Abstract
The ongoing pandemic of COVID-19 (Coronavirus Disease-2019), a respiratory disease caused by the novel coronavirus strain, SARS-CoV-2, has affected more than 42 million people already, with more than one million deaths worldwide (as of October 25, 2020). We are in urgent need of therapeutic interventions that target the host-virus interface, which requires a molecular understanding of the SARS-CoV-2 life-cycle. Like other positive-sense RNA viruses, coronaviruses remodel intracellular membranes to form specialized viral replication compartments, including double-membrane vesicles (DMVs), where viral RNA genome replication takes place. Here we review the current knowledge of the structure, lipid composition, function, and biogenesis of coronavirus-induced DMVs, highlighting the druggable viral and cellular factors that are involved in the formation and function of DMVs.
Keywords: Coronavirus, Double membrane vesicles, Lipid metabolism
1. Introduction
Prior to the emergence of Coronavirus Disease-2019 (COVID-19) caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the outbreak of highly pathogenic SARS-CoV (2003) and Middle East respiratory syndrome coronavirus (MERS-CoV) (2012) [1] in the past few years had already raised awareness towards the urgent need to develop effective therapeutic options to combat coronavirus infections.
Like other positive-sense (+) RNA viruses, including hepatitis C virus (HCV), dengue virus, Zika virus, and polioviruses, coronaviruses share the feature of establishing specialized membranous replication organelles with unique lipid compositions to enable robust viral replication [2]. Depending on the groups of the viruses and the time course of the infection, the structure, composition, and formation of (+) RNA virus replication organelles appear to be varied and dynamic [3]. The accumulating knowledge concerning the involvement and requirement of cellular membranes for viral RNA synthesis specify the establishment of these viral replication organelles as an evolutionarily conserved and essential step in the early stages of the viral life cycle. The replication organelles serve multiple purposes: first of all, they provide an optimal microenvironment specialized for the synthesis of viral RNA by concentrating viral components (RNA and proteins) and host factors (specific proteins and lipid species) required for viral RNA synthesis. Because host factors are actively recruited to appropriate membrane sites in and around replication organelles, viruses can interact with them within microenvironments (thereby generating higher local concentrations) and with less interference from host membrane protein traffic or the possibility of diffusion of soluble factors. Furthermore, viral replication very likely takes place in the inner membrane facing the cytosolic interior of the replication organelles. Therefore, replication intermediates (e.g. double-stranded (ds)RNA) could be physically shielded from the host innate immune defenses [4]. In addition, formation of replication organelles allows spatial orchestration of the different steps of viral replication and assembly. Hence, membrane remodeling plays a crucial role in the (+) RNA virus life cycle.
(+) RNA virus-induced replication organelles can be classified into single-membrane spherules, double-membrane vesicles (DMVs), cubic membranes/membranous webs, and planar oligomeric arrays according to their morphological features [3,5]. Identifying both viral and cellular factors involved in the formation of these replication organelles is anticipated to facilitate the development of novel therapeutics or enable repurposing of existing antiviral drugs [6].
This review focuses particularly on coronaviral DMVs, which are structurally complex and frequently accompanied by other membrane rearrangements [4], with a spot light on the viral and cellular factors involved in DMV formation that may serve as druggable targets.
2. Characteristics of DMVs generated during coronavirus replication
2.1. Morphological features of coronavirus-induced DMVs
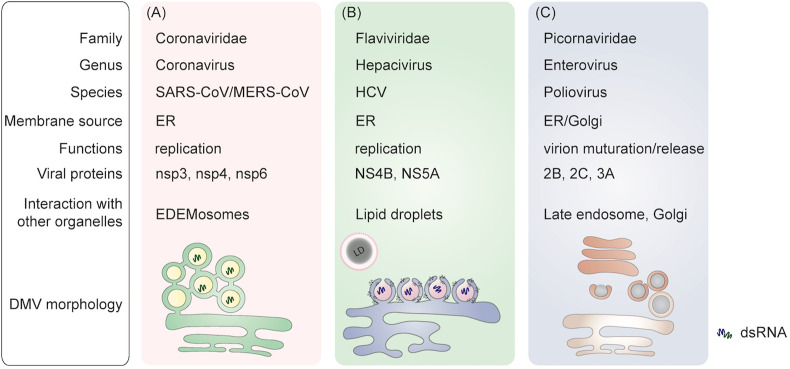
The morphological features of DMVs vary between different (+) RNA viruses (Fig. 1 ). DMVs are typically generated via bending of the donor membrane into the cytoplasm, resulting in the induction of positive membrane curvature [7]. One of the most studied examples is HCV-induced membrane rearrangements [3,5,8]. HCV infection induces significant rearrangements of intracellular membranes in a time-dependent manner. At early stages of infection, HCV-induced DMVs appear as protrusions from the ER into the cytosol, often connected to the ER membrane via a neck-like structure (Fig. 1B) [9]. Electron microscopy analyses have revealed a diameter of approximately 50–250 nm of these DMVs as described in detail elsewhere [9,10]. The remodeled membranes in poliovirus-infected cells, however, show a distinctive morphology with single-walled, connected and branched tubular compartments at early stages of infection, which later transform into double-membrane structures with a diameter of 100–300 nm via extending the membranous walls and/or collapsing of the lumenal cavity of the single-walled structures [11] (Fig. 1C).
Fig. 1.
A brief comparation of typical DMVs generated by (+) RNA viruses. A) Coronavirus-induced DMVs are interconnected by their outer membrane via a narrow neck. These DMVs are part of an elaborate network that is connected with the rough ER. B) HCV-induced DMVs emerge as protrusions from the ER into the cytosol, connected to the ER membrane via a neck-like structure and in a close proximity to lipid droplets (LDs). This is different from the membrane invagination induced during replication of members of the Flavivirus genus [19]. C) Poliovirus-induced DMVs are generated from single-walled connecting and branching tubular compartments, which later transform into double-membrane structures by extending the membranous walls and/or collapsing of the lumenal cavity of the single-walled structures.
Two highly pathogenic human coronaviruses, MERS-CoV and SARS-CoV, are well documented to induce ER-derived DMVs [12,13]. In SARS-CoV-infected Vero E6 cells, DMVs that are distributed throughout the cytoplasm can be observed as early as 2 h post-infection (p.i.), with a diameter of 150–300 nm. The number of DMVs increases dramatically at 4 h p. i., accompanied by their connection to ER structures. The DMVs become more clustered in the perinuclear area of the cell as the infection develops [12,14]. SARS-CoV-induced DMVs are interconnected by their outer membrane via a narrow 8 nm neck. In addition, they form a part of an elaborate network that remains connected with the rough ER. Distinct from HCV-induced DMVs, in-depth analysis of 3D tomographic reconstructions of SARS-CoV-induced DMVs failed to reveal any membrane openings between DMVs and the cytoplasm [12] (Fig. 1A). MERS-CoV-induced DMVs also range from 150 to 320 nm. Similar to SARS-CoV, these DMV clusters also change in number and distribution in the cytosol along with the progress of the infection [13]. Furthermore, there are no clear openings connecting the interior of the MERS-CoV-induced DMVs and the cytosol. In addition to DMVs, double-membrane spherules (DMS) were also detected in MERS-CoV-infected cells. These DMSs are smaller in size (80 nm in diameter) and without a clear opening to the cytosol. DMSs are linked to convoluted membranes (CM), and CM to ER, while ER membranes are continuous with DMVs [15]. Hence, MERS-CoV infection also creates a network of largely interconnected modified ER membrane structures as its replication organelle (Fig. 1A). The generation of DMSs was also observed in murine hepatitis virus (MHV)-infected and human coronavirus 229E (HCoV-299E)-infected cells, suggesting that inducing the same replication organelle elements is a shared feature by coronaviruses [15].
A very recent report unveiled a molecular pore complex that spans both membranes of the DMVs induced by MHV or SARS-CoV-2 [16]. Using cellular electron cryo-microscopy, 6-fold symmetrized subtomogram average of the pore complexes in MHV-induced DMVs were observed. A cytosolic structure with a crown-like morphology extends approximately 13 nm into the cytosol and is based on a ∼24 nm wide platform embedded between the two layers of DMV membranes, which maintain the typical inter-membrane spacing found in DMVs. The complex towards the cytosol side has an opening of ∼2 3 nm, allowing the transition of RNA strands that later would be encapsidated in the cytosol [16]. This discovery provides more insights on the detailed roles that coronavirus-induced DMVs play during viral replication and assembly.
By comparing the morphology of DMVs induced by different (+) RNA viruses (Fig. 1), it is clear that despite significant similarities shared by DMVs, the architecture of membrane rearrangements induced by different (+) RNA viruses are unique, which may be due to the specific roles they serve in the viral life cycle. In the case of coronavirus infection, it remains to be elucidated why and how the DMVs form an interconnected network, a unique feature of coronaviral DMVs, that may relate to its biological functions.
2.2. Lipid composition of coronavirus-induced DMVs
As the major components of cell membranes, host lipids play indispensable roles in membrane flexibility and rigidity, which are important for multiple morphological transformation-related membrane functions, including those of DMVs during the replication of (+) RNA viruses [17]. Each lipid molecule has a specific shape (cylindrical, conical, or inverted conical). The formation of the viral replication organelles requires the bending of membranes, hence, enrichment of certain types of lipids are necessary to achieve these membrane structures [17]. Therefore, specific lipid compositions are preferred by different viruses for the development of their optimal replication organelles [18]. (+) RNA viruses, including coronaviruses, manipulate host cellular lipid metabolism, as well as lipid trafficking pathways, to ensure certain types of lipids are available [17,19].
To cope with the rapidly elevated anabolic demands during viral replication, and to provide building blocks while maintaining the specific composition of cellular lipids required for DMV formation, coronaviruses often employ multiple strategies to reprogram the host lipids metabolic network. One strategy is to up-regulate lipid biosynthesis pathways. Sterol regulatory element-binding proteins (SREBPs) are a group of transcription factors that regulate the biosynthesis of cellular cholesterol and fatty acids [20], and have been reported to be activated during Flavivirus infections [21]. As one of the major regulators for lipid biosynthesis, the ER-resident SREBP is activated through proteolytic processing to release its N-terminal domain (nSREBP), which subsequently translocates to the nucleus and binds to a nonpalindromic sterol response elements (SRE). Binding between nSREBP and SRE activates the transcription of lipid biosynthesis pathways [20,22]. Yuan et al discovered that SREBPs play an essential role in MERS-CoV viral replication, particularly in DMV formation and viral protein palmitoylation. Pharmacological inhibition of SREBP activation significantly inhibits viral replication and DMV formation. Moreover, targeting SREBP downstream pathways, such as fatty acid and cholesterol synthesis pathways, can also suppress MERS-CoV replication [23].
The extensive membrane rearrangements induced during coronavirus replication necessitates that besides significantly enhanced lipid biosynthesis, lipid metabolic enzymes also regulate DMV formation. Cytosolic phospholipase A2α (cPLA2α) is a lipolytic enzyme that catalyzes the hydrolysis of membrane phospholipids at the sn2-position and releases fatty acid, lysophospholipid (LPL), and arachidonic acid (AA). Several mass spectra-based lipidomic studies have shown selective upregulation of downstream products of cPLA2α activation, including glycerophospholipids, LPL, and fatty acid in MERS-CoV-infected or HCoV-229E-infected cells [18,23,24], and long-chain polyunsaturated fatty acids (PUFA) in SARS-CoV-2 infected patient sera [25]. As a non-bilayer lipid, LPL can introduce curvature stress to the bio-membranes and induce their fission and fusion [26]. Treatment of a low-molecular-weight nonpeptidic inhibitor, pyrrolidine-2, which inhibits production of LPL, significantly reduced formation of DMVs and DMV-associated replication/transcription complexes (RTC) in HCoV-299E-infected cells, indicating that LPL is a crucial component of HCoV-299E-induced DMVs and the virus may manipulate LPL levels through activating cPLA2α [24].
2.3. Functions of coronavirus-induced DMVs
Like other types of viral replication compartments, DMVs provide a favorable environment for viral RNA replication by creating an appropriate replicase topology and a barrier between viral RNA replication compartments and the innate immune sensors, as well as RNA degradation machinery that are present in the cytosol [4].
The HCV-induced DMVs fit in this classic scenario. The co-purification of HCV-induced DMVs together with essential viral/cellular factors for de novo synthesis of HCV RNA suggests that the major role of the HCV-induced DMVs is to support viral RNA replication [10]. Moreover, the HCV-induced membranous web structure protects the viral genome from being recognized by cytoplasmic pattern recognition receptors and therefore avoids activation of cellular innate immune responses [27].
In contrast, polioviral RNA does not replicate inside the DMVs [28]. Nevertheless, polioviral DMVs, which resemble autophagosomes, can fuse with late endosomes to form amphisomes [29]. Interfering with this fusion process impairs the final maturation step in viral particle production, indicating the importance of DMVs in virion maturation and the non-lytic release of progeny poliovirus [30].
Coronavirus replicase proteins are mostly detected in CMs but not in DMVs. However, dsRNA, which is an intermediate of viral RNA replication, and a marker for this process, is predominantly found in coronaviral DMVs [12]. A previous study reported the existence of a pore in the DMVs generated during infection by MHV [72], which could potentially enable transport of viral RNA, and was corroborated in a more recent electron cryo-microscopy study [16]. Another recent study by Klein and colleagues using cryo-transmission electron microscopy clearly showed the presence of strands reminiscent of RNA inside the DMVs, suggesting that they may be the site of coronavirus RNA synthesis [70]. Additionally, SARS-CoV viral capsid proteins were found at the budding sites in close proximity to the DMVs [31], and SARS-CoV virions budded into membranes directly adjacent to and continuous with the outer DMVs membranes at the later stage of infection cycle [12]. The observation of double-membrane-spanning molecular pore in MHV-infected cells further elaborated how viral genomic RNA from the putative site of RNA synthesis inside DMVs gets transported via the channel of the pore into the cytoplasm, where it would be encapsidated [16]. These emerging data reinforce the hypothesis that coronavirus-induced DMVs participate in viral replication and budding. However, the size and number of DMVs in infected cells are not correlated with competitive fitness of viral progenies. Viral mutants that generate smaller DMVs or less DMVs replicated as efficiently as wild-type virus [32]. In addition, coronavirus-induced membrane rearrangements are not determinants of viral pathogenicity [33]. Collectively, these evidences suggest that coronaviruses have evolved to have tremendous plasticity in the capability to form membrane-associated replication complexes for efficient viral replication [32,33].
3. The interaction between coronavirus non-structural proteins and DMVs
The striking similarities in morphology and architecture of DMVs across different genera of coronavirus, including the emerging SARS-CoV-2 [15], suggests a conservative mechanism behind the formation of these organelles. The biogenesis of coronavirus-induced DMVs relies on the interactions between nonstructural viral proteins and the host factors. The replication of two thirds of the human coronaviruses is initiated by translation of the two overlapping open reading frames (ORF1a and ORF1b) to generate two polyproteins, pp1a and pp1ab. The polyproteins are further processed by several viral proteases to generate 16 nonstructural proteins (nsp1–16) which are involved in different stages of viral life cycle. The other ORFs encode four structural proteins (spike, membrane, nucleocapsid, and envelope). A variable number of accessory proteins are also encoded by different coronaviruses [71]. Among the nonstructural proteins, nsp3, 4, and 6 are demonstrated to play fundamental roles in the replication of coronaviruses, particularly the biogenesis of DMVs [34]. Co-transfection of plasmids encoding the full-length nsp3, 4, and 6 can induce formation of CMs and DMVs in the ER region that resemble viral replication organelles observed in SARS-CoV infected cells [35]. In addition to their functions in host membrane rearrangement, these nsps also interact with host and viral proteins to ensure the successful replication process. Their distinctive impact in DMV formation and functions will be discussed in the following sections.
3.1. Non-structural proteins as structural scaffolds for DMV formation
3.1.1. Nsp3
Coronavirus nsp3 is considered to be the central scaffolding protein involved in the formation of DMVs. Despite the different genera of coronavirus, all nsp3 proteins broadly contain six functional domains, including a ubiquitin-like (Ubl1) domain that binds to single-stranded RNA (ssRNA) and nucelocapsid; macrodomain 1 (X domain) that possess ADP-ribose binding activity; macrodomain 2 which binds to ssRNA; a papain-like viral protease; a nucleic acid-binding (NAB) domain; a transmembrane domain, and seven-transmembrane G protein-coupled receptor C [[36], [37], [38]].
Formation of distinctive double-membrane structures of DMVs requires 1) excessive membrane materials; 2) folding and rearrangement of the membrane to form paired membrane structures. Nsp3 alone is capable of inducing ER membrane proliferation and expansion to provide membrane materials for the formation of DMVs [35].
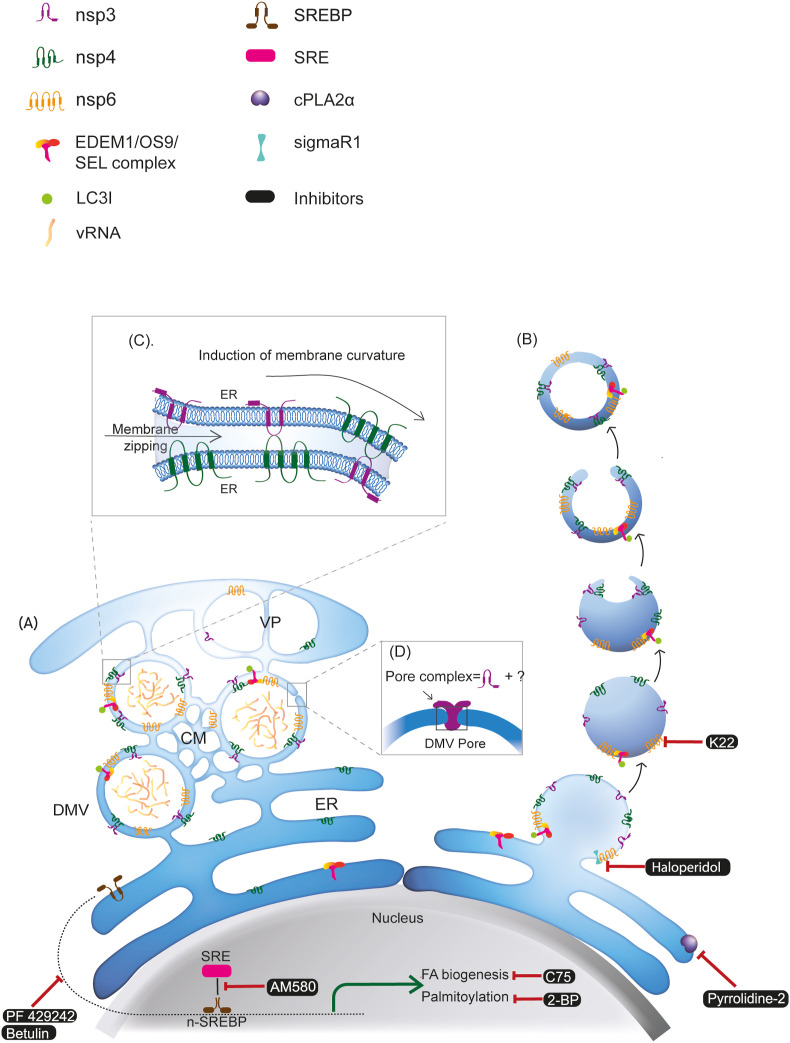
Wolff et al. identified nsp3 as one of the major constituents of the double-membrane spanning pore complex on MHV-induced DMVs, with its Ubl1 domain residing in the cytosolic prongs of the complex (Fig. 2 D). Other membrane spanning nsps (i.e. nsp4 and 6) and host factors are also potential candidates for the pore complex formation, although further investigation is required [16].
Fig. 2.
Biogenesis and architecture of coronavirus-induced DMVs with druggable targets and their pharmacological inhibitors. Formation of DMV is induced by viral nonstructural proteins (nsp3, nsp4, and nsp6) and host proteins involved in the ERAD machinery (EDEM1/OS9/SEL1). Two models have been proposed for the biogenesis of the coronavirus-induced DMVs: A) Model 1: viral infection triggers the generation of reticulovesicular network (RVN) of modified ER that integrates convoluted membranes (CMs) and numerous interconnected DMVs, which also connect to the ER. B) Model 2: by the action of nsp3, nsp4, and nsp6, formation of DMVs starts with exvaginations of the ER membrane, which then pinch off to form single-membraned EDEMsomes. These vesicles then undergo partial invagination to form cup-like structures that are then sealed to form DMVs. C) Heterotypical interaction between nsp3-nsp4 via their lumenal domains can induce membrane zipping and curvature which are essential for the formation of DMVs. D) nsp3 is one of the major constituents of the double-membrane spanning pore complex on coronavirus induced DMVs. VP: vesicle packets; SRE: sterol response elements; SREBP: sterol regulatory element-binding protein; vRNA: viral RNA.
In addition to its central role in DMV formation, nsp3 also serves as part of the viral immune defense machinery by promoting cytokine expression through its ADP-ribose binding, de-ubiquitylation, and de-ISGylation activities [38]. Moreover, nsp3 can also bind to cognate nucleocapsid protein mainly via its Ubl1 domain and tether the infecting nucleocapsid to the newly translated RTC at early stage of infection [13].
3.1.2. Nsp4
Nsp4 of coronavirus is a tetraspanning membrane protein [39,40]. Nsp4 binds to the transmembrane domain of nsp3 through its first large lumenal loop, whereas two amino acids H120 and F121 are identified to be essential for the binding, and the subsequent membrane rearrangement functions [41].
The importance of coronavirus nsp4 in biogenesis of DMVs has been observed in different studies. MHV mutant carrying nsp4 N258T caused a temperature-sensitive phenotype, accompanied with decreased numbers of DMVs. Moreover, nsp4-N258T, together with other replicase components (e.g. nsp3) localized on mitochondria, which were abnormally enlarged in size and extensively vacuolated that may due to fusion of DMVs [42]. Abolition of the N-glycosylation sites of MHV nsp4 resulted in suppressed viral RNA synthesis in addition to the appearance of aberrant DMVs and elevated numbers of CMs [43].
3.1.3. Nsp6
Coronavirus Nsp6 is an ER-located integral membrane protein with multiple transmembrane domains. Nsp6 itself can induce membrane proliferation and rearrangements. Overexpression of SARS-CoV nsp6 alone resulted in formation of vesicles named microtubule organizing center vesiculation (MTOCV) [35]. Nsp6 of avian coronavirus, infectious bronchitis virus (IBV), SARS-CoV, and MERS-CoV is capable of generating Atg5- and LC3ІІ-positive autophagosomes via an omegasome intermediate from the ER [44]. Interestingly, nsp6 appeared to limit the expansion of endogenous autophagosome and autolysosome, possibly providing a means to compromise the ability of autophagosomes to deliver viral components to lysosomes for degradation [45].
A recent bioinformatic and evolutionary study showed that SARS-CoV-2 nsp6 contains mutations in the outer membrane binding region that may favor a more stable binding of nsp6 to the ER membrane. However, whether the mutation would further enhance the ability of nsp6 to limit the antiviral autophagy responses requires more experimental evidence [46].
3.1.4. Heterotypical interactions between the nsps and other viral proteins
Although nsp3, 4, and 6 of coronavirus have their unique roles in viral replication, paralleled membrane structure formation involves different membranes and interactions from different sides of the membranes. Therefore, heterotypical interactions between nsp3-nsp4-nsp6 is necessary for the formation of DMVs.
The protein-protein binding between nsp3 and nsp4 is indispensable for the re-localization of this nsp3-4 complex from ER to RTC and the membrane pairing required during the formation of DMVs in SARS-CoV [35,39], MHV [47], and MERS-CoV [48] -infected cells (Fig. 2C). Loss of this nsp3-nsp4 interaction abolishes viral replication in SARS-CoV replicon systems [35,39,41]. Interaction between nsp3 and nsp4 is, however, insufficient to induce the formation of DMVs in the case of SARS-CoV and MHV. Rather, nsp6 and multiple host factors from the ER-turning machineries are involved in DMV formation with an exception of MERS-CoV, where co-expression of nsp3 and nsp4 is sufficient to form MERS-CoV-induced DMVs [48].
Coronavirus accessary protein ORF6 interacts with nsp3 and nsp8 [[49], [50], [51]]. As a multifunctional Golgi - ER membrane-associated protein, the N-terminal region of ORF6 induces the formation of membranous structures, which partially resemble DMVs that are observed in native virus replication [51]. Given the critical function of nsp3 in DMV formation and of nsp8 in viral RNA synthesis, it is possible that coronaviral ORF6 is also involved in the formation/function of DMVs via its interaction with these two nsps which may have an impact on virulence and pathogenicity [50].
Another accessory protein of coronavirus, ORF9b, has a central hydrophobic cavity that binds lipid molecules. ORF9b colocalizes with intracellular vesicles, suggesting a possible role in virus assembly via membrane association [52]. Despite the fact that ORF9b is dispensable for viral replication, its interactions with nsp8, nsp14, and ORF6 still strongly indicate the involvement of this lipid-binding viral protein in viral replication and assembly [53,54].
3.2. Non-structural proteins and biogenesis of DMVs
As discussed in the previous sections, the membrane-spanning nsp 3, 4, and 6 provide a structural scaffold for coronavirus-induced DMVs. Their lumenal domains are N-glycosylated -- a posttranslational modification that takes place on the ER. Moreover, when expressed alone intracellularly, ER resident nsp4 re-localized to the DMVs upon infection [40]. In addition, ultrastructure analysis of SARS-CoV-induced DMVs showed that they are part of a unique reticulovesicular network which connects to the rough ER [12]. Multiple lines of findings strongly suggest that coronaviruses hijack ER or ER-derived membranes to generate the lipid bilayer of the DMVs [12,[55], [56], [57]]. Two models have been proposed by which the coronavirus-induced DMVs are assembled and modified.
3.2.1. Via ER-spanning
In an early proposed model by Knoops et al, coronavirus infection leads to the formation of reticulovesicular network with multiple interconnected DMVs, which remain connected to ER membranes via their outer membrane [12] (Fig. 1A). The hydrophobic domains of the nsp 3, 4, and 6 anchored to the ER membrane and target the viral RTC to the unmodified ER. Particularly, heterotypical interaction between nsp3-nsp4 via their lumenal domains can induce membrane zipping and curvature that are essential for the formation of DMVs (Fig. 2C). As the viral replication proceeds, locally accumulated transmembrane nsps may induce further expansion, distortion, and rearrangement of ER membrane, while recruiting host factors required for viral replication (Fig. 2A).
In this model, apart from the hydrophobic transmembrane domains, the occurrence of non-membrane-spanning hydrophobic/amphipathic regions in nsp3 and nsp6 may also contribute to their membrane bending ability, and this feature seems to be common among DMV-inducing RNA viruses [58].
3.2.2. Via autophagy-independent LC3 engagement
Both macro and selective forms of autophagy have been described for (+) RNA viruses for various steps in the viral lifecycle [59,60]. Given the distinctive zipper membrane architecture of coronavirus-induced DMVs, it was speculated initially that the virus may hijack double-membraned autophagosome to generate DMVs. However, up to date, the role of autophagy in coronavirus replication remains controversial. Two earlier studies from the same group showed a decreased replication of MHV in autophagy essential gene Atg5 deleted cells and later co-localization of nsp 2, 3 and 8 with endogenous LC3, suggesting a pro-viral role of autophagy [55,61]. However, these findings could not be reproduced in later studies with MHV and SARS-CoV models by other groups, in which deletion of ATG5 and ATG7 had no significant impact on viral replication [62,63]. It seems more plausible that autophagy is not directly involved in either the replication or DMV formation, rather, single components of the autophagy pathways (e.g. LC3) are likely utilized by coronaviruses.
As the central hub for host membrane and protein synthesis, maturation and secretion, ER-associated degradation (ERAD) pathway is at the core of ER homeostasis. The level of ERAD is fine-tuned by selectively discarding ERAD effectors (such as mannosidase alpha-like 1 (EDEM1), osteosarcoma amplified 9 (OS9), and ERAD component SEL1) from the ER via ER-derived vesicles named EDEMosomes [63]. Unlike autophagosomes, which contain the lipidated form of LC3 (LC3ІІ), EDEMosomes are coated with LC3І. Surprisingly, MHV-induced DMVs contain multiple components of EDEMosomes, including LC3І, EDEM1, and OS-9 [62, 64]. In addition, siRNA-mediated LC3І knockdown led to severe block of viral replication and defective DMV production, indicating that LC3І is directly involved in the replication of the virus and formation of DMVs. These results suggest a mechanism by which coronaviruses hijack the LC3І-positive EDEMosomes for the biogenesis of DMVs. In this scenario, coronaviruses could recruit either EDEMosome-cargo receptors or directly LC3І as well as other ERAD effectors to the site of replication by one or more of the transmembrane nsps (nsp3, 4, and/or 6). The formation of DMVs starts with evaginations of the ER membrane, which then pinches off to form single-membraned EDEMsomes. These vesicles then undergo partial invagination to form a cup-like structure that is then sealed to form a DMV [63,65] (Fig. 2B). Although the direct interactions between coronavirus nsps and EDEMsome components have not yet been identified, EDEM1, OS9, and LC3І were detected to co-localize with nsp2/3. Moreover, EDEM1 is involved in the ER N-glycosylation processing [66]. It is possible that the interaction between EDEM1 and glycoprotein nsp6 is essential for the formation of DMVs [65].
4. Druggable targets of coronaviral DMVs
As elaborated above, the importance of DMVs during (+) RNA viruses replication makes this type of membrane compartments a very attractive target for drug development. The formation of DMVs involves 1) induction of membrane curvature via viral and cellular factors (e.g., insertion of transmembrane proteins and/or irregularly shaped lipids); 2) modification of DMV lipid composition, either via lipid transfer proteins or de novo lipid synthesis [4]. The identification of pharmacological inhibitors or dominant-negative mutants of viral or host proteins involved in the formation of coronavirus-induced DMVs may facilitate the development of antiviral treatment of related infectious diseases, including the highly pathogenic SARS-CoV-2 (Fig. 2).
4.1. Targeting viral factors
Anti-HCoV-229E compound K22 inhibits early yet post entry phase of the HCoV-229E replication cycle via diminishing the formation of DMVs. This antiviral effect of K22 is probably due to its interaction with nsp6. Moreover, K22 inhibits a broad range of coronaviruses, including MERS-CoV, making K22 a promising anti-coronavirus candidate for further development [67,68].
A very recent SARS-CoV-2 drug screening project using affinity-purification mass spectrometry successfully identified more than three hundred cellular factors that interact with viral proteins, among which there are 66 druggable human factors targeted by 69 compounds. Amongst the identified hits, the authors found an interaction between SARS-CoV-2 nsp6 and human sigmaR1 [6]. SigmaR1 is a unique multifunctional ER membrane protein that is involved in lipid transport, ER stress, and autophagy [69]. The interaction between nsp6 and sigmaR1 can be targeted by four approved drugs (e.g. Haloperidol, E−52862), one drug that is in clinical trial, and three preclinical agents [6].
4.2. Targeting cellular factors
Targeting SREBP and its downstream fatty acid synthesis pathways provide a broad spectrum of potential antivirals against coronaviruses. Small molecular inhibitor AM580, which blocks SREBP activation by disrupting the binding of n-terminal SREBP1 and SRE, significantly abolishes the production of DMVs in MERS-CoV-infected cells and SARS-CoV-infected cells [23]. In line with the antiviral effect of AM580, several pharmacological inhibitors of the SREBP-associated pathways, such as PF 429242 and Betulin that inhibit SREBP cleavage and maturation, C75 that inhibits fatty acid biosynthesis, and 2-BP that inhibits palmitoylation, all effectively reduce MERS-CoV production [23].
As discussed above, cPLA2α is the crucial component of HCoV-299E-induced DMVs. Inhibition of cPLA2α with a nonpeptidic inhibitor, pyrrolidine-2, significantly decrease the formation of DMVs as well as the production of infectious virus progeny [24].
5. Conclusion
The replication organelles formed by (+) RNA viruses are remarkably sophisticated and complex, particularly the DMVs induced by HCV, coronaviruses, and poliovirus, that are intricate labyrinths of modified host membranes with specific composition of lipids as well as viral and host proteins. In this review, we have summarized the current knowledge on the architecture, function, and biogenesis of DMVs with a focus on those induced by coronaviruses.
Author Contributions
JZ and YL drafted the manuscript and contributed equally to this work. SS supervised, evaluated, and edited the manuscript.
Declaration of competing interest
The authors declare no commercial or financial conflicts of interest.
Acknowledgement
This work was partially supported by Health and Medical Research Funds (17161022, 17161032 and 17161202) and Medical Research Council, UK (MC_PC_19063).
References
- 1.Paules C.I., Marston H.D., Fauci A.S. Coronavirus infections-more than just the common cold. J. Am. Med. Assoc. 2020;323(8):707–708. doi: 10.1001/jama.2020.0757. [DOI] [PubMed] [Google Scholar]
- 2.Nagy P.D., Strating J.R., van Kuppeveld F.J. Building viral replication organelles: close encounters of the membrane types. PLoS Pathog. 2016;12 doi: 10.1371/journal.ppat.1005912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Risco C. Three-Dimensional imaging of viral infections. Annu. Rev. Virol. 2014;1:453–473. doi: 10.1146/annurev-virology-031413-085351. [DOI] [PubMed] [Google Scholar]
- 4.Shulla A., Randall G. (+) RNA virus replication compartments: a safe home for (most) viral replication. Curr. Opin. Microbiol. 2016;32:82–88. doi: 10.1016/j.mib.2016.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harak C., Lohmann V. Ultrastructure of the replication sites of positive-strand RNA viruses. Virology. 2015;479–480:418–433. doi: 10.1016/j.virol.2015.02.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gordon D.E. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583:459–468. doi: 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Strating J.R., van Kuppeveld F.J. Viral rewiring of cellular lipid metabolism to create membranous replication compartments. Curr. Opin. Cell Biol. 2017;47:24–33. doi: 10.1016/j.ceb.2017.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Paul D., Madan V., Bartenschlager R. Hepatitis C virus RNA replication and assembly: living on the fat of the land. Cell Host Microbe. 2014;16:569–579. doi: 10.1016/j.chom.2014.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Romero-Brey I. Three-dimensional architecture and biogenesis of membrane structures associated with hepatitis C virus replication. PLoS Pathog. 2012;8 doi: 10.1371/journal.ppat.1003056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Paul D., Hoppe S., Saher G., Krijnse-Locker J., Bartenschlager R. Morphological and biochemical characterization of the membranous hepatitis C virus replication compartment. J. Virol. 2013;87:10612–10627. doi: 10.1128/JVI.01370-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Belov G.A. Complex dynamic development of poliovirus membranous replication complexes. J. Virol. 2012;86:302–312. doi: 10.1128/JVI.05937-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Knoops K. SARS-coronavirus replication is supported by a reticulovesicular network of modified endoplasmic reticulum. PLoS Biol. 2008;6 doi: 10.1371/journal.pbio.0060226. e226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Wilde A.H. MERS-coronavirus replication induces severe in vitro cytopathology and is strongly inhibited by cyclosporin A or interferon-alpha treatment. J. Gen. Virol. 2013;94:1749–1760. doi: 10.1099/vir.0.052910-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Snijder E.J. Ultrastructure and origin of membrane vesicles associated with the severe acute respiratory syndrome coronavirus replication complex. J. Virol. 2006;80:5927–5940. doi: 10.1128/JVI.02501-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Snijder E.J. A unifying structural and functional model of the coronavirus replication organelle: tracking down RNA synthesis. PLoS Biol. 2020;18 doi: 10.1371/journal.pbio.3000715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wolff G. A molecular pore spans the double membrane of the coronavirus replication organelle. Science. 2020;369:1395–1398. doi: 10.1126/science.abd3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang Z. Host lipids in positive-strand RNA virus genome replication. Front. Microbiol. 2019;10 doi: 10.3389/fmicb.2019.00286. 286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yan B. Characterization of the lipidomic profile of human coronavirus-infected cells: implications for lipid metabolism remodeling upon coronavirus replication. Viruses. 2019;11 doi: 10.3390/v11010073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang J., Lan Y., Sanyal S. Modulation of lipid droplet metabolism-A potential target for therapeutic intervention in flaviviridae infections. Front. Microbiol. 2017;8 doi: 10.3389/fmicb.2017.02286. 2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Horton J.D., Goldstein J.L., Brown M.S. SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J. Clin. Invest. 2002;109:1125–1131. doi: 10.1172/JCI15593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pombo J.P., Sanyal S. Perturbation of intracellular cholesterol and fatty acid homeostasis during flavivirus infections. Front. Immunol. 2018;9:1276. doi: 10.3389/fimmu.2018.01276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ye J., DeBose-Boyd R.A. Regulation of cholesterol and fatty acid synthesis. Cold Spring Harb Perspect Biol. 2011;3 doi: 10.1101/cshperspect.a004754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yuan S. SREBP-dependent lipidomic reprogramming as a broad-spectrum antiviral target. Nat. Commun. 2019;10 doi: 10.1038/s41467-018-08015-x. 120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Muller C. Inhibition of cytosolic phospholipase A2alpha impairs an early step of coronavirus replication in cell culture. J. Virol. 2018;92 doi: 10.1128/JVI.01463-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thomas T. COVID-19 infection results in alterations of the kynurenine pathway and fatty acid metabolism that correlate with IL-6 levels and renal status. JCI Insight. 2020;5 doi: 10.1172/jci.insight.140327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fuller N., Rand R.P. The influence of lysolipids on the spontaneous curvature and bending elasticity of phospholipid membranes. Biophys. J. 2001;81:243–254. doi: 10.1016/S0006-3495(01)75695-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Neufeldt C.J. The hepatitis C virus-induced membranous web and associated nuclear transport machinery limit access of pattern recognition receptors to viral replication sites. PLoS Pathog. 2016;12 doi: 10.1371/journal.ppat.1005428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Egger D., Pasamontes L., Bolten R., Boyko V., Bienz K. Reversible dissociation of the poliovirus replication complex: functions and interactions of its components in viral RNA synthesis. J. Virol. 1996;70:8675–8683. doi: 10.1128/jvi.70.12.8675-8683.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jackson W.T. Subversion of cellular autophagosomal machinery by RNA viruses. PLoS Biol. 2005;3 doi: 10.1371/journal.pbio.0030156. e156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Richards A.L., Jackson W.T. Intracellular vesicle acidification promotes maturation of infectious poliovirus particles. PLoS Pathog. 2012;8 doi: 10.1371/journal.ppat.1003046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stertz S. The intracellular sites of early replication and budding of SARS-coronavirus. Virology. 2007;361:304–315. doi: 10.1016/j.virol.2006.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Al-Mulla H.M. Competitive fitness in coronaviruses is not correlated with size or number of double-membrane vesicles under reduced-temperature growth conditions. mBio. 2014;5:e01107–e01113. doi: 10.1128/mBio.01107-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maier H.J. Extensive coronavirus-induced membrane rearrangements are not a determinant of pathogenicity. Sci. Rep. 2016;6 doi: 10.1038/srep27126. 27126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.J Alsaadi E.A., Jones I.M. Membrane binding proteins of coronaviruses. Future Virol. 2019;14:275–286. doi: 10.2217/fvl-2018-0144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Angelini M.M., Akhlaghpour M., Neuman B.W., Buchmeier M.J. Severe acute respiratory syndrome coronavirus nonstructural proteins 3, 4, and 6 induce double-membrane vesicles. mBio. 2013;4 doi: 10.1128/mBio.00524-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kanjanahaluethai A., Chen Z., Jukneliene D., Baker S.C. Membrane topology of murine coronavirus replicase nonstructural protein 3. Virology. 2007;361:391–401. doi: 10.1016/j.virol.2006.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Claverie J.-M. The likely role of de-Mono-ADP-ribosylation of STAT1 by the SARS-CoV-2 nsp3 protein in the cytokine storm syndrome of COVID-19. Viruses. 2020;12:646–654. doi: 10.3390/v12060646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lei J., Kusov Y., Hilgenfeld R. Nsp3 of coronaviruses: structures and functions of a large multi-domain protein. Antivir. Res. 2018;149:58–74. doi: 10.1016/j.antiviral.2017.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hagemeijer M.C. Mobility and interactions of coronavirus nonstructural protein 4. J. Virol. 2011;85:4572–4577. doi: 10.1128/JVI.00042-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Oostra M. Localization and membrane topology of coronavirus nonstructural protein 4: involvement of the early secretory pathway in replication. J. Virol. 2007;81:12323–12336. doi: 10.1128/JVI.01506-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sakai Y. Two-amino acids change in the nsp4 of SARS coronavirus abolishes viral replication. Virology. 2017;510:165–174. doi: 10.1016/j.virol.2017.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Clementz M.A., Kanjanahaluethai A., O’Brien T.E., Baker S.C. Mutation in murine coronavirus replication protein nsp4 alters assembly of double membrane vesicles. Virology. 2008;375:118–129. doi: 10.1016/j.virol.2008.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gadlage M.J. Murine hepatitis virus nonstructural protein 4 regulates virus-induced membrane modifications and replication complex function. J. Virol. 2010;84:280–290. doi: 10.1128/JVI.01772-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cottam E.M. Coronavirus nsp6 proteins generate autophagosomes from the endoplasmic reticulum via an omegasome intermediate. Autophagy. 2011;7:1335–1347. doi: 10.4161/auto.7.11.16642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cottam E.M., Whelband M.C., Wileman T. Coronavirus NSP6 restricts autophagosome expansion. Autophagy. 2014;10:1426–1441. doi: 10.4161/auto.29309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Benvenuto D. Evolutionary analysis of SARS-CoV-2: how mutation of Non-Structural Protein 6 (NSP6) could affect viral autophagy. J. Infect. 2020;81:e24–e27. doi: 10.1016/j.jinf.2020.03.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hagemeijer M.C. Membrane rearrangements mediated by coronavirus nonstructural proteins 3 and 4. Virology. 2014;458–459:125–135. doi: 10.1016/j.virol.2014.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Oudshoorn D. Expression and cleavage of middle east respiratory syndrome coronavirus nsp3-4 polyprotein induce the formation of double-membrane vesicles that mimic those associated with coronaviral RNA replication. mBio. 2017;8 doi: 10.1128/mBio.01658-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kumar P. The nonstructural protein 8 (nsp8) of the SARS coronavirus interacts with its ORF6 accessory protein. Virology. 2007;366:293–303. doi: 10.1016/j.virol.2007.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tangudu C., Olivares H., Netland J., Perlman S., Gallagher T. Severe acute respiratory syndrome coronavirus protein 6 accelerates murine coronavirus infections. J. Virol. 2007;81:1220–1229. doi: 10.1128/JVI.01515-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhou H. The N-terminal region of severe acute respiratory syndrome coronavirus protein 6 induces membrane rearrangement and enhances virus replication. J. Virol. 2010;84:3542–3551. doi: 10.1128/JVI.02570-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Meier C. The crystal structure of ORF-9b, a lipid binding protein from the SARS coronavirus. Structure. 2006;14:1157–1165. doi: 10.1016/j.str.2006.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Calvo E. Severe acute respiratory syndrome coronavirus accessory proteins 6 and 9b interact in vivo. Virus Res. 2012;169:282–288. doi: 10.1016/j.virusres.2012.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.von Brunn A. Analysis of intraviral protein-protein interactions of the SARS coronavirus ORFeome. PloS One. 2007;2 doi: 10.1371/journal.pone.0000459. e459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Prentice E., Jerome W.G., Yoshimori T., Mizushima N., Denison M.R. Coronavirus replication complex formation utilizes components of cellular autophagy. J. Biol. Chem. 2004;279:10136–10141. doi: 10.1074/jbc.M306124200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Romero-Brey I., Bartenschlager R. Endoplasmic reticulum: the favorite intracellular niche for viral replication and assembly. Viruses. 2016;8(6) doi: 10.3390/v8060160. 160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bagchi P. Endoplasmic reticulum in viral infection. Int Rev Cell Mol Biol. 2020;350:265–284. doi: 10.1016/bs.ircmb.2019.10.005. [DOI] [PubMed] [Google Scholar]
- 58.Oostra M. Topology and membrane anchoring of the coronavirus replication complex: not all hydrophobic domains of nsp3 and nsp6 are membrane spanning. J. Virol. 2008;82:12392–12405. doi: 10.1128/JVI.01219-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang J. Flaviviruses exploit the lipid droplet protein AUP1 to trigger lipophagy and drive virus production. Cell Host Microbe. 2018;23:819–831. doi: 10.1016/j.chom.2018.05.005. e815. [DOI] [PubMed] [Google Scholar]
- 60.Wong H.H., Sanyal S. Manipulation of autophagy by (+) RNA viruses. Semin. Cell Dev. Biol. 2020;101:3–11. doi: 10.1016/j.semcdb.2019.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Prentice E., McAuliffe J., Lu X., Subbarao K., Denison M.R. Identification and characterization of severe acute respiratory syndrome coronavirus replicase proteins. J. Virol. 2004;78:9977–9986. doi: 10.1128/JVI.78.18.9977-9986.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Reggiori F., de Haan C.A.M., Molinari M. Unconventional use of LC3 by coronaviruses through the alleged subversion of the ERAD tuning pathway. Viruses. 2011;3:1610–1623. doi: 10.3390/v3091610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Noack J., Bernasconi R., Molinari M. How viruses hijack the ERAD tuning machinery. J. Virol. 2014;88:10272–10275. doi: 10.1128/JVI.00801-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Reggiori F. Coronaviruses Hijack the LC3-I-positive EDEMosomes, ER-derived vesicles exporting short-lived ERAD regulators, for replication. Cell Host Microbe. 2010;7:500–508. doi: 10.1016/j.chom.2010.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.de Haan C.A., Molinari M., Reggiori F. Autophagy-independent LC3 function in vesicular traffic. Autophagy. 2010;6:994–996. doi: 10.4161/auto.6.7.13309. [DOI] [PubMed] [Google Scholar]
- 66.Olivari S. EDEM1 regulates ER-associated degradation by accelerating de-mannosylation of folding-defective polypeptides and by inhibiting their covalent aggregation. Biochem. Biophys. Res. Commun. 2006;349:1278–1284. doi: 10.1016/j.bbrc.2006.08.186. [DOI] [PubMed] [Google Scholar]
- 67.Lundin A. Targeting membrane-bound viral RNA synthesis reveals potent inhibition of diverse coronaviruses including the middle East respiratory syndrome virus. PLoS Pathog. 2014;10 doi: 10.1371/journal.ppat.1004166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Li G., De Clercq E. Therapeutic options for the 2019 novel coronavirus (2019-nCoV) Nat. Rev. Drug Discov. 2020;19:149–150. doi: 10.1038/d41573-020-00016-0. [DOI] [PubMed] [Google Scholar]
- 69.Yang H., Shen H., Li J., Guo L.W. SIGMAR1/Sigma-1 receptor ablation impairs autophagosome clearance. Autophagy. 2019;15:1539–1557. doi: 10.1080/15548627.2019.1586248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Klein S. SARS-CoV-2 structure and replication characterized by in situ cryo-electron tomography. bioRxiv. 2020 doi: 10.1101/2020.06.23.167064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Forni D., Cagliani R., Clerici M., Sironi M. Molecular evolution of human coronavirus genomes. Trends Microbiol. 2017;25:35–48. doi: 10.1016/j.tim.2016.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Barcena M. Cryo-electron tomography of mouse hepatitis virus: insights into the structure of the coronavirion. Proc. Natl. Acad. Sci. Unit. States Am. 2009;106:582–587. doi: 10.1073/pnas.0805270106. [DOI] [PMC free article] [PubMed] [Google Scholar]