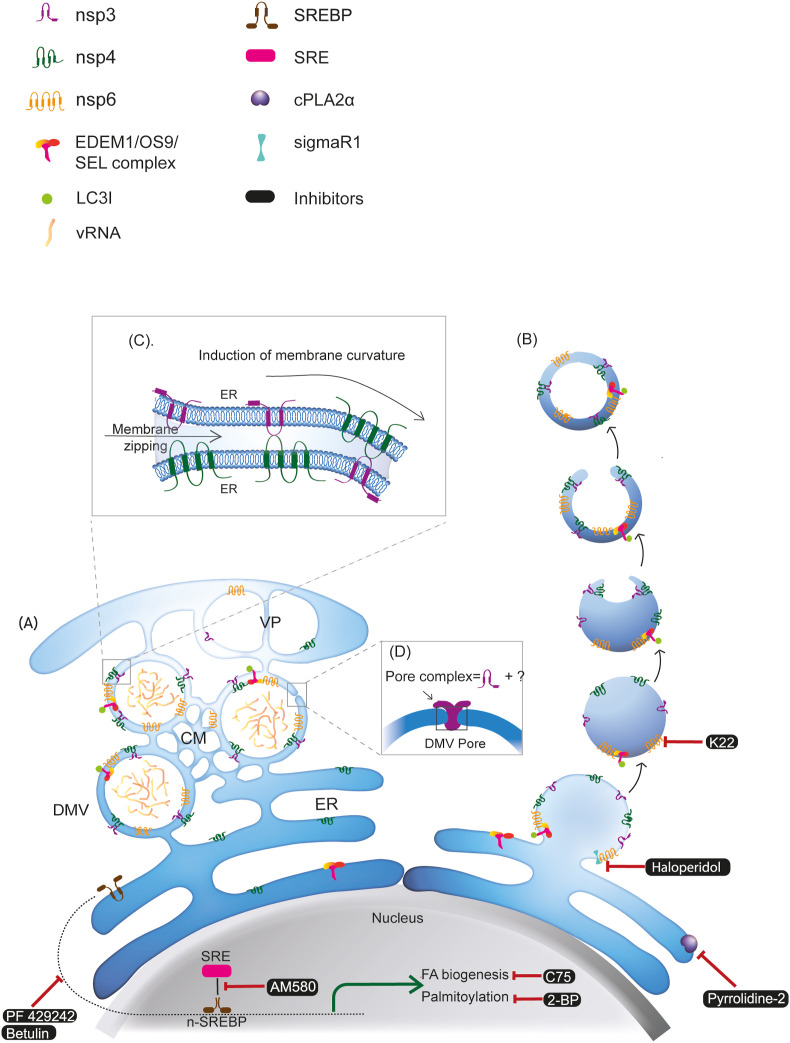
Fig. 2.
Biogenesis and architecture of coronavirus-induced DMVs with druggable targets and their pharmacological inhibitors. Formation of DMV is induced by viral nonstructural proteins (nsp3, nsp4, and nsp6) and host proteins involved in the ERAD machinery (EDEM1/OS9/SEL1). Two models have been proposed for the biogenesis of the coronavirus-induced DMVs: A) Model 1: viral infection triggers the generation of reticulovesicular network (RVN) of modified ER that integrates convoluted membranes (CMs) and numerous interconnected DMVs, which also connect to the ER. B) Model 2: by the action of nsp3, nsp4, and nsp6, formation of DMVs starts with exvaginations of the ER membrane, which then pinch off to form single-membraned EDEMsomes. These vesicles then undergo partial invagination to form cup-like structures that are then sealed to form DMVs. C) Heterotypical interaction between nsp3-nsp4 via their lumenal domains can induce membrane zipping and curvature which are essential for the formation of DMVs. D) nsp3 is one of the major constituents of the double-membrane spanning pore complex on coronavirus induced DMVs. VP: vesicle packets; SRE: sterol response elements; SREBP: sterol regulatory element-binding protein; vRNA: viral RNA.