Abstract
Background
RNA splicing defects are emerging molecular hallmarks of cancer. The gene encoding splicing factor RNA binding motif protein 10 (RBM10) has been found frequently mutated in various types of cancer, particularly lung adenocarcinoma (LUAD), but how RBM10 affects cancer pathogenesis remains to be determined. Moreover, the functional roles and clinical significance of RBM10 mutation-associated splicing events in LUAD are largely unknown.
Methods
RBM10 mutations and their functional impacts were examined in LUAD patients from a Chinese patient cohort and The Cancer Genome Atlas (TCGA). Alternative splicing (AS) changes induced by RBM10 mutations in LUAD were identified by RNA sequencing and correlated with patient survival. Functions of RBM10 and the splice variants of eukaryotic translation initiation factor 4H containing or lacking exon 5 (EIF4H-L and EIF4H-S respectively) in LUAD development and progression were examined by cellular phenotypic assays and xenograft tumour formation.
Findings
RBM10 mutations in LUAD generally lead to loss-of-function and cause extensive alterations in splicing events that can serve as prognostic predictors. RBM10 suppresses LUADprogression largely by regulating alternative splicing of EIF4H exon 5. Loss of RBM10 in LUAD enhances the expression of EIF4H-L in LUAD. EIF4H-L, but not EIF4H-S, is critical for LUAD cell proliferation, survival and tumourigenesis.
Interpretation
Our study demonstrates a new molecular mechanism underlying RBM10 suppressive functions in lung cancer and the therapeutic value of RBM10-regulated AS events, providing important mechanistic and translational insights into splicing defects in cancer.
Keywords: RBM10, Lung cancer, Alternative splicing, EIF4H
Research in context.
Evidence before this study
Aberrant RNA splicing plays significant roles in tumour development and progression and can be used as promising therapeutic targets. The RNA binding motif protein 10 (RBM10) encodes an important regulator of RNA splicing. RBM10 mutations frequently occur in lung adenocarcinoma (LUAD), the most common subtype of lung cancer. Previous studies reported that RBM10 mutations associated with splicing changes in LUAD. However, the functions, molecular mechanisms and clinical significance of RBM10-regulated splicing events in LUAD pathogenesis remains largely unexplored. In addition, inconsistent functions of RBM10 in LUAD have been proposed.
Added value of this study
We systematically identified and confirmed RNA splicing changes induced by RBM10 mutations in LUAD. Moreover, we demonstrated that RBM10 suppresses LUAD progression by inhibiting alternative splicing of an important translation initiation regulator EIF4H. Importantly, EIF4H splice variant containing exon 5 (EIF4H-L) is significantly upregulated in RBM10 deficient LUAD, critical for LUAD cell proliferation and tumourigenesis, and can be used as a new therapeutic target.
Implications of the all the available evidence
Our study demonstrates that RBM10 exerts suppressive functions in LUAD by controlling alternative splicing of the mRNA translation regulator EIF4H and that RBM10-regulated splicing events can serve as new prognostic markers and therapeutic targets for lung cancer, providing critical insights into cancer-associated splicing aberrations.
Alt-text: Unlabelled box
1. Introduction
Lung cancer is the leading cause of cancer-related death worldwide [1], with lung adenocarcinoma (LUAD) being the most common histological subtype [2]. During the past two decades, important advancements have been achieved in characterizing mutational spectrum and molecular subtypes of LUAD, which provide grounds for targeted therapies that dramatically improved clinical outcomes for LUAD patients [3, 4]. However, a large proportion of LUAD patients lack clinically actionable oncogenic mutations, and for those who received targeted therapies, responses are generally incomplete or temporary [2, 3]. For these reasons, identifying new driver genes and elucidating their functional roles in LUAD development and progression are pressing issues for better management of this disease.
The majority (>90%) of human genes undergo alternative splicing (AS) to produce multiple variants with distinct functions [5]. Dysregulation of AS is one of the major causes of human diseases including cancer [6, 7]. A dynamically assembled complex comprising of >100 core spliceosomal proteins and five snRNAs, spliceosome, catalyzes the step-wise splicing reactions [8, 9]. In addition, various auxiliary splicing factors specifically recognize regulatory cis-elements in pre-mRNAs to regulate AS by mainly affecting spliceosomal assembly [5, 8].
Mutations or aberrant expression of splicing factors and abnormal splicing of cancer-associated genes are frequently observed in cancer patients [10], [11], [12], [13]. The resulting splicing defects are considered to be key contributors to carcinogenesis and may serve as promising prognostic markers and/or therapeutic targets for various types of cancer [12], [13], [14], [15]. To date, splicing defects are best characterized in hematopoietic malignancies where the core spliceosomal genes splicing factor 3b subunit 1 (SF3B1), U2 small nuclear RNA auxiliary factor 1 (U2AF1) and serine and arginine rich splicing factor 2 (SRSF2) are frequently mutated and act as oncogenic drivers [16]. In addition, auxiliary splicing factors were also found to be mutated or aberrantly expressed in cancer and contribute to carcinogenesis [17], [18], [19]. However, the functional roles and the underlying molecular mechanisms of splicing factors mutated or deregulated in cancer, particularly in solid tumours, remain largely elusive.
RNA binding motif protein 10 (RBM10) is located on the X chromosome (Xp11.3) and encodes an RNA binding protein. Previous studies revealed that RBM10 functions as an auxiliary splicing factor to promote exon skipping by binding to adjacent intronic regions [20], [21], [22]. RBM10 germline mutations are the cause of TARP syndrome (OMIM #311900), a severe developmental disorder inherited in an X-linked recessive manner [23]. Somatic mutations of RBM10 frequently occur in several types of solid tumours, including LUAD, colorectal carcinomas [24], pancreatic ductal adenocarcinoma [25] and bladder cancer [26]. The RBM10 mutation frequency is ~8% in The Cancer Genome Atlas (TCGA) LUAD patients [27, 28], but is less well-defined in other populations with distinct oncogenic mutational spectrums [29]. Notably, RBM10 mutations have been shown to be enriched [30] and clonal in early stage LUADs [31], suggesting a role in driving the development of LUAD. The analyses of LUAD samples in TCGA indicate that RBM10 mutations may contribute to LUAD progression [32] and associate with a number of splicing changes [33]. Previous functional studies using lung cancer cell lines showed contradictory roles of RBM10 in LUAD progression [34], [35], [36], [37]. Therefore, the exact function and underlying molecular mechanism of RBM10 in lung cancer remain to be determined. Moreover, the function, regulation and clinical significance of RBM10 mutation-associated splicing events are largely unclear.
In this study, we demonstrate that RBM10 suppresses LUAD by regulating alternative splicing of the eukaryotic translation initiation factor 4H (EIF4H), and that RBM10-regulated splicing events can serve as new prognostic and therapeutic targets.
2. Materials and methods
2.1. RBM10 mutation detection and immunohistochemistry analysis
RBM10 mutations were detected in LUAD tissues by amplifying RBM10 exons and their flanking intronic regions from genomic DNA using a set of PCR primers (Supplementary Table S6). The amplified products were then purified and sent for Sanger sequencing (BGI genomics). Mutations were identified by manual examination of the sequencing chromatograms and comparison with reference sequences using VectorNT1 software. Candidate mutations were confirmed by sequencing the corresponding PCR products from the opposite direction.
Tumour blocks were cut into 4 µm-thick sections. All sections were stained with hematoxylin-eosin. At least two sections from different areas of each tumour sample were examined. Sections and slides were prepared by pathologists in Department of Pathology at the Fudan University Shanghai Cancer centre (FUSCC). Slides were treated with 3% hydrogen peroxide for 15 min at room temperature to block endogenous peroxidase activity. Antigen retrieval was performed using sodium citrate buffer (10 mM, pH = 6.0). Primary antibody against RBM10 (1:150, cat No. HPA034972, Sigma) was applied and incubated for 1 h at room temperature. The DAB Envision Kit (Gene Tech) was used to detect the immunoreactivity according to the manufacturer's manual. For RBM10 staining, score was graded on the basis of intensity and percentage of immunoreactive tumour cells as following: 3+, strong staining intensity in more than 10% of tumour cells; 2+, moderate staining intensity in more than 10% of tumour cells; 1+, faint or weak staining intensity in more than 10% of tumour cells; and 0, no staining or any staining in less than 10% of tumour cells. Tumours with a score 2 or 3 were classified as moderate or high RBM10 expression and RBM10 positive, whereas those with a score 0 or 1 were classified as no or low RBM10 expression and RBM10 negative, respectively. The scores were independently reviewed by two experienced pathologists at the FUSCC. For a few cases with inconsistent results, the two pathologists discussed with each other to reach an agreement.
2.2. Cell culture
Cell lines were purchased from and authenticated by COBIOER or the Cell Bank of Chinese Academy of Sciences. A549, PC9, H1975 and H1944 (human lung adenocarcinoma cell lines) were cultured in 1640 medium supplemented with 10% FBS and 6 mM l-glutamine. BEAS-2B (human bronchial epithelium cell line) and HEK293 cells were cultured in DMEM (Invitrogen) supplemented with 10% FBS. 293FT cells were cultured in DMEM medium containing 10% FBS, 6 mM l-glutamine, 1 mM sodium pyruvate (Sigma-Aldrich) and 0.1 mM non-essential amino acids (NEAA, Invitrogen). All cultures were maintained under standard culture conditions (37 °C, 5% CO2). Cells were regularly checked for absence of mycoplasma infection by using PCR with primers targeting mycoplasma.
Stable cell lines inducibly expressing RBM10 were obtained using the Lenti-X Tet-On Advanced Inducible Expression System (Clontech) as previously described [38]. Stably transfected cells were maintained in normal culture medium and FLAG-RBM10 expression was induced by exposing the cells to 100 or 1000 ng/ml doxycycline (Dox, Sigma-Aldrich). Other stable cell lines were obtained by infecting with lentiviral particles and selecting the infected cells with 2 µg/mL puromycin.
2.3. Construct
A tet-on lentiviral plasmid encoding FLAG-RBM10 (Tet-FLAG-RBM10) was constructed by subcloning pFRT-TO-RBM10 [21] into NheI and EcoRI linearized pLVX-Tight-Puro (Clontech) using the ClonExpress II One Step Cloning Kit (Vazyme). RBM10-EGFP mutants were constructed using Mut Express II Fast Mutagenesis Kit V2 (Vazyme) as described previously [32].
Lentiviral plasmids encoding wild type (WT) or mutant KRAS or EGFR were constructed by inserting corresponding coding sequence into NotI and BamHI linearized pLVX-IRES-Neo vector (Clontech) using ClonExpress II One Step Cloning Kit (Vazyme). Wild type KRAS and EGFR coding sequences were amplified using cDNA template derived from BEAS-2B cells. KRASG12D and EGFRE19del coding sequences were amplified using cDNA template derived from A549 and PC9 cells, respectively. Vector expressing EGFRL858R was derived from WT EGFR plasmid using Mut Express II Fast Mutagenesis Kit V2 (Vazyme). Empty pLVX-IRES-Neo vector was used as a control.
EIF4H-L/S constructs were generated by cloning the coding region of EIF4H isoform 1 (NM_022170.1) or isoform 2 (NM_031992.1) into pLVX-IRES-Neo plasmid using ClonExpress II One Step Cloning Kit (Vazyme). The EIF4H-E5M minigene reporter was generated by amplifying genomic segments including the EIF4H exon 4, intron 4, exon 5, intron 5 (truncated) and exon 6 (chr7:73,604,152–73,605,652; 73,608,505–73,609,208) and ligating into pcDNA3.1 plasmid using ClonExpress II One Step Cloning Kit (Vazyme).
Lentiviral vectors expressing small hairpin RNAs (shRNA) were constructed by ligating annealed oligos into AgeI and EcoRI linearized pLKO vector (Clontech) using T4 DNA ligase. Sequences of primers and oligos are listed in Supplementary Table S6.
2.4. Cellular phenotypic assays
Cellular phenotypic assays were conducted as previously described [18, 39]. Briefly, cell proliferation rates were measured by determining viable cell numbers within continuous days using Cell Counting Kit-8 (Dojindo Laboratories) according to the manufacturer's instruction. Oncogenic capacities of cells were estimated by anchorage-dependant and -independent colony formation assays. In vitro cell migration and invasion were determined by Trans-well (Falcon, BD) assays. Cell cycle was assessed by propidium iodine (PI) staining and subsequent Flow Cytometer (BD Accuri C6) analysis. Apoptotic cells were detected by using Annexin V-FITC Apoptosis Detection Kit (Vazyme) and Flow Cytometer.
2.5. Xenograft tumour formation
5 × 106 cells resuspended in PBS were injected subcutaneously into lower flanks of BALB/c-nu mice. RBM10 overexpression in PC9 cells was induced by feeding the mice with drinking water containing 2 mg/mL doxycycline (Dox, Sigma-Aldrich). H1944 cells were collected 48 h after transfection with 20 nM siRNAs targeting the EIF4H long splice variant (siEIF4H-L) and non-targeting control (siCtrl) respectively and injected into nude mice. Viability difference between siEIF4H-L- and siCtrl-treated cells was not observed until 72 h after siRNA transfection in cell culture plate. Tumour volume (V) was calculated using the equation V (mm3) = a × b2/2, where a is the largest diameter and b is the smallest diameter. Tumours were dissected at the end of experiments and used for RNA and protein analyses.
2.6. RNA extraction and RT-PCR
Total RNA extraction and reverse transcription followed by PCR (RT-PCR) or quantitative PCR (RT-qPCR) were performed as previously described [38]. Briefly, total RNAs from cells were extracted by TRIzol reagent (Invitrogen), and tissue samples were first homogenized and total RNAs were then extracted by TRIzol (Invitrogen) or DNA/RNA/protein extraction kit (TIANGEN). Total RNAs after genomic DNA removal with TURBO DNA-free Kit (Thermo Fisher Scientific) were reverse transcribed using HiScript II Reverse Transcriptase (Vazyme). Splicing outcomes were determined by estimating percent-spliced-in (PSI) values based on band intensities of RT-PCR products as described previously [38].
2.7. Western blot
Protein levels were detected by Western blot analysis as previously described(38). The following primary antibodies were used: RBM10 (cat No. HPA034972, 1:10,000, Sigma), KRAS (cat No.sc-30,1:200, Santa Cruz Biotechnologies Inc.), EGFR (cat No.D38B1,1:1000,Cell signalling Technology), β-actin (cat No.66009–1-Ig,1:10,000,Proteintech), cleaved caspase 3 (cat No. 9661, Asp175, 1:500, Cell signalling Technology), cleaved PARP (cat No.D64E10,Asp214,1:500, Cell signalling Technology), EIF4H (cat No. 3469,D85F2,1:1000, Cell signalling Technology), c-Myc (cat No.ab32072,1:5000,abcam), cyclin D1 (cat No.ab134175,1:1000,abcam). The following secondary antibodies were used: Goat anti-rabbit IgG HRP (cat. No. 458, 1:5000, MBL), Goat anti-mouse IgG HRP (cat No.M21001,1:5000, Abmart).
2.8. siRNA-mediated silencing
We used small interfering RNAs (siRNAs) to knockdown RBM10, and selectively knockdown the long or short splice variant of EIF4H and both variants. Cells were transfected with siRNA oligos at final concentration of 20 nM using Lipofectamine RNAiMAX (Thermo Fisher Scientific) according to the manufacturer's instructions. The cells were harvested for RNA and protein analyses as well as phenotypic assays 48 h after transfection.
2.9. Antisense oligonucleotide (ASO)-mediated splicing blockade
ASOs were synthesized with a full-length phosphorothioate backbone and 2′-O-methyl modified ribose molecules (GenScript Biotech Corp). ASO was transfected at final concentration of 100 nM using Lipofectamine RNAiMAX (Thermo Fisher Scientific) according to the manufacturer's instructions. The cells were harvested for RNA and protein analyses as well as phenotypic assays 48 h after transfection.
2.10. Minigene splicing reporter assays
To study EIF4H exon 5 splicing following RBM10 overexpression or knockdown, HEK293 cells were co-transfected with minigene splicing reporters and the pcDNA3.1-RBM10 expression plasmid or siRNA oligos targeting the RBM10 coding sequence (siRBM10), respectively. The splicing outcomes were determined as previously described [38].
2.11. mRNA-Seq and data analysis
RNA integrity was assessed using RNA Nano 6000 Kit and the Agilent 2100 Bioanalyzer (Agilent). 500 ng total RNA with Bioanalyzer RNA integrity number > 7.0 for each sample was used for mRNA-Seq library preparation. The sequencing library was prepared using Stranded mRNA-seq Library Prep Kit (Vazyme) following the manufacturer's manual. The prepared libraries were quantified by Qubit spectrometry (Life Technologies), and assessed by DNA 1000 Kit and the Agilent 2100 Bioanalyzer (Agilent). The assessed libraries labelled with different sequence indexes were pooled at equal molar and subsequently sequenced on Illumina X-ten using standard protocol.
The RNA-seq reads were mapped to the human genome reference (UCSC genome browser hg19) using mapsplice [40] with default parameters. Differential splicing events were identified using the MISO pipeline [41]. To complement for the MISO method, we also estimated the PSI (Percent-Spliced-In) value for each AS event based on the number of splice junction reads. MISO method cutoffs: delta PSI > 0.1 or <−0.1 and Bayes factor > 5 between compared groups; PSI method cutoffs: delta PSI > 0.1 or < −0.1 and P value < 0.05 (Student's t-test). The final results are presented as the combination of the two methods. In case splicing changes were identified by both methods, we reported the PSI changes obtained from MISO. For TCGA level 3 data analysis, splicing changes were obtained using the PSI method. Gene expression levels were estimated using RSEM [42]. Differentially expressed genes were identified using R package ‘DESeq2’ with the cutoff: |log2FoldChange| > 1 and P value < 0.05. Gene ontology (GO) analysis was performed using the DAVID online tool (http://david.abcc.ncifcrf.gov/) [43, 44].
2.12. CLIP-PCR and PAR-CLIP data analysis
CLIP-PCR experiment was performed as previously described [38] to confirm the interaction between RBM10 and its binding regions in EIF4H pre-mRNA.
The PAR-CLIP data for RBM10 was downloaded from GEO database [21]. The sequencing reads were trimmed with fastx clipper to cut the adaptor sequence before alignment. The adaptor-trimmed reads were aligned to the human genome (UCSC genome browser hg19) using bowtie [45] with the parameter -v 1 -m 10. The aligned reads were processed with the PARalyzer [46] to call peaks and define RBM10 binding site. Putative RBM10 motifs within the strong binding sites in EIF4H were predicted according to motifs identified by our and other studies [20, 21, 47, 48]
2.13. Primer and oligonucleotide
All primer and oligonucleotide sequences are listed in Supplementary Table S6.
2.14. Statistics and plots
For all experimental results, statistical tests were carried out using GraphPad Prism 6 (GraphPad software). For comparisons between sample pairs, two-tailed Student's t-tests were used. For experiments with more than two conditions, one-way or two-way ANOVA followed by multiple comparisons tests were used.
For all results obtained from data analysis, plots were generated and the related statistics were performed using R version 3.4.1. The co-mutation plot for LUAD patients was generated by R package ‘GenVisR’. Differential gene expression analysis was conducted using ‘DDESeq2’. Principal component analysis (PCA) was performed and the plot were generated using R package ‘mixOmics’. Survival analysis was conducted using the R package ‘survival’ and ‘survminer’. A Cox regression was used to estimate the P value and the regression coefficients, and the risk score was estimated using multivariable cox analysis for every patient. Other plots were generated using R package ‘ggplot2’.
2.15. BioID assay
BioID assays were performed as previously described [49]. Briefly, pcDNA3.1-BirA-HA plasmid or pcDNA3.1-EIF4H-L/S-BirA-HA plasmids were transiently transfected into 5 × 106 HEK293T cells in 15 cm dishes with Lipofectamine 3000 reagent (Thermo Fisher Scientific). Then cells were incubated in complete media supplemented with 50 μM biotin (Sigma-Aldrich, cat No. B4639) for 36 h. Subsequently, cells were collected and lysed in 1 ml lysis buffer (50 mM Tris–HCl, pH = 7.4, 150 mM NaCl, 1% Triton X-100, 1% deoxycholate, and 0.1% SDS) with 1 × Complete protease inhibitor cocktail (Roche). After sonication and centrifugation, supernatants were incubated with 400 μL Dynabeads (M-280 Streptavidin, Invitrogen) at 4 °C overnight. The beads were collected and washed in very stringent wash buffer, and then kept in 50 mM NH4HCO3 for mass spectrometry analysis. For the assay of biotin labelling and pulldown efficiency, 5% of the beads were taken out and assessed by western blot with Streptavidin-HRP antibody (CST, cat No. 3999). Another 5% of beads were used for SDS-PAGE and silver staining analysis.
The remaining beads were eluted with 50 µL denature buffer (8 M urea in 100 mM Tris–HCl, pH 8.5) and sonicated for 30 min. The eluted proteins were analysed by nanoACQUITY UPLC (Waters) and mass spectrometry (MS) at MS platform of National centre for Protein Science Shanghai using standard procedures. To identify candidate interacting proteins of eIF4H-L/S, the acquired MS/MS data were compared to the UniProt database (released on Oct 22, 2015) using Integrated Proteomics Pipeline (IP2, http://integratedproteomics.com/). A decoy database containing the reversed sequences of all the proteins was appended to the target database to accurately estimate peptide probabilities and false discovery rate (FDR), and FDR threshold was set at 0.01. We quantified each protein using the spectrum counts obtained from MS and calculated the fold change of spectrum counts for EIF4H-L/S-BirA vs BirA ((EIF4H-L/S-BirA+0.0001)/(BirA+0.0001)) respectively. We selected proteins with fold change > 1.5 as candidates.
To confirm the interaction between eIF4H-L and eIF4E2, we co-transfected plasmids expressing Flag-eIF4E2 and eIF4H-L/S-HA in HEK293 cells in 10 cm culture dish using Lipofectamine 3000 (Thermo Fisher Scientific). Cells were harvested 48 h after transfection, and lysed by 1 mL coIP lysis buffer (50 mM Tris–HCl, pH=7.4, 150 mM NaCl, 1 mM EDTA, 0.5% NP40, protease inhibitor (bimake, cat No. B14001)) on ice for 30mins, followed by 12,000 g centrifugation at 4 °C for 20mins. After centrifugation, the supernatant was collected and cell debris was discarded. 10% of the lysate was saved as input for Western blot analysis. The remaining supernatant was mixed with 100 µL anti-Flag conjugated beads (Smart-Lifesciences, cat No. SM00901) or anti-IgG affinity beads (Smart-Lifesciences, cat No. SA068C), rotated overnight at 4 °C. The beads were then washed by 1xTBS buffer (50 mM Tris–HCl, pH=7.4, 150 mM NaCl) three times after removing the supernatant. The washed beads were re-suspended in 50 µL 1xSDS-PAGE sample buffer and boiled at 100 °C for 5 mins. 10µL protein sample was used for Western blot analysis.
2.16. Ethics statement
Patient tissue samples. Human lung adenocarcinoma (LUAD) specimens were obtained with the approval by the institutional review board of Shanghai Cancer Hospital, Fudan University, Shanghai, China. All patients underwent surgery and provided informed consent. Samples were snap-frozen in liquid nitrogen at the time of resection and stored at −80 °C until use. All cases were re-reviewed by pathologists for confirmation of tumour histology and tumour content.
Mice studies. All the animal experiments were performed in compliance with the NIH Guide for the Care and Use of Laboratory Animals (National Academies Press, 2011) and were approved by the Animal Ethics committee of School of Basic Medical Sciences at Fudan University.
2.17. Data availability
All RNA sequencing raw data has been deposited in NODE (http://www.biosino.org/node) and can be accessed via the accession number OEP001100.
2.18. Role of funding source
Funders provide financial support for this study, and do not participate in study design, data collection, data analyses, interpretation, or writing of report.
3. Results
3.1. RBM10 loss-of-function mutations frequently occur in LUAD patients
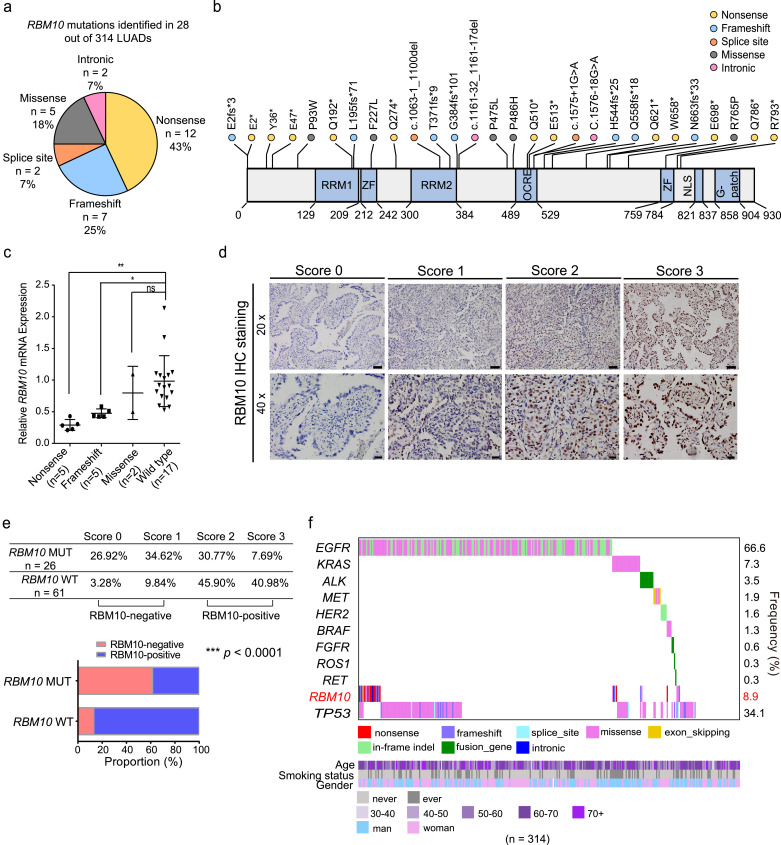
To further define RBM10 mutations in LUAD and determine their pathological roles and clinical relevance, we characterized the RBM10 mutational spectrum in a cohort of Chinese LUAD patients (Supplementary Fig. S1A, B and Table S1). The mutations of known driver genes, including EGFR, KRAS, ALK, MET, HER2, BRAF, FGFR, ROS1, RET1 and TP53, have been previously determined in these patients [29,50,51]. We found that approximately 8.9% (28 out of 314) LUAD patients harbour RBM10 mutations, which locate across the coding sequence of RBM10 and the majority (75%) are protein-truncating mutations, including nonsense, frameshift or splice site mutations (Fig. 1a, b and Supplementary Table S2). This RBM10 mutational spectrum is similar to that observed in LUAD patients from The Cancer Genome Atlas (TCGA, 7.3% (41 out of 561) with RBM10 mutations) [28,32] and in canonical tumour suppressor genes [52]. These protein-truncating mutations in RBM10 are predicted to introduce premature termination codon (PTC) and subsequently trigger nonsense-mediated mRNA decay (NMD), resulting in the decreased expression of RBM10 and loss-of-function. Consistently, we found that representative protein-truncating mutations disrupted RBM10 protein expression and led to loss-of-function in modulating splicing of target genes (Supplementary Fig. S1C, D). In addition, both RBM10 mRNA and protein levels in LUAD tissues harbouring RBM10 mutations were significantly lower compared to RBM10 wild type LUAD, as judged by qPCR and immunohistochemistry analysis respectively (Fig. 1c–e).
Fig. 1.
Characterization of RBM10 mutations in lung adenocarcinoma (LUAD). (a) Categories of RBM10 mutations identified in a cohort of Chinese LUAD patients. (b) Locations of mutations within RBM10 protein. RRM: RNA recognition motif; ZF: zinc finger; OCRE, Octamer Repeat; NLS: nuclear localization signal; G-patch: Glycine-patch. (c) RT-qPCR analysis of relative RBM10 RNA levels in LUAD tissue samples harbouring RBM10 nonsense, frameshift or missense mutations or lacking RBM10 mutations (wild type) compared to their adjacent non-tumour tissues. * P < 0.05, ** P < 0.01, ns: not significant, one-way ANOVA followed by Dunnett's tests for comparisons with wild type. (d) Representative images of RBM10 immunohistochemistry analysis of LUAD tissue samples with mutated or wild type RBM10 (RBM10 MUT and RBM10 WT). Scale bar: 50 µm for 20 × magnification, and 20 µm for 40 × magnification. Relative RBM10 staining intensities from low to high were scored 0, 1, 2, and 3. (e) Summary and quantification of immunohistochemistry results in (d). *** P < 0.0001, Fisher's exact tests. (f) Co-mutation plot for RBM10 and known oncogenic mutations identified in the Chinese LUAD patient cohort, with their smoking status, age and gender shown at the bottom. Known oncogenic mutations had previously been identified in these patients [29,50,51].
Co-mutation analysis revealed that 93% (26/28) and 78% (32/41) RBM10 mutations co-occurred with known oncogene mutations, mostly with EGFR and KRAS mutations, in the Chinese and TCGA LUAD cohorts respectively (Fig. 1f and Supplementary Fig. S1E). These observations suggest that RBM10 mutations contribute to pathogenesis of LUAD patients with distinct genetic backgrounds. On the other hand, there are 7% (2/28) and 22% (9/41) LUAD patients in the Chinese and TCGA LUAD cohorts harbouring RBM10 mutations but no known oncogene mutations, indicating that the RBM10 mutation may function as oncogenic driver in LUAD development (Fig. 1f and Supplementary Fig. S1E). In support of the functional role of RBM10 loss, survival analysis using TCGA data revealed that LUAD patients with low RBM10 expression level correlated with reduced survival rates (Supplementary Fig. S1F), indicating a critical role of RBM10 in LUAD progression.
3.2. RBM10 mutations lead to extensive splicing alterations in LUAD patients
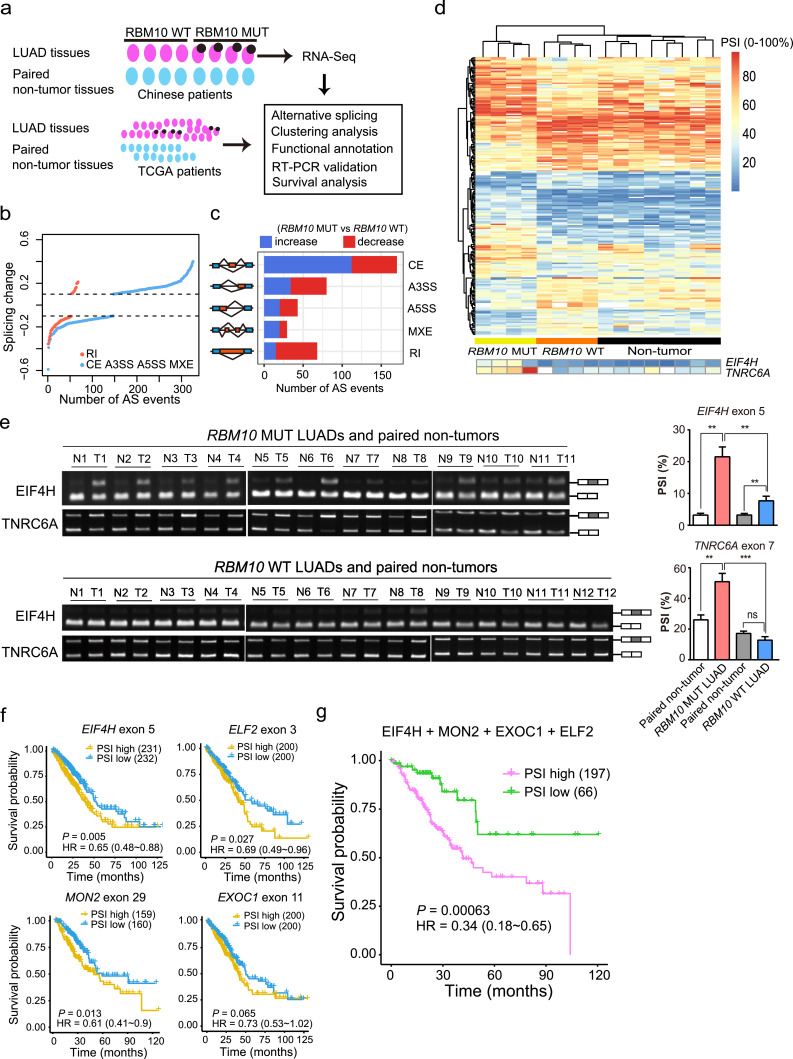
RBM10 regulates splicing of hundreds of target genes [20, 21] and may affect LUAD progression by modulating key cancer-associated AS events [20,32]. To gain new molecular insights into RBM10 mutations in LUAD pathogenesis, we performed RNA-Seq for LUAD samples with RBM10 loss-of-function mutations (Supplementary Table S2) or wild type RBM10 (4 samples in each category) together with their matched adjacent non-tumour tissues to systematically identify AS events associated with RBM10 mutations (Fig. 2a). We found that 394 AS alterations were significantly associated with LUAD samples harbouring RBM10 mutations (Fig. 2b, c and Supplementary Table S3; P<0.05, |△PSI (percent-spliced-in)| > 0.1), of which 43% are cassette exons. The majority of these cassette exons showed increased PSI values in RBM10 mutated LUAD samples (Fig. 2c), suggesting that RBM10 primarily suppresses inclusion of cassette exons.
Fig. 2.
RBM10 mutations lead to extensive alternative splicing (AS) changes in LUAD tissues. (a) Experimental design for identifying RBM10 mutation-associated AS events in LUAD patient tissues. LUAD tissues with RBM10 nonsense or frameshift mutations (RBM10 MUT), with wild type RBM10 expression (RBM10 WT) and their matched adjacent non-tumour tissues from Chinese patients were used for RNA-Seq experiments. (b) Splicing changes between RBM10 MUT and RBM10 WT LUAD samples (P < 0.05, |splicing change| > 0.1). CE: cassette exon; A3SS: alternative 3′ splice site; A5SS: alternative 5′ splice site; MEX: mutually exclusive exon; RI: retained intron. (c) The numbers of AS events associated with RBM10 mutations in each AS category. (d) Hierarchical clustering of LUAD tissue samples with or without RBM10 mutation and their adjacent non-tumour tissue samples based on AS events affected by RBM10 mutations. Each row represents an AS event, and each column represents a tissue sample. Two representative AS events were enlarged and shown at the bottom. (e) RT-PCR validation of AS changes in RBM10 mutated, RBM10 wild type LUAD and their adjacent non-tumour tissues from Chinese patients. Left panel: agarose gel image; right panel: quantification result. PSI: percent-spliced-in; T: LUAD tissues, N: non-tumour tissues. Shown are two representative AS events. (f, g) Kaplan-Meier survival curves of TCGA LUAD patients stratified by exon inclusion levels of individual (f) and combination (g) of RBM10 mutation-associated AS events. HR: hazard ratio. P values were calculated by Log-rank tests.
The unsupervised hierarchical clustering using these AS events revealed that the LUAD samples with RBM10 mutations are well separated from the tumours with wild type RBM10, and both LUAD samples are separated from the non-tumour tissues (Fig. 2d). Interestingly, the LUADs with wild type RBM10 are more similar to the non-tumour tissues. Such results suggest that these AS events may potentially be used as molecular markers for classification of LUAD. We selected several AS events with significantly higher PSI levels in RBM10 mutated samples and confirmed their splicing changes by RT-PCR (Fig. 2e and Supplementary Fig. S2A, B). The ontology analysis showed that genes harbouring RBM10 mutation-associated AS events are enriched in cancer-related molecular functions, including protein phosphorylation and acetylation (Supplementary Fig. S2C).
To further corroborate the results obtained from the Chinese LUAD samples, we also analysed TCGA data for splicing differences between LUAD samples harbouring mutated and wild type RBM10. We identified 49 AS alternations correlated with RBM10 mutations (Supplementary Fig. S2D, E and Table S3), and found that the common AS events between the Chinese and TCGA samples exhibited highly consistent changes (Supplementary Fig. S2F), supporting the credibility of AS events obtained in each cohort. In the rest of this study, we defined the union of AS events identified from both datasets as “RBM10 mutation-associated AS events”.
3.3. RBM10 mutation-associated AS events are predictive of LUAD survival
Increasing lines of evidences have suggested the clinical significance of cancer-associated AS events as prognostic markers [11,14]. To investigate whether RBM10 mutation-associated AS events in LUAD are predictive of patient survival, we performed Kaplan-Meier analysis of LUAD patients grouped by the PSI values of those AS events using the data from TCGA. We found that PSI values of EIF4H exon 5, ELF2 exon 3, MON2 exon 29 and EXOC1 exon 11 are all negatively correlated with survival rates of LUAD patients (Fig. 2f, P < 0.05 for all events), indicating that the RBM10 mutation-associated AS events are predictive of LUAD patient survival and thus may be used as potential prognostic markers. Importantly, combining the set of above four AS events using multivariable cox analysis provided a stronger prognostic predictor (Fig. 2g).
3.4. RBM10 suppresses LUAD development and progression
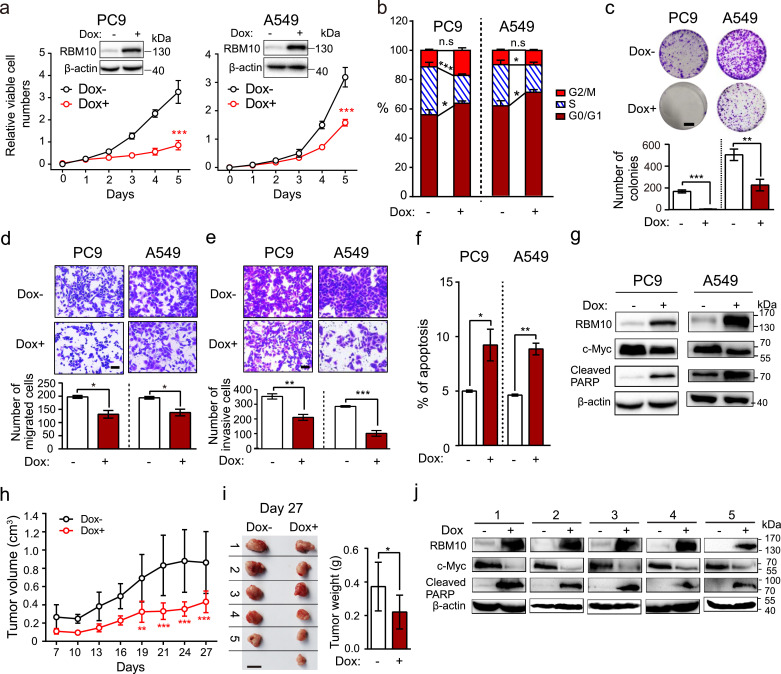
RBM10 mutations mostly co-occur with oncogenic EGFR or KRAS mutations (Fig. 1f and Supplementary Fig. S1E), suggesting that RBM10 may function in LUAD harbouring either one of the two most common oncogenic drivers. To directly examine RBM10 effects on LUAD progression in both contexts, we selected two commonly used LUAD cell lines, PC9 and A549, which respectively contain EGFRE19del and KRASG12D mutations and thus are representative of the oncogene mutations in LUAD. These two cell lines have lower expression of RBM10 compared to the immortalized lung bronchial epithelium-derived BEAS-2B cells (Supplementary Fig. S3A, B). Using a doxycycline-inducible system, we ectopically express RBM10 in LUAD cells in a controlled manner (Supplementary Fig. S3C, D). In both LUAD cell lines, we observed significantly reduced cell proliferation (Fig. 3a), cell cycle progression (Fig. 3b), anchorage-dependant colony formation (Fig. 3c), and cell migration and invasion (Fig. 3d, e). These processes were accompanied by the increased apoptosis (Fig. 3f) and apoptotic PARP and caspase 3 cleavage as well as the decreased c-Myc expression (Fig. 3g and Supplementary Fig. S3E). Importantly, RBM10 inducible overexpression significantly suppressed xenograft tumour growth in the immuno-compromised mice (Fig. 3h, i), accompanied by dramatic decrease of c-Myc expression and increased apoptotic PARP cleavage in xenograft tumours (Fig. 3j). Consistent with the effects of RBM10 overexpression, knockdown of RBM10 in LUAD A549 and PC9 cells increased cell proliferation (Supplementary Fig. S3F, G). Taken together, these data indicate that RBM10 can suppress LUAD development and progression.
Fig. 3.
RBM10 suppresses LUAD development and progression. (a-f) Cellular proliferation rates (a; n = 3; inserts are Western blot analysis of RBM10 protein levels, loading control: β-actin, Dox: doxycycline.), cell cycle (b; n = 5), anchorage-dependant colony formation (c; n = 3 for PC9, n = 4 for A549; scale bar: 1 cm), in vitro migration (d; n = 3; scale bar: 50 µm) and invasion (e; n = 4 for PC9, n = 3 for A549; scale bar: 50 µm) and apoptosis (f; n = 3) under inducible overexpression of RBM10 in LUAD cells. (g) Western blot analysis of indicated marker proteins under RBM10 overexpression. Loading control: β-actin. (h) Growth curve of xenograft tumours in nude mice following subcutaneous injection of PC9 cells with or without RBM10 overexpression. Error bar: ±SD. (i) Xenograft tumours were removed at day 27 and weighed. n = 5 for Dox- group, n = 6 for Dox+ group; scale bar: 1 cm; error bar: ±SD. (j) Western blot analysis of indicated marker proteins in xenograft tumours described in (i). Loading control: β-actin.
Error bars represent ±SEM unless indicated. * P < 0.05, ** P < 0.01, *** P < 0.001, ns: not significant. Two-way ANOVA followed by Bonferroni's tests were used in a and h; Student's t-tests were used in b, c, d, e, f and i.
We also examined the effects of RBM10 loss on oncogenic potential using BEAS-2B cells, and found that RBM10 silencing led to increased cell proliferation and anchorage-independent colony formation (Supplementary Fig. S3H-J). In addition, we observed additive effects of RBM10 silencing in BEAS-2B cells co-expressing oncogenic mutations KRASG12D, EGFRE19del or EGFRL858R (Supplementary Fig. S3I, J). These data suggest that loss of RBM10 can potentially induce the oncogenic transformation of lung epithelial cells.
3.5. RBM10 regulates splicing of cancer-related genes
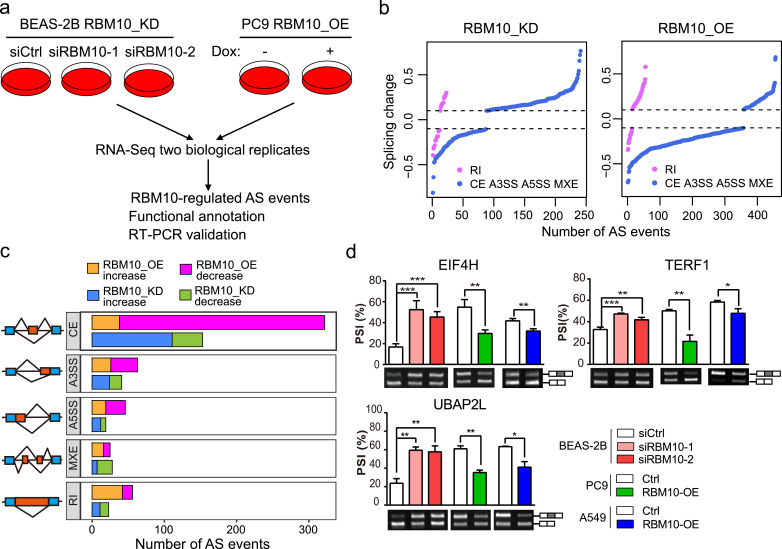
To further delineate the molecular mechanisms underlying RBM10 functions in LUAD, we performed RNA-Seq analysis in lung bronchial epithelium-derived BEAS-2B cells with RBM10 silencing or LUAD PC9 cells with RBM10 overexpression to identify the RBM10 targets transcriptome-wide (Fig. 4a). With two biological replicates, we identified 264 and 512 RBM10-regulated AS events in BEAS-2B and PC9 cells induced by RBM10 knockdown and overexpression respectively (Fig. 4b, c and Supplementary Table S4). The majority of RBM10-regulated AS events are the cassette exons, where the PSI values for most exons increased upon RBM10 silencing and decreased upon RBM10 overexpression (Fig. 4c). Gene ontology analysis revealed that RBM10-regulated AS events are enriched in genes with cancer-related cellular functions, including ubiquitin protein ligase activity and the cell-cell adhesion junction (Supplementary Fig. S4A).
Fig. 4.
RBM10 regulates alternative splicing (AS) of cancer-associated genes. (a) Experimental design for identifying AS changes induced by RBM10 knockdown (RBM10_KD) in lung epithelial BEAS-2B cells and overexpression (RBM10_OE) in LUAD PC9 cells. (b) Splicing changes following RBM10 KD in BEAS-2B cells or RBM10 OE in PC9 cells (P < 0.05, |splicing change| > 0.1). AS types were indicated in Fig. 2b. (c) The numbers of altered splicing events in each AS category. (d) RT-PCR validation of RBM10-regulated AS events under the indicated conditions. Agarose gel images (lower panel) and quantification of exon inclusion levels (upper panel) are shown for 3 representative genes. PSI: percent-spliced-in. Error bars represent +SEM, n = 3–4 biological replicates; * P < 0.05, ** P < 0.01, *** P < 0.001; One-way ANOVA followed by Dunnett's tests were used for comparisons with control (siCtrl) in BEAS-2B cells; Student's t-tests were used for comparisons with control (Ctrl) in PC9 and A549 cells.
Twenty-nine of these RBM10-regulated AS events were significantly altered in both RBM10 knockdown and overexpression cells, amongst which 25 exhibited inverse changes in the two conditions (Supplementary Fig. S4B and Table S4). We then selected 13 candidate AS events for validation using RT-PCR (Fig. 4d and Supplementary Fig. S4C), and found highly correlated splicing changes between RT-PCR and RNA-Seq (Supplementary Fig. S4D).
We further determined to what extent RBM10 mutation-associated AS events identified in LUAD patients are directly regulated by RBM10. By comparing changes of all AS events identified in the Chinese and TCGA LUAD samples and those identified in two cell lines, we found that AS changes obtained in two cohorts of LUAD tissue samples positively correlated with each other and with those induced by RBM10 knockdown in BEAS-2B cells, whereas they negatively correlated with those induced by RBM10 overexpression in PC9 cells (Supplementary Fig. S4E). These data suggest that RBM10 mutation-associated AS events identified in LUADs are largely induced by RBM10 loss.
In addition to splicing changes, we also analysed gene expression alterations and found that only a small number of genes (13 genes) had significant expression changes upon knockdown of RBM10, and hundreds of genes showed altered expression upon RBM10 overexpression (Supplementary Fig. S4F and Table S5; P < 0.05 and |log2FoldChange| ≥ 1 in both cases).
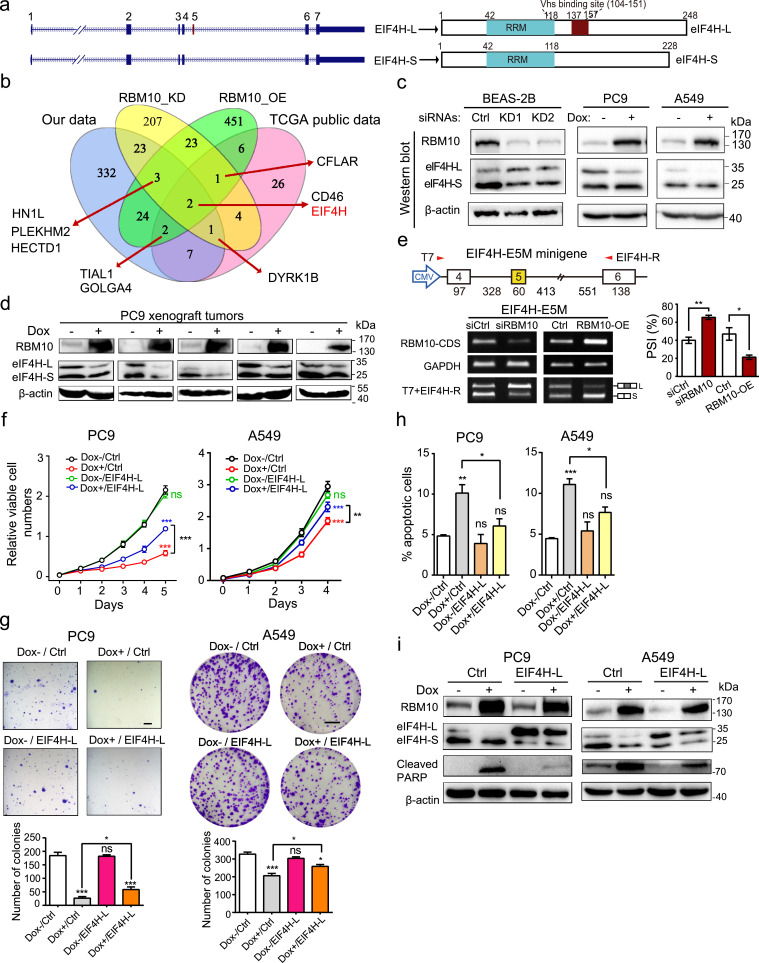
3.6. RBM10 suppresses LUAD progression by regulating EIF4H splicing
To elucidate the molecular mechanism and functional consequence of RBM10-regulated AS events in more detail, we focused on a key RBM10 target, the 5th exon of EIF4H (Fig. 5a). EIF4H encodes a regulator of translation initiation, the rate-limiting step of translation that has been closely linked with cancer [53, 54], and alternative splicing of its exon 5 produces two splice variants that we named as EIF4H-L and EIF4H-S (Fig. 5a). We selected this particular splicing event due to the following reasons. First, EIF4H exon 5 is one of the two AS events that were significantly affected by RBM10 mutations in the Chinese and TCGA LUAD samples and regulated by RBM10 in BEAS-2B and PC9 cells (Fig 5b). For the two AS events, only EIF4H exon 5 showed PSI increase > 20% in RBM10 mutated versus wild type LUAD samples, strongly indicating that it is a key target of RBM10. Second, EIF4H exon 5 inclusion level is low in LUAD adjacent non-tumour tissues and normal lung epithelial cells (BEAS-2B), but is dramatically increased in RBM10 mutated LUAD tissues and in BEAS-2B cells with RBM10 knockdown (Figs. 2e, 4d and Supplementary S5A). In addition, it is negatively correlated with LUAD patient survival (Fig. 2f, g). These observations indicate a specific function of EIF4H-L in promoting LUAD development and progression. Third, RBM10-mediated regulation of EIF4H exon 5 splicing led to consistent changes at protein levels (Fig. 5c, d) and RNA levels (Fig. 4d).
Fig. 5.
RBM10 suppresses LUAD progression by regulating EIF4H splicing. (a) Exon/Intron structures and encoded proteins of EIF4H splice variants containing or lacking exon 5. RRM: RNA recognition motif. Vhs: virion host shutoff (Vhs) protein binding region. (b) Venn diagram of genes whose splicing was affected by RBM10 mutations in Chinese and TCGA LUAD samples (our data and TCGA public data respectively) and regulated by RBM10 knockdown (RBM10_KD) in BEAS-2B and RBM10 overexpression (RBM10_OE) in PC9 cells. (c) Protein expression of two EIF4H splice variants following RBM10 knockdown in BEAS-2B cells or RBM10 overexpression in A549 and PC9 cells examined by Western blot analysis. Shown are the representative results of three biological replicates. Dox: doxycycline. (d) Protein levels of EIF4H splice variants in xenograft tumours shown in Fig. 3i. Loading control: β-actin. (e) Effects of RBM10 silencing and overexpression on EIF4H exon 5 splicing were examined using minigene splicing reporter assays. Upper panel: scheme of minigene reporter. Lower panel: representative agarose gel images (left) and quantification of results (right). (f, g, h) Cell proliferation rates (f; n = 3) and anchorage independent (PC9) or dependant (A549) colony formation (g; Upper panel: representative images, scale bar: 1 mm for PC9 and 1 cm for A549 cells, respectively; Lower panel: quantification of results; n = 4) and apoptosis (h; n = 3) of LUAD cells under conditions of control (Dox-/Ctrl), overexpression of RBM10 alone (Dox+/Ctrl), overexpression of EIF4H-L alone (Dox-/EIF4H-L), or both proteins together (Dox+/EIF4H-L). (i) Expression of indicated proteins under conditions in (f) were examined by Western blot. Loading control: β-actin.
Error bars represent ±SEM or +SEM; * P < 0.05, ** P < 0.01, *** P < 0.001, ns: not significant; Student's t-tests in e; One-way ANOVA followed by Turkey's tests for multiple comparisons in g, h; two-way ANOVA followed by Bonferroni's tests in f.
To validate the direct effect of RBM10 on splicing of EIF4H exon 5, we analysed previously reported RBM10-RNA interaction datasets [21] and found strong RBM10 binding signals in the intronic regions adjacent to the splice sites of EIF4H exon 5 (Supplementary Fig. S5B). The specific interactions of RBM10 with these sites in EIF4H pre-mRNA were further confirmed by CLIP-PCR (Supplementary Fig. S5C). In agreement with the splicing changes of endogenous EIF4H (Fig. 4d), we observed that RBM10 knockdown promoted EIF4H exon 5 inclusion, while overexpression of RBM10 significantly inhibited its inclusion, as judged by minigene splicing reporter containing a genomic segment spanning EIF4H exon 4 and exon 6 (Fig. 5e). Collectively, these results indicate that RBM10 directly binds to EIF4H pre-mRNA and promotes the skipping of exon 5.
To directly test if the suppression of LUAD by RBM10 is through regulation of EIF4H splicing, we conducted rescue experiments. We ectopically re-expressed EIF4H-L in LUAD PC9 and A549 cells where the doxycycline-induced RBM10 expression can arrest cell proliferation and induce apoptosis (Figs. 3 and 5f-h). While overexpressing EIF4H-L alone had an undetectable effect, overexpression of EIF4H-L together with RBM10 partially reversed the anti-proliferative and pro-apoptotic effects of RBM10, as judged by the results of cell proliferation (Fig. 5f), colony formation (Fig. 5g), apoptosis assays (Fig. 5h) and apoptotic marker cleaved-PARP measurement (Fig. 5i). To exclude the confounding effects of EIF4H-S expression promoted by RBM10 (Figs. 4d and 5c), we silenced EIF4H-S under RBM10 overexpression in LUAD PC9 cells and found no discernable differences in cell proliferation (Supplementary Fig. S5D). In line with the results of EIF4H-L overexpression (Fig. 5f-h), silencing EIF4H-L counteracted the pro-proliferation effects of RBM10 knockdown in LUAD A549 cells (Supplementary Fig. S5E). Taken together, these results indicate that RBM10 suppresses LUAD progression, at least in part, by regulating EIF4H splicing.
3.7. EIF4H-L is critical for LUAD cell proliferation and a valuable therapeutic target
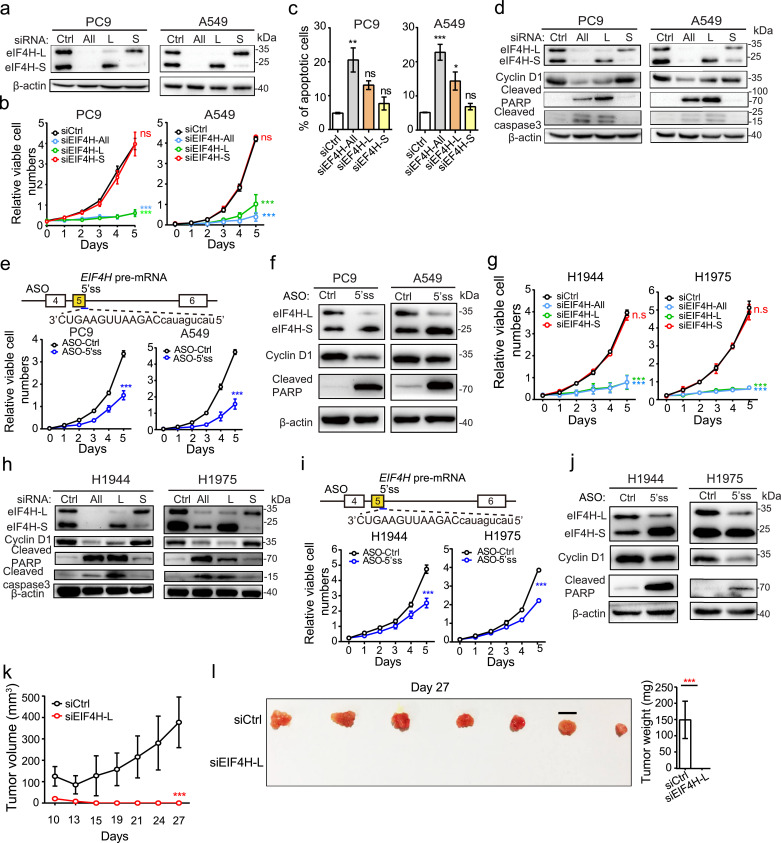
We next sought to investigate the functions of different EIF4H splice variants by selectively silencing the EIF4H splice variants with/without exon 5 (EIF4H-L, EIF4H-S) or both variants in LUAD cells (PC9 and A549) using siRNAs. The specificity and efficiency of these siRNAs were first confirmed by RT-PCR and Western blotting (Fig. 6a). We found that the silencing of EIF4H-L significantly suppressed cell proliferation and promoted apoptosis accompanied by concordant changes of corresponding molecular markers (cyclin D1, cleaved caspase 3 and cleaved PARP), whereas silencing of EIF4H-S had virtually no effect compared to cells transfected with control siRNAs (Fig. 6b-d). Interestingly, silencing of both EIF4H splice variants produced similar effect of EIF4H-L silencing (Fig. 6b-d). In addition, we used antisense oligonucleotide (ASO) to shift the splicing of EIF4H from EIF4H-L to EIF4H-S, and observed similar anti-proliferative and pro-apoptotic effects in LUAD cells and consistent expression changes in cyclin D1 and cleaved-PARP (Fig. 6e, f). These results suggest that EIF4H-L, but not EIF4H-S, plays a critical role in LUAD cell proliferation and survival.
Fig. 6.
EIF4H exon 5 inclusion is crucial for LUAD cell proliferation and a potential therapeutic target for LUAD. (a) Western blot analysis of EIF4H protein expression in indicated LUAD cells transfected with control siRNA (siCtrl), siRNAs targeting total EIF4H (si-All) or selectively targeting the long (si-L) or short (si-S) EIF4H variants. (b, c, g) Cell proliferation rates (b, g) and apoptosis (c) of indicated LUAD cells under conditions described in (a). (d, h) Western blot analysis of cell cycle and apoptotic markers under conditions described in (a). Loading control: β-actin. (e, i) Cell proliferation rates dramatically decreased following antisense oligonucleotide (ASO)-mediated blockade of EIF4H exon 5 inclusion in indicated LUAD cells. Targeting location and sequence of the ASO were shown in the upper panel. (f, j) Western blot analysis of indicated proteins under conditions in (e, i) respectively. Loading control: β-actin. (k) Growth curves of xenograft tumours formed by H1944 cells transfected with siRNAs against EIF4H-L or control. The sizes of xenograft tumours were measured at indicated time points. n = 6 per condition. Error bar: ±SD. (l) Xenograft tumours were removed at day 27 and weighed. Error bar: ±SD.
Error bars represent ±SEM unless indicated; n = 3 biological replicates unless indicated; * P < 0.05, ** P < 0.01, *** P < 0.001; two-way ANOVA followed by Bonferroni's tests compared to siCtrl in b, e, g, i and k; One-way ANOVA followed by Dunnett's tests compared to siCtrl in c; and Student's t-test in l.
To further confirm the effects of EIF4H splicing in RBM10-deficient LUADs, we used two additional LUAD cells lines with RBM10 loss-of-function mutations (H1944 and H1975, carrying a frameshift mutation respectively). As expected, both cell lines exhibited higher levels of EIF4H exon 5 inclusion compared to normal lung epithelial BEAS-2B cells (Supplementary Fig. S6A, B). In agreement with results from A549 and PC9 cells, the siRNA-mediated EIF4H-L silencing and ASO-mediated splicing shift dramatically inhibited cell proliferation and promoted apoptosis of both RBM10-mutated LUAD cell lines (Fig. 6g-j and Supplementary Fig. 6C). These results demonstrate that the RBM10-mediated splicing switch of EIF4H plays a critical role in regulating LUAD progression, and thus may be used as a promising therapeutic target for RBM10-deficient LUADs.
To determine the effectiveness of EIF4H-L silencing in suppressing tumourigenesis in vivo, we used xenograft tumour model with LUAD H1944 cells treated with siRNAs against EIF4H-L. Strikingly, we found that, compared to control siRNAs, treatment with siRNA against EIF4H-L almost completely abolished xenograft tumour formation when subcutaneously injected into the immuno-compromised mice (Fig. 6k, l). Collectively, these results demonstrate that EIF4H-L is indispensable for LUAD cell proliferation and survival in vitro and in vivo, providing a potentially new therapeutic route to suppress LUAD through splicing manipulation.
4. Discussion
Splicing factors are frequently mutated or dysregulated in cancer, and thus elucidating their functional roles and clinical significance is of great importance for managing cancers. RBM10 is one of the most commonly mutated splicing factor genes in solid tumours, particularly LUAD. Here we report that RBM10 suppresses lung cancer primarily by altering the splicing of the key target EIF4H, and demonstrate that the inclusion of EIF4H exon 5 could serve as a valuable therapeutic target in lung cancer.
RBM10 mutations frequently occur in LUAD patients from the Chinese (~9%) and TCGA (~7%) cohorts, and mostly co-occur with EGFR and KRAS mutations respectively (Fig. 1f and Supplementary S1E). Notably, two very recent studies showed that RBM10 is the second most frequently mutated gene compared to commonly mutated tumour suppressor genes (e.g. TP53, STK11, RB1, and APC etc.) in LUAD patients from East Asia [55, 56] and enriched in female and younger non-smoking LUADs [55], highlighting the broad functional and clinical significance of RBM10 mutations in LUAD. Besides mutations, we examined the RBM10 expression in TCGA LUAD samples without RBM10 mutation and found that 12.8% of RBM10 wild type LUAD samples had lower RBM10 expression than the median of RBM10-mutant LUADs (Supplementary Fig. S6D). Interestingly, RBM10 wild type LUADs with low RBM10 expression (termed as RBM10-low LUADs) correlated with splicing changes of RBM10 target genes in the same direction with RBM10-mutant LUADs, although to a lesser extent, compared to LUADs with high RBM10 expression (Supplementary Fig. S6E). These results indicate that RBM10 downregulation may also be functionally significant in LUAD, expanding the frequency and clinical significance of RBM10 loss in LUAD.
RBM10 mutations in LUADs generally result in loss-of-function, making RBM10 itself a poor therapeutic target. Our study demonstrates that targeting RBM10-regulated AS events may provide a valuable alternative route for RBM10-mutant/low LUADs. There are several advantages in this strategy. Firstly, most RBM10-regulated AS events are significantly upregulated in RBM10-deficient LUADs compared to non-tumour tissues (Fig. 2b), providing a diverse repertoire of potential targets. In addition, exon inclusion of several RBM10-regulated AS events negatively correlate with patient survival (Fig. 2f, g), offering prognostic markers for future treatment. Finally, exciting advances have recently been achieved in disease treatment based on splicing modulation. For example, antisense oligonucleotides (ASOs) that alter splicing of disease-causing genes have recently been approved by FDA to treat Duchenne muscle dystrophy and spinal muscular atrophy, and the ASO and siRNA-based strategies have also shown promises in treatment of cancer [14].
An important RBM10-regulated target is the translation initiation factor EIF4H. We, for the first time, demonstrate that the inclusion of EIF4H exon 5 is significantly enhanced by RBM10 loss and can be used as a promising therapeutic target for RBM10-mutant/low LUADs (Figs. 5 and 6). In addition to lung cancer, analysis of TCGA data revealed that the inclusion of EIF4H exon 5 is significantly increased in various other cancers compared to the adjacent normal tissues (Supplementary Fig. S6F), including the previously reported oesophageal and colorectal cancer [57], suggesting its general role as a cancer-specific splicing alteration. Moreover, the analyses of the Genotype-Tissue Expression (GTEx) data demonstrated that the inclusion of EIF4H exon 5 is relatively low in almost all normal tissues except for testis (Supplementary Fig. S6G), implying a molecular function similar to cancer/testis antigens [58].
The eIF4H is an auxiliary translation initiation factor that has been proposed to promote translation by stimulating the core translation initiation factor eIF4A [59, 60]. However, the molecular mechanisms underlying the different functions of the two isoforms as shown in our study are unknown. The two eIF4H isoforms, eIF4H-L and eIF4H-S, differ by 20 amino acids (aa) within their virion host shutoff (Vhs) region [61]. We hypothesize that the 20 amino acids difference between the two eIF4H isoforms may lead to distinct interacting protein partners and/or binding capacities to target mRNAs, thereby resulting in different regulatory and physio-pathological functions. To support this hypothesis, we attempted to characterize the interacting proteins for eIF4H-L and eIF4H-S using BioID (Supplementary Fig.7A, B). Our preliminary data showed that, in addition to a common set of proteins involved in translation regulation, eIF4H-L and eIF4H-S may interact with specific proteins (Supplementary Fig.7C, D). Substantial future efforts will be needed to elucidate the exact molecular mechanisms underlying distinct functions between eIF4H-L and eIF4H-S. Such efforts include identifying the protein interacting partners of eIF4H-L and eIF4H-S by using immunoprecipitation and mass spectrometry, and examining their global effects on mRNA translation by using polysome profiling. In polysome profiling experiments, siRNAs can be used to specifically knockdown EIF4H-L and EIF4H-S respectively. Due to the transient and possible off-target effects of siRNA, the results need to be confirmed by using independent siRNAs and/or other genetic perturbation methods (e.g. ASO and CRISPR-CAS9). Other than involved in mRNA translation regulation, eIF4H-L and/or eIF4H-S may also have currently unrevealed functions that underscore their different roles in cancer and warrant more investigations.
Besides EIF4H-L, we identified splicing alterations caused by RBM10 deficiency in a number of genes that are known to play key roles in cancer, including genes involved in cell proliferation (e.g. TERF1) and RNA processing (e.g. PCBP2). This may explain why eIF4H-L overexpression partially rescued the anti-proliferation and pro-apoptotic effect of RBM10 overexpression in LUAD cells (Fig. 5f-i). The splicing of NUMB exon 9 was previously reported to be inhibited by RBM10 and linked with RBM10 suppressive functions in LUAD [20], and we found that this splicing event exhibited small changes below the cutoff (P< 0.05, |△PSI| > 0.1) between RBM10 mutated and wild type LUAD tissue samples, which was confirmed by RT-PCR (Supplementary Fig. S2A, B). Nevertheless, functional splicing events with relatively small changes may also contribute to the impacts of dysregulated splicing factors in cancer. Therefore, further studies will be required to elucidate functional roles and contributions of various specific splice variants affected by mutations of RBM10.
In summary, our study demonstrates a new molecular mechanism underlying the tumour suppressive functions of RBM10, and the critical role as well as the therapeutic value of EIF4H splicing in LUAD, providing important insights for future mechanistic and translational studies of splicing defects in cancer.
Declaration of Competing Interest
A patent application by authors Y.B.W., Z.F.W., Y.F.B, X.F.S., and M.X. using the splicing of EIF4H exon 5 as a new cancer therapeutic target has been submitted to National Intellectual Property Administration, PRC. Other authors declare no competing interests.
Acknowledgments
Acknowledgements
We thank Prof. Wei Chen at Department of Biology, Southern University of Science and Technology, Shenzhen, China, for helpful discussions on conceiving this project.
Funding Sources
This work was supported by the National Natural Science Foundation of China [81871878, 31371299 to Y.B.W.; 31730110, 31661143031 and 31570823 to Z.F.W.; 81572264 to Y.H.S; 81571090 to D.S.], and by the Shanghai Municipal Natural Science Fundation [20ZR1406500 to Y.B.W]. Z.F.W. is also supported by the type A CAS Pioneer 100-Talent program.
Data sharing statement
The RNA sequencing data has been deposited in NODE (http://www.biosino.org/node) and can be accessed via the accession number OEP001100.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.ebiom.2020.103067.
Contributor Information
Yihua Sun, Email: sun_yihua76@hotmail.com.
Zefeng Wang, Email: wangzefeng@picb.ac.cn.
Yongbo Wang, Email: wangyongbo@fudan.edu.cn.
Appendix. Supplementary materials
References
- 1.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68(1):7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 2.Herbst R.S., Morgensztern D., Boshoff C. The biology and management of non-small cell lung cancer. Nature. 2018;553:446. doi: 10.1038/nature25183. [DOI] [PubMed] [Google Scholar]
- 3.Rotow J., Bivona T.G. Understanding and targeting resistance mechanisms in NSCLC. Nat Rev Cancer. 2017;17:637. doi: 10.1038/nrc.2017.84. [DOI] [PubMed] [Google Scholar]
- 4.Swanton C., Govindan R. Clinical implications of genomic discoveries in lung cancer. N Engl J Med. 2016;374(19):1864–1873. doi: 10.1056/NEJMra1504688. [DOI] [PubMed] [Google Scholar]
- 5.Lee Y., Rio D.C. Mechanisms and regulation of alternative Pre-mRNA Splicing. Annu. Rev. Biochem. 2015;84(1):291–323. doi: 10.1146/annurev-biochem-060614-034316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Scotti M.M., Swanson M.S. RNA mis-splicing in disease. Nat Rev Genet. 2016;17(1):19–32. doi: 10.1038/nrg.2015.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Daguenet E., Dujardin G., Valcárcel J. The pathogenicity of splicing defects: mechanistic insights into pre‐mRNA processing inform novel therapeutic approaches. EMBO Rep. 2015;16(12):1640–1655. doi: 10.15252/embr.201541116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Matera A.G., Wang Z. A day in the life of the spliceosome. Nat Rev Mol Cell Biol. 2014;15:108. doi: 10.1038/nrm3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shi Y. Mechanistic insights into precursor messenger RNA splicing by the spliceosome. Nat Rev Mol Cell Biol. 2017;18:655. doi: 10.1038/nrm.2017.86. [DOI] [PubMed] [Google Scholar]
- 10.Lee S.C.-.W., Abdel-Wahab O. Therapeutic targeting of splicing in cancer. Nat Med. 2016;22(9):976–986. doi: 10.1038/nm.4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Urbanski L.M., Leclair N., Anczuków O. Alternative-splicing defects in cancer: splicing regulators and their downstream targets, guiding the way to novel cancer therapeutics. Wiley Interdisc Rev: RNA. 2018;9(4):e1476. doi: 10.1002/wrna.1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bonnal S.C., López-Oreja I., Valcárcel J. Roles and mechanisms of alternative splicing in cancer — Implications for care. Nat Rev Clin Oncol. 2020 doi: 10.1038/s41571-020-0350-x. [DOI] [PubMed] [Google Scholar]
- 13.Wang Y., Bao Y., Zhang S., Wang Z. Splicing dysregulation in cancer: from mechanistic understanding to a new class of therapeutic targets. Sci China Life Sci. 2020;63(4):469–484. doi: 10.1007/s11427-019-1605-0. [DOI] [PubMed] [Google Scholar]
- 14.Agrawal A.A., Yu L., Smith P.G., Buonamici S. Targeting splicing abnormalities in cancer. Curr Opin Genet Dev. 2018;48:67–74. doi: 10.1016/j.gde.2017.10.010. [DOI] [PubMed] [Google Scholar]
- 15.Song X., Zeng Z., Wei H., Wang Z. Alternative splicing in cancers: from aberrant regulation to new therapeutics. Semin Cell Dev Biol. 2018;75:13–22. doi: 10.1016/j.semcdb.2017.09.018. [DOI] [PubMed] [Google Scholar]
- 16.Dvinge H., Kim E., Abdel-Wahab O., Bradley R.K. RNA splicing factors as oncoproteins and tumour suppressors. Nat Rev Cancer. 2016;16(7):413–430. doi: 10.1038/nrc.2016.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Anczuków O., Krainer A.R. Splicing-factor alterations in cancers. RNA. 2016;22(9):1285–1301. doi: 10.1261/rna.057919.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang Y., Chen D., Qian H., Tsai Yihsuan S., Shao S., Liu Q. The splicing factor RBM4 controls apoptosis, proliferation, and migration to suppress tumor progression. Cancer Cell. 2014;26(3):374–389. doi: 10.1016/j.ccr.2014.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rahman M.A., Krainer A.R., Abdel-Wahab O. SnapShot: splicing alterations in cancer. Cell. 2020;180(1) doi: 10.1016/j.cell.2019.12.011. 208-.e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bechara Elias G., Sebestyén E., Bernardis I., Eyras E., Valcárcel J. RBM5, 6, and 10 differentially regulate NUMB alternative splicing to control cancer cell proliferation. Mol Cell. 2013;52(5):720–733. doi: 10.1016/j.molcel.2013.11.010. [DOI] [PubMed] [Google Scholar]
- 21.Wang Y., Gogol-Döring A., Hu H., Fröhler S., Ma Y., Jens M. Integrative analysis revealed the molecular mechanism underlying RBM10-mediated splicing regulation. EMBO Mol Med. 2013;5(9):1431–1442. doi: 10.1002/emmm.201302663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rodor J., FitzPatrick D.R., Eyras E., Cáceres J.F. The RNA-binding landscape of RBM10 and its role in alternative splicing regulation in models of mouse early development. RNA Biol. 2017;14(1):45–57. doi: 10.1080/15476286.2016.1247148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnston J.J., Teer J.K., Cherukuri P.F., Hansen N.F., Loftus S.K., Chong K. Massively parallel sequencing of exons on the X chromosome identifies RBM10 as the gene that causes a syndromic form of cleft palate. A J Hum Genetics. 2010;86(5):743–748. doi: 10.1016/j.ajhg.2010.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Giannakis M., Mu Xinmeng J., Shukla Sachet A., Qian Zhi R., Cohen O., Nishihara R. Genomic correlates of immune-cell infiltrates in colorectal carcinoma. Cell Rep. 2016;15(4):857–865. doi: 10.1016/j.celrep.2016.03.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bailey P., Chang D.K., Nones K., Johns A.L., Patch A.-.M., Gingras M.-.C. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature. 2016;531(7592):47–52. doi: 10.1038/nature16965. [DOI] [PubMed] [Google Scholar]
- 26.Nordentoft I., Lamy P., Birkenkamp-Demtröder K., Shumansky K., Vang S., Hornshøj H. Mutational context and diverse clonal development in early and late bladder cancer. Cell Rep. 2014;7(5):1649–1663. doi: 10.1016/j.celrep.2014.04.038. [DOI] [PubMed] [Google Scholar]
- 27.Imielinski M., Berger A.H., Hammerman P.S., Hernandez B., Pugh T.J., Hodis E. Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing. Cell. 2012;150(6):1107–1120. doi: 10.1016/j.cell.2012.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Network T. Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511(7511):543–550. doi: 10.1038/nature13385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang R., Hu H., Pan Y., Li Y., Ye T., Li C. RET fusions define a unique molecular and clinicopathologic subtype of non–small-cell lung cancer. J Clin Oncol. 2012;30(35):4352–4359. doi: 10.1200/JCO.2012.44.1477. [DOI] [PubMed] [Google Scholar]
- 30.Chen H., Carrot-Zhang J., Zhao Y., Hu H., Freeman S.S., Yu S. Genomic and immune profiling of pre-invasive lung adenocarcinoma. Nat Commun. 2019;10(1):5472. doi: 10.1038/s41467-019-13460-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jamal-Hanjani M., Wilson G.A., McGranahan N., Birkbak N.J., Watkins T.B.K., Veeriah S. Tracking the evolution of non–small-cell lung cancer. N Engl J Med. 2017;376(22):2109–2121. doi: 10.1056/NEJMoa1616288. [DOI] [PubMed] [Google Scholar]
- 32.Zhao J., Sun Y., Huang Y., Song F., Huang Z., Bao Y. Functional analysis reveals that RBM10 mutations contribute to lung adenocarcinoma pathogenesis by deregulating splicing. Sci Rep. 2017;7:40488. doi: 10.1038/srep40488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Seiler M., Peng S., Agrawal A.A., Palacino J., Teng T., Zhu P. Somatic mutational landscape of splicing factor genes and their functional consequences across 33 cancer types. Cell Rep. 2018;23(1):282–296. doi: 10.1016/j.celrep.2018.01.088. .e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hernández J., Bechara E., Schlesinger D., Delgado J., Serrano L., Valcárcel J. Tumor suppressor properties of the splicing regulatory factor RBM10. RNA Biol. 2016;13(4):466–472. doi: 10.1080/15476286.2016.1144004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ji Y., Xie S., Jiang L., Liu L., Li L., Luo L. Increased cell apoptosis in human lung adenocarcinoma and in vivo tumor growth inhibition by RBM10, a tumor suppressor gene. Oncol Lett. 2017;14(4):4663–4669. doi: 10.3892/ol.2017.6765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sun X., Jia M., Sun W., Feng L., Gu C., Wu T. Functional role of RBM10 in lung adenocarcinoma proliferation. Int J Oncol. 2019;54(2):467–478. doi: 10.3892/ijo.2018.4643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Loiselle J.J., Sutherland L.C. RBM10: harmful or helpful-many factors to consider. J Cell Biochem. 2018;119(5):3809–3818. doi: 10.1002/jcb.26644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sun Y., Bao Y., Han W., Song F., Shen X., Zhao J. Autoregulation of RBM10 and cross-regulation of RBM10/RBM5 via alternative splicing-coupled nonsense-mediated decay. Nucl Acids Res. 2017;45(14):8524–8540. doi: 10.1093/nar/gkx508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang J.-.Z., Chen M., Chen D., Gao X.-.C., Zhu S., Huang H. A peptide encoded by a putative lncRNA HOXB-AS3 suppresses colon cancer growth. Mol Cell. 2017;68(1):171–184. doi: 10.1016/j.molcel.2017.09.015. .e6. [DOI] [PubMed] [Google Scholar]
- 40.Wang K., Singh D., Zeng Z., Coleman S.J., Huang Y., Savich G.L. MapSplice: accurate mapping of RNA-seq reads for splice junction discovery. Nucl Acids Res. 2010;38(18) doi: 10.1093/nar/gkq622. e178-e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Katz Y., Wang E.T., Airoldi E.M., Burge C.B. Analysis and design of RNA sequencing experiments for identifying isoform regulation. Nat Methods. 2010;7:1009. doi: 10.1038/nmeth.1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li B., Dewey C.N. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformat. 2011;12(1):323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Huang D.W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 44.Huang D.W., Sherman B.T., Lempicki R.A. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucl Acids Res. 2009;37(1):1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Langmead B., Trapnell C., Pop M., Salzberg S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10(3):R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Corcoran D.L., Georgiev S., Mukherjee N., Gottwein E., Skalsky R.L., Keene J.D. PARalyzer: definition of RNA binding sites from PAR-CLIP short-read sequence data. Genome Biol. 2011;12(8):R79. doi: 10.1186/gb-2011-12-8-r79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Collins K.M., Kainov Y.A., Christodolou E., Ray D., Morris Q., Hughes T. An RRM-ZnF RNA recognition module targets RBM10 to exonic sequences to promote exon exclusion. Nucl Acids Res. 2017 doi: 10.1093/nar/gkx225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Maaskola J., Rajewsky N. Binding site discovery from nucleic acid sequences by discriminative learning of hidden Markov models. Nucl Acids Res. 2014;42(21):12995–13011. doi: 10.1093/nar/gku1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Roux K.J., Kim D.I., Raida M., Burke B. A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J Cell Biol. 2012;196(6):801–810. doi: 10.1083/jcb.201112098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pan Y., Zhang Y., Li Y., Hu H., Wang L., Li H. ALK, ROS1 and RET fusions in 1139 lung adenocarcinomas: a comprehensive study of common and fusion pattern-specific clinicopathologic, histologic and cytologic features. Lung Cancer. 2014;84(2):121–126. doi: 10.1016/j.lungcan.2014.02.007. [DOI] [PubMed] [Google Scholar]
- 51.Wang R., Pan Y., Li C., Hu H., Zhang Y., Li H. The use of quantitative real-time reverse transcriptase PCR for 5′ and 3′ Portions of ALK transcripts to detect ALK rearrangements in lung cancers. Clin Cancer Res. 2012;18(17):4725–4732. doi: 10.1158/1078-0432.CCR-12-0677. [DOI] [PubMed] [Google Scholar]
- 52.Vogelstein B., Papadopoulos N., Velculescu V.E., Zhou S., Diaz L.A., Kinzler K.W. Cancer genome. Landscapes. 2013;339(6127):1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Grzmil M., Hemmings B.A. Translation regulation as a therapeutic target in cancer. Cancer Res. 2012;72(16):3891–3900. doi: 10.1158/0008-5472.CAN-12-0026. [DOI] [PubMed] [Google Scholar]
- 54.Truitt M.L., Ruggero D. New frontiers in translational control of the cancer genome. Nat Rev Cancer. 2016;16:288. doi: 10.1038/nrc.2016.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen Y.-.J., Roumeliotis T.I., Chang Y.-.H., Chen C.-.T., Han C.-.L., Lin M.-.H. Proteogenomics of non-smoking lung cancer in East Asia delineates molecular signatures of pathogenesis and progression. Cell. 2020;182(1):226–244. doi: 10.1016/j.cell.2020.06.012. .e17. [DOI] [PubMed] [Google Scholar]
- 56.Chen J., Yang H., Teo A.S.M., Amer L.B., Sherbaf F.G., Tan C.Q. Genomic landscape of lung adenocarcinoma in East Asians. Nat. Genet. 2020;52(2):177–186. doi: 10.1038/s41588-019-0569-6. [DOI] [PubMed] [Google Scholar]
- 57.Di W., Kazuyuki M., Hisahiro M., Fumio N., Takeshi T. An alternative splicing isoform of eukaryotic initiation factor 4H promotes tumorigenesis in vivo and is a potential therapeutic target for human cancer. Int J Cancer. 2011;128(5):1018–1030. doi: 10.1002/ijc.25419. [DOI] [PubMed] [Google Scholar]
- 58.Simpson A.J.G., Caballero O.L., Jungbluth A., Chen Y.-.T., Old L.J. Cancer/testis antigens, gametogenesis and cancer. Nat Rev Cancer. 2005;5(8):615–625. doi: 10.1038/nrc1669. [DOI] [PubMed] [Google Scholar]
- 59.Rozovsky N., Butterworth A.C., Moore M.J. Interactions between eIF4AI and its accessory factors eIF4B and eIF4H. RNA. 2008;14(10):2136–2148. doi: 10.1261/rna.1049608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sun Y., Atas E., Lindqvist L., Sonenberg N., Pelletier J., Meller A. The eukaryotic initiation factor eIF4H facilitates loop-binding, repetitive RNA unwinding by the eIF4A DEAD-box helicase. Nucl Acids Res. 2012;40(13):6199–6207. doi: 10.1093/nar/gks278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Feng P., Everly D.N., Read G.S. mRNA Decay during Herpesvirus Infections: interaction between a putative viral nuclease and a cellular translation factor. J. Virol. 2001;75(21):10272–10280. doi: 10.1128/JVI.75.21.10272-10280.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All RNA sequencing raw data has been deposited in NODE (http://www.biosino.org/node) and can be accessed via the accession number OEP001100.