promising ways to improve generalizability and leverage large-related datasets to train networks (Kouw and Loog, 2019). They can also adapt the network to work better on different data—e.g. MRI scans with different quality/resolutions, or different scanners, or from under-represented patient groups. A degree of adaptation is possible even in the extreme case where no training labels are available in the new dataset, by comparing unlabelled data in the new context to the original dataset. This can be important for generalizing, or harmonizing, the network to work with data from different hospitals, using different scanners for example, where there may be insufficient data to perform transfer learning.
Augmented Intelligence: the interplay between human expertise and AI algorithms
Although AI has the potential to transform healthcare as we know it, its success will depend on how successful we are at developing a symbiotic relationship between human domain-specific expertise and predictive AI algorithms, also optimized and fine-tuned by human experts. The concept of Augmented Intelligence emphasizes the assistive role of AI in advancing human capabilities and decision-making [see Gennatas et al. (2020) for more information]. An AI programme can provide a decision or prediction after learning patterns from data, but the interpretation and real-world implementation of AI models requires human expertise. Humans ultimately must decide how AI models should be integrated into clinical practice (Bærøe et al., 2020; Reddy et al., 2020).
Furthermore, understanding of the decisions made by complex AI models is a critical element of confidence in the advice they provide (Israelsen and Ahmed, 2019). This builds on trust in the models (‘AI assurance’), and being able to explain the decisions that they make (‘explainability’)—distinguishing here between explaining decisions and explaining the mechanisms by which they arrive at those decisions (Adadi and Berrada, 2018; Guidotti et al., 2018; Miller, 2019). The advantage of adhering to the concept of Augmented Intelligence in a clinical and research setting is that human experts can use less time on automatable tasks such as identifying abnormal imaging features and focus on the tasks that demand uniquely human skills, including asking contextually appropriate questions about a patient’s condition, interpreting and critically analysing data, and discussing individual needs and values that may determine the best treatment decision for a given patient. Human experts may do better at understanding unusual and rare cases with uncommon pathologies, where it is not possible to get adequate training data for AI analysis—this is something that makes Augmented Intelligence important now and in the future.
The performance of an AI model must be benchmarked against a known clinical outcome that provides an appropriate target label for AI prediction (e.g. seizure versus no seizure; drug response versus no drug response; depression versus no depression). Accurate identification of these target labels requires clinical knowledge, and we are dependent on people with extensive clinical experience and expertise to provide reliable outcome measures in our patients. Humans and machines need to work together to ensure that the outputs of AI models are robust enough for clinical prediction (Elshafeey et al., 2019).
In terms of identifying and prioritizing the problems and questions where AI methods can be most useful, the clinicians may assist in monitoring the use of algorithms in particular clinical situations—to understand at some level what the limitations of the algorithms might be, and to flag when a decision does not seem to be correct (either because it does not align with a subjective clinical intuition, or when a patient outcome is contrary to a prediction) to support further refinement and improvement of algorithms and general safety monitoring of the algorithms in practice. A common scenario in the AI community is that different research groups—with different AI algorithms—compete to produce the best predictive result to a specific clinical problem or question. This competition or crowd-sourcing approach is embodied in platforms such as Kaggle, supported by Google (www.kaggle.com). Here, researchers explore and build predictive models in a web-based data-science environment. This encourages collaboration between researchers and engineers to solve intricate data-science problems or questions that can be fed back to the clinicians for further refinement or implementation.
AI to assist prognosis avoids potential overdiagnosis
Improvements in the sensitivity of diagnostic technology, whether or not driven by AI, have the potential to result in overdiagnosis. A classic example is the availability in South Korea of an inexpensive yet sensitive test for the presence of thyroid cancers. Its introduction and popularity resulted in an order of magnitude increase in the detection rate of thyroid cancers over a decade, entirely attributable to the detection of papillary thyroid cancer—yet over the same period, there was virtually no change in mortality (Ahn et al., 2014). The ‘improved’ testing was essentially detecting an increase in benign cases, resulting in unnecessary treatment and anxiety, and wasting precious healthcare dollars. AI predictive tools trained on patient outcome measures, rather than diagnostic surrogates, prospectively avoids this problem. An outcomes-trained predictive tool provides clinicians and patients with the prognostic information they really need—for example helping to answer questions such as those indicated in Fig. 4.
Ethical principles are imperative in the fast-changing field of AI
At present, the rapid advances in precision medicine technologies, large data and AI-led analysis are outstripping societal and regulatory response. As the pace of AI technology continues to drive transformation in health, it is imperative to consider the ethical and safety implications of AI systems for research and practice. As AI pushes the boundaries of what we can do with data, we face a responsibility to ensure that the highest standards for data management and AI development are upheld, while also ensuring the continuing development of AI tools to improve diagnosis and treatment of disease (Topol, 2019).
Public trust and confidence in AI are crucial to its success in medicine. Recent ethical frameworks promote understanding of AI ethics and regulations in medicine (Bryson and Winfield, 2017; Floridi et al., 2018; Jobin et al., 2019), including the Royal Australian and New Zealand College of Radiologists and the EU’s initiative to develop a trustworthy ethical framework (see Box 4).
Box 4.
Ethical principles for AI
EU’s initiative to develop a trustworthy ethical AI framework includes human agency and oversight; technical robustness and safety; privacy and data governance; transparency and diversity; non-discrimination and fairness; societal and environmental well-being as well as human accountability (https://ec.europa.eu/futurium/en/ai-alliance-consultation). The Royal Australian and New Zealand College of Radiologists ethical AI framework includes safety; privacy and protection of data; avoidance of bias; transparency and exploitability and application of human values; decision-making on diagnosis and treatment; teamwork; responsibility for decisions made and governance (https://www.ranzcr.com/college/document-library/ethical-principles-for-ai-in-medicine).
These two ethical AI frameworks are overlapping and reinforce the importance of clear and safe ethics guidelines for AI algorithms, as they are becoming routinely used as a clinical support tool.
The US Food and Drug Administration has also called on AI researchers to provide expert input on how to ensure sound governance and ethical responsibility in the field of AI in medicine (https://www.fda.gov/files/medical%20devices/published/US-FDA-Artificial-Intelligence-and-Machine-Learning-Discussion-Paper.pdf). They have proposed a set of rules intended to provide regulatory oversights of AI algorithms used in healthcare. For example, there is a low risk of using AI if its purpose is to inform clinical management in non-critical healthcare situations. But AI algorithms are of high risk when they are a driver of clinical decision-making in acute disease. Requirements for AI-based software will need to: carefully review of the safety and effectiveness of such software; address the allowable post-approval modifications to the software; and manage unanticipated divergence in the software’s eventual performance from the original product which was approved (Hwang et al., 2019). Regulatory agencies, institutions and industries will need to formulate guidelines and policies regarding the use of patient data to underpin commercialization of algorithms developed using patient data.
Despite the apprehension of how AI can be misused, the Commonwealth Scientific and Industrial Research Organisation recently released an AI roadmap and alluded to the point that we need to build trust in the field of AI (https://data61.csiro.au/en/Our-Research/Our-Work). Integral to building trust in AI is quality assurance, safety, security and traceability of data and its platforms. As discussed above, AI models are superfluous without human expertise to tune and clinically interpret AI results—and clinicians and scientists need to come together to build interpretable AI models, to improve treatment and care in neurology. Ethical, privacy and security considerations are paramount in any advance of precision medicine and the use of large data sets and AI. These concerns, however, can be managed and should not lead to inertia as AI has the potential to change lives (Topol, 2019).
Concluding remarks: large-scale projects are needed to unlock AI’s clinical potential
Precision medicine and AI is likely to be a big part of the future of medical practice (Collins and Varmus, 2015). AI has the potential to create a paradigm shift in the diagnosis, treatment, prediction and economics of neurological disease. People living with a neurological disease yearn for such precision—Will I have another seizure? Will this medication work for me? Should I have surgery? Am I depressed? Advancements in AI technology have the potential to reduce the uncertainty surrounding diagnosis and treatment of all neurological disease. But to achieve this, a deep effort is needed to fund large-scale studies with data derived from realistic clinical documentation that includes participant outcome measures. This will create an invaluable asset to drive advances in the future of healthcare.
Acknowledgements
The Florey Institute of Neuroscience and Mental Health acknowledges the strong support from the Victorian Government and in particular the funding from the Operational Infrastructure Support Grant. We also acknowledge the facilities, and the scientific and technical assistance of the National Imaging Facility (NIF), an Australian Government National Collaborative Research Infrastructure Strategy (NCRIS) capability, at the Florey node, and The Victorian Biomedical Imaging Capability (VBIC).
Funding
This work was supported by an Australian Government Medical Research Future Fund Frontier Health and Medical Research Program Stage One grant (MRFF75908). D.F.A. acknowledges fellowship funding from the Australian National Imaging Facility. M.J. is supported by the National Institute for Health Research (NIHR) Oxford Biomedical Research Centre (BRC), and this research was funded by the Wellcome Trust (215573/Z/19/Z). The Wellcome Centre for Integrative Neuroimaging is supported by core funding from the Wellcome Trust (203139/Z/16/Z).
Competing interests
The authors report no competing interest.
Glossary
- AI
= artificial intelligence
Contributor Information
Mangor Pedersen, The Florey Institute of Neuroscience and Mental Health, The University of Melbourne, Heidelberg, VIC 3084, Australia; Department of Psychology, Auckland University of Technology (AUT), Auckland, 0627, New Zealand.
Karin Verspoor, School of Computing and Information Systems, The University of Melbourne, Parkville, VIC 3010, Australia.
Mark Jenkinson, Wellcome Centre for Integrative Neuroimaging, FMRIB, Nuffield Department of Clinical Neurosciences, University of Oxford, Oxford, OX3 9DU, UK; South Australian Health and Medical Research Institute (SAHMRI), Adelaide, SA 5000, Australia; Australian Institute for Machine Learning (AIML), The University of Adelaide, Adelaide, SA 5000, Australia.
Meng Law, Department of Radiology, Alfred Hospital, Melbourne, VIC 3181, Australia; Department of Electrical and Computer Systems Engineering, Monash University, Melbourne, VIC 3181, Australia; Department of Neuroscience, Monash School of Medicine, Nursing and Health Sciences, Melbourne, VIC 3181, Australia.
David F Abbott, The Florey Institute of Neuroscience and Mental Health, The University of Melbourne, Heidelberg, VIC 3084, Australia; Department of Medicine Austin Health, The University of Melbourne, Heidelberg, VIC 3084, Australia.
Graeme D Jackson, The Florey Institute of Neuroscience and Mental Health, The University of Melbourne, Heidelberg, VIC 3084, Australia; Department of Medicine Austin Health, The University of Melbourne, Heidelberg, VIC 3084, Australia; Department of Neurology, Austin Health, Heidelberg, VIC 3084, Australia.
References
- Adadi A, Berrada M. Peeking inside the black-box: a survey on explainable artificial intelligence (XAI). IEEE Access 2018; 6: 52138–60. [Google Scholar]
- Agatonovic-Kustrin S, Beresford R. Basic concepts of artificial neural network (ANN) modeling and its application in pharmaceutical research. J Pharm Biomed Anal 2000; 22: 717–27. [DOI] [PubMed] [Google Scholar]
- Ahn HS, Kim HJ, Welch HG. Korea’s thyroid-cancer ‘epidemic’—screening and overdiagnosis. N Engl J Med 2014; 371: 1765–7. [DOI] [PubMed] [Google Scholar]
- Akkus Z, Galimzianova A, Hoogi A, Rubin DL, Erickson BJ. Deep learning for brain MRI segmentation: state of the art and future directions. J Digit Imaging 2017; 30: 449–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardila D, Kiraly AP, Bharadwaj S, Choi B, Reicher JJ, Peng L, et al. End-to-end lung cancer screening with three-dimensional deep learning on low-dose chest computed tomography. Nat Med 2019; 25: 954–61. [DOI] [PubMed] [Google Scholar]
- Badrinarayanan V, Kendall A, Cipolla R. SegNet: a deep convolutional encoder-decoder architecture for image segmentation. IEEE Trans Pattern Anal Mach Intell 2017; 39: 2481–95. [DOI] [PubMed] [Google Scholar]
- Bærøe K, Miyata-Sturm A, Henden E. How to achieve trustworthy artificial intelligence for health. Bull World Health Organ 2020; 98: 257–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baltrušaitis T, Ahuja C, Morency L-P. Multimodal Machine Learning: A Survey and Taxonomy. ArXiv170509406 Cs. 2017. Available at: http://arxiv.org/abs/1705.09406 (2 April 2020, date last accessed). [DOI] [PubMed]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B Methodol 1995; 57: 289–300. [Google Scholar]
- Brosch T, Yoo Y, Li DKB, Traboulsee A, Tam R, Modeling the variability in brain morphology and lesion distribution in multiple sclerosis by deep learning. In: Golland P, Hata N, Barillot C, Hornegger J, Howe R, editors. Medical image computing and Computer-assisted intervention—MICCAI 2014. Cham: Springer International Publishing; 2014. p. 462–9. [DOI] [PubMed] [Google Scholar]
- Bryson J, Winfield A. Standardizing ethical design for artificial intelligence and autonomous systems. Computer 2017; 50: 116–9. [Google Scholar]
- Chilamkurthy S, Ghosh R, Tanamala S, Biviji M, Campeau NG, Venugopal VK, et al. Deep learning algorithms for detection of critical findings in head CT scans: a retrospective study. Lancet 2018; 392: 2388–96. [DOI] [PubMed] [Google Scholar]
- Choi J-H, Lee J-S. EmbraceNet: a robust deep learning architecture for multimodal classification. Inf Fusion 2019; 51: 259–70. [Google Scholar]
- Codella NCF, Nguyen Q-B, Pankanti S, Gutman DA, Helba B, Halpern AC, et al. Deep learning ensembles for melanoma recognition in dermoscopy images. IBM J Res Dev 2017; 61: 5:1–15. [Google Scholar]
- Cohen KB, Glass B, Greiner HM, Holland-Bouley K, Standridge S, Arya R, et al. Methodological issues in predicting pediatric epilepsy surgery candidates through natural language processing and machine learning. Biomed Inform Insights 2016; 8: BII.S38308–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins FS, Varmus H. A new initiative on precision medicine. N Engl J Med 2015; 372: 793–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortes C, Vapnik V. Support-vector networks. Mach Learn 1995; 20: 273–97. [Google Scholar]
- Crevier DA. The tumultuous history of the search for artificial intelligence. 1st edn. New York: Basic Books; 1993. [Google Scholar]
- Cui B, Tung AKH, Zhang C, Zhao Z. Multiple feature fusion for social media applications. In: Proceedings of the 2010 ACM SIGMOD International Conference on Management of data (SIGMOD ’10). 2010. New York: Association for Computing Machinery, pp. 435–446. doi: 10.1145/1807167.1807216. [DOI]
- Dahl GE, Sainath TN, Hinton GE. Improving deep neural networks for LVCSR using rectified linear units and dropout. In: 2013 IEEE International Conference on Acoustics, Speech and Signal Processing. IEEE, 2013. p. 8609–8613.
- Dai W, Chen Y, Xue G, Yang Q, Yu Y. Translated learning: transfer learning across different feature spaces. In: Koller D, Schuurmans D, Bengio Y, Bottou L, editors. Advances in neural information processing systems 21. New York: Curran Associates, Inc.; 2009. p. 353–60. [Google Scholar]
- Daubechies I, DeVore R, Foucart S, Hanin B, Petrova G. Nonlinear Approximation and (Deep) ReLU Networks. ArXiv190502199 Cs. 2019. Available at: http://arxiv.org/abs/1905.02199 (2 April 2020, date last accessed).
- Dibbens LM, de Vries B, Donatello S, Heron SE, Hodgson BL, Chintawar S, et al. Mutations in DEPDC5 cause familial focal epilepsy with variable foci. Nat Genet 2013; 45: 546–51. [DOI] [PubMed] [Google Scholar]
- D’Mello SK, Westlund JK. A review and meta-analysis of multimodal affect detection systems. ACM Computing Surveys (CSUR); 2015; 47: 1–36. [Google Scholar]
- Dreiseitl S, Ohno-Machado L. Logistic regression and artificial neural network classification models: a methodology review. J Biomed Inform 2002; 35: 352–9. [DOI] [PubMed] [Google Scholar]
- Duong CT, Lebret R, Aberer K. Multimodal Classification for Analysing Social Media. ArXiv170802099 Cs. 2017. Available at: http://arxiv.org/abs/1708.02099 (2 April 2020, date last accessed).
- Eitel F, Soehler E, Bellmann-Strobl J, Brandt AU, Ruprecht K, Giess RM, et al. Uncovering convolutional neural network decisions for diagnosing multiple sclerosis on conventional MRI using layer-wise relevance propagation. NeuroImage Clin 2019; 24: 102003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elshafeey N, Kotrotsou A, Hassan A, Elshafei N, Hassan I, Ahmed S, et al. Multicenter study demonstrates radiomic features derived from magnetic resonance perfusion images identify pseudoprogression in glioblastoma. Nat Commun 2019; 10: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteva A, Kuprel B, Novoa RA, Ko J, Swetter SM, Blau HM, et al. Dermatologist-level classification of skin cancer with deep neural networks. Nature 2017; 542: 115–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everitt BS, Skrondal A. The Cambridge Dictionary of Statistics (9780521766999). Cambridge: Cambridge University Press; 2002.
- Faust O, Hagiwara Y, Hong TJ, Lih OS, Acharya UR. Deep learning for healthcare applications based on physiological signals: a review. Comput Methods Programs Biomed 2018; 161: 1–13. [DOI] [PubMed] [Google Scholar]
- Floridi L, Cowls J, Beltrametti M, Chatila R, Chazerand P, Dignum V, et al. AI4People—an ethical framework for a good AI society: opportunities, risks, principles, and recommendations. Minds Machines 2018; 28: 689–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friston KJ, Worsley KJ, Frackowiak RS, Mazziotta JC, Evans AC. Assessing the significance of focal activations using their spatial extent. Hum Brain Mapp 1994; 1: 210–20. [DOI] [PubMed] [Google Scholar]
- Gennatas ED, Friedman JH, Ungar LH, Pirracchio R, Eaton E, Reichmann LG, et al. Expert-augmented machine learning. Proc Natl Acad Sci USA 2020; 117: 4571–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs JW. Elementary Principles in Statistical Mechanics: Developed with Especial Reference to the Rational Foundation of Thermodynamics (Cambridge Library Collection - Mathematics). Cambridge: Cambridge University Press. 2010. doi: 10.1017/CBO9780511686948. [DOI]
- Goodfellow I, Bengio Y, Courville A. Deep learning. Cambridge: MIT Press; 2016. [Google Scholar]
- Guidotti R, Monreale A, Ruggieri S, Turini F, Pedreschi D, Giannotti F. A Survey Of Methods For Explaining Black Box Models. ArXiv180201933 Cs. 2018. Available at: http://arxiv.org/abs/1802.01933 (2 April 2020, date last accessed).
- Hannun AY, Rajpurkar P, Haghpanahi M, Tison GH, Bourn C, Turakhia MP, et al. Cardiologist-level arrhythmia detection and classification in ambulatory electrocardiograms using a deep neural network. Nat Med 2019; 25: 65–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassoun MH. Fundamentals of artificial neural networks. Cambridge: MIT Press; 1995. [Google Scholar]
- Hawkins DM. The problem of overfitting. J Chem Inf Comput Sci 2004; 44: 1–12. [DOI] [PubMed] [Google Scholar]
- He K, Zhang X, Ren S, Sun J. Deep Residual Learning for Image Recognition. ArXiv151203385 Cs. 2015. Available at: http://arxiv.org/abs/1512.03385 (2 April 2020, date last accessed).
- Hestness J, Narang S, Ardalani N, Diamos G, Jun H, Kianinejad H, et al. Deep Learning Scaling is Predictable, Empirically. ArXiv171200409 Cs Stat 2017. Available from: http://arxiv.org/abs/1712.00409
- Hosseini M-P, Tran TX, Pompili D, Elisevich K, Soltanian-Zadeh H. Multimodal data analysis of epileptic EEG and rs-fMRI via deep learning and edge computing. Artif Intell Med 2020; 104: 101813. [DOI] [PubMed] [Google Scholar]
- Hwang TJ, Kesselheim AS, Vokinger KN. Lifecycle regulation of artificial intelligence- and machine learning-based software devices in medicine. JAMA 2019; 322: 2285. [DOI] [PubMed] [Google Scholar]
- Israelsen BW, Ahmed NR. “Dave…I can assure you …that it’s going to be all right …” a definition, case for, and survey of algorithmic assurances in human-autonomy trust relationships. ACM Comput Surv 2019; 51: 1–37. [Google Scholar]
- Jackson GD. New techniques in magnetic resonance and epilepsy. Epilepsia 1994; 35: S2–13. [DOI] [PubMed] [Google Scholar]
- Jackson GD, Pedersen M, Harvey AS. How small can the epileptogenic region be? A case in point. Neurology 2017; 88: 2017–9. [DOI] [PubMed] [Google Scholar]
- Jafari MH, Karimi N, Nasr-Esfahani E, Samavi S, Soroushmehr SMR, Ward K, et al. Skin lesion segmentation in clinical images using deep learning. In: 2016 23rd International Conference on Pattern Recognition (ICPR). IEEE, 2016. p. 337–42.
- Janocha K, Czarnecki WM. On Loss Functions for Deep Neural Networks in Classification. ArXiv170205659 Cs. 2017. Available at: http://arxiv.org/abs/1702.05659 (8 April 2020, date last accessed).
- Jobin A, Ienca M, Vayena E. The global landscape of AI ethics guidelines. Nat Mach Intell 2019; 1: 389–99. [Google Scholar]
- Kohavi R. A study of cross-validation and bootstrap for accuracy estimation and model selection. In: Proceedings of the 14th international joint conference on artificial intelligence. Vol. 2. Montreal, Quebec, Canada: Morgan Kaufmann Publishers Inc.; 1995. p. 1137–43. [Google Scholar]
- Korfiatis P, Kline TL, Erickson BJ. Automated segmentation of hyperintense regions in FLAIR MRI using deep learning. Tomography 2016; 2: 334–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouw WM, Loog M. A review of domain adaptation without target labels. IEEE Trans Pattern Anal Mach Intell 2019; 1. doi: 10.1109/tpami.2019.2945942. [DOI] [PubMed] [Google Scholar]
- Kuzniecky RI, Bilir E, Gilliam F, Faught E, Palmer C, Morawetz R, et al. Multimodality MRI in mesial temporal sclerosis: relative sensitivity and specificity. Neurology 1997; 49: 774–8. [DOI] [PubMed] [Google Scholar]
- Le QV, Ngiam J, Coates A, Lahiri A, Prochnow B, Ng AY. On optimization methods for deep learning. 2011.Available at: https://icml.cc/2011/papers/210_icmlpaper.pdf (8 April 2020, date last accessed).
- LeCun Y, Bengio Y, Hinton G. Deep learning. Nature 2015; 521: 436–444. [DOI] [PubMed] [Google Scholar]
- Lee E-J, Kim Y-H, Kim N, Kang D-W. Deep into the brain: artificial intelligence in stroke imaging. J Stroke 2017; 19: 277–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lillicrap TP, Santoro A, Marris L, Akerman CJ, Hinton G. Backpropagation and the brain. Nat Rev Neurosci 2020; 21: 335–12. [DOI] [PubMed] [Google Scholar]
- Liu H, Cocea M. Semi-random partitioning of data into training and test sets in granular computing context. Granul Comput 2017; 2: 357–386. [Google Scholar]
- Makin JG, Moses DA, Chang EF. Machine translation of cortical activity to text with an encoder-decoder framework. Nat Neurosci 2020; 23: 575–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marini C, Harkin LA, Wallace RH, Mulley JC, Scheffer IE, Berkovic SF. Childhood absence epilepsy and febrile seizures: a family with a GABAA receptor mutation. Brain 2003; 126: 230–40. [DOI] [PubMed] [Google Scholar]
- Miller T. Explanation in artificial intelligence: insights from the social sciences. Artif Intell 2019; 267: 1–38. [Google Scholar]
- Minsky M, Papert SA, Bottou L. Perceptrons. Reissue edn. Cambridge, MA: MIT Press; 2017. [Google Scholar]
- Motsinger AA, Ritchie MD. Multifactor dimensionality reduction: an analysis strategy for modelling and detecting gene–gene interactions in human genetics and pharmacogenomics studies. Hum Genomics 2006; 2: 318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mwangi B, Tian TS, Soares JC. A review of feature reduction techniques in neuroimaging. Neuroinformatics 2014; 12: 229–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngiam J, Khosla A, Kim M, Nam J, Lee H, Ng AY. Multimodal Deep Learning. 2011. Available at: https://people.csail.mit.edu/khosla/papers/icml2011_ngiam.pdf (8 April 2020, date last accessed).
- Obermeyer Z, Emanuel EJ. Predicting the future—big data, machine learning, and clinical medicine. N Engl J Med 2016; 375: 1216–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen M, Omidvarnia AH, Walz JM, Jackson GD. Increased segregation of brain networks in focal epilepsy: an fMRI graph theory finding. NeuroImage Clin 2015; 8: 536–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen RC, Aisen PS, Beckett LA, Donohue MC, Gamst AC, Harvey DJ, et al. Alzheimer’s Disease Neuroimaging Initiative (ADNI). Neurology 2010; 74: 201–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Premaladha J, Ravichandran KS. Novel approaches for diagnosing melanoma skin lesions through supervised and deep learning algorithms. J Med Syst 2016; 40: 96. [DOI] [PubMed] [Google Scholar]
- Rahhal MMA, Bazi Y, AlHichri H, Alajlan N, Melgani F, Yager RR. Deep learning approach for active classification of electrocardiogram signals. Inf Sci 2016; 345: 340–54. [Google Scholar]
- Reddy S, Allan S, Coghlan S, Cooper P. A governance model for the application of AI in health care. J Am Med Inform Assoc JAMIA 2020; 27: 491–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuben C, Karoly P, Freestone DR, Temko A, Barachant A, Li F, et al. Ensembling crowdsourced seizure prediction algorithms using long-term human intracranial EEG. Epilepsia 2020; 61: e7–12. [DOI] [PubMed] [Google Scholar]
- Ripley BD. Pattern Recognition and Neural Networks. Cambridge: Cambridge University Press; 1996. doi: 10.1017/CBO9780511812651. [DOI] [Google Scholar]
- Rojas R. The backpropagation algorithm. In: Rojas R, editor. Neural networks: a systematic introduction. Berlin, Heidelberg: Springer; 1996. p. 149–82. [Google Scholar]
- Rosenblatt F. The perceptron: a probabilistic model for information storage and organization in the brain. Psychol Rev 1958; 65: 386–408. [DOI] [PubMed] [Google Scholar]
- Sainath TN, Vinyals O, Senior A, Sak H. Convolutional, Long Short-Term Memory, fully connected Deep Neural Networks. In: 2015 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP). IEEE, 2015. p. 4580–4
- Sarle WS. Neural Networks and Statistical Models. In: Proceedings of the Nineteenth Annual SAS Users Group International Conference, Cary, NC: SAS Institute, 1994, pp. 1538–1550.
- Savage N. How AI and neuroscience drive each other forwards. Nature 2019; 571: S15–7. [DOI] [PubMed] [Google Scholar]
- Scheffer IE, Berkovic SF. Generalized epilepsy with febrile seizures plus. A genetic disorder with heterogeneous clinical phenotypes. Brain 1997; 120: 479–90. [DOI] [PubMed] [Google Scholar]
- Schmidhuber J. Deep learning in neural networks: an overview. Neural Netw 2015; 61: 85–117. [DOI] [PubMed] [Google Scholar]
- Sejnowski TJ. The unreasonable effectiveness of deep learning in artificial intelligence. Proc Natl Acad Sci USA 2020. doi: 10.1073/pnas.1907373117. [DOI] [PMC free article] [PubMed]
- Smith SL, Kindermans P-J, Ying C, Le QV. Don’t Decay the Learning Rate, Increase the Batch Size. ArXiv171100489 Cs Stat. 2018. Available at: http://arxiv.org/abs/1711.00489 (8 April 2020, date last accessed).
- Tan C, Sun F, Kong T, Zhang W, Yang C, Liu C. A survey on deep transfer learning. In: Kůrková V, Manolopoulos Y, Hammer B, Iliadis L, Maglogiannis I, editors. Artificial neural networks and machine learning—ICANN 2018. Cham: Springer International Publishing; 2018. p. 270–9. [Google Scholar]
- Topol EJ. High-performance medicine: the convergence of human and artificial intelligence. Nat Med 2019; 25: 44–56. [DOI] [PubMed] [Google Scholar]
- Torrey L, Shavlik J. Transfer learning. In: Soria E, Martin J, Magdalena R, Martinez M, Serrano A, editors. Handbook of research on machine learning applications and trends: algorithms, methods and technique. Pennsylvania, IGI Global, 2010. p. 242–64.
- Weiss K, Khoshgoftaar TM, Wang D. A survey of transfer learning. J Big Data 2016; 3: 9. [Google Scholar]
- Whittington JCR, Bogacz R. Theories of error back-propagation in the brain. Trends Cogn Sci 2019; 23: 235–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing Y, Ge Z, Zeng R, Mahapatra D, Seah J, Law M, et al. Adversarial pulmonary pathology translation for pairwise chest X-ray data augmentation. ArXiv191004961 Cs Eess 2019; 11769: 757–65. [Google Scholar]
- Xu Y, Goodacre R. On splitting training and validation set: a comparative study of cross-validation, bootstrap and systematic sampling for estimating the generalization performance of supervised learning. J Anal Test 2018; 2: 249–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaharchuk G, Gong E, Wintermark M, Rubin D, Langlotz CP. Deep learning in neuroradiology. Am J Neuroradiol 2018; 39: 1776–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeppelzauer M, Schopfhauser D. Multimodal classification of events in social media. Image Vis Comput 2016; 53: 45–56. [Google Scholar]
- Zhang Q, Zhou D, Zeng X. HeartID: a multiresolution convolutional neural network for ECG-based biometric human identification in smart health applications. IEEE Access 2017; 5: 11805–16. [Google Scholar]
- Zhou Z-H, Jiang Y, Yang Y-B, Chen S-F. Lung cancer cell identification based on artificial neural network ensembles. Artif Intell Med 2002; 24: 25–36. [DOI] [PubMed] [Google Scholar]
- Zhu W, Liu C, Fan W, Xie X. DeepLung: Deep 3D Dual Path Nets for Automated Pulmonary Nodule Detection and Classification. In: 2018 IEEE Winter Conference on Applications of Computer Vision (WACV). IEEE, 2018. p. 673–81.