Abstract
Preeclampsia is believed to be caused by impaired placentation with insufficient trophoblast invasion, leading to impaired uterine spiral artery remodeling and angiogenesis. However, the underlying molecular mechanism remains unknown. We recently carried out transcriptome profiling of placental long noncoding RNAs (lncRNAs) and identified 383 differentially expressed lncRNAs in early-onset severe preeclampsia. Here, we are reporting our identification of lncRNA INHBA-AS1 as a potential causal factor of preeclampsia and its downstream pathways that may be involved in placentation. We found that INHBA-AS1 was upregulated in patients and positively correlated with clinical severity. We systematically searched for potential INHBA-AS1-binding transcription factors and their targets in databases and found that the targets were enriched with differentially expressed genes in the placentae of patients. We further demonstrated that the lncRNA INHBA-AS1 inhibited the invasion and migration of trophoblast cells through restraining the transcription factor CENPB from binding to the promoter of TNF receptor-associated factor 1 (TRAF1). Therefore, we have identified the dysregulated pathway “INHBA-AS1-CENPB-TRAF1” as a contributor to the pathogenesis of preeclampsia through prohibiting the proliferation, invasion, and migration of trophoblasts during placentation.
Keywords: placenta, transcriptome, long noncoding RNA, preeclampsia, trophoblast, cell invasion, cell migration
Graphical Abstract
Preeclampsia is believed to be caused by impaired placentation. Xinping Yang and colleagues report the identification of lncRNA INHBA-AS1 as a potential causal factor of preeclampsia. They found that INHBA-AS1 is positively correlated with clinical severity and involved in the pathogenesis through inhibiting the invasion and migration of trophoblast cells.
Introduction
Preeclampsia (PE) is a pregnancy-specific hypertensive syndrome, with onset of hypertension and proteinuria after 20 weeks’ gestation.1 The prevalence of PE is 3%–5% of pregnancies,2 accounting for 10% to 15% of maternal mortality.3 The grave consequences of PE on maternal health are mainly attributed to cerebral edema, intracranial hemorrhage, and eclampsia.4,5 PE also has significant consequences on fetal development and growth, often leading to perinatal and infant morbidity or mortality,6 and it contributes to preterm births7,8 and ∼15% cases of fetal growth restriction (FGR).8,9 In the long term, PE affects brain development and functions and has profound consequences on the offspring in later life.10 Because the etiology is unclear, there is still a lack of both early diagnosis and radical treatment for PE.
Recent studies suggest that the root cause of PE lies with the placenta. The impairment of uterine spiral artery remodeling caused by insufficient trophoblast cell infiltration might be critical in the early stage of PE development,11 characterized by placental endothelial cell damage, caused by shallow invasion of trophoblasts into the endometrium and the placental vascular recasting barriers.6,12, 13, 14, 15 Genetic factors play a role in its pathogenesis, and the heritability of PE is about 50%.16,17 Genome-wide linkage studies on pedigrees have identified risk loci on chromosomes 2p13, 2q23, 11q23, 10q22, 22q12, 2p25, 9p13, 4q32, and 9p11.18, 19, 20, 21, 22 Genome-wide association studies (GWASs) have found some variants associated with PE.23,24 Numerous placental microarray studies have identified some differentially expressed genes (DEGs) and related pathways involved in placentation, angiogenesis, immune function, and hypoxia,25, 26, 27, 28, 29, 30, 31, 32 pointing to placental mechanisms underlying PE. Recent studies suggest that some long noncoding RNAs (lncRNAs) may be involved in placental gene dysregulation. For example, H19 is reported to be reduced in the placentae of pregnancies with FGR through altering trophoblast cell invasion and migration regulated by the H19/TβR3 pathway.33 It is reported that MEG3 and MALAT1 are downregulated in placental tissues from preeclamptic patients, and MEG3 inhibits apoptosis and promotes migration of trophoblast cells.34,35 SPRY4-IT1 is found to suppress the migration and invasion of trophoblast.36,37 HOTAIR is well known as a master regulator of chromatin status in cancer38 and is found to promote invasion of trophoblast cells.39 These studies suggest that lncRNAs may play a critical role in placenta development, and the dysregulation of some lncRNAs may lead to placenta-origin diseases, such as PE.
One of the major functions of lncRNAs is to recruit/inhibit transcription factors (TFs) to the promoter of their targeting genes.40,41 Although the transcriptome sequencing has revealed a large number of lncRNAs in normal human placenta,42 we do not know to what extent the lncRNAs might be involved in PE. In order to search for lncRNAs involved in PE, our group has recently carried out a transcriptome sequencing on placentae from patients with early-onset severe PE (EOSPE) and normal controls and identified 383 lncRNAs differentially expressed. We found that the lncRNA INHBA-AS1 was upregulated in PE patients and positively correlated with severity of the clinical presentation of the pregnant women. We performed a systematic search for interacting proteins of INHBA-AS1, including TFs and their targets. We found significant enrichment of EOSPE-associated genes in INHBA-AS1 targeting genes, suggesting the potential involvement of INHBA-AS1 in the dysregulated gene expression in the placentae of patients with EOSPE. Here, we demonstrated that INHBA-AS1 may be involved in EOSPE through recruiting the TFs to the promoter of TRAF1 (TNF receptor-associated factor 1) that are important for the proliferation, invasion, and migration of trophoblast cells, which are considered to be the key cellular functions for placentation.
Results
INHBA-AS1 Is Upregulated in the Placentae of Patients with EOSPE
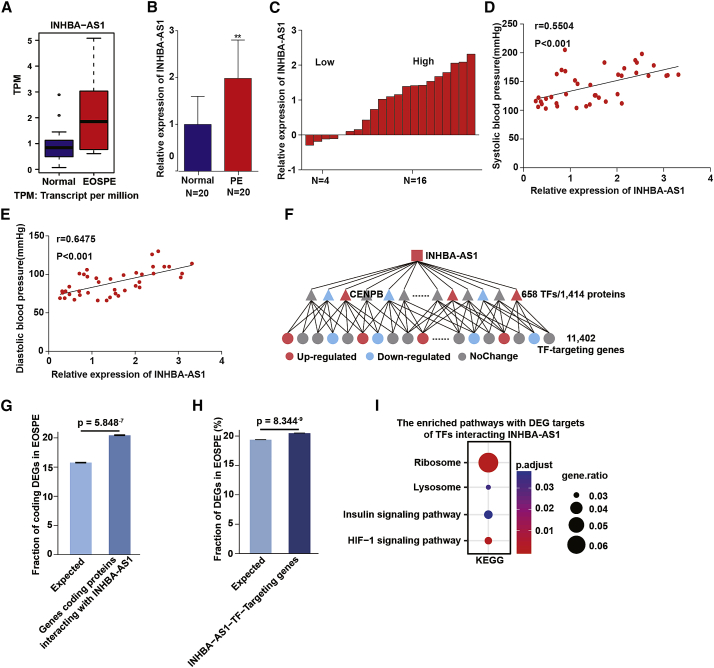
The lncRNA INHBA-AS1 was among the top lncRNAs with significant upregulation in the placental transcriptomes of EOSPE patients (Figure 1A). To further confirm the association of INHBA-AS1 with EOSPE, we collected placentae from 20 cases of PE, which showed significant difference in blood pressure and proteinuria compared to 20 cases of normal controls (Table 1), and found that INHBA-AS1 was significantly upregulated in 80% (16/20) of PE cases (p < 0.01) (Figures 1B and 1C) using qRT-PCR. Interestingly, the expression levels of INHBA-AS1 in placentae were positively correlated with the severity of clinical characteristics, such as systolic and diastolic blood pressures (Figures 1D and 1E). Since lncRNAs usually exert their functions through interacting with proteins, we obtained empirical DElncRNA-protein interactions from the RNA-protein interaction databases starBase, RNA Association Interaction Database (RAID), and NPInter and predicted DElncRNA-protein interactions using catRAPID omics. From these data we extracted INHBA-AS1-protein interactions, 658 of which were TFs (Figure 1F; Table S1). The genes encoding proteins interacting with INHBA-AS1 are significantly enriched with differentially expressed coding genes in EOSPE (Figure 1G; Fisher’s exact test, p = 5.848−7). These TFs target 11,402 genes (including 2,336 DEGs in EOSPE; Figure 1F), which are significantly enriched with DEGs in EOSPE (Figure 1H; Fisher’s exact test, p = 8.344−9), suggesting that INHBA-AS1 might be involved in gene dysregulation of EOSPE. We further carried out pathway enrichment analysis on the targets dysregulated in EOSPE and found some enriched pathways (Figure 1I): the hypoxia-inducible factor 1 (HIF−1) signaling pathway,43, 44, 45 lysosome pathway,46,47 and insulin signaling pathway,48,49 which are all known to be associated with PE.
Figure 1.
Expression of INHBA-AS1 in Placentae of Patients with Preeclampsia (PE)
(A) Boxplot of INHBA-AS1 expression levels in EOSPE placentae (n = 9) compared with normal placentae (n = 32). (B) The relative expression level of INHBA-AS1 measured by qRT-PCR. y axis, the fold change; red bar, placentae of patients with PE, n = 20; blue bar, normal placentae, n = 20. The values are shown as the mean ± SD; ∗∗p < 0.01. (C) The relative expression level of INHBA-AS1 in PE samples measured by qRT-PCR. Average expression level in normal samples was set at 0; red bars represent relative expression levels in placentae of PE patients. (D) Positive correlation (r = 0.5504, p < 0.001) between systolic blood pressure and the expression levels of lncRNA INHBA-AS1 (Pearson one-tail correlation analysis). (E) Positive correlation (r = 0.6475, p < 0.001) between diastolic blood pressure and the expression of lncRNA INHBA-AS1 (Pearson one-tail correlation analysis). (F) Hierarchical “INHBA-AS1-transcription factor (TF)-Target” interaction network. (G) Gene-coding proteins interacting with INHBA-AS1 are enriched with coding differentially expressed genes (DEGs) in EOSPE. (H) The targets in the INHBA-AS1-TF-Target interaction network are enriched with DEGs in EOSPE. (I) The enriched pathways with DEGs as targets of the INHBA-AS1-TF-Target interaction network. The values are shown as the mean ± SD of three independent experiments; ∗∗p < 0.01, ∗p < 0.05. See also Table S1.
Table 1.
Clinical Characteristics of Patients with PE and Normal Pregnancies
| Variable | PE (n = 20) | Control (n = 20) | p Value |
|---|---|---|---|
| Maternal age (year) | 31.40 ± 5.879 | 31.00 ± 3.974 | >0.05 |
| Maternal weight (kg) | 69.85 ± 7.289 | 65.28 ± 8.131 | >0.05 |
| Systolic blood pressure (mm Hg) | 169.00 ± 15.980 | 115.65 ± 8.248 | <0.01 |
| Diastolic blood pressure (mm Hg) | 103.60 ± 10.619 | 74.90 ± 5.857 | <0.01 |
| Proteinuria (g/day) | >0.3 | <0.3 | <0.05 |
The values are shown as the mean ± SD.
INHBA-AS1 Inhibits the Invasion, Migration, and Proliferation and Promotes Apoptosis of Trophoblast Cells
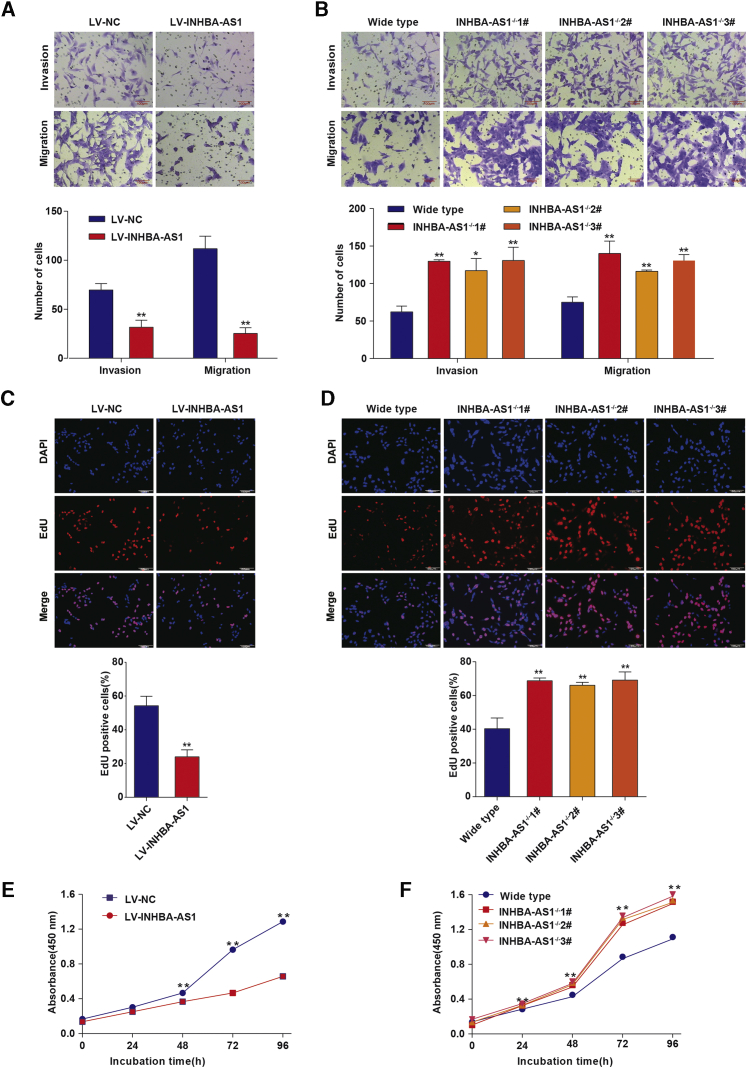
The invasion and migration of trophoblasts are essential for placentation, dysregulation of which is thought to play a critical role in the pathogenesis of PE. Therefore, we tested the effect of INHBA-AS1 on the invasion and migration of trophoblast cells using overexpression or knockout. The overexpression of INHBA-AS1 in a human villous trophoblast cell line, HTR-8/SVneo cells, was established by transfecting the full-length cDNA of INHBA-AS1 into the cells (Figure S1A). Multiple INHBA-AS1-knockout monoclonal cell lines were established using CRISPR/Cas9 method (Figure S1B). We evaluated the invasion and migration using a Transwell assay (Corning, USA) and found that overexpression of INHBA-AS1 inhibited invasion and migration of HTR-8/SVneo cells (Figure 2A), whereas its knockout promoted invasion and migration (Figure 2B). We tested cell proliferation using a 5-ethynyl-2′-deoxyuridine (EdU) assay and found that overexpression of INHBA-AS1 promoted proliferation (Figure 2C), whereas knockout inhibited proliferation (Figure 2D). We also measured cell proliferation using cell counting kit-8 (CCK-8; Dojindo) and obtained similar results (Figures 2E and 2F).
Figure 2.
The Effect of INHBA-AS1 on Invasion, Migration, and Proliferation in Trophoblast Cell Line
(A and B) Invasion and migration of HTR-8/SVneo cells were detected by a Transwell assay after INHBA-AS1 overexpression (A) or knockout (B). Upper panel: pictures of cells stained with 0.1% crystal violet solution; lower panel: statistics of cells. Scale bars, 100 μm. (C and D) The proliferation of HTR-8/SVneo cells was detected by an EdU assay after INHBA-AS1 overexpression (C) or knockout (D). Upper panel: pictures of cells stained with 4′,6-diamidino-2-phenylindole (DAPI) and EdU; lower panel: statistics of EdU-positive cells. (E and F) Cell Counting Kit-8 was used to evaluate the cell proliferation of HTR-8/SVneo cells after INHBA-AS1 overexpression (E) or knockout (F). LV, lentivirus; NC, normal control. The values are shown as the mean ± SD of three independent experiments; ∗∗p < 0.01, ∗p < 0.05. See also Figure S1.
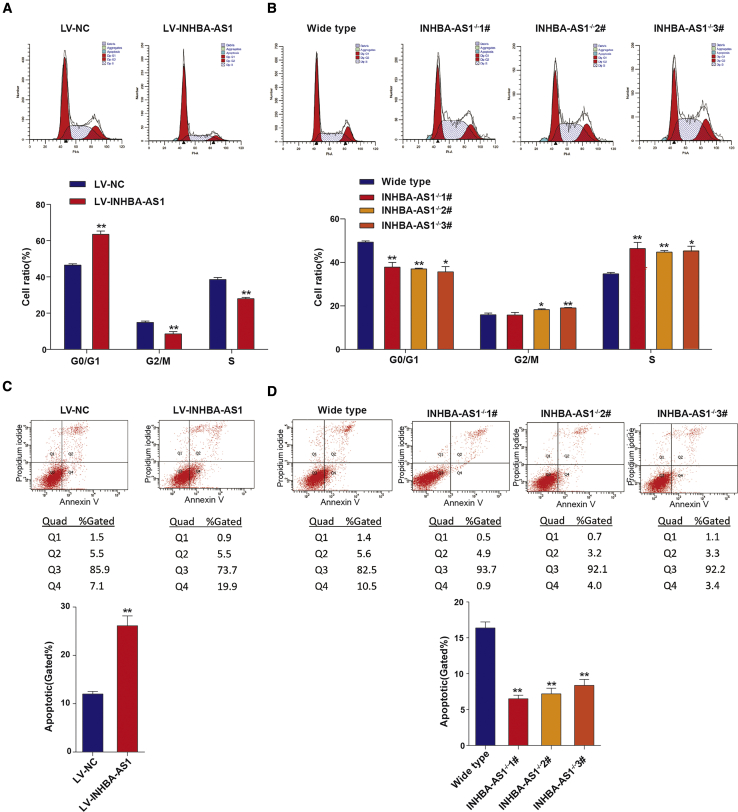
Consistent with the results on cell proliferation, overexpression of INHBA-AS1 arrested cell cycle and reduced the number of cells entering S phase (Figure 3A), whereas knockout of it promoted the transition from G0/G1 phase to S phase (Figure 3B). We further found that overexpression of INHBA-AS1 promoted apoptosis (Figure 3C), whereas its knockout inhibited apoptosis (Figure 3D). These results demonstrate that INHBA-AS1 inhibits invasion, migration, and proliferation and promotes apoptosis of trophoblast cells, suggesting that the upregulation of INHBA-AS1 might lead to poor placentation due to impaired trophoblast invasion and migration.
Figure 3.
INHBA-AS1 Prohibits Cell Cycle and Promotes Apoptosis in HTR8/SVneo Cells
(A) Cell-cycle analysis by flow cytometry on cells transfected by LV harboring human full-length INHBA-AS1 or empty vector. Upper panels: frequency histograms of cell-cycle phases; lower panels: statistics of cell-cycle phases. (B) Cell-cycle analysis by flow cytometry on cells with INHBA-AS1 knockout. Upper panels: frequency histograms of cell-cycle phases; lower panels: statistics of cell-cycle phases. (C and D) Apoptosis analysis using Annexin-V assay coped with flow cytometry on cells with INHBA-AS1 overexpression (C) or knockout (D). Upper panels: histograms of apoptotic cells; lower panels: statistics of apoptotic cells. The apoptotic cells include cells in quad 2 (Q2) and Q4. Cells in Q2 are dead apoptotic cells (PI positive and Annexin-V positive), and cells in Q4 are ongoing apoptotic cells (PI negative but Annexin-V positive). The values are shown as the mean ± SD of three independent experiments; ∗∗p < 0.01, ∗p < 0.05. See also Figure S1.
CENPB as a Downstream Effector of INHBA-AS1
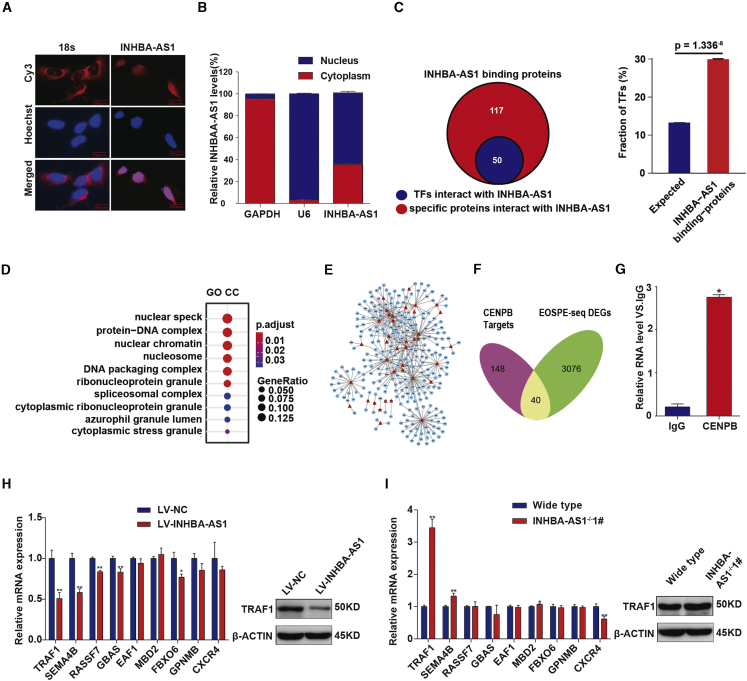
With the use of FISH (fluorescence in situ hybridization) and cell fractionation, we found that INHBA-AS1 was mainly located in the nucleus (Figures 4A and 4B), suggesting that INHBA-AS1 might participate in transcriptional regulation. To search for proteins binding to INHBA-AS1, we carried out an RNA-pulldown assay (Figure S2A) and detected 167 proteins in the INHBA-AS1-protein complexes using mass spectrometry (Figure S2B; Table S2). Of the 167 proteins binding to INHBA-AS1, 50 were TFs, showing significant enrichment (29.94%, p = 1.336−8) (Figure 4C). We performed enrichment analysis for Gene Ontology (GO) terms (cellular components) and found that INHBA-AS1-binding proteins might be associated with protein complexes in the nucleus (Figure 4D).
Figure 4.
INHBA-AS1 Interacts with CENPB and Inhibits TRAF1 Expression
(A) The subcellular localization of INHBA-AS1 in HTR-8/SVneo detected by FISH assays. Red, INHBA-AS1; blue, nucleus. Scale bar, 20 μm. (B) The subcellular localization of INHBA-AS1 in the HTR-8/SVneo cell detected by cell fractionation assays. U6, nucleus marker; GAPDH, cytoplasm marker. (C) INHBA-AS1-binding proteins detected by pulldown and mass spectrometry. (D) Enrichment analysis of GO term cellular component (GO-CC) on INHBA-AS1-binding proteins. (E) TF-target network (TFTNet) for EOSPE DEGs. Red triangles, TFs; blue nodes, targets of the TFs. (F) Overlaps between CENPB target genes (188) and DEGs (3,116) in EOSPE detected in transcriptome sequencing. (G) Binding of INHBA-AS1 with CENPB detected by RIP assays and qRT-PCR. (H) Transcription level of CENPB target genes in cells with INHBA-AS1 overexpression. Left panel shows qPCR results; blue bars, cells transfected with empty vector; red bars, cells transfected with INHBA-AS1; right panel shows western blotting results for TRAF1. (I) Transcription level detected of CENPB target genes in cells with INHBA-AS1 knockout. Left panel shows qPCR results; blue bars, cells without knockout; red bars, cells with INHBA-AS1 knockout; right panel shows western blotting result of TRAF1. The values are shown as the mean ± SD of three independent experiments; ∗∗p < 0.01, ∗p < 0.05. See also Figure S2 and Tables S2, S3, and S4.
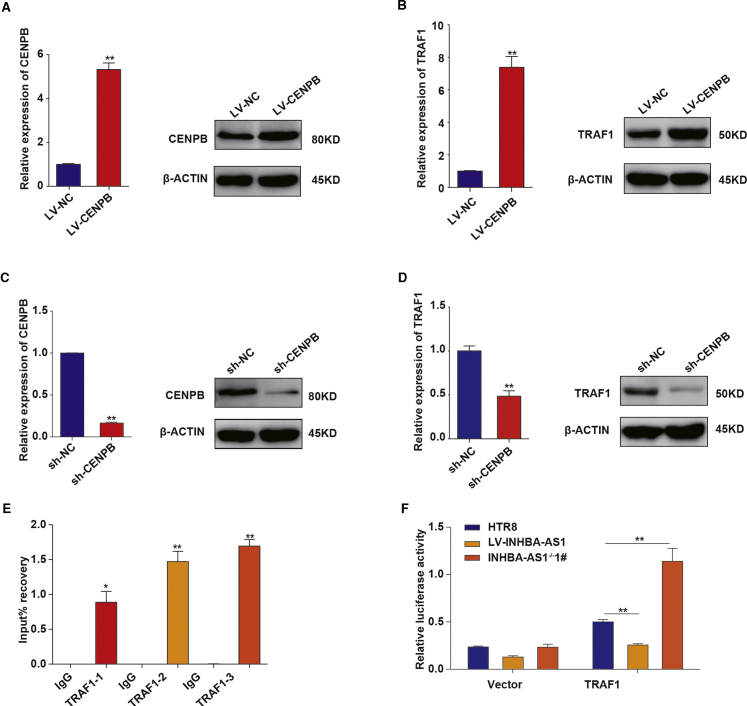
We obtained 1,440 targets of these 50 TFs (Table S3) by searching databases TRED,50 ITFP,51 ENCODE,52 Neph2012,53 TRRUST2,54 and Marbach2016,54 and 262 of the 1,440 genes were dysregulated in EOSPE (Table S3). We constructed a TF-target regulatory network, which contains the 50 TFs and their differentially expressed targets in EOSPE (Figure 4E). CENPB has the highest betweenness centrality (Table S4), suggesting that CENPB might play a central role in this regulatory network. The TF CENPB targets 188 genes, and about 21.28% (40/188) are differentially expressed in EOSPE (Figures 2C and 4F). We verified the physical interaction between INHBA-AS1 and CENPB using an RNA immunoprecipitation (RIP) assay (Figure 4G).
To test INHBA-AS1-mediated expression changes of these targets, we established overexpression or knockout of INHBA-AS1 in HTR-8/SVneo cells and performed qRT-PCR on the top 9 CENPB target genes, which showed the largest fold change in EOSPE transcriptome sequencing. We found that the target TRAF1 was most significantly downregulated by INHBA-AS1 in the cells (Figures 4H and 4I). The regulation of TRAF1 by INHBA-AS1 was confirmed at the protein level using western blot (Figures 4H and 4I). However, the overexpression or the deletion of INHBA-AS1 in HTR-8/SVneo cells did not have significant effects on the expression of CENPB (Figure S2D). These results demonstrated that INHBA-AS1 may bind to CENPB to regulate expression of the target genes, such as TRAF1.
INHBA-AS1 Prohibits the Binding of CENPB to the Promoter Region of TRAF1 (PTRAF1–3)
To investigate the effect of the TF CENPB on TRAF1, we established overexpression of CENPB in HTR-8/SVneo cells (Figure 5A) and found that overexpression of CENPB upregulated the mRNA and protein levels of TRAF1 (Figure 5B), whereas knockdown (Figure 5C), conversely, downregulated mRNA levels and protein levels of TRAF1 (Figure 5D).
Figure 5.
CENPB Binds the Promoter Region of TRAF1 and Promotes Its Transcription
(A) The expression of CENPB in HTR8/SVneo cells transfected with full-length CENPB. Left panel: results of qPCR; right panel: results of western blot. (B) Increased expression of TRAF1 in HTR8/SVneo cells after CENPB overexpression. Left panel: results of qPCR; right panel: results of western blot. (C) The expression of CENPB in HTR8/SVneo cells transfected with shRNA of CENPB. Left panel: results of qPCR; right panel: results of western blot. (D) Decreased expression of TRAF1 in HTR8/SVneo cells after CENPB overexpression. Left panel: results of qPCR; right panel: results of western blot. (E) Binding of CENPB to the promoter of TRAF1 detected using ChIP assays and qPCR in HTR8/SVneo cells. (F) Luciferase reporter assay on the transcription activity of the TRAF1 promoter in HTR8/SVneo cells after INHBA-AS1 overexpression or knockout. The values are shown as the mean ± SD of three independent experiments; ∗∗p < 0.01, ∗p < 0.05. See also Table S5.
We detected the binding of CENPB to the promoter region of TRAF1 using a chromatin immunoprecipitation (ChIP) assay (Figure 5E). With the use of luciferase as reporter, we tested the effect of INHBA-AS1 on the promoter activity of the PTRAF1–3 in HTR-8/SVneo cells with INHBA-AS1 overexpression or INHBA-AS1 knockout. The activity of the luciferase showed that INHBA-AS1 reduced the transcriptional effects of CENPB on the TRAF1 promoter compared to the empty controls (Figure 5F). These results demonstrate that INHBA-AS1 inhibits the binding of CENPB to PTRAF1–3 and thus inactivates its transcription.
INHBA-AS1 Prohibits Invasion and Migration of Trophoblast through TRAF1
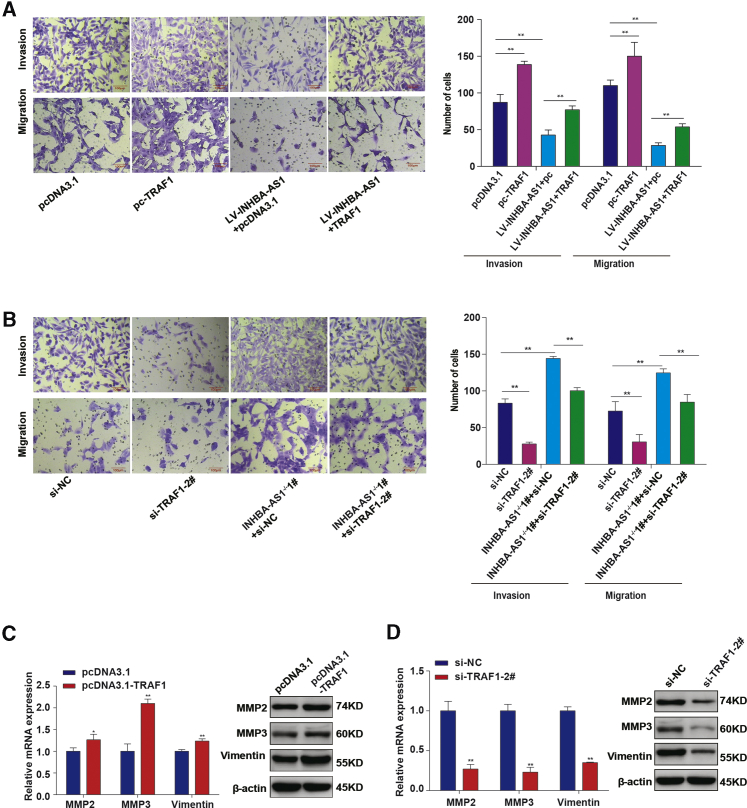
We have demonstrated that INHBA-AS1 inhibited invasion and migration of placental trophoblast cells through binding to CENPB (Figures 2A, 2B, and 4G) and that CENPB upregulated TRAF1 through binding to the promoter (Figure 5). To further confirm the function of the pathway INHBA-AS1-CENPB-TRAF1 in HTR-8/SVneo cells, we used a strategy to overexpress INHBA-AS1, TRAF1 separately, and in combination (Figures 6A, S1A, and S3A). The results showed that overexpression of TRAF1 could partially rescue the invasion and migration inhibited by overexpression of INHBA-AS1 (Figure 6A). We also performed knockout of INHBA-AS1 and knockdown of TRAF1 separately and in combination (Figures 6B, S1B, and S3B). Knockout of INHBA-AS1 promoted invasion and migration of HTR-8/SVneo cells (Figure 6B), whereas knockdown of TRAF1, on the contrary, could partially inhibit the invasion and migration induced by INHBA-AS1 knockout (Figure 6B).
Figure 6.
TRAF1 Promoted the Invasion and Migration of Trophoblast Cells and Weakened the Effect of INHBA-AS1
(A) Overexpression of TRAF1 partially rescued the invasion and migration of HTR-8/SVneo cells inhibited by INHBA-AS1. Scale bars, 100 μm. (B) INHBA-AS1 knockout partially rescued invasion and migration caused by knockdown of TRAF1. Scale bars, 100 μm. (C) Overexpression of TRAF1 upregulated the expression of MMP2, MMP3, and vimentin at mRNA level and protein level. (D) Knockdown of TRAF1 in HTR-8/SVneo cells downregulated the expression of MMP2, MMP3, and vimentin at the mRNA level and protein level. The values are shown as the mean ± SD of three independent experiments; ∗∗p < 0.01, ∗p < 0.05. See also Figure S3 and Table S5.
Since matrix metalloproteinase 2 (MMP2), MMP3, and vimentin are molecular markers for the pathways involved in cell invasion and migration,55, 56, 57, 58 we looked at their expression in cells overexpressing/knocking down gene TRAF1. We found that TRAF1 overexpression upregulated these markers (Figure 6C), whereas knockdown of TRAF1 downregulated these markers (Figure 6D). These results suggest that INHBA-AS1 may inhibit the invasion and migration of trophoblast cells through hindering the binding of TF CENPB to the promoter of TRAF1.
Discussion
It is thought that the root cause of PE is located in the placenta. During early gestation, poor trophoblast invasion and incomplete vascular formation of spiral arteries lead to placenta dysfunctions. This view on the pathogenesis of PE is supported by the fact that the patients are often characterized by placental tissue ischemia hypoxia and endothelial cell damage, caused by shallow invasion of the trophoblasts into the endometrium and the placental vascular recasting barriers.6,12, 13, 14, 15 Numerous lncRNAs, such as H19,33 MEG3,34 MALAT-1,35 SPRY4-IT1,36,37 and HOTAIR,38,39 are reported to be involved in the invasion and migration of trophoblast cells. A systematic search for PE-associated lncRNAs will enable the discovery of regulatory molecules key to placenta development.
INHBA-AS1 is among the top upregulated lncRNAs (Figure 1A) in transcriptome profiling on placentae of patients with EOSPE. The INHBA-AS1 expression level was positively correlated with severity of the symptoms (Figures 1B–1E), suggesting that INHBA-AS1 might contribute substantially to PE. We have constructed a DElncRNA-protein interaction network using interaction data from databases and prediction. INHBA-AS1 interacts with 658 TFs, which target 11,402 genes (Figure 1F). These TF targets are enriched with DEGs in EOSPE (Figure 1H), suggesting that INHBA-AS1 might play a role in gene dysregulation in this disease.
A number of studies have shown that insufficient invasion of placental trophoblast cells leads to impaired placental function and the occurrence of PE.6,12, 13, 14, 15 Several immortal trophoblast cell lines are available for the study of trophoblast biology.59 We chose HTR-8SV/neo cells for cellular functional studies because this cell line is widely used in previous studies on the pathogenesis of PE.60, 61, 62, 63, 64, 65, 66, 67 We overexpressed or knocked out this gene in this placenta-derived trophoblast cell line and found that INHBA-AS1 prohibited the invasion, migration, and proliferation (Figures 2A–2D, 3A, and 3B) and promoted apoptosis (Figures 3C and 3D). These results demonstrated that INHBA-AS1 may participate in the pathogenesis of PE through inhibiting trophoblast invasion, migration, and proliferation, which may lead to shallow implantation of the placenta. The placental shallow implantation caused by insufficient trophoblastic infiltration during early pregnancy is thought to be crucial in the development of PE.68 It has also been reported that the apoptosis of extravillous trophoblast cells in the preeclamptic placenta is increased.69
The nuclear localization of INHBA-AS1 (Figures 4A and 4B) suggested that INHBA-AS1 might be involved in transcriptional regulation. We identified 167 proteins binding to INHBA-AS1 using an RNA-pulldown assay and mass spectrometry (Figures S2A and S2B; Table S2). The INHBA-AS1-binding proteins are enriched with TFs (50/167, p = 1.336−8) (Figure 4C), confirming our hypothesis that INHBA-AS1 may participate in transcriptional regulation. Among the 1,440 targets of these 50 TFs from databases TRED, ITFP, ENCODE, Neph2012, TRRUST2, and Marbach2016 (Table S3), 262 genes were dysregulated in EOSPE (Table S3), suggesting that these TFs may contribute substantially to gene dysregulation in EOSPE. CENPB has the highest betweenness centrality in the regulatory network (Table S4), suggesting a central role in the transcriptional regulation in EOSPE. Of the 188 CENPB target genes, about 21.28% (40/188) are differentially expressed in EOSPE (Figures 4F and S2C). We detected the physical interaction between CENPB and INHBA-AS1 using a RIP assay (Figure 4G), and we also detected the binding of CENPB to PTRAF1–3 using a ChIP assay (Figure 5E). Although CENPB is well known to bind a 17-bp sequence (CENPB box) in the centromeric alpha satellite DNA and facilitates the centromere formation,70 it has recently been reported to regulate gene transcription.71 We verified the regulation of CENPB on the transcription of TRAF1 by overexpression or knockout of CENPB (Figures 5B and 5D) and showed an inhibitory effect of INHBA-AS1 on the promoter activity of PTRAF1–3 using luciferase as a reporter (Figure 5F).
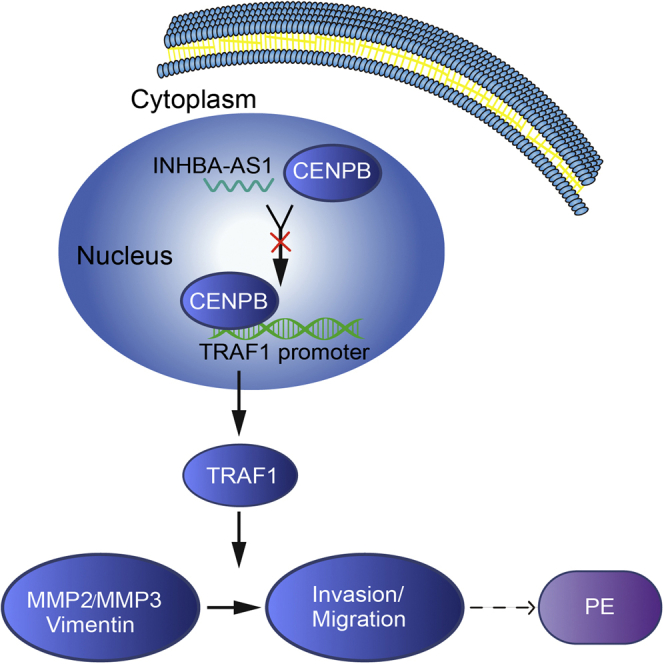
We have confirmed the function of the pathway INHBA-AS1-CENPB-TRAF1 on invasion and migration, using overexpression/knockdown of INHBA-AS1, TRAF1 in different combinations (Figure 6). First, we have identified the lncRNA INHBA-AS1 as a potential PE candidate gene using transcriptome profiling on placentae from EOSPE patients. Second, we have demonstrated that INHBA-AS1 prohibits CENPB from binding to the promoter of TRAF1 and thus downregulates its expression and inhibits invasion and migration of trophoblasts. All of these findings have illustrated a potential underlying pathway for PE, INHBA-AS1-CENPB-TRAF1, which may regulate trophoblast invasion and migration during placentation.
Materials and Methods
Sample Collection and Patient Information
All samples were collected at the Department of Obstetrics & Gynecology of Nanfang Hospital in China. The 40 placental samples for qRT-PCR were collected between December 2016 and May 2018. The placental tissue samples were collected from the midsection between the chorionic and maternal basal surfaces at four different positions of the placenta within 5 min after delivery. The tissues were washed immediately with PBS buffer and preserved in RNA later at −80°C for later RNA isolation. The clinical characteristics of each patient were extracted from the medical records, strictly following the guidelines from the American Board of Obstetrics and Gynecology, Williams Obstetrics, 24th edition. The clinical information for the patients and controls is shown in Table 1. This research has been approved by the Ethnics Board of Nanfang Hospital of Southern Medical University, and all patients have signed the informed consent.
Identifying lncRNA INHBA-AS1 as a PE Candidate Gene from Transcriptome Sequencing Data
We have carried out transcriptome sequencing and identified a total number of 3,116 DEGs, including 383 differentially expressed lncRNAs. INHBA-AS1 was among the top DElncRNAs based on fold change. From DElncRNA-protein interaction data (Databases: RNA-protein interaction databases starBase, RAID) and NPInter and predicted DElncRNA-protein interactions using catRAPID omics, we obtained INHBA-AS1 658 interacting TFs, which target 11,402 genes (Table S1). Because these targets are enriched with DEGs in EOSPE, we chose INHBA-AS1 as a PE candidate gene for further study.
RNA Isolation and qRT-PCR
Total RNA was isolated using the RNeasy Plus Universal Mini Kit (QIAGEN), according to the manufacturer’s instruction. RNA (500 ng) was reverse transcribed using the PrimeScript RT Reagent Kit (Takara, Japan), and qRT-PCR was performed with the SYBR Premix Ex Taq Kit (Takara, Japan) in a LightCycler 480 (Roche, Swiss) system to detect expression of genes, following the manufacturer’s instruction. The sequences of specific primers were shown in Table S5. The relative gene expression was calculated using the 2−ΔΔCT method and converted to fold changes using β-actin (ACTB) as internal controls.
Cell Culture
The cells used in this study were human villous trophoblasts HTR-8/SVneo, purchased from the ATCC Cell Bank. The culture conditions were RPMI-1640 medium (Cellgro), supplemented with 10% fetal bovine serum (FBS; Gibco) and placed in a cell incubator at 37°C, 5% carbon dioxide concentration, and humidified air. Fresh medium was replaced every 2 days, depending on the cell status.
Lentiviral Expression Constructs and Transfection
The full length of lncRNA INHBA-AS1 and CENPB was synthesized and cloned into a pGC-FU vector (Genechem, China). The short hairpin RNA (shRNA) specific for CENPB was designed and cloned into a pGC-FU vector (Genechem, China). HTR8/SVneo cells were transfected following the manufacturer’s instruction. Stable cell lines of lncRNA INHBA-AS1 and CENPB were selected by puromycin (Gibco, USA). The interference sequence of shRNA for CENPB was 5′-GAGCACGATCCTGAAGAACAA-3′.
Plasmid and Small Interfering RNA (siRNA) Transfection
The plasmids pcDNA3.1, pcDNA-3.1-TRAF1 were purchased from Genechem (China). The transfection was done using Lipofectamine 3000 (Invitrogen), according to the manufacturer’s instruction. After 48 h transfection, HTR8/SVneo cells were harvested for further experiments. The sequences of siRNAs were listed in Table S5.
CRISPR/Cas9-Mediated INHBA-AS1 Knockout
The CRISPR/Cas9 targeting was performed as previously published.72 Guide RNA (gRNA) for INHBA-AS1 was designed on the website (https://zlab.bio/guide-design-resources) and inserted into the pX330 plasmid. After being treated with puromycin for 1 week, HTR8/SVneo cells were plated at 96-well plates to select monoclonal cells for further experiments. The sequences of specific primers were shown in Table S5.
Transwell Assays
The 24-well Transwell chambers with 8 mm pore size polycarbonate membranes (Corning, USA) were used to test invasion and migration of HTR8/SVneo cells. 200 μL cell suspension (40,000–60,000 cells) in medium without FBS was added in the upper side of the membrane coated with/without Matrigel (Beckman Dickinson [BD], USA), and 600 μL culture medium RPMI 1640 containing 10% FBS was added in the lower chamber and placed in the cell culture incubator. After 24 h (for migration) or 48 h (for invasion), the upper chambers were removed with cotton swabs. The lower chambers were fixed by methanol for 30 min and stained with 0.1% crystal violet solution for 20 min. After the chambers were washed with PBS 3 times, the number of cells that penetrated the filter membrane was observed with an optical microscope. Nine randomly selected visual fields were photographed and counted.
EdU Assays
The cell proliferation was determined by EdU assays using the EdU Labeling/Detection Kit (RiboBio, China). Each confocal dish inoculated with cells was added with 200 μL diluent A and incubated in the cell incubator for 2 h. Cells were washed with PBS buffer and then fixed by 4% paraformaldehyde for 30 min and decolorized with 200 μL 2 mg/mL glycine for 5 min in a shaker at room temperature. Then, 200 μL of 0.5% Triton X-100 was added to each well and put at room temperature for 10 min. After one wash with PBS buffer, each well was incubated for 30 min in the dark with 200 μL of pre-prepared 1× Apollo stain at room temperature. 0.5% Triton X-100 was added to each well following 2 washes with PBS buffer and incubated for 10 min. Each well was stained with 200 μL diluent F and incubated in the dark at room temperature for 30 min and then again twice washed with PBS buffer. After staining, the dishes were observed under a fluorescence microscope.
Cell Viability Assay
Cell viability was detected using the CCK-8 (Dojindo). First, cells were seeded in 96-well plates at a density of 10,000 cells per well, and they were given 2 h incubation to attach to the dishes. The CCK-8 was used to calculate the number of viable cells by measuring the absorbance at 450 nm using the BMG microplate reader (CLARIOstar; BMG Labtech). The measurements were performed every 24 h until 96 h.
Flow Cytometric Analysis of Cell Cycle and Apoptosis
Cell cycle was evaluated using the RNase A/Propidium Iodide (PI) Detection kit (KeyGEN, China). The staining buffer of PI/RNase A was prepared at a 9:1 dilution. After precipitation by centrifugation, the cell pellets were suspended using 500 μL of 70% cold ethanol and then fixed for 2 h to overnight at 4°C. After washing with PBS, the cell pellets were incubated with staining solution in the dark for 30–60 min. The percentage of G0/G1, S, and G2/M phases was estimated using flow cytometry (BD LSRFortessa, USA). Apoptosis was determined with the Annexin-V/PI Detection kit (KeyGEN, China). The suspended cells in the culture were collected first, and then the attached cells were collected after being digested with Trypsin. After centrifugation, the cells were resuspended with 500 μL binding buffer. 5 μL of Annexin-V-FITC and 5 μL of PI were added and gently mixed. The reaction was conducted in the dark at room temperature for 5–15 min, and the percentages of early and late apoptosis, as well as viable and dead cells, was detected by flow cytometry (BD LSRFortessa, USA) within 1 h.
RNA-Pulldown and Mass Spectrometry Assay
INHBA-AS1 and its antisense RNA were transcribed with the biotin RNA labeling mix (Roche, USA) and T7 RNA polymerase (Roche, USA). Biotinylated RNA was incubated with HTR8/SVneo cell nuclear extracts. The pulldown proteins were run on SDS-PAGE gels (Sigma, USA) and stained with silver staining solution (Beyotime, China), followed by mass spectrometry.
FISH
The localization and distribution of lncRNA INHBA-AS1 were detected with a FISH kit (RiboBio, China), according to the manufacturer’s instruction.
Subcellular Fractionation
The PARIS Kit (Life Technologies, USA) was used to separate cytosolic and nuclear fractions of the HTR8/SVneo cell following the manufacturer’s instruction. The levels of INHBA-AS1, glyceraldehyde 3-phosphate dehydrogenase (GAPDH), and U6 were examined by qRT-PCR. U6 was used as nuclear control and GAPDH as a cytoplasm control.
RIP Assays
RIP assays were performed following the protocol of the EZ-Magna RIP Kit (Millipore, USA). Anti-CENPB antibody and immunoglobulin G (IgG; negative control) were purchased from Santa Cruz and Millipore. After being collected by centrifugation, HTR-8/SVneo cells were lysed in RIP lysis buffer. Cell lysates were used for immunoprecipitation, then the immunoprecipitated RNA was examined by qRT-PCR, and the data were normalized to the input. The sequences of specific primers were listed in Table S5.
Western Blot Assays
Total cellular proteins were extracted with the Whole Protein Extraction Kit (KeyGEN, China). Protein concentration was detected by the BCA (Bicinchoninic Acid) Protein Assay Kit (KeyGEN, China). The following primary antibodies were used: anti-TRAF1 (Cell Signaling Technology, USA), anti-CENPB (Santa Cruz), anti-MMP2, anti-MMP3, and anti-vimentin (Cell Signaling Technology, USA). Anti-β-actin (Cell Signaling Technology, USA) was used as control. The antibody was diluted according to the manufacturer’s instruction.
ChIP Assays
ChIP assays were performed using the Agarose ChIP Kit (Pierce, USA), according to the manufacturer’s instructions. Anti-CENPB was purchased from Santa Cruz. Immunoprecipitated DNA was analyzed by qRT-PCR, and the data were normalized to the input. Promoter primers are listed in Table S5.
Luciferase Reporter Assays
Luciferase Assay Kit (Promega, USA) was used to measure luciferase activity, according to the manufacturer’s instruction. We cloned the TRAF1 site 3 (Table S5) into PGL3-Basic vector. These 2 plasmids and PGL3 empty vector were transfected into the HTR8/SV/neo cells, cells with INHBA-AS1 overexpression, and cells with INHBA-AS1 knockout, respectively. pRL-TK vector was transfected into the three kinds of cells simultaneously. After incubating for 48 h, the cells were lysed in 1× Passive lysis, and the firefly luciferase activity was examined using Renilla luciferase as control.
Statistical Analysis
Statistical analyses were performed using SPSS20.0 (IBM), and the data were presented as the mean ± SD in three independently repeated experiments. The comparison between the two independent groups was conducted by Student’s t test. Statistical significance was described as ∗p <0.05 or ∗∗p <0.01.
Author Contributions
XinpingY. designed the project. Y.Y. provided some constructive ideas and support. S.J., Q.C., and L.X. carried out the experiments. S.J., Q.C., and M.Z. collected samples. Yunfei G., S.J., and Q.C. collected and analyzed the clinical information. S.J., Q.C., H.L., Yue G., Xiaoxue.Y., and Z.R. analyzed the data. S.J. and Xinping.Y. prepared the manuscript.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
We thank the patients for their generous support for this study and also Profs. Guangjin Pan and Yongli Shan at Guangzhou Institutes of Biomedicine and Health, Chinese Academy of Sciences, for providing plasmids. This study was supported by National Key Programs for R&D grants 2018YFA0507800/2018YFA0507803 (to X.Y.); National Natural Science Foundation of China grants 31771434 (to X.Y.), 81771609 (to Y.Y.), and 81971413 (to M.Z.); and Science and Technology Projects of Guangdong Province grant 2015B050501006 (to M.Z.).
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtn.2020.09.033.
Supplemental Information
References
- 1.Chaiworapongsa T., Chaemsaithong P., Yeo L., Romero R. Pre-eclampsia part 1: current understanding of its pathophysiology. Nat. Rev. Nephrol. 2014;10:466–480. doi: 10.1038/nrneph.2014.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ananth C.V., Keyes K.M., Wapner R.J. Pre-eclampsia rates in the United States, 1980-2010: age-period-cohort analysis. BMJ. 2013;347:f6564. doi: 10.1136/bmj.f6564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Khan K.S., Wojdyla D., Say L., Gülmezoglu A.M., Van Look P.F. WHO analysis of causes of maternal death: a systematic review. Lancet. 2006;367:1066–1074. doi: 10.1016/S0140-6736(06)68397-9. [DOI] [PubMed] [Google Scholar]
- 4.World Health Organization . World Health Organization; 2011. WHO Recommendations for Prevention and Treatment of Pre-Eclampsia and Eclampsia. [PubMed] [Google Scholar]
- 5.Zeeman G.G. Neurologic complications of pre-eclampsia. Semin. Perinatol. 2009;33:166–172. doi: 10.1053/j.semperi.2009.02.003. [DOI] [PubMed] [Google Scholar]
- 6.Mol B.W.J., Roberts C.T., Thangaratinam S., Magee L.A., de Groot C.J.M., Hofmeyr G.J. Pre-eclampsia. Lancet. 2016;387:999–1011. doi: 10.1016/S0140-6736(15)00070-7. [DOI] [PubMed] [Google Scholar]
- 7.Davies E.L., Bell J.S., Bhattacharya S. Preeclampsia and preterm delivery: A population-based case-control study. Hypertens. Pregnancy. 2016;35:510–519. doi: 10.1080/10641955.2016.1190846. [DOI] [PubMed] [Google Scholar]
- 8.Mitani M., Matsuda Y., Makino Y., Akizawa Y., Ohta H. Clinical features of fetal growth restriction complicated later by preeclampsia. J. Obstet. Gynaecol. Res. 2009;35:882–887. doi: 10.1111/j.1447-0756.2009.01120.x. [DOI] [PubMed] [Google Scholar]
- 9.Weiler J., Tong S., Palmer K.R. Is fetal growth restriction associated with a more severe maternal phenotype in the setting of early onset pre-eclampsia? A retrospective study. PLoS ONE. 2011;6:e26937. doi: 10.1371/journal.pone.0026937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu X., Zhao W., Liu H., Kang Y., Ye C., Gu W., Hu R., Li X. Developmental and Functional Brain Impairment in Offspring from Preeclampsia-Like Rats. Mol. Neurobiol. 2016;53:1009–1019. doi: 10.1007/s12035-014-9060-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cui Y., Wang W., Dong N., Lou J., Srinivasan D.K., Cheng W., Huang X., Liu M., Fang C., Peng J. Role of corin in trophoblast invasion and uterine spiral artery remodelling in pregnancy. Nature. 2012;484:246–250. doi: 10.1038/nature10897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.El-Sayed A.A.F. Preeclampsia: A review of the pathogenesis and possible management strategies based on its pathophysiological derangements. Taiwan. J. Obstet. Gynecol. 2017;56:593–598. doi: 10.1016/j.tjog.2017.08.004. [DOI] [PubMed] [Google Scholar]
- 13.Hod T., Cerdeira A.S., Karumanchi S.A. Molecular Mechanisms of Preeclampsia. Cold Spring Harb. Perspect. Med. 2015;5:a023473. doi: 10.1101/cshperspect.a023473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sircar M., Thadhani R., Karumanchi S.A. Pathogenesis of preeclampsia. Curr. Opin. Nephrol. Hypertens. 2015;24:131–138. doi: 10.1097/MNH.0000000000000105. [DOI] [PubMed] [Google Scholar]
- 15.Jim B., Karumanchi S.A. Preeclampsia: Pathogenesis, Prevention, and Long-Term Complications. Semin. Nephrol. 2017;37:386–397. doi: 10.1016/j.semnephrol.2017.05.011. [DOI] [PubMed] [Google Scholar]
- 16.Salonen Ros H., Lichtenstein P., Lipworth L., Cnattingius S. Genetic effects on the liability of developing pre-eclampsia and gestational hypertension. Am. J. Med. Genet. 2000;91:256–260. [PubMed] [Google Scholar]
- 17.Cnattingius S., Reilly M., Pawitan Y., Lichtenstein P. Maternal and fetal genetic factors account for most of familial aggregation of preeclampsia: a population-based Swedish cohort study. Am. J. Med. Genet. A. 2004;130A:365–371. doi: 10.1002/ajmg.a.30257. [DOI] [PubMed] [Google Scholar]
- 18.Arngrímsson R., Sigurõardóttir S., Frigge M.L., Bjarnadóttir R.I., Jónsson T., Stefánsson H., Baldursdóttir A., Einarsdóttir A.S., Palsson B., Snorradóttir S. A genome-wide scan reveals a maternal susceptibility locus for pre-eclampsia on chromosome 2p13. Hum. Mol. Genet. 1999;8:1799–1805. doi: 10.1093/hmg/8.9.1799. [DOI] [PubMed] [Google Scholar]
- 19.Moses E.K., Lade J.A., Guo G., Wilton A.N., Grehan M., Freed K., Borg A., Terwilliger J.D., North R., Cooper D.W., Brennecke S.P. A genome scan in families from Australia and New Zealand confirms the presence of a maternal susceptibility locus for pre-eclampsia, on chromosome 2. Am. J. Hum. Genet. 2000;67:1581–1585. doi: 10.1086/316888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Laivuori H., Lahermo P., Ollikainen V., Widen E., Häivä-Mällinen L., Sundström H., Laitinen T., Kaaja R., Ylikorkala O., Kere J. Susceptibility loci for preeclampsia on chromosomes 2p25 and 9p13 in Finnish families. Am. J. Hum. Genet. 2003;72:168–177. doi: 10.1086/345311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lachmeijer A.M., Arngrímsson R., Bastiaans E.J., Frigge M.L., Pals G., Sigurdardóttir S., Stéfansson H., Pálsson B., Nicolae D., Kong A. A genome-wide scan for preeclampsia in the Netherlands. Eur. J. Hum. Genet. 2001;9:758–764. doi: 10.1038/sj.ejhg.5200706. [DOI] [PubMed] [Google Scholar]
- 22.van Dijk M., Mulders J., Poutsma A., Könst A.A., Lachmeijer A.M., Dekker G.A., Blankenstein M.A., Oudejans C.B. Maternal segregation of the Dutch preeclampsia locus at 10q22 with a new member of the winged helix gene family. Nat. Genet. 2005;37:514–519. doi: 10.1038/ng1541. [DOI] [PubMed] [Google Scholar]
- 23.Zhao L., Bracken M.B., DeWan A.T. Genome-wide association study of pre-eclampsia detects novel maternal single nucleotide polymorphisms and copy-number variants in subsets of the Hyperglycemia and Adverse Pregnancy Outcome (HAPO) study cohort. Ann. Hum. Genet. 2013;77:277–287. doi: 10.1111/ahg.12021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnson M.P., Brennecke S.P., East C.E., Göring H.H., Kent J.W., Jr., Dyer T.D., Said J.M., Roten L.T., Iversen A.C., Abraham L.J., FINNPEC Study Group Genome-wide association scan identifies a risk locus for preeclampsia on 2q14, near the inhibin, beta B gene. PLoS ONE. 2012;7:e33666. doi: 10.1371/journal.pone.0033666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Enquobahrie D.A., Meller M., Rice K., Psaty B.M., Siscovick D.S., Williams M.A. Differential placental gene expression in preeclampsia. Am. J. Obstet. Gynecol. 2008;199:566.e1–566.e11. doi: 10.1016/j.ajog.2008.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tsai S., Hardison N.E., James A.H., Motsinger-Reif A.A., Bischoff S.R., Thames B.H., Piedrahita J.A. Transcriptional profiling of human placentas from pregnancies complicated by preeclampsia reveals disregulation of sialic acid acetylesterase and immune signalling pathways. Placenta. 2011;32:175–182. doi: 10.1016/j.placenta.2010.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sitras V., Paulssen R.H., Grønaas H., Leirvik J., Hanssen T.A., Vårtun A., Acharya G. Differential placental gene expression in severe preeclampsia. Placenta. 2009;30:424–433. doi: 10.1016/j.placenta.2009.01.012. [DOI] [PubMed] [Google Scholar]
- 28.Kang J.H., Song H., Yoon J.A., Park D.Y., Kim S.H., Lee K.J., Farina A., Cho Y.K., Kim Y.N., Park S.W. Preeclampsia leads to dysregulation of various signaling pathways in placenta. J. Hypertens. 2011;29:928–936. doi: 10.1097/HJH.0b013e328344a82c. [DOI] [PubMed] [Google Scholar]
- 29.Nishizawa H., Pryor-Koishi K., Kato T., Kowa H., Kurahashi H., Udagawa Y. Microarray analysis of differentially expressed fetal genes in placental tissue derived from early and late onset severe pre-eclampsia. Placenta. 2007;28:487–497. doi: 10.1016/j.placenta.2006.05.010. [DOI] [PubMed] [Google Scholar]
- 30.Junus K., Centlow M., Wikström A.K., Larsson I., Hansson S.R., Olovsson M. Gene expression profiling of placentae from women with early- and late-onset pre-eclampsia: down-regulation of the angiogenesis-related genes ACVRL1 and EGFL7 in early-onset disease. Mol. Hum. Reprod. 2012;18:146–155. doi: 10.1093/molehr/gar067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pang Z.J., Xing F.Q. Expression profile of trophoblast invasion-associated genes in the pre-eclamptic placenta. Br. J. Biomed. Sci. 2003;60:97–101. doi: 10.1080/09674845.2003.11783682. [DOI] [PubMed] [Google Scholar]
- 32.Nishizawa H., Ota S., Suzuki M., Kato T., Sekiya T., Kurahashi H., Udagawa Y. Comparative gene expression profiling of placentas from patients with severe pre-eclampsia and unexplained fetal growth restriction. Reprod. Biol. Endocrinol. 2011;9:107. doi: 10.1186/1477-7827-9-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zuckerwise L., Li J., Lu L., Men Y., Geng T., Buhimschi C.S., Buhimschi I.A., Bukowski R., Guller S., Paidas M., Huang Y. H19 long noncoding RNA alters trophoblast cell migration and invasion by regulating TβR3 in placentae with fetal growth restriction. Oncotarget. 2016;7:38398–38407. doi: 10.18632/oncotarget.9534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Y., Zou Y., Wang W., Zuo Q., Jiang Z., Sun M., De W., Sun L. Down-regulated long non-coding RNA MEG3 and its effect on promoting apoptosis and suppressing migration of trophoblast cells. J. Cell. Biochem. 2015;116:542–550. doi: 10.1002/jcb.25004. [DOI] [PubMed] [Google Scholar]
- 35.Chen H., Meng T., Liu X., Sun M., Tong C., Liu J., Wang H., Du J. Long non-coding RNA MALAT-1 is downregulated in preeclampsia and regulates proliferation, apoptosis, migration and invasion of JEG-3 trophoblast cells. Int. J. Clin. Exp. Pathol. 2015;8:12718–12727. [PMC free article] [PubMed] [Google Scholar]
- 36.Zuo Q., Huang S., Zou Y., Xu Y., Jiang Z., Zou S., Xu H., Sun L. The Lnc RNA SPRY4-IT1 Modulates Trophoblast Cell Invasion and Migration by Affecting the Epithelial-Mesenchymal Transition. Sci. Rep. 2016;6:37183. doi: 10.1038/srep37183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zou Y., Jiang Z., Yu X., Sun M., Zhang Y., Zuo Q., Zhou J., Yang N., Han P., Ge Z. Upregulation of long noncoding RNA SPRY4-IT1 modulates proliferation, migration, apoptosis, and network formation in trophoblast cells HTR-8SV/neo. PLoS ONE. 2013;8:e79598. doi: 10.1371/journal.pone.0079598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bhan A., Mandal S.S. LncRNA HOTAIR: A master regulator of chromatin dynamics and cancer. Biochim. Biophys. Acta. 2015;1856:151–164. doi: 10.1016/j.bbcan.2015.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang Y., Jin F., Li X.C., Shen F.J., Ma X.L., Wu F., Zhang S.M., Zeng W.H., Liu X.R., Fan J.X. The YY1-HOTAIR-MMP2 Signaling Axis Controls Trophoblast Invasion at the Maternal-Fetal Interface. Mol. Ther. 2017;25:2394–2403. doi: 10.1016/j.ymthe.2017.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 40.Long Y., Wang X., Youmans D.T., Cech T.R. How do lncRNAs regulate transcription? Sci. Adv. 2017;3:eaao2110. doi: 10.1126/sciadv.aao2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xue Z., Hennelly S., Doyle B., Gulati A.A., Novikova I.V., Sanbonmatsu K.Y., Boyer L.A. A G-Rich Motif in the lncRNA Braveheart Interacts with a Zinc-Finger Transcription Factor to Specify the Cardiovascular Lineage. Mol. Cell. 2016;64:37–50. doi: 10.1016/j.molcel.2016.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Majewska M., Lipka A., Paukszto L., Jastrzebski J.P., Gowkielewicz M., Jozwik M., Majewski M.K. Preliminary RNA-Seq Analysis of Long Non-Coding RNAs Expressed in Human Term Placenta. Int. J. Mol. Sci. 2018;19:E1894. doi: 10.3390/ijms19071894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang Y., Zhao H.J., Xia X.R., Diao F.Y., Ma X., Wang J., Gao L., Liu J., Gao C., Cui Y.G., Liu J.Y. Hypoxia-induced and HIF1α-VEGF-mediated tight junction dysfunction in choriocarcinoma cells: Implications for preeclampsia. Clin. Chim. Acta. 2019;489:203–211. doi: 10.1016/j.cca.2017.12.010. [DOI] [PubMed] [Google Scholar]
- 44.Tal R. The role of hypoxia and hypoxia-inducible factor-1alpha in preeclampsia pathogenesis. Biol. Reprod. 2012;87:134. doi: 10.1095/biolreprod.112.102723. [DOI] [PubMed] [Google Scholar]
- 45.Korkes H.A., De Oliveira L., Sass N., Salahuddin S., Karumanchi S.A., Rajakumar A. Relationship between hypoxia and downstream pathogenic pathways in preeclampsia. Hypertens. Pregnancy. 2017;36:145–150. doi: 10.1080/10641955.2016.1259627. [DOI] [PubMed] [Google Scholar]
- 46.Jackson D.W., Sciscione A., Hartley T.L., Haynes A.L., Carder E.A., Blakemore K.J., Idrisa A., Glew R.H. Lysosomal enzymuria in preeclampsia. Am. J. Kidney Dis. 1996;27:826–833. doi: 10.1016/s0272-6386(96)90520-x. [DOI] [PubMed] [Google Scholar]
- 47.Sharma S. Autophagy-Based Diagnosis of Pregnancy Hypertension and Pre-Eclampsia. Am. J. Pathol. 2018;188:2457–2460. doi: 10.1016/j.ajpath.2018.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Scioscia M., Gumaa K., Kunjara S., Paine M.A., Selvaggi L.E., Rodeck C.H., Rademacher T.W. Insulin resistance in human preeclamptic placenta is mediated by serine phosphorylation of insulin receptor substrate-1 and -2. J. Clin. Endocrinol. Metab. 2006;91:709–717. doi: 10.1210/jc.2005-1965. [DOI] [PubMed] [Google Scholar]
- 49.Khaliq O.P., Murugesan S., Moodley J., Mackraj I. Differential expression of miRNAs are associated with the insulin signaling pathway in preeclampsia and gestational hypertension. Clin. Exp. Hypertens. 2018;40:744–751. doi: 10.1080/10641963.2018.1431257. [DOI] [PubMed] [Google Scholar]
- 50.Jiang C., Xuan Z., Zhao F., Zhang M.Q. TRED: a transcriptional regulatory element database, new entries and other development. Nucleic Acids Res. 2007;35:D137–D140. doi: 10.1093/nar/gkl1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zheng G., Tu K., Yang Q., Xiong Y., Wei C., Xie L., Zhu Y., Li Y. ITFP: an integrated platform of mammalian transcription factors. Bioinformatics. 2008;24:2416–2417. doi: 10.1093/bioinformatics/btn439. [DOI] [PubMed] [Google Scholar]
- 52.The ENCODE (ENCyclopedia Of DNA Elements) Project Science. 2004;306:636–640. doi: 10.1126/science.1105136. [DOI] [PubMed] [Google Scholar]
- 53.Neph S., Stergachis A.B., Reynolds A., Sandstrom R., Borenstein E., Stamatoyannopoulos J.A. Circuitry and dynamics of human transcription factor regulatory networks. Cell. 2012;150:1274–1286. doi: 10.1016/j.cell.2012.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Han H., Cho J.W., Lee S., Yun A., Kim H., Bae D., Yang S., Kim C.Y., Lee M., Kim E. TRRUST v2: an expanded reference database of human and mouse transcriptional regulatory interactions. Nucleic Acids Res. 2018;46(D1):D380–D386. doi: 10.1093/nar/gkx1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Denys H., De Wever O., Nusgens B., Kong Y., Sciot R., Le A.T., Van Dam K., Jadidizadeh A., Tejpar S., Mareel M. Invasion and MMP expression profile in desmoid tumours. Br. J. Cancer. 2004;90:1443–1449. doi: 10.1038/sj.bjc.6601661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Uraoka N., Oue N., Sakamoto N., Sentani K., Oo H.Z., Naito Y., Noguchi T., Yasui W. NRD1, which encodes nardilysin protein, promotes esophageal cancer cell invasion through induction of MMP2 and MMP3 expression. Cancer Sci. 2014;105:134–140. doi: 10.1111/cas.12316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jimenez L., Lim J., Burd B., Harris T.M., Ow T.J., Kawachi N., Belbin T.J., Angeletti R., Prystowsky M.B., Childs G., Segall J.E. miR-375 Regulates Invasion-Related Proteins Vimentin and L-Plastin. Am. J. Pathol. 2017;187:1523–1536. doi: 10.1016/j.ajpath.2017.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang H., Wu X., Xiao Y., Wu L., Peng Y., Tang W., Liu G., Sun Y., Wang J., Zhu H. Coexpression of FOXK1 and vimentin promotes EMT, migration, and invasion in gastric cancer cells. J. Mol. Med. (Berl.) 2019;97:163–176. doi: 10.1007/s00109-018-1720-z. [DOI] [PubMed] [Google Scholar]
- 59.Renaud S.J. Reproductive and Developmental Toxicology. Second Edition. Academic Press; 2017. Strategies for investigating hemochorial placentation; pp. 1259–1273. [Google Scholar]
- 60.Wu D., Xu Y., Zou Y., Zuo Q., Huang S., Wang S., Lu X., He X., Wang J., Wang T., Sun L. Long Noncoding RNA 00473 Is Involved in Preeclampsia by LSD1 Binding-Regulated TFPI2 Transcription in Trophoblast Cells. Mol. Ther. Nucleic Acids. 2018;12:381–392. doi: 10.1016/j.omtn.2018.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang N., Li R., Xue M. Potential regulatory network in the PSG10P/miR-19a-3p/IL1RAP pathway is possibly involved in preeclampsia pathogenesis. J. Cell. Mol. Med. 2019;23:852–864. doi: 10.1111/jcmm.13985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xu Y., Wu D., Liu J., Huang S., Zuo Q., Xia X., Jiang Y., Wang S., Chen Y., Wang T., Sun L. Downregulated lncRNA HOXA11-AS Affects Trophoblast Cell Proliferation and Migration by Regulating RND3 and HOXA7 Expression in PE. Mol. Ther. Nucleic Acids. 2018;12:195–206. doi: 10.1016/j.omtn.2018.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Xu Y., Lian Y., Zhang Y., Huang S., Zuo Q., Yang N., Chen Y., Wu D., Sun L. The long non-coding RNA PVT1 represses ANGPTL4 transcription through binding with EZH2 in trophoblast cell. J. Cell. Mol. Med. 2018;22:1272–1282. doi: 10.1111/jcmm.13405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zou Y., Li Q., Xu Y., Yu X., Zuo Q., Huang S., Chu Y., Jiang Z., Sun L. Promotion of trophoblast invasion by lncRNA MVIH through inducing Jun-B. J. Cell. Mol. Med. 2018;22:1214–1223. doi: 10.1111/jcmm.13400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Niu Z.R., Han T., Sun X.L., Luan L.X., Gou W.L., Zhu X.M. MicroRNA-30a-3p is overexpressed in the placentas of patients with preeclampsia and affects trophoblast invasion and apoptosis by its effects on IGF-1. Am. J. Obstet. Gynecol. 2018;218:249.e1–249.e12. doi: 10.1016/j.ajog.2017.11.568. [DOI] [PubMed] [Google Scholar]
- 66.Canfield J., Arlier S., Mong E.F., Lockhart J., VanWye J., Guzeloglu-Kayisli O., Schatz F., Magness R.R., Lockwood C.J., Tsibris J.C.M. Decreased LIN28B in preeclampsia impairs human trophoblast differentiation and migration. FASEB J. 2019;33:2759–2769. doi: 10.1096/fj.201801163R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang Y., Cao L., Jia J., Ye L., Wang Y., Zhou B., Zhou R. CircHIPK3 is decreased in preeclampsia and affects migration, invasion, proliferation, and tube formation of human trophoblast cells. Placenta. 2019;85:1–8. doi: 10.1016/j.placenta.2019.07.010. [DOI] [PubMed] [Google Scholar]
- 68.Meekins J.W., Pijnenborg R., Hanssens M., McFadyen I.R., van Asshe A. A study of placental bed spiral arteries and trophoblast invasion in normal and severe pre-eclamptic pregnancies. Br. J. Obstet. Gynaecol. 1994;101:669–674. doi: 10.1111/j.1471-0528.1994.tb13182.x. [DOI] [PubMed] [Google Scholar]
- 69.Whitley G.S., Dash P.R., Ayling L.J., Prefumo F., Thilaganathan B., Cartwright J.E. Increased apoptosis in first trimester extravillous trophoblasts from pregnancies at higher risk of developing preeclampsia. Am. J. Pathol. 2007;170:1903–1909. doi: 10.2353/ajpath.2007.070006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ohzeki J., Nakano M., Okada T., Masumoto H. CENP-B box is required for de novo centromere chromatin assembly on human alphoid DNA. J. Cell Biol. 2002;159:765–775. doi: 10.1083/jcb.200207112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lambert S.A., Jolma A., Campitelli L.F., Das P.K., Yin Y., Albu M., Chen X., Taipale J., Hughes T.R., Weirauch M.T. The Human Transcription Factors. Cell. 2018;172:650–665. doi: 10.1016/j.cell.2018.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shan Y., Liang Z., Xing Q., Zhang T., Wang B., Tian S., Huang W., Zhang Y., Yao J., Zhu Y. PRC2 specifies ectoderm lineages and maintains pluripotency in primed but not naïve ESCs. Nat. Commun. 2017;8:672. doi: 10.1038/s41467-017-00668-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.