Abstract
Objectives
This study was performed during the early outbreak period of coronavirus disease 2019 (COVID-19) and the seasonal epidemics of other respiratory viral infections, in order to describe the extent of co-infections of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) with other respiratory viruses. It also compared the diagnostic performances of upper respiratory tract (URT) and lower respiratory tract (LRT) samples for SARS-CoV-2 infection.
Methods
From 25 January to 29 March 2020, all URT and LRT samples collected from patients with suspected COVID-19 received in the virology laboratory of Pitié-Salpêtrière University Hospital (Paris, France) were simultaneously tested for SARS-CoV-2 and other respiratory viruses.
Results
A total of 1423 consecutive patients were tested: 677 (47.6%) males, 746 (52.4%) females, median age 50 (range, 1–103) years. Twenty-one (1.5%) patients were positive for both SARS-CoV-2 and other respiratory viruses. The detection rate of SARS-CoV-2 was significantly higher in LRT than in URT (53.6% vs. 13.4%; p < 0.0001). The analysis of paired samples from 117 (8.2%) patients showed that SARS-CoV-2 load was lower in URT than in LRT samples in 65% of cases.
Conclusion
The detection of other respiratory viruses in patients during this epidemic period could not rule out SARS-CoV-2 co-infection. Furthermore, LRT samples increased the accuracy of diagnosis of COVID-19.
Keywords: SARS-CoV-2, Other respiratory viruses, Co-infection, Lower respiratory tract
Introduction
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) responsible for coronavirus disease 2019 (COVID-19) emerged in Wuhan, China, in December 2019 (Zhu et al., 2020). A few studies have reported proportions of SARS-CoV-2 co-infections with other respiratory viruses (range, 0–20%) (Chen et al., 2020, Kim et al., 2020, Leuzinger et al., 2020, Lin et al., 2020a, Wee et al., 2020). Furthermore, lower respiratory tract (LRT) samples have been found to significantly improve the efficiency of diagnosis compared with upper respiratory tract (URT) samples for non-SARS-CoV-2 respiratory infections (Branche et al., 2014, Falsey et al., 2012).
This study, performed during the early outbreak period of COVID-19 and the seasonal epidemics of other respiratory viral infections, aimed to describe the extent of co-infections of SARS-CoV-2 with other respiratory viruses. A second objective of this study was to compare the diagnostic performances of URT and LRT samples for SARS-CoV-2 infection. From 25 January to 29 March 2020, all URT and LRT samples collected from patients with suspected COVID-19 received in the virology laboratory of Pitié-Salpêtrière University Hospital (Paris, France) were tested for SARS-CoV-2 and other respiratory viruses. The study population consisted of hospitalised symptomatic adults. All patients had respiratory symptoms, as mentioned in the definition of possible COVID-19 cases at that time in France. SARS-CoV-2 RNA was detected by real-time RT-PCR (Corman et al., 2020). SARS-CoV-2 loads were estimated with cycle threshold (Ct) values. Other respiratory viruses were tested with the Filmarray® RP2plus (BIOMERIEUX): influenza virus, respiratory syncytial virus, metapneumovirus, adenovirus, parainfluenza virus, non-SARS-CoV-2 coronavirus, and rhinovirus/enterovirus. For statistical analyses, continuous variables were expressed as median (range) and categorical variables as numbers (percentages). Comparisons were performed using Mann-Whitney U test (continuous variables) and Chi-squared test (categorical variables). P < 0.05 was considered to be statistically significant.
During the study period, 1423 consecutive hospitalised patients (677 males, 746 females, median age 50 years) were included. Patients originated from Pitié-Salpêtrière hospital (n = 1167) or from other close hospitals (n = 256). Most of patients from Pitié-Salpêtrière hospital were hospitalised within 10 days after the onset of symptoms and were tested for SARS-CoV-2 and other respiratory viruses within 24 h of admission. Among patients from Pitié-Salpêtrière hospital, 233/1167 (20.0%) were hospitalised in the intensive care unit (ICU). Detection rates of respiratory viruses among patients were: 724 (50.9%) negative for respiratory viruses, 398 (28.0%) positive for other respiratory viruses, 280 (19.7%) positive for SARS-CoV-2, and 21 (1.5%) positive for SARS-CoV-2 and other respiratory viruses (Table 1 ). Among patients negative for SARS-CoV-2 (n = 1122), those who were positive for other respiratory viruses were statistically younger than those who were negative (median, 39 vs. 52 years, p < 0.0001). Conversely, among patients who were positive for SARS-CoV-2 (n = 301), no significant age difference was observed between those who were positive for other respiratory viruses and those who were negative (median, 56 vs. 54 years, not significant). Among the 21 patients co-infected with SARS-CoV-2, other respiratory viruses that were detected were non-SARS-CoV-2 coronavirus (n = 6), influenza virus (n = 5), adenovirus (n = 3), rhinovirus/enterovirus (n = 3), parainfluenza virus (n = 3), and adenovirus + rhinovirus/enterovirus (n = 1). Four of 21 (19.0%) co-infected patients were hospitalised in ICU, which was significantly lower than the 113/280 (46.4%) patients infected with SARS-CoV-2 alone (p = 0.020) (Table 1).
Table 1.
Characteristics of suspected COVID-19 patients, according to respiratory virological findings.
| SARS-CoV-2 (−) Other respiratory viruses (−) |
SARS-CoV-2 (−) Other respiratory viruses (+) |
SARS-CoV-2 (+) Other respiratory viruses (−) |
SARS-CoV-2 (+) Other respiratory viruses (+) |
|
|---|---|---|---|---|
| No. (%) of patients | 724 (50.9%) | 398 (28.0%) | 280 (19.7%) | 21 (1.5%) |
| Demographic characteristics | ||||
| Male | 328 (45.3%) | 178 (44.7%) | 160 (57.1%) | 11 (52.4%) |
| Female | 396 (54.7%) | 220 (55.3%) | 120 (42.9%) | 10 (47.6%) |
| Median age (y) (range) | 52 (0–103)a | 39 (0–101)a | 56 (12–94) | 54 (30–94) |
| Hospitalisation in ICUb | ||||
| Yes | 89 (15.3%) | 27 (8.4%) | 113 (46.7%)c | 4 (19.0%)c |
| No | 493 (84.7%) | 295 (91.6%) | 129 (53.3%) | 17 (81.0%) |
Abbreviations: (−), negative; (+), positive; ICU, intensive care unit; y, years.
p < 0.0001 (Mann–Whitney U test).
Rates of hospitalisation in ICU only for the patients from Pitié-Salpêtrière University Hospital (n = 1167).
p = 0.020 (Chi-squared test).
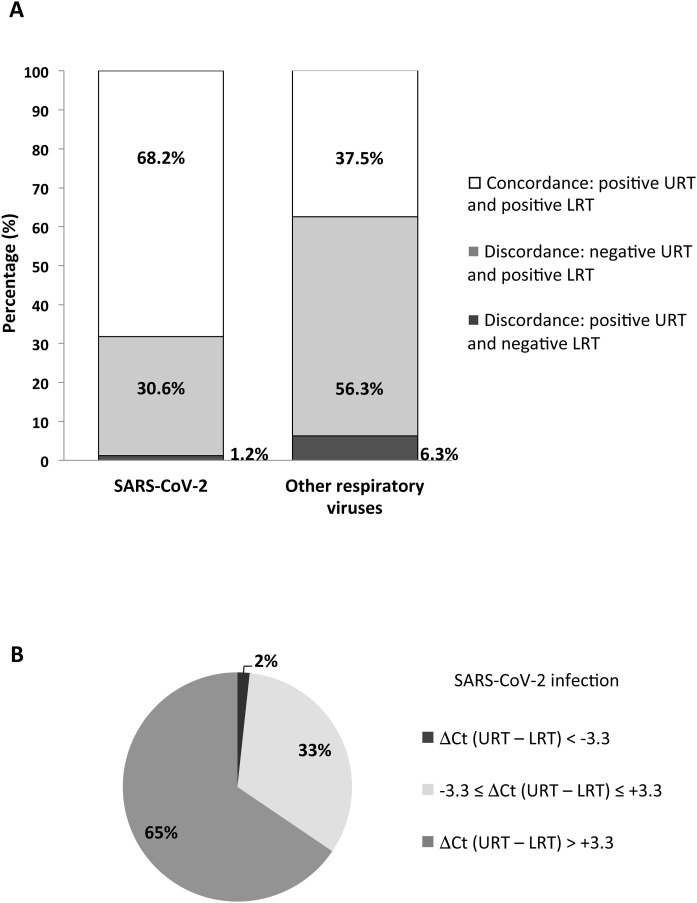
A total of 1160 URT and 379 LRT samples were collected. The detection rate of SARS-CoV-2 was significantly higher in LRT (203, 53.6%) than in URT (1160, 13.4%; p < 0.0001). Paired samples from URT and LRT were obtained from 117 (8.2%) patients. Among the 85 patients who were positive for SARS-CoV-2, 52 (68.2%) showed concordant positive results in URT and LRT samples (Figure 1A), but SARS-CoV-2 load was at least 1 log higher in LRT than in URT samples for 38 patients (65%) (Figure 1B). Moreover, 26 (30.6%) patients showed discordant SARS-CoV-2 results, with negative URT and positive LRT samples (Figure 1A). A similar profile was observed for the 32 patients infected with other respiratory viruses: concordant positive results in URT and LRT for 12 (37.5%), while 18 (56.3%) showed discordant results, with negative URT and positive LRT samples (Figure 1A). The proportion of patients positive for SARS-CoV-2 with discordant results was significantly lower than the one of patients positive for other respiratory viruses with discordant results: 30.6% vs. 56.3% (p = 0.012).
Figure 1.
Comparison of upper respiratory tract (URT) and lower respiratory tract (LRT) for diagnosing infections with SARS-CoV-2 and other respiratory viruses, using paired samples from COVID-19 suspected patients. (A) Distribution of paired respiratory samples according to the concordance of detection of SARS-CoV-2 and other respiratory viruses in URT and LRT. (B) Distribution of paired respiratory samples according to the difference in SARS-CoV-2 load between URT and LRT. SARS-CoV-2 loads were estimated with cycle threshold (Ct) values and the difference of Ct values obtained between URT and LRT (ΔCt URT – LRT) was calculated. Paired samples were then distributed according to the ΔCt value of 3.3, given that this difference approximately corresponds with a SARS-CoV-2 load of 1 log.
In the present study, 7% (21/301) of SARS-CoV-2-positive patients were co-infected with other respiratory viruses. This co-infection rate is similar to some previously reported rates (range, 0–6.5%) (Chen et al., 2020, Leuzinger et al., 2020, Lin et al., 2020a), but lower than others (up to 20%) (Kim et al., 2020). This might be due to different study populations or potential spatiotemporal variations in viral epidemiology. In particular, it is very likely that the lockdown in France, which started on 17 March and lasted until 11 May 2020, had no (or very little) influence on the circulation of respiratory viruses during the present study performed from 25 January to 29 March 2020. In line with previous studies, different types of other respiratory viruses were detected together with SARS-CoV-2 among co-infected patients, including non-SARS-CoV-2 coronavirus, influenza virus, adenovirus, rhinovirus/enterovirus, and parainfluenza virus (Kim et al., 2020, Leuzinger et al., 2020, Lin et al., 2020a, Wee et al., 2020). Patients co-infected with SARS-CoV-2 and other respiratory viruses did not significantly differ in age and gender from those infected with SARS-CoV-2 alone, as previously described (Kim et al., 2020). Among ICU patients, the proportion of co-infected patients was significantly lower than the one infected with SARS-CoV-2 alone, possibly indicating that co-infection with other respiratory viruses might not worsen the severity of SARS-CoV-2-associated respiratory disease, which was in accordance with a previous study (Wee et al., 2020). The current study found a higher efficiency of LRT than URT samples for COVID-19 diagnosis, with a significantly higher rate of detection of SARS-CoV-2 and a 1 log higher SARS-CoV-2 load for the majority of infected patients. This discrepancy could be partly explained by the variability in the delay between the onset of symptoms and the sampling among patients. However, this higher diagnostic performance of LRT samples for respiratory infections, including COVID-19, has been previously reported (Branche et al., 2014, Falsey et al., 2012, Lin et al., 2020b, Wang et al., 2020). In the current study, the proportion of patients positive for SARS-CoV-2 with discordant results was significantly lower than the one of patients who were positive for other respiratory viruses with discordant results. Those different profiles of compartmentalisation of PCR positivity within the respiratory tract may suggest some differences in pathophysiology of SARS-CoV-2 infection compared with infections by other respiratory viruses.
In conclusion, the detection of other respiratory viruses in patients during this epidemic period could not rule out SARS-CoV-2 co-infection, and LRT samples increased the accuracy of diagnosis of viral respiratory infections, including COVID-19.
Conflict of interest
DB and PH received lecture honorarium and fees for congress transportation and registration from Biomerieux. MD received honorarium for fees for travel expenses from Lungpacer. CEL received lecture honorarium from Biomerieux. Other authors have no conflict of interest to declare.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial or not-for-profit sectors.
Ethical approval
Due to the observational and retrospective nature of the study, the need for specific informed consent from individual patients was not required. All data were generated from routine standard clinical management of the patients. An informed consent to the treatment of personal data was acquired upon hospital admission from the patients or their legal representatives.
Acknowledgements
We thank all healthcare workers involved in the diagnosis and treatment of COVID-19 patients in Pitié-Salpêtrière University Hospital (Paris, France).
References
- Branche A.R., Walsh E.E., Formica M.A., Falsey A.R. Detection of respiratory viruses in sputum from adults by use of automated multiplex PCR. J Clin Microbiol. 2014;52:3590–3596. doi: 10.1128/JCM.01523-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N., Zhou M., Dong X., Qu J., Gong F., Han Y. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395:507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K.W. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25:1–8. doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falsey A.R., Formica M.A., Walsh E.E. Yield of sputum for viral detection by reverse transcriptase PCR in adults hospitalized with respiratory illness. J Clin Microbiol. 2012;50:21–24. doi: 10.1128/JCM.05841-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Quinn J., Pinsky B., Shah N.H., Brown I. Rates of co-infection between SARS-CoV-2 and other respiratory pathogens. JAMA. 2020;323:2085–2086. doi: 10.1001/jama.2020.6266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leuzinger K., Roloff T., Gosert R., Sogaard K., Naegele K., Rentsch K. Epidemiology of Severe Acute Respiratory Syndrome Coronavirus 2 emergence amidst community-acquired respiratory viruses. J Infect Dis. 2020;222:1270–1279. doi: 10.1093/infdis/jiaa464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin D., Liu L., Zhang M., Hu Y., Yang Q., Guo J. Co-infections of SARS-CoV-2 with multiple common respiratory pathogens in infected patients. Sci China Life Sci. 2020;63:606–609. doi: 10.1007/s11427-020-1668-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C., Xiang J., Yan M., Li H., Huang S., Shen C. Comparison of throat swabs and sputum specimens for viral nucleic acid detection in 52 cases of novel coronavirus (SARS-Cov-2)-infected pneumonia (COVID-19) Clin Chem Lab Med. 2020;58:1089–1094. doi: 10.1515/cclm-2020-0187. [DOI] [PubMed] [Google Scholar]
- Wang W., Xu Y., Gao R., Lu R., Han K., Wu G. Detection of SARS-CoV-2 in different types of clinical specimens. JAMA. 2020;323:1843–1844. doi: 10.1001/jama.2020.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wee L.E., Ko K.K.K., Ho W.Q., Kwek G.T.C., Tan T.T., Wijaya L. Community-acquired viral respiratory infections amongst hospitalized inpatients during a COVID-19 outbreak in Singapore: co-infection and clinical outcomes. J Clin Virol. 2020;128:104436. doi: 10.1016/j.jcv.2020.104436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]