Abstract
Background
Many studies found that VPS53, one of the subunits of the golgi-associated retrograde protein (GARP) complexes, was aberrantly expressed in human diseases.
Aim
This study investigated the functions and molecular mechanisms of VPS53 in colorectal cancer (CRC).
Methods
Expression and correlation of Beclin 1 and VPS53 were analyzed by RT-qPCR and Pearson’s correlation in CRC tissues, and VPS53 expression was also determined in CRC cells. The changes of proliferation, migration, invasion, apoptosis, and autophagy of CRC cells were examined by a succession of functional experiments including CCK-8, flow cytometry, transwell assay, and electron microscopy. The levels of autophagy related proteins were evaluated by Western blotting analysis.
Results
RT-qPCR results found that VPS53 was downregulated in CRC tissues and cells, and Beclin 1 expression was also decreased in CRC tissues. There was a positive correlation between VPS53 and Beclin 1. Functional results showed that overexpression of VPS53 could suppress proliferation, migration, and invasion, and accelerate apoptosis and autophagy of CRC cells. Also, VPS53 could upregulate Beclin 1 and LC3BII, suggesting the inductive effect of VPS53 on CRC cell autophagy. Furthermore, it was found that the autophagy inhibitor (Inhb) could attenuate the inhibition of VPS53 on CRC progression.
Conclusion
VPS53 repressed CRC progression by regulating the autophagy signaling pathway, suggesting that VPS53 might be a promising therapeutic target for CRC.
Keywords: colorectal cancer, VPS53, autophagy, cell migration, cell invasion
Introduction
Colorectal cancer (CRC) is a malignant tumor transformed from the epithelial tissue of the colorectal mucosa under the comprehensive actions of various pathogenic factors such as genes, diet, and the environment.1,2 Its incidence and mortality are the third and fourth, respectively, among malignant tumors in the world.3 In developed countries, the incidence of CRC ranks second in malignant tumors.4 At present, the treatment of CRC is mainly comprehensive surgical treatment, including surgical resection of tumor tissues, chemotherapy, radiotherapy, biological therapy, and immunotherapy.5,6 However, the 5-year survival rate of CRC is not high due to its complex etiology, high malignancy, rapid tumor growth, and strong invasive ability.7
Vesicular transport mainly consists of budding, directional transport, tethering, anchoring, and membrane fusion.8 The initial connection between the transport vesicle and membrane receptor is mediated by tethering factors, which recognize the transport vesicle and narrowed the distance between it and the membrane receptor.9 Vps53 might be involved in intracellular cholesterol transport and lysosomal sphingolipid homeostasis.10 Also, Vps53 was found to be differentially expressed in a variety of tumors.11–14 Specially, Gan et al found that VPS53 serves as a tumor suppressor in CRC, by inhibiting cell proliferation, attenuating tumor growth.15 However, the mechanism of VPS53 in CRC was still unclear.
In this study, we further confirmed expression of Vps53 and the regulatory relationship between Vps53 and the autophagy pathway in CRC. Functionally, we also determined the influence of Vps53 on CRC cell proliferation, migration, invasion, apoptosis, and autophagy by mediating the autophagy pathway. Therefore, we speculated that Vps53 might be of great clinical significance for the early diagnosis and treatment of CRC.
Materials and Methods
Patient Tissue Samples
CRC and adjacent normal tissues were collected from 15 patients with CRC who underwent surgical resection in Nanchong Central Hospital between November 2017 and February 2018. No patient had received chemotherapy before the operation. Written informed consent was obtained from each participant. Our study was also approved by the ethics committee of The Second clinical Medical College. Samples were placed in cryopreservation tubes, quick-frozen with liquid nitrogen, and transferred to a −80°C refrigerator.
Cell Culture
Normal colon FHC cells (BNCC338003) and LoVo (BNCC338601) cells were obtained from BeNa Culture Collection (Suzhou, China). SW620 (CCL-227), HT29 (HTB-38), SW480 (CCL-228), and HCT116 (CCL-247) cells were purchased from ATCC. FHC and HCT116 cells were maintained in RPMI-1640 medium (ATCC, cat. no. 30–2001) with 10% fetal bovine serum (FBS; Invitrogen Cat. no. 10,099–141) and glutamine. SW620 and SW480 cells were cultured in L-15 medium (Gibco, cat. no. 41,300,039) with 10% FBS, HT29 cells were grown in McCoy's 5A (Sigma, cat. no. M4892) with 10% FBS, and LoVo cells were maintained in F-12K medium (Gibco) with 10% FBS. All cells were cultured at 37°C with 5% CO2.
Vector Construction and Transduction
PrimerSTAR Max DNA Polymerase Mix (Takara, Japan) was used to amplify the full-length VPS53, and the products were cloned into the pcDNA3.1+vector (Invitrogen). SW620 and LoVo cells (2×105 cells/well) were spread onto 6 well plates and incubated at 37°C for 8 hrs. After incubation, cells in each well were transfected with empty vector (NC) and pcDNA-VPS53 using Lipofectamine 3000 (Invitrogen) according to the instruction provided by the supplier. SW620 and LoVo cells were also treated with an autophagy inhibitor (Inhb).
RNA Extraction and Quantitative Real-Time PCR (RT-qPCR) Assay
Total RNA was extracted using TRIzol reagent (Invitrogen) from the tissue homogenates and cells. RNA was used to synthesize cDNA according to the instructions of the reverse transcription kit (Takara, Japan). Expressions of the target genes and housekeeping gene (GAPDH) were examined using the SYBR-Green PCR kit (Qiagen, Hilden, Germany) on the ABI 7500 Fast Real-Time PCR system. Relative expression levels of the target genes were analyzed using the 2−ΔΔCt calculation method.16 All primers used in this experiment are listed in Table 1.
Table 1.
Primers Used in Realtime PCR in This Study
| ID | Sequence (5ʹ- 3ʹ) | Length (bp) |
|---|---|---|
| GAPDH F | TGTTCGTCATGGGTGTGAAC | 154 |
| GAPDH R | ATGGCATGGACTGTGGTCAT | |
| Beclin-1 F | AGGTTGAGAAAGGCGAGACA | 133 |
| Beclin-1 R | GTCCACTGCTCCTCAGAGTT | |
| VPS53 F | TGTTCCCAACCGAGCAATCTC | 115 |
| VPS53 R | ACGTTCGTCTGACCTCTTACA |
Abbreviations: F, forward primer; R, reversed primer.
Western Blot Assay
The tissues were ground and 500μL of RIPA lysate (Beyotime, cat. no. P0013B) was added to the processed cells in order to extract the total proteins. The Pierce BCA Protein Assay kit (Thermo Fisher Scientific) was used to quantify the concentrations of the extracted proteins. Thirty microgram protein was added into each of the holes in 10% SDS-PAGE, and the different sizes of proteins were separated by electrophoresis and transferred to PVDF membranes (Bio-Rad, cat. no. 170–4157). After being sealed for 1h using 5% skim milk, the membranes were treated with primary antibodies at 4°C overnights. After treatment with secondary antibodies (Santa Cruz), the ECL system (Amersham Pharmacia Biotech) was used to colorize the membranes. The primary antibodies were: anti-GAPDH antibody (1:3000; Abcam, ab9485), anti-VPS53 antibody (0.4 µg/mL; Abcam, ab251759), anti-LC3B antibody (1:2000; Abcam, ab51520), and anti-Beclin 1 antibody (1.0 µg/mL; Abcam, ab62557).
Proliferation Assay
SW620 and LoVo cells (1×104 cells/well) were plated onto 96-well plates. After transfection with the VPS53-overexpressed plasmid or/and treatment with the autophagy inhibitor (Inhb), 3-MA for 0, 24, 48, and 72 hrs, 15 µL CCK-8 reagent was added to each well. After incubation for 3 hrs, the OD values were measured using a microplate reader (Thermo Fisher) at 450 nm.
Flow Cytometry Analysis
The treated SW620 and LoVo cells were harvested by cell dissociation and centrifuged (1000 ×g for 5 mins). Cell deposits were suspended using PBS buffer. Apoptosis was assessed using the Annexin V-FITC/PI apoptosis detection kit (cat. no. MA0220). Following the instructions, 500µL binding buffer, 5µL Annexin V-FITC, and 10µL PI were added to the cells for 10 mins. The number of apoptotic cells was monitored using flow cytometry (BD Biosciences).
Transwell Assay
Cell migration and invasion were examined by utilizing Transwell chambers (Costar, cat. no. 3422) with and without Matrigel matrix (Cat. no. 356,230). Two hundred microliters of treated SW620 and LoVo cells (1×105 cells/mL) in serum-free medium were placed in the upper chamber, and the medium with 20% FBS was put in the lower chamber. After incubation at 37 °C for 24 hrs, the migrated and invaded cells were fixed for 10 mins, stained with 0.5% crystal violet (Cat. no. C3886) for 20 mins, and observed using a microscope.
Transmission Electron Microscope
The treated SW620 and LoVo cells were collected and centrifuged (500 ×g for 5 mins). Cell deposits at 4°C had precooled 5% glutaraldehyde solution added for 5 mins. After suspension and centrifugation, cells were treated with 5% glutaraldehyde and 10% bovine serum albumin (BSA; cat. no. BP1605) for 4 hrs on ice. After washing, cells were fixed with 8% potassium pyrophosphate and 1% osmium tetroxide. After dehydration with acetone, the cells were soaked in epoxy resin for 2 days. Sections were obtained using a Leica EM UC7 Ultramicrotome. The sections were stained with 2% uranyl acetate and observed using a transmission electron microscope.
Statistical Analysis
All the experiments were repeated three times, and the data were expressed as mean ± standard deviation (SD). All results were analyzed using SPSS 19.0 (IBM, Chicago, IL, USA). The statistical graphs were drawn using GraphPad Software (Version 5.0). The differences were evaluated using Student’s t-tests. The correlation between Beclin 1 and VPS53 was analyzed by Pearson’s correlation. P<0.05 indicated that the difference was statistically significant.
Results
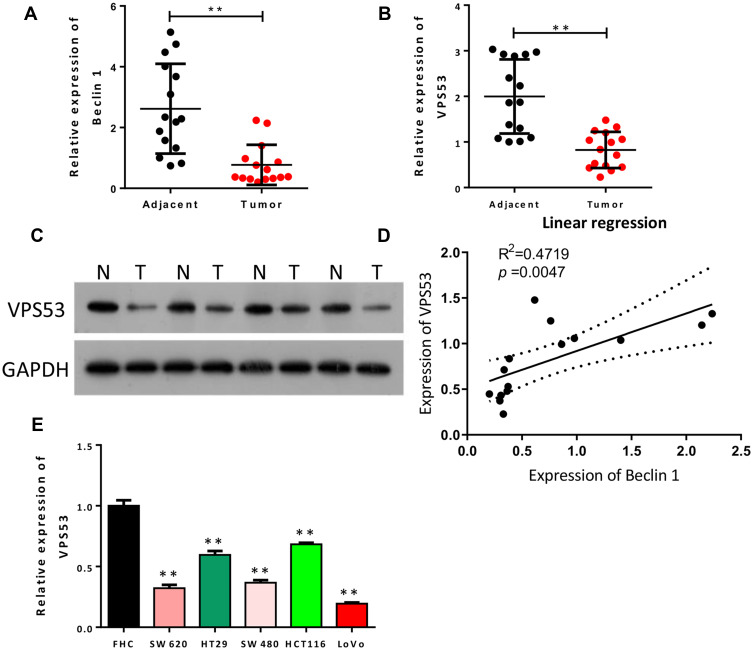
Decreased VPS53 Was Found to Be Positively Correlated with Beclin 1 in CRC
To investigate the relationship between autophagy and CRC, we first identified the level of the autophagy-related gene (Beclin 1) in CRC tissues. Analysis showed that Beclin 1 expression was dramatically reduced in CRC tissues with respect to adjacent normal tissues (P<0.01, Figure 1A). Also, to determine the potential contribution of VPS53 in CRC, we focused on and examined the level of VPS53 in CRC. As indicated by Figure 1B and C, the level of VPS53 was lower in CRC tissues than in adjacent normal tissues (P<0.01). In addition, the results showed that expression of Beclin 1 was positively correlated with that of VPS53 in CRC (R2=0.4719, Figure 1D). Furthermore, VPS53 expression was confirmed by RT-qPCR assay in human CRC cells, and the results determined that the level of VPS53 showed a significant decline in CRC cells, especially LoVo cells, in relation to normal colon FHC cells, and SW620 and SW480 cells displayed moderate VPS53 expressions (P<0.01, Figure 1E). Therefore, SW620 and LoVo were used in vitro experiments subsequently.
Figure 1.
Downregulated VPS53 was positively correlated with Beclin 1 in CRC. (A) RT-qPCR results of expression of Beclin 1 in adjacent normal tissues (n=15) and CRC tissues (n=15), **P < 0.01. (B) The level of VPS53 as also examined by RT-qPCR analysis in adjacent normal tissues (n=15) and CRC tissues (n=15), **P < 0.01. (C) Western blotting analysis of VPS53 in adjacent normal and CRC tissues. (D) The correlation analysis of Beclin 1 and VPS53 in CRC tissues (R2=0.4719, p=0.0047). (E) The level of VPS53 as determined by RT-qPCR assay in normal colon FHC cells and human CRC cells (SW620, HT29, SW480, HCT116, and LoVo), **P < 0.01.
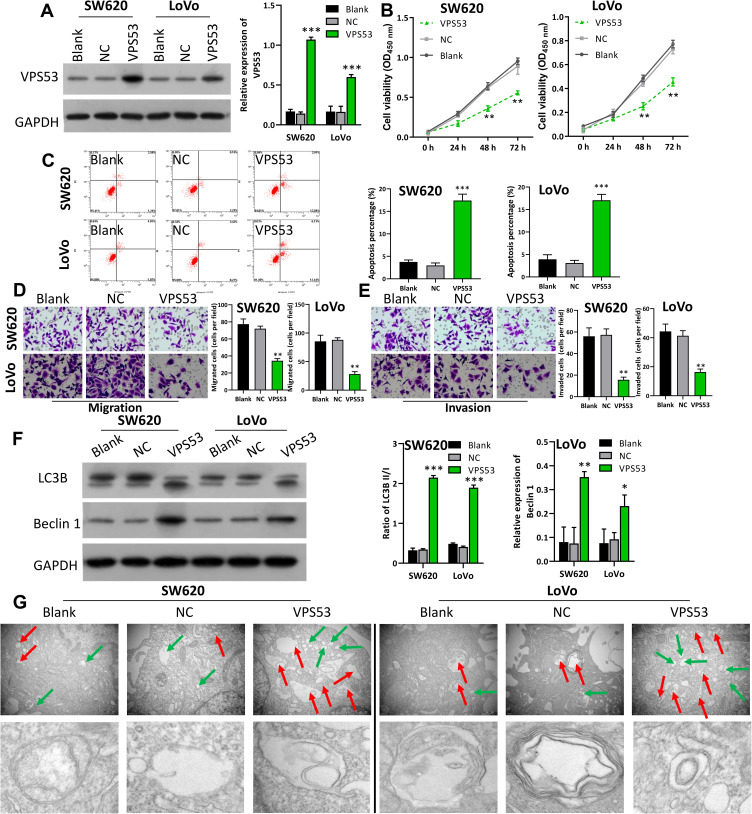
Overexpression of VPS53 Suppressed Proliferation, Migration, and Invasion, and Facilitated Apoptosis and Autophagy of CRC Cells
In order to further determine the probably function of VPS53 on the proliferation, migration, invasion, apoptosis, and autophagy of CRC cells, we transfected SW620 and LoVo cells with the pcDNA-VPS53 plasmid. Western blotting analysis found that VPS53 expression was notably raised in SW620 and LoVo cells after transfection with pcDNA3.0-VPS53 relative to cells transfected with the pcDNA3.0 vector (NC), indicating that there was a significant transfection effect of VPS53 in SW620 and LoVo cells (P<0.001, Figure 2A). We further detected the effect of VPS53 overexpression on proliferation of SW620 and LoVo cells. According to the CCK-8 assay, cell viability was significantly reduced in SW620 and LoVo cells transfected with the pcDNA-VPS53 plasmid versus cells with NC (P<0.001, Figure 2B). A characteristic of cancer is its capacity to evade apoptosis. So, we next examined the influence of VPS53 on CRC cell apoptosis using flow cytometry. As exhibited in Figure 2C, overexpression of VPS53 significantly accelerated apoptosis in SW620 and LoVo cells (P<0.001). Also, the roles of VPS53 on the migration and invasion of SW620 and LoVo cells were further confirmed. As indicated in Figure 2D and E, transwell assay determined that overexpression of VPS53 markedly inhibited the migration and invasion of SW620 and LoVo cells (P<0.01). Moreover, the effects of VPS53 overexpression on expressions of autophagy-related proteins including Beclin 1 and LC3B in SW620 and LoVo cells were verified. Western blotting analysis showed that overexpression of VPS53 increased the protein expressions of Beclin 1 and LC3BII (P<0.001, Figure 2F). Simultaneously, it was observed that VPS53 could prominently increase autophagosomes in SW620 and LoVo cells (Figure 2G).
Figure 2.
Overexpression of VPS53 suppressed proliferation, migration, and invasion, and facilitated apoptosis and autophagy of CRC cells. SW620 and LoVo cells were transfected with the pcDNA-VPS53 plasmid and NC, respectively. (A) VPS53 expression was determined by Western blotting analysis and the VPS53 level was normalized to GAPDH, ***P < 0.001. (B) CCK-8 assay investigated the effects of VPS53 overexpression on SW620 and LoVo cells, **P < 0.01. (C) Apoptotic analysis of SW620 and LoVo cells was evaluated by flow cytometry, and then the percentage of apoptosis was calculated, ***P < 0.001. (D and E) Transwell assays were used, and the results show the number of migrated and invaded cells per field, magnification, ×200; **P < 0.01. (F) Western blotting analysis displays expressions of Beclin 1 and LC3B, and the ratio of LC3BII/I and Beclin 1 level were counted based on the grayscale, *P < 0.05, **P < 0.01, ***P < 0.001. (G) Electron microscopy shows the number of autophagosomes in each group. The red arrow denotes autophagosomes and the green arrow denotes the lysosomes.
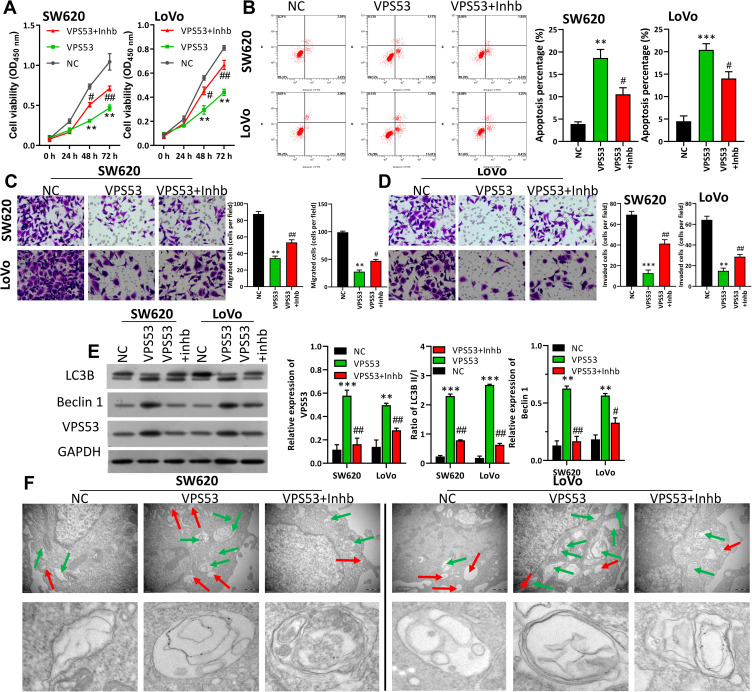
VPS53 Inhibited CRC Cell Carcinogenesis by Regulating Autophagy
The downstream regulation mechanism of VPS53 in CRC cells was further investigated. In order to confirm if the effect of VPS53 on the CRC process was achieved by regulating autophagy, the autophagy inhibitor (Inhb) was used to treat VPS53-overexpressed SW620 and LoVo cells, and functional experiments were performed. After a range of functional restoration assays, the results from CCK-8 assay certified that overexpression of VPS53 dramatically suppressed CRC viability, while Inhb attenuated the suppression mediated by VPS53 in SW620 and LoVo cells (P<0.001, Figure 3A). Flow cytometry assay also demonstrated that overexpression of VPS53 notably accelerated apoptosis of SW620 and LoVo cells, while Inhb weakened this acceleration induced by VPS53 (P<0.001, Figure 3B). Also, it was found that Inhb treatment could reverse the inhibition of CRC cell migration and invasion mediated by VPS53 overexpression (P<0.05, P<0.01, P<0.001, Figure 3C and D). More importantly, it was determined that expressions of LC3BII, Beclin 1, and VPS53 were markedly increased in VPS53-overexpressed SW620 and LoVo cells, whereas they were attenuated by Inhb treatment (P<0.05, P<0.01, P<0.001, Figure 3E). As shown in Figure 3F, autophagosome was enhanced in SW620 and LoVo cell by VPS53 overexpression, while reversed by Inhb treatment (Figure 3F). Collectively, these data showed that VPS53 inhibited proliferation, migration, and invasion, and induced apoptosis and autophagy by directly regulating autophagy pathways.
Figure 3.
VPS53 inhibited CRC cell carcinogenesis by regulating autophagy. SW620 and LoVo cells were treated with the VPS53-overexpressed plasmid and the autophagy inhibitor (Inhb). (A) CCK-8 assay indicates the influence of Inhb on the VPS53-inhibited proliferation of SW620 and LoVo cells, **P < 0.01 vs NC group; #P < 0.05, ##P < 0.01 vs VPS53 group. (B) The role of Inhb on the VPS53-induced apoptosis analyzed by flow cytometry, and the apoptosis percentage is shown, **P < 0.01, ***P < 0.001 vs NC group; #P < 0.05 vs VPS53 group. (C and D) VPS53 plasmid and Inhb-treated SW620 and LoVo cells were subjected to the migration and invasion assays, and the migrated and invaded cells were counted, **P < 0.01, ***P < 0.001 vs NC group; #P < 0.05, ##P < 0.01 vs VPS53 group. (E) Western blot analysis results show expressions of LC3B and Beclin 1, and the quantitative results were analyzed, **P < 0.01, ***P < 0.001 vs NC group; #P < 0.05, ##P < 0.01 vs VPS53 group. (F) Ultrastructure of autophagic bodies observed by an electron microscope. The red arrow denotes autophagosomes and the green arrow denotes the lysosomes.
Discussion
The pathogenesis of CRC has not been fully elucidated. GARP can facilitate retrograde transport vector fusion into TGN, and loss of the GARP function can impair the growth, reproduction or viability of defective organisms.17 VPS53 as a subunit of GARP complexes, can participate in the reverse transport of proteins from endosomes to late golgi apparatus, and transport and accelerate the differentiation of pluripotent cells.17 Moreover, a large number of studies confirmed that VPS53 is downregulated in multiple cancers, such as medullary thyroid cancer,11 prostate cancer,12,18 and cervical cancer.13 Also, VPS53 can induce apoptosis of ectopic endometrial stromal cells.1414 More importantly, several studies disclosed that VPS53 was screened for low expression in CRC,19–21 and HCCS1, as a homologous gene of VPS53, has a strong anti-tumor effect on human colon cancer and liver cancer cells.15 In our study, we demonstrated that VPS53 was down-expressed in CRC tissues and cells relative to normal adjacent tissues or normal colon cells.
Autophagy, as highly-conserved lysosome-dependent auto-degradation pathways, has significant effect in maintaining cell homeostasis.22,23 Numerous studies also certified that autophagy can make a significant contribution in cancer,24,25 cardiovascular disease,26 and neurodegenerative diseases by removing the damaged organelles and misfolded proteins.27 In addition, more and more evidences showed that autophagy is vital to the development and treatment of CRC, and is also correlated with its prognosis.28,29 In autophagy, LC3 is processed into cytoplasmic LC3-I, and subsequently modified into membrane-bound LC3-II, which can be bound to an autophagosome membrane, and become a vital symbol of autophagosomes.30 Autophagy levels can be evaluated by the LC3-II/LC3-I ratio.31 Microtubule-associated protein 1 light chain 3B (LC3B), as a homologue of yeast ATG8, is a crucial autophagy marker in mammals.32 Beclin-1 is essential for the initial steps of autophagy, which indirectly implicates its important role in numerous biological cellular processes, including aging and cell death.33 Autophagy induction can facilitate CRC cell death, while autophagy inhibition can accelerate CRC cell survival.34 Therefore, autophagy-related pathways might be potential new targets for prevention and treatment of CRC. mTORC1-mediated autophagy can affect receptor tyrosine kinase phosphorylation in CRC by the mTORC2-regulated mechanism.35 Stimulation of tumor‑associated macrophage (TAM) autophagy can enhance the radio-sensitivity of CRC.36 Besides, in this study, the expression of Beclin 1 was upregulated with increasing of LC3B II and cell apoptosis. This might be due to the interaction between Beclin 2 and Bcl-2. The combination of Bcl-2 and Beclin 1 can mediate cell survival and autophagy.37 In our preliminary experiment, we found that Beclin-1 was lowly expressed in CRC tissues, and its expression was positively correlated with VPS53. We also found that VPS53 could markedly improve the LC3-II/LC3-I ratio and upregulate Beclin 1 in CRC. Moreover, functional experiments showed that the autophagy inhibitor (Inhb) could attenuate the inhibition of proliferation, migration, and invasion, and the promotion effects of apoptosis and autophagy in CRC. Therefore, it was demonstrated that VPS53 can suppress tumorigenesis and progression of CRC by regulating autophagy pathways.
Conclusions
It was determined that VPS53 is a critical regulator of CRC progression. VPS53 expression is prominently reduced in CRC, and overexpression of VPS53 can suppress the proliferation, migration, and invasion, and accelerate apoptosis and autophagy of CRC cells through induction of autophagy pathways. Therefore, our results suggested that VPS53 has a significant role in the developmental process of CRC.
Funding Statement
This work was supported by North Sichuan Medical College City school cooperation program (18SXHZ0364, 18SXHZ0408, 19SXHZ0297); Science and Technology Research Project of Sichuan Province (2017JY0116); School Grade Research Key Project of North Sichuan Medical College (CBY18-A-ZD13), and Basic Scientific and Technological Strategic Cooperation of Nanchong Municipal Schools (NSMC20170311).
Disclosure
The authors declared no conflicts of interest.
References
- 1.Saltz LB. Colorectal cancer. Hematol Oncol Clin North America. 2015;29:ix. doi: 10.1016/j.hoc.2014.10.012 [DOI] [PubMed] [Google Scholar]
- 2.Dulai PS, Sandborn WJ, Gupta S. Colorectal cancer and dysplasia in inflammatory bowel disease: a review of disease epidemiology, pathophysiology, and management. Cancer Prevent Res. 2016;9:887–894. doi: 10.1158/1940-6207.CAPR-16-0124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arnold M, Sierra MS, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global patterns and trends in colorectal cancer incidence and mortality. Gut. 2017;66:683–691. doi: 10.1136/gutjnl-2015-310912 [DOI] [PubMed] [Google Scholar]
- 4.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262 [DOI] [PubMed] [Google Scholar]
- 5.Roncucci L, Mariani F. Prevention of colorectal cancer: how many tools do we have in our basket? Eur J Internal Med. 2015;26:752–756. doi: 10.1016/j.ejim.2015.08.019 [DOI] [PubMed] [Google Scholar]
- 6.Svestka T, Krechler T. [Preventing colorectal cancer]. Casopis Lekaru Ceskych. 2016;155:27–29. [PubMed] [Google Scholar]
- 7.Siegel RL, Miller KD, Fedewa SA, et al. Colorectal cancer statistics, 2017. CA Cancer J Clin. 2017;67::177–193. doi: 10.3322/caac.21395 [DOI] [PubMed] [Google Scholar]
- 8.Best JT, Xu P, Graham TR. Phospholipid flippases in membrane remodeling and transport carrier biogenesis. Curr Opin Cell Biol. 2019;59:8–15. doi: 10.1016/j.ceb.2019.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kummel D, Heinemann U. Diversity in structure and function of tethering complexes: evidence for different mechanisms in vesicular transport regulation. Curr Protein Pept Sci. 2008;9:197–209. doi: 10.2174/138920308783955252 [DOI] [PubMed] [Google Scholar]
- 10.Hausman-Kedem M, Ben-Shachar S, Menascu S, Geva K, Sagie L, Fattal-Valevski A. VPS53 gene is associated with a new phenotype of complicated hereditary spastic paraparesis. Neurogenetics. 2019;19. [DOI] [PubMed] [Google Scholar]
- 11.Shi L, Zhao SM, Luo Y, et al. MiR-375: A prospective regulator in medullary thyroid cancer based on microarray data and bioinformatics analyses. Pathol Res Pract. 2017;213::1344–1354. doi: 10.1016/j.prp.2017.09.024 [DOI] [PubMed] [Google Scholar]
- 12.Rebbeck TR. Prostate cancer genetics: variation by race, ethnicity, and geography. Semin Radiat Oncol.2017;27:3–10. doi: 10.1016/j.semradonc.2016.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim TE, Kim YW, Hwang SY, et al. Candidate tumor suppressor, HCCS‐1, is downregulated in human cancers and induces apoptosis in cervical cancer. Int j Cancer. 2002;97::780–786. doi: 10.1002/ijc.10124 [DOI] [PubMed] [Google Scholar]
- 14.Zhang W, Zhang X, Zheng Y, et al. Preparation of polyaniline@MoS2@Fe3O4 nanowires with a wide band and small thickness toward enhancement in microwave absorption. ACS Appl Nano Mater. 2018;1:5865–5875. doi: 10.1021/acsanm.8b01452 [DOI] [Google Scholar]
- 15.Gan Y, Gu J, Cai X, Hu J, Liu XY, Zhao X. Adenovirus-mediated HCCS1 overexpression elicits a potent antitumor efficacy on human colorectal cancer and hepatoma cells both in vitro and in vivo. Cancer Gene Ther. 2008;15:808–816. doi: 10.1038/cgt.2008.52 [DOI] [PubMed] [Google Scholar]
- 16.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- 17.Vasan N, Hutagalung A, Novick P, Reinisch KM. Structure of a C-terminal fragment of its Vps53 subunit suggests similarity of Golgi-associated retrograde protein (GARP) complex to a family of tethering complexes. Proc Nat Acad Sci USA. 2010;107:14176–14181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goh CL, Eeles RA. Germline genetic variants associated with prostate cancer and potential relevance to clinical practice. 2014. Prostate Cancer Prev. 9–26. City; Springer. [DOI] [PubMed] [Google Scholar]
- 19.Qi L, Ding Y. Screening of tumor suppressor genes in metastatic colorectal cancer. Bio Med Res Int. 2017;2017:1–7. doi: 10.1155/2017/2769140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nakao M, Kawauchi S, Uchiyama T, et al. DNA copy number aberrations associated with the clinicopathological features of colorectal cancers: identification of genomic biomarkers by array-based comparative genomic hybridization. Oncol Rep. 2011;25::1603–1611.doi: 10.3892/or.2011.1246 [DOI] [PubMed] [Google Scholar]
- 21.Li H, Yang X, Wang G, et al. KDM4B plays an important role in mitochondrial apoptosis by upregulating HAX1 expression in colorectal cancer. Oncotarget. 2016;7:57866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smith M, Wilkinson S. ER homeostasis and autophagy. Essays Biochem. 2017;61:625–635. doi: 10.1042/EBC20170092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yan Y, Finkel T. Autophagy as a regulator of cardiovascular redox homeostasis. Free Radic Biol Med. 2016;109:108–113. doi: 10.1016/j.freeradbiomed.2016.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen C, Lu L, Yan S, et al. Autophagy and doxorubicin resistance in cancer. Anti-Cancer Drugs. 2018;29:1–9. doi: 10.1097/CAD.0000000000000572 [DOI] [PubMed] [Google Scholar]
- 25.Amaravadi R, Kimmelman AC, White E. Recent insights into the function of autophagy in cancer. Genes Dev. 2016;30:1913–1930. doi: 10.1101/gad.287524.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bravo-San Pedro JM, Kroemer G, Galluzzi L. Autophagy and mitophagy in cardiovascular disease. Circ Res. 2017;120:1812–1824. doi: 10.1161/CIRCRESAHA.117.311082 [DOI] [PubMed] [Google Scholar]
- 27.Guo F, Liu X, Cai H, Le W. Autophagy in neurodegenerative diseases: pathogenesis and therapy. Brain Pathol. 2018;28:3–13. doi: 10.1111/bpa.12545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu T, Guo F, Yu Y, et al. Fusobacterium nucleatum promotes chemoresistance to colorectal cancer by modulating autophagy. Cell. 2017;170:548–563.e516. doi: 10.1016/j.cell.2017.07.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mokarram P, Albokashy M, Zarghooni M, et al. New frontiers in the treatment of colorectal cancer: autophagy and the unfolded protein response as promising targets. Autophagy. 2017;13:781–819. doi: 10.1080/15548627.2017.1290751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dikic I, Elazar Z. Mechanism and medical implications of mammalian autophagy. Nat Rev Mol Cell Biol. 2018;19:349. doi: 10.1038/s41580-018-0003-4 [DOI] [PubMed] [Google Scholar]
- 31.Aparicio I, Munoz PM, Salido G, Pena F, Tapia J. The autophagy-related protein LC3 is processed in stallion spermatozoa during short-and long-term storage and the related stressful conditions. Animal. 2016;10:1182–1191. doi: 10.1017/S1751731116000240 [DOI] [PubMed] [Google Scholar]
- 32.Xu WN, Chen DH, Liu WB, Xu JX, Yang SS. Molecular characterization of microtubule-associated protein 1-light chain 3B in megalobrama amblycephala fed with high fat/berberine diets. J Appl Gene. 2018;59:345–355. doi: 10.1007/s13353-018-0451-8 [DOI] [PubMed] [Google Scholar]
- 33.QJ W, Z Y. The Beclin 1-VPS34 complex–at the crossroads of autophagy and beyond.%A Funderburk SF. Trends Cell Biol. 2010;20::355–362. doi: 10.1016/j.tcb.2010.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu L, Wang HJ, Meng T, et al. lncRNA GAS5 inhibits cell migration and invasion and promotes autophagy by targeting miR-222-3p via the GAS5/PTEN-signaling pathway in CRC. Mol Therapy Nucleic Acids. 2019;17:644–656. doi: 10.1016/j.omtn.2019.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 35.Lampada A, O’Prey J, Szabadkai G, Ryan KM, Hochhauser D, Salomoni P. mTORC1-independent autophagy regulates receptor tyrosine kinase phosphorylation in colorectal cancer cells via an mTORC2-mediated mechanism. Cell Death Differ. 2017;24:1045–1062. doi: 10.1038/cdd.2017.41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shao LN, Zhu BS, Xing CG, Yang XD, Young W, Cao JP. Effects of autophagy regulation of tumor-associated macrophages on radiosensitivity of colorectal cancer cells. Mol Med Rep. 2016;13:2661–2670. doi: 10.3892/mmr.2016.4820 [DOI] [PubMed] [Google Scholar]
- 37.Fu LL, Cheng Y, Liu B. Beclin-1: autophagic regulator and therapeutic target in cancer. Int J Biochem Cell Biol. 2013;45:921–924. doi: 10.1016/j.biocel.2013.02.007 [DOI] [PubMed] [Google Scholar]