Abstract
Objective
To investigate the role of fucoxanthin, reported to have significant anticancer effects, and histone Cluster 1 H3 Family Member D (HIST1H3D; implicated in tumorigenesis) in cervical cancer.
Methods
The half maximal inhibitory concentration (IC50) of fucoxanthin against HeLa and SiHa cervical cancer cells was determined. Differentially expressed genes (DEGs) in SiHa cells treated with IC50 fucoxanthin were screened by high-throughput techniques and subjected to signal enrichment. Following identification of HIST1H3D as a candidate gene, HIST1H3D-knockdown models were created via transfection with a short hairpin HIST1H3D payload. Impacts on cell proliferation, cell-cycle distribution, colony formation, and apoptosis were studied.
Results
The fucoxanthin IC50 was 1 445 and 1 641 µM (Hela and SiHa cells, respectively). Chip results revealed 2 255 DEGs, including 943 upregulated and 1 312 downregulated genes, in fucoxanthin-treated versus untreated SiHa cells. Disease and function analysis indicated that these DEGs are primarily associated with cancer and organismal injuries and abnormalities, and online integrated pathway analysis showed that the DEGs were mainly enriched in p53 signalling. HIST1H3D was significantly downregulated in response to fucoxanthin. Inhibition of HIST1H3D mRNA significantly reduced cell proliferation and colony formation, significantly augmented the percentage of apoptotic HeLa and SiHa cells, and cells were arrested in G0/G1 cell cycle phase.
Conclusion
The results suggest that HIST1H3D may be an oncogene in cervical carcinogenesis and a potential fucoxanthin target in treating cervical cancer.
Keywords: Fucoxanthin, cervical cancer, proliferation, colony formation
Introduction
Cervical cancer is the fourth most common female malignancy in the world,1,2 accounting for 4% of all tumours diagnosed globally. About 50.3% of patients with cervical cancer die each year from the disease.3 Nearly 84% of cervical cancer cases occur in developing countries, and cervical cancer is the third most common cause of death in women in these regions.1,4,5
Infection with human papillomavirus (HPV), herpes simplex virus 2 and/or chlamydia, smoking, oral contraceptives, and genetic factors may be associated with increased cervical cancer risk.6–15 In addition, a lack of prevention, screening and treatment are risk factors for disease onset.16,17
The Papanicolaou test helps to detect precancerous lesions, resulting in effective treatment or even cure, making the 5-year survival rate of patients with cervical cancer close to 100%.18,19 However, women in developing countries are prone to cervical cancer because they fail to receive screening for precancerous lesions or HPV immunotherapy in time.18,20 Therefore, in addition to the above strategies, clarifying the pathogenesis of cervical cancer will help to improve survival rates in patients with cervical cancer, and also help to provide potential novel targets for cervical cancer treatment.
Fucoxanthin is a non‐provitamin A carotenoid that is widely found in brown algae,21 and its structure is shown in Figure 1a. Fucoxanthin has shown a wide range of biological effects, including antitumour, antioxidant and antidiabetic activities.22 In addition, fucoxanthin has an antiangiogenic effect that prevents tumour growth.23 Studies have shown that the antiproliferative effects of fucoxanthin against tumour cells is achieved through phosphatidylinositol 3-kinase (PI3K)/Akt and/or nuclear factor (NF)‐κB signalling pathways.24,25 However, the effect of fucoxanthin on cervical cancer remains to be elucidated.
Figure 1.
Inhibitory effect of fucoxanthin on human cervical cancer cell lines: (a) the structure of fucoxanthin; (b) HeLa cell inhibition curve and (c) SiHa cell inhibition curve showing the half maximal inhibitory concentration (IC50) of fucoxanthin.
In eukaryotic cells, histones are a class of nuclear proteins involved in chromatin condensation, and accumulated evidence suggests that histone regulation of gene expression is achieved through regulation of chromatin states.26–28 Abnormal expression of histones, or histone mutation, is associated with tumorigenesis.29 For example, glioblastoma is associated with histone mutations,30 and abnormal expression of histone H2A.Z has been observed in prostate cancer.31
The human H3 clustered histone 6 (H3C6) gene (also known as HIST1H3D) encodes histone H3.1.32 Methylated H3.1 has been shown to be associated with gene-silencing and heterochromatin formation,33 and HIST1H3D is reported to be elevated in tumour necrosis factor (TNF)-α-induced endothelial cell inflammation and lung disease involving pseudomonas infection.34,35 Furthermore, HIST1H3D expression is shown to be upregulated in tumour tissues, such as lung cancer and primary gastric cancer tissues.36,37
The aim of the present study was to investigate the roles of fucoxanthin and HIST1H3D in cervical cancer through investigation of genes that are differentially expressed between cervical cancer cells treated with fucoxanthin and untreated cells, and the effect of HIST1H3D expression on cervical cancer cell activity.
Materials and methods
Cell lines
The HeLa and SiHa human cervical cancer cell lines were obtained from the Shanghai Cell Bank of China (Shanghai, China). Both cell lines were maintained in Dulbecco’s modified Eagle’s medium (DMEM, catalogue No. 11965; Gibco™, Shanghai, China) supplemented with 10% fetal bovine serum (catalogue No. 10099141; Gibco™) and 0.1% gentamicin sulphate (A100304; Sangon Biotech, Shanghai, China) at 37°C in a 5% CO2 humidified incubator. All experiments were performed using cells grown to >75% confluence.
IC50 assay
To assess the half maximal inhibitory concentration (IC50) of fucoxanthin, HeLa and SiHa cells in logarithmic growth phase were trypsinized to detach from the culture vessel, then resuspended in complete DMEM and plated into 96-well plates (4 × 103 cells/100 μl/well). After 24 h of incubation at 37 °C/5% CO2, different concentrations of fucoxanthin (0, 0.1, 0.5, 1, 5, 10, and 25 μM, each in triplicate) were added and cells were cultured for a further 48 h. Following fucoxanthin treatment, 10 μl/well of cell counting kit (CCK)-8 reagent (catalogue No. E606335, Sangon Biotech, Shanghai, China) was added, and the plates were incubated for 4 h at 37 °C. The optical density (OD) value at 490 nm wavelength was then measured using an Elx-800 microplate reader (Biotek® Instruments, Winooski, VT, USA).
Screening of differentially expressed genes (DEGs) using microarray
The expression status of genes was determined within cervical cancer SiHa cells treated with 0.5 μM/l fucoxanthin or untreated negative controls (NC) using GeneChip Primeview human gene expression array (catalogue No. 901838; Affymetrix, Santa Clara, CA, USA). After treatment of SiHa cells with fucoxanthin or NC for 48 h, total RNA was extracted using TRIzol® reagent (Invitrogen, Shanghai, China), and inspected for subsequent microarray analysis. For gene expression profiling, cDNA was synthesized using 0.5 μg of RNA per sample as a template and AffinityScript QPCR synthesis kit (catalogue No. 600559; Stratagene, La Jolla, CA, USA). Biotin-labelled amplified RNA was synthesized from double-stranded cDNA using the GeneChip® 3′ IVT labelling kit (Affymetrix). The microarrays were washed and stained with GeneChip® hybridization wash and stain kit (Affymetrix) according to the manufacturer's instructions. Finally, the probe arrays were scanned directly using a GeneChip® scanner 3000 (Affymetrix) post hybridization.
Microarray data were normalized using GeneSpring software, version 11 (Agilent Technologies, Santa Clara, CA, USA), and generated lists of DEGs (at least ± 2.0-fold, P ≤ 0.05, relative to the negative control). The newly generated list of differential expression transcripts was used for clustering hierarchically based on certain correlations and signalling pathway enrichment. Pathways were enriched using an online integrated pathway analysis database.38 Finally, genes of interest (for example, HIST1H3D) with meaningful and significantly different expression, were utilized to identify molecular functions using real-time quantitative polymerase chain reaction (qPCR).
Real-time qPCR
Total RNA was extracted from SiHa cells using TRIzol® reagent (Invitrogen, Shanghai, China), and used as a template for cDNA synthesis with M-MLV reverse transcriptase (Promega, Shanghai, China), according to the manufacturer's instructions. Real-time qPCR of cDNA was then performed. The GAPDH internal control primer sequences were 5′-TGA CTT CAA CAG CGA CAC CCA-3′ (forward) and 5′-CAC CCT GTT GCT GTA GCC AAA-3′ (reverse), and provided a 121-bp product; and the primer sequences for HIST1H3D were 5′-TTC GCA AAC TGC CAT TCC-3′ (forward) and 5′-GAG CCT TTG GGT TTT GGT T-3′ (reverse), with a 264-bp PCR product. Each real-time qPCR reaction was performed using a Platinum® qPCR SuperMix-UDG (Invitrogen, Carlsbad, CA, USA) and the following thermal cycling procedure: 95°C for 30 s, then 40 cycles of denaturation at 95°C for 5 s and extension at 60°C for 30 s. All samples were assayed in triplicate, and the relative level of HIST1H3D expression was calculated using the 2-ΔΔCT method.
Lentiviral recombinant plasmid and cell transfection
Small interfering (si)RNA and short hairpin (sh)RNA for specifically knocking down human HIST1H3D expression was synthesized by GeneChem Co. Ltd (Shanghai, China) according to the full-length HIST1H3D sequence (GenBank no. NM_003530). The siRNA sequence was 5′-GCA ACT CAA AGA CCT GGA A-3′ and the shRNA sequence was 5′-CCG GGC TGA TTC GCA AAC TGC CAT TCT CGA GAA TGG CAG TTT GCG AAT CAG CTT TTT G-3′. To assess knockdown efficiency, stem-loop-stem oligonucleotides were synthesized and inserted into a pFV055-GFP vector (GeneChem). Lentivirus particle packaging, collection, and titre determination were performed as previously described.39
For cell transfection, HeLa and SiHa cells (5 × 104 cells per well) were seeded into 6-well plates and transfected with the shHIST1H3D or shCtrl lentivirus at a multiplicity of infection (MOI) of 20. Then, cells were incubated at 37°C in a 5% CO2 humidified environment. After 72 h, the growth state of the cells was observed under a fluorescence microscope (MicroPublisher™ 3.3, Olympus, Japan). After 2 days, normally growing cells were determined for knockdown efficiency by real-time qPCR (as described above).
CCK-8 assay
Proliferation of cells transfected with either an HIST1H3D knockdown (shHIST1H3D) or empty plasmid (shCtrl) was determined with the CCK-8 assay. Transfected HeLa and SiHa cervical cancer cells (1 × 103 cells per well), within logarithmic growth phase, were reseeded in 96-well plates and incubated at 37°C with 5% CO2 for 5 consecutive days. On each day, the CCK-8 reagent (10 μl per well) was added to a portion of the wells in each treatment group, cells were incubated for 4 h at 37 °C, then the OD value at 490 nm was measured using an Elx-800 microplate reader (Biotek® Instruments).
Colony formation assay
The effect of HIST1H3D silencing on colony formation was determined in parallel with a negative control. Transfected HeLa and SiHa cervical cancer cells (1 × 103 cells per well) were seeded in 6-well plates and incubated at 37°C in 5% CO2 for 14 days to form cell colonies. Cell colonies were then washed 3 times with phosphate buffered saline (PBS, pH 7.2) and fixed with 4% paraformaldehyde (Sangon Biotech) for 60 min. The fixed cells were washed with PBS to remove unreacted paraformaldehyde and then stained with 100 µl of Giemsa staining solution (G5637, Sigma-Aldrich®, Shanghai, China) for 20 min. The number of colonies (>50 cells per colony) were counted under light microscopy.
Flow cytometric analysis for cell cycle and apoptosis
The effect of HIST1H3D knockdown on cell cycle progression and apoptosis was determined by assessing the proportion of cells at each cell cycle stage or undergoing apoptosis using flow cytometry. At 4 days following lentiviral infection, cells (1 × 106 cells per dish) were reseeded into 6-cm dishes. Cells were harvested when coverage reached >80%, and fixed with 70% pre-cooled ethanol for 1 h at 4 °C. For cell cycle analysis, cells were washed 3 times with PBS (pH 7.2), and incubated with 1.5 ml PBS containing 50 μg/ml propidium iodide (PI; catalogue No. P4170; Sigma-Aldrich, St. Louis, MO, USA) and 100 μg/ml RNase A (catalogue No. EN0531; Fermentas®, Shanghai, China) to stain the DNA, in the dark for 30 min at room temperature. The cell suspension was filtered through a 300 mesh and the proportion of cells at each cell cycle phase was determined using a FACSCalibur flow cytometer (BD Biosciences, San Jose, CA, USA) according to the manufacturer’s instructions. For apoptosis analysis, the HeLa and SiHa cell concentrations were adjusted to 1 × 106/ml with 1 × staining buffer (catalogue No. 00-0055; ThermoFisher Scientific, Shanghai, China). Cells in suspension (100 µl) were stained with 5 μl of eBioscience™ Annexin V-APC (catalogue No. 88-8007; ThermoFisher Scientific), and incubated in the dark at room temperature for 15 min. The proportion of apoptotic cells were then analysed by FACSCalibur flow cytometry (BD Biosciences) within 1 h. All experiments were performed in triplicate.
Statistical analyses
Data are presented as mean ± SD, and were compared using Student’s t-test. All statistical analyses were conducted within GraphPad Prism software, version 8.0.1 (GraphPad Software, San Diego, CA, USA). The threshold of statistical significance was set to a P value < 0.05.
Results
Fucoxanthin significantly alters gene expression in SiHa cells
In order to clarify the optimal concentration of fucoxanthin for inhibition of HeLa and SiHa cervical cancer cell lines, an IC50 test was performed. The IC50 of fucoxanthin for inhibition of Hela and SiHa cells was found to be 1 445 and 1 641 µM, respectively (Figure 1b and 1c).
To analyse the effects of fucoxanthin on downstream gene expression, DEGs were assessed in SiHa cells following treatment with fucoxanthin. Relative to untreated control SiHa cells, a total of 2 255 DEGs (±2.0-fold change; P < 0.05), including 943 upregulated and 1 312 downregulated, were obtained. Hierarchical clustering of the DEGs is presented in Figure 2a. Disease and functions analysis showed that these 2 255 DEGs were primarily associated with cancer and organismal injuries and abnormalities (Figure 2b). Kyoto encyclopaedia of genes and genomes (KEGG) pathway enrichment of these DEGs identified pathways mainly involved in cancer regulation, including p53 and cell cycle signalling pathways (Figure 2c and 2d). Within the list of DEGs, HIST1H3D was shown to be significantly downregulated in fucoxanthin-treated SiHa cells (approximately 2.5 fold down), using qPCR analysis (P = 0.007; Figure 3). Because the gene is abnormally expressed in several tumours and participates in tumorigenesis, the biological role of HIST1H3D in cervical cancer was explored in subsequent experiments.
Figure 2.
Effect of fucoxanthin treatment on SiHa cells: (a) volcano map of differentially expressed genes (DEGs); (b) gene ontology enrichment of DEGs in fucoxanthin-treated SiHa cells compared with untreated controls; (c) Kyoto encyclopaedia of genes and genomes (KEGG) pathway enrichment of DEGs in fucoxanthin-treated SiHa cells compared with controls; and (d) integrated pathway analysis (IPA) showing the top 10 networks influenced by fucoxanthin treatment.
Figure 3.
Analysis of H3 clustered histone 6 (HIST1H3D) mRNA levels using reverse transcription-quantitative polymerase chain reaction showed that HIST1H3D gene expression in SiHa cells was inhibited by fucoxanthin, with an inhibition efficiency of 62.5%. Data presented as mean ± SD; **P < 0.01 versus untreated controls.
Decreased HIST1H3D expression in cervical cancer cells transfected with shHIST1H3D
To elucidate the biological function of HIST1H3D in cervical cancer, HIST1H3D expression was silenced in HeLa and SiHa cervical cancer cells by transfection with a lentivirus delivering a shRNA payload specifically targeting human HIST1H3D (shHIST1H3D). The efficacy of this transfection was >80% (Figure 4a). To evaluate the knockdown efficacy, mRNA was isolated from HeLa and SiHa cervical cancer cells transfected with either an shHIST1H3D or shCtrl payload, and HIST1H3D mRNA levels were then determined by RT-qPCR. As shown in Figure 4b, the levels of HIST1H3D mRNA were markedly lower in the shHIST1H3D group than in the shCtrl group. Thus, shHIST1H3D was shown to significantly downregulate HIST1H3D expression in HeLa and SiHa cervical cancer cells, suggesting that HIST1H3D knockdown is specific to the target gene and its loss-of-function outcome is possibly not due to an off-target effect.
Figure 4.
H3 clustered histone 6 (HIST1H3D) mRNA levels after transfection with shHIST1H3D or shCtrl lentiviruses in human HeLa and SiHa cervical cancer cells: (a) representative photomicrographs obtained at 72 h following shHIST1H3D or shCtrl lentiviral infection; and (b) levels of HIST1H3D mRNA measured by reverse transcription-quantitative PCR. Data presented as mean ± SD; **P < 0.01 versus shCtrl group.
HIST1H3D knockdown inhibits proliferation of cervical cancer cells
To examine the effect of HIST1H3D on cell growth, HeLa and SiHa cervical cancer cells transfected with either an shHIST1H3D or control lentivirus were reseeded into 96-well plates and incubated for 5 days. Proliferation was assessed by CCK-8 assay on each day. Cells transfected with the control lentivirus greatly expanded during the 5 days of observation, while shHIST1H3D-transfected cells proliferated more slowly (P < 0.01; Figures 5a and 5b). Thus, HIST1H3D knockdown was shown to significantly inhibit the proliferation of HeLa and SiHa cells.
Figure 5.
Cell proliferation analysis using cell counting kit-8 assay, showing: (a) effect of H3 clustered histone 6 (HIST1H3D) knockdown on (a) HeLa cells; and (b) SiHa cells. Data presented as mean ± SD; **P < 0.01 versus shCtrl group.
HIST1H3D knockdown arrests cervical cancer cells at G0/G1 phase
To determine the contribution of cell cycle arrest to the observed growth inhibition by HIST1H3D silencing, the proportion of HeLa and SiHa cells at different phases of the cell cycle were analysed using flow cytometry (Figure 6a). The knockdown of HIST1H3D significantly increased the proportion of cells in the G0/G1 phase compared with the control group in both cell lines (both P < 0.05; Figures 6b and 6c). Knockdown of HIST1H3D also reduced the proportion of HeLa cells in S phase (P < 0.05; Figure 6b). Collectively, these results indicate that shHIST1H3D is able to arrest the cell cycle of HeLa and SiHa cells at G0/G1 phase, leading to slower cell growth.
Figure 6.
H3 clustered histone 6 (HIST1H3D) silencing arrested cell cycle at the G0/G1 phase in human cervical cancer cells: (a) representative flow cytometry images showing the distribution of HeLa and SiHa cells in cell cycle phases; and the proportion of (b) HeLa cells and (c) SiHa cells in each cell cycle phase, as determined by flow cytometry. Data presented as mean ± SD; *P < 0.05 versus shCtrl group.
HIST1H3D knockdown increases apoptosis in cervical cancer cells
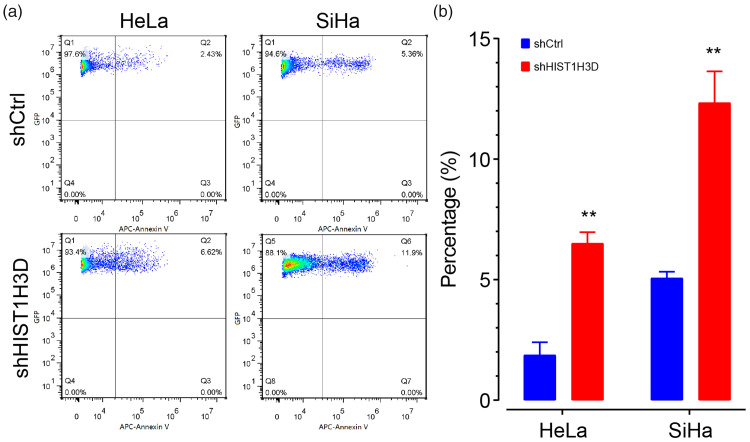
To determine if HIST1H3D inactivation induces apoptosis in cervical cell lines, HIST1H3D was knocked down in HeLa and SiHa cells, and the proportion of apoptotic cells was assessed using Annexin-V staining and flow cytometry (Figure 7a). The proportion of apoptotic cells was significantly higher in cells with knockdown HIST1H3D than controls with normal HIST1H3D expression (both cell lines P < 0.01; Figure7b). These results suggest that HIST1H3D expression is a determinant affecting apoptosis in HeLa and SiHa cells.
Figure 7.
H3 clustered histone 6 (HIST1H3D) silencing increased the proportion of apoptotic cells in human HeLa and SiHa cervical cancer cells: (A) representative flow cytometry plot showing apoptotic cells detected by Annexin V staining; and (B) quantification of flow cytometry results. Data presented as mean ± SD; **P < 0.01 versus shCtrl group.
HIST1H3D silencing restricts colony formation of cervical cancer cells
The influence of HIST1H3D silencing on the ability of HeLa and SiHa cells to form colonies was determined. There was a visible impairment of colony formation within shHIST1H3D-transfected cells compared with colony formation in control cells (Figure 8a). Quantification of colony formation showed that colony numbers in HIST1H3D-knockdown cells were significantly lower than control cells with functional HIST1H3D (P < 0.05 and P < 0.01, HeLa and SiHa cells, respectively; Figure 8b).
Figure 8.
H3 clustered histone 6 (HIST1H3D) silencing suppresses the colony formation ability of Hela and SiHa human cervical cancer cells: (A) representative photomicrographs of HeLa and SiHa cell colonies at 14 days after transfection with short hairpin (sh)HIST1H3D or shCtrl lentivirus; and (B) comparison of colony numbers between HeLa and SiHa cells. Data presented as mean ± SD, *P < 0.05 or **P < 0.01 versus shCtrl group.
Discussion
Fucoxanthin has been shown to have a wide range of antitumour biological activities,40 but its biological role in cervical cancer cells remains unclear. In the present study, DEGs from fucoxanthin-treated SiHa cells were first determined. The enrichment of KEGG signalling pathways indicated that p53 signalling is induced following fucoxanthin treatment. These results raised the hypothesis that fucoxanthin may be effective in the therapy of cervical cancer through its effects on p53 signalling, and this required further investigation.
Interestingly, HIST1H3D was found to be significantly downregulated by fucoxanthin, and studies have shown that this gene is downregulated in TNF-α-activated HUVEC endothelial cells.34 In addition, HIST1H3D may be involved in the process of cystic fibrosis.35
Histone family members play an important role in chromatin condensation and cell cycle progression.28 Histones are abnormally expressed in tumours and contribute to the pathogenesis of various malignancies.41,42 However, as a member of the histone family, there are few studies on the development of tumorigenesis involving HIST1H3D. Iwaya et al.37 found that HIST1H3D is highly expressed in advanced gastric cancer; and Rui et al.36 reported that HIST1H3D is highly expressed in lung cancer and can promote the pathogenesis of lung cancer. Its biological function in cervical cancer, however, remains unclear.
To assess the biological role of HIST1H3D in cervical cancer, the expression of HIST1H3D in human HeLa and SiHa cervical cancer cells was specifically silenced via a lentiviral system. After confirming that HIST1H3D was silenced in HeLa and SiHa cells, the proliferation and colony forming ability of cervical cancer cells was found to be decreased, while the proportion of apoptotic cells increased. These results suggest that increased expression of HIST1H3D may contribute to the development of cervical cancer.
The cell cycle is an important event in cell proliferation. To investigate the effect of HIST1H3D on proliferation of cervical cancer cells, the cell cycle was analysed in HeLa and SiHa cells using flow cytometry. HIST1H3D silencing was found to arrest cervical cancer cells in the G0/G1 phase, suggesting that HIST1H3D promotes cell proliferation and colony formation in human cervical cancer cells by regulating cell cycle progression. However, its detailed mechanism has yet to be unveiled.
The present results may be limited by several shortcomings. For example, whether the inhibition of HIST1H3D by fucoxanthin is direct or indirect requires further clarification. If the effect is direct, in what way does fucoxanthin act on HIST1H3D? In addition, what is the role of p53 signalling in its effect? All of these factors need to be confirmed by further studies.
In summary, the present study demonstrates that fucoxanthin inhibits HIST1H3D expression. Lentiviral-mediated silencing of HIST1H3D inhibits cell proliferation and colony formation, promotes apoptosis, and leads to cell cycle arrest in the G0/G1 phase in cervical cancer cells. These results suggest that HIST1H3D plays an important role in the development of cervical cancer. Further studies are required to elucidate the specific effects of fucoxanthin on HIST1H3D in cervical cancer cells.
Declaration of conflicting interest
The authors declare that there is no conflict of interest.
Funding
This research was funded by the Natural Science Research Project of Anhui Educational Committee (KJ2019A0343), and the Science Research Project of Bengbu Medical College (BYKF1861).
ORCID iD
Guoliu Ye https://orcid.org/0000-0003-4634-7942
References
- 1.Bermudez A, Bhatla N, Leung E. Cancer of the cervix uteri. Int J Gynaecol Obstet 2015; 131 Suppl 2: S88–S95. [DOI] [PubMed] [Google Scholar]
- 2.Berek JS, Matsuo K, Grubbs BH, et al. Multidisciplinary perspectives on newly revised 2018 FIGO staging of cancer of the cervix uteri. J Gynecol Oncol 2019; 30: e40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Skinner SR, Apter D, De Carvalho N, et al. Human papillomavirus (HPV)-16/18 AS04-adjuvanted vaccine for the prevention of cervical cancer and HPV-related diseases. Expert Rev Vaccines 2016; 15: 367–387. [DOI] [PubMed] [Google Scholar]
- 4.Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018; 68: 394–424. [DOI] [PubMed] [Google Scholar]
- 5.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin 2019; 69: 7–34. [DOI] [PubMed] [Google Scholar]
- 6.Tomkins A, White C, Higgins SP. Primary herpes simplex virus infection mimicking cervical cancer. BMJ Case Rep 2015; 2015: bcr2015210194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cao S, Gan Y, Dong X, et al. Herpes simplex virus type 2 and the risk of cervical cancer: a meta-analysis of observational studies. Arch Gynecol Obstet 2014; 290: 1059–1066. [DOI] [PubMed] [Google Scholar]
- 8.Veijalainen O, Kares S, Kujala P, et al. Implementation of HPV-based cervical cancer screening in an organised regional screening programme: 3 years of experience. Cytopathology 2019; 30: 150–156. [DOI] [PubMed] [Google Scholar]
- 9.Barra F, Leone Roberti Maggiore U, Bogani G, et al. New prophylactics human papilloma virus (HPV) vaccines against cervical cancer. J Obstet Gynaecol 2019; 39: 1–10. [DOI] [PubMed] [Google Scholar]
- 10.Hoover DS, Spears CA, Vidrine DJ, et al. Smoking cessation treatment needs of low SES cervical cancer survivors. Am J Health Behav 2019; 43: 606–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sugawara Y, Tsuji I, Mizoue T, et al. Cigarette smoking and cervical cancer risk: an evaluation based on a systematic review and meta-analysis among Japanese women. Jpn J Clin Oncol 2019; 49: 77–86. [DOI] [PubMed] [Google Scholar]
- 12.Mignot S, Ringa V, Vigoureux S, et al. Pap tests for cervical cancer screening test and contraception: analysis of data from the CONSTANCES cohort study. BMC Cancer 2019; 19: 317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Niu F, Wang T, Li J, et al. The impact of genetic variants in IL1R2 on cervical cancer risk among Uygur females from China: a case–control study. Mol Genet Genomic Med 2019; 7: e00516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Du GH, Wang JK, Richards JR, et al. Genetic polymorphisms in tumor necrosis factor alpha and interleukin-10 are associated with an increased risk of cervical cancer. Int Immunopharmacol 2019; 66: 154–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Babapour N, Mehramiz M, Rastgar Moghadam A, et al. Association of TNF-308 G>A polymorphism located in tumor necrosis factor a with the risk of developing cervical cancer and results of pap smear. J Cell Biochem 2019; 120: 5444–5448. [DOI] [PubMed] [Google Scholar]
- 16.Michelow P, Firnhaber C. HPV vaccination in Southern Africa: a jab of hope in the fight against cervical cancer. Cancer Cytopathol 2016; 124: 695–698. [DOI] [PubMed] [Google Scholar]
- 17.Saleem A, Tristram A, Fiander A, et al. Prophylactic HPV vaccination: a major breakthrough in the fight against cervical cancer? Minerva Med 2009; 100: 503–523. [PubMed] [Google Scholar]
- 18.Kessler TA. Cervical cancer: prevention and early detection. Semin Oncol Nurs 2017; 33: 172–183. [DOI] [PubMed] [Google Scholar]
- 19.Carvallo-Michelena A, Rojas-Dominguez JL, Piscoya A. Early prevention and screening of cervical cancer in a developing country. Am J Prev Med 2015; 48: e1. [DOI] [PubMed] [Google Scholar]
- 20.Foran C, Brennan A. Prevention and early detection of cervical cancer in the UK. Br J Nurs 2015; 24: S22–24, S26, S28–29. [DOI] [PubMed] [Google Scholar]
- 21.D'Orazio N, Gemello E, Gammone MA, et al. Fucoxantin: a treasure from the sea. Mar Drugs 2012; 10: 604–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jung HA, Ali MY, Choi RJ, et al. Kinetics and molecular docking studies of fucosterol and fucoxanthin, BACE1 inhibitors from brown algae Undaria pinnatifida and Ecklonia stolonifera. Food Chem Toxicol 2016; 89: 104–111. [DOI] [PubMed] [Google Scholar]
- 23.Ganesan P, Matsubara K, Sugawara T, et al. Marine algal carotenoids inhibit angiogenesis by down-regulating FGF-2-mediated intracellular signals in vascular endothelial cells. Mol Cell Biochem 2013; 380: 1–9. [DOI] [PubMed] [Google Scholar]
- 24.Kumar SR, Hosokawa M, Miyashita K. Fucoxanthin: a marine carotenoid exerting anti-cancer effects by affecting multiple mechanisms. Mar Drugs 2013; 11: 5130–5147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang J, Ma Y, Yang J, et al. Fucoxanthin inhibits tumour-related lymphangiogenesis and growth of breast cancer. J Cell Mol Med 2019; 23: 2219–2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Turner BM. Cellular memory and the histone code. Cell 2002; 111: 285–291. [DOI] [PubMed] [Google Scholar]
- 27.Berger SL. Histone modifications in transcriptional regulation. Curr Opin Genet Dev 2002; 12: 142–148. [DOI] [PubMed] [Google Scholar]
- 28.Jenuwein T, Allis CD. Translating the histone code. Science 2001; 293: 1074–1080. [DOI] [PubMed] [Google Scholar]
- 29.Cruft HJ, Mauritzen CM, Stedman E. Abnormal properties of histones from malignant cells. Nature 1954; 174: 580–585. [DOI] [PubMed] [Google Scholar]
- 30.Rheinbay E, Louis DN, Bernstein BE, et al. A tell-tail sign of chromatin: histone mutations drive pediatric glioblastoma. Cancer cell 2012; 21: 329–331. [DOI] [PubMed] [Google Scholar]
- 31.Dryhurst D, Ausio J. Histone H2A.Z deregulation in prostate cancer. Cause or effect? Cancer Metastasis Rev 2014; 33: 429–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Albig W, Kioschis P, Poustka A, et al. Human histone gene organization: nonregular arrangement within a large cluster. Genomics 1997; 40: 314–322. [DOI] [PubMed] [Google Scholar]
- 33.Peters AH, Schubeler D. Methylation of histones: playing memory with DNA. Curr Opin Cell Biol 2005; 17: 230–238. [DOI] [PubMed] [Google Scholar]
- 34.Knyazev EN, Mal'tseva DV, Zakharyants AA, et al. Expression of microRNA genes MIR221, MIR222, and MIR181B1 increases during induction of inflammation in the endothelial barrier model. Bull Exp Biol Med 2018; 164: 749–752. [DOI] [PubMed] [Google Scholar]
- 35.Fesen K, Silveyra P, Fuentes N, et al. The role of microRNAs in chronic pseudomonas lung infection in cystic fibrosis. Respir Med 2019; 151: 133–138. [DOI] [PubMed] [Google Scholar]
- 36.Rui Y, Peng WJ, Wang M, et al. HIST1H3D: a promising therapeutic target for lung cancer. Int J Oncol 2017; 50: 815–822. [DOI] [PubMed] [Google Scholar]
- 37.Iwaya T, Fukagawa T, Suzuki Y, et al. Contrasting expression patterns of histone mRNA and microRNA 760 in patients with gastric cancer. Clin Cancer Res 2013; 19: 6438–6449. [DOI] [PubMed] [Google Scholar]
- 38.Zhang F, Drabier R. IPAD: the integrated pathway analysis database for systematic enrichment analysis. BMC Bioinformatics 2012; 13 Suppl 15: S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lois C, Hong EJ, Pease S, et al. Germline transmission and tissue-specific expression of transgenes delivered by lentiviral vectors. Science 2002; 295: 868–872. [DOI] [PubMed] [Google Scholar]
- 40.Satomi Y. Antitumor and cancer-preventative function of fucoxanthin: a marine carotenoid. Anticancer Res 2017; 37: 1557–1562. [DOI] [PubMed] [Google Scholar]
- 41.Esteller M. Cancer epigenomics: DNA methylomes and histone-modification maps. Nat Rev Genet 2007; 8: 286–298. [DOI] [PubMed] [Google Scholar]
- 42.Berdasco M, Esteller M. Aberrant epigenetic landscape in cancer: how cellular identity goes awry. Dev Cell 2010; 19: 698–711. [DOI] [PubMed] [Google Scholar]
- 43.Nahand JS, Taghizadeh‐Boroujeni S, Karimzadeh M, et al. microRNAs: new prognostic, diagnostic, and therapeutic biomarkers in cervical cancer. J Cell Physiol 2019; 234: 17064–17099. [DOI] [PubMed] [Google Scholar]
- 44.Shafabakhsh R, Reiter RJ, Mirzaei H, et al. Melatonin: a new inhibitor agent for cervical cancer treatment. J Cell Physiol 2019; 234: 21670–21682. [DOI] [PubMed] [Google Scholar]
- 45.Ghasemi F, Shafieeb M, Banikazemic Z, et al. Curcumin inhibits NF-kB and Wnt/β-catenin pathways in cervical cancer cells. Pathol Res Pract 2019; 215: 152556. [DOI] [PubMed] [Google Scholar]
- 46.Sadri Nahand J, Moghoofe M, Salmaninejad A, et al. Pathogenic role of exosomes and microRNAs in HPV-mediated inflammation and cervical cancer: a review. Int J Cancer 2020; 146: 305–320. [DOI] [PMC free article] [PubMed] [Google Scholar]