Abstract
Friedreich ataxia (FRDA) is a neurodegenerative disorder characterized by neuromuscular and neurological manifestations. It is caused by mutations in the FXN gene, which results in loss of the mitochondrial protein frataxin. Endoplasmic Reticulum-mitochondria associated membranes (MAMs) are inter-organelle structures involved in the regulation of essential cellular processes, including lipid metabolism and calcium signaling. In the present study, we have analyzed in both, unicellular and multicellular models of FRDA, calcium management and integrity of MAMs. We observed that function of MAMs is compromised in our cellular model of FRDA, which was improved upon treatment with antioxidants. In agreement, promoting mitochondrial calcium uptake was sufficient to restore several defects caused by frataxin deficiency in Drosophila Melanogaster. Remarkably, our findings describe for the first time frataxin as a member of the protein network of MAMs, where interacts with two of the main proteins implicated in endoplasmic reticulum-mitochondria communication. These results suggest a new role of frataxin, indicate that FRDA goes beyond mitochondrial defects and highlight MAMs as novel therapeutic candidates to improve patient's conditions.
Keywords: MAMs, Frataxin, Calcium, Lipid peroxidation, Vitamin E, N-acetylcysteine
Graphical abstract
List of abbreviations
- FRDA
Friedreich's Ataxia
- MAMs
Endoplasmic-reticulum-mitochondria associated membranes
- Ca2+
Calcium
- VDAC
Voltage-dependent anion channel
- IP3R
Inositol-1,4,5-triphosphate receptor
- GRP75
Glucose response protein 75
- FXN
Frataxin
- DRG
Dorsal Root Ganglia
- ROS
Reactive Oxygen Species
- ER
Endoplasmic Reticulum
- MCU
Mitochondrial Calcium Uniporter
- Mfn
Mitofusin
- mtDNA
mitochondrial DNA
- ROI
Region of Interest
- La
Lamina
- Me
Medulla
- OC
Outer Chiasm
- PLA
Proximity Ligation Assay
- NAC
N-Acetylcysteine
- AD
Alzheimer's disease
- PD
Parkinson's disease
- HD
Huntington's disease
- OMM
Outer mitochondrial membrane
- IMM
Inner mitochondrial membrane
- PDC
Pyruvate dehydrogenase complex
- Nrf2
Nuclear factor E2-related factor 2
1. Introduction
Friedreich Ataxia (FRDA) is a neurodegenerative disorder mainly caused by homozygous GAA repeat expansion mutations within intron 1 of the FXN gene, which encodes frataxin (FXN), a protein associated with the mitochondrial inner membrane [1]. The length of GAA expansions decreases the expression of frataxin, which involves neurological and neuromuscular manifestations in patients including progressive trunk and limb ataxia, dysarthria, scoliosis, cardiomyopathy and diabetes mellitus [2]. FRDA is characterized by the degeneration of the large sensory neurons at the dorsal root ganglia (DRG), in charge of proprioception and sense of positioning. This neurodegenerative process affects both, the central and peripheral nervous systems, including spinocerebellar tracts, corticospinal tracts, posterior columns and cerebellum [3].
Frataxin has been proposed to play a role in many physiological functions, mainly related to iron metabolism [4], such as the assembly of the iron-sulfur clusters in the mitochondria, acting as a chaperone [[5], [6], [7]], as an iron-storage protein regulating mitochondrial iron transport [8,9] and participating in heme groups maturation [10,11]. Other processes like mitochondrial energy conversion [12] and oxidative phosphorylation [13] have been described regarding frataxin, as well as its participation in oxidative stress regulation by reducing the production of reactive oxygen species (ROS) [[14], [15], [16]]. Moreover, frataxin has an important role in proper calcium (Ca2+) handling [17,18], whose effect in neurons is the formation of multiple axonal spheroids mainly caused by Ca2+ imbalance.
Mitochondria plays a key role in energy metabolism, specifically in ATP production through oxidative phosphorylation. It is considered the main source of ROS in most cells, which could generate an imbalance in cellular redox state in circumstances of alteration of the electron transport chain [19]. As frataxin is ligated to different mitochondrial processes, its deficiency leads to mitochondrial dysfunction associated with redox imbalance and reduced mitochondrial energy production [20].
Mitochondrial dysfunction also affects its communication with other cellular compartments and the processes they regulate [20]. In relation to this, we and others have observed increased endoplasmic reticulum (ER) stress in different FRDA models [18,21]. The physical interaction between these compartments termed ER-mitochondria associated membranes (MAMs) [22] requires specific protein networks. The wide variety of cellular processes in which MAMs are involved in, including lipid metabolism [23], autophagy [24], mitochondrial morphology and cell death [25] are usually altered in several neurodegenerative disorders [[26], [27], [28]], as well as in FRDA [18,[29], [30], [31]].
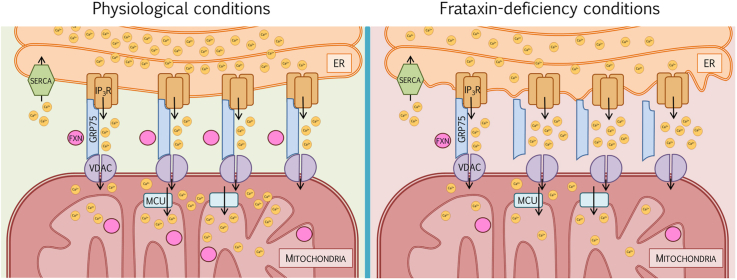
One of the better characterized functions of MAMs is the rapid exchange of Ca2+ between both organelles [32]. The main route for Ca2+ stored in the ER into mitochondria is throughout the channels VDAC1/porin and mitochondrial calcium uniporter (MCU), located at the outer and inner mitochondrial membrane, respectively [33,34]. Ca2+ homeostasis is crucial to maintain a proper physiology in different metabolic processes and signaling pathways, specifically mitochondrial regulation of cell survival and oxidative phosphorylation [35]. Importantly, we have also previously reported that frataxin-silenced cells show an impairment in Ca2+ buffering, as a consequence of reduced mitochondrial Ca2+ uptake capacity [18]. The involvement of MAMs in FRDA pathophysiology has been recently suggested in a Drosophila model of the disease in which downregulation of fly mitofusin (Mfn) was sufficient to counteract some frataxin-deficient phenotypes via reduction of ER stress [21]. However, undoubtful and definitive evidences regarding the intimate link between frataxin and MAMs are still elusive.
In this work, we assess MAMs architecture and integrity in a well stablished FRDA model in which frataxin is depleted in the SH-SY5Y neuroblastoma cell line by means of FXN gene silencing with two independent short hairpins (FXN-138.1 and FXN-138.2). We confirm that the interactions between the ER and mitochondria are reduced in frataxin-depleted cells, which can be reverted by treating cells with the antioxidants Trolox and N-acetyl cysteine. Concomitantly, both compounds also improve mitochondrial Ca2+ uptake and alleviate oxidative stress effects in membrane lipids. In agreement with all this, promotion of Ca2+ transport into the mitochondria was sufficient to restore several defects triggered by frataxin silencing in the fruit fly, Drosophila melanogaster. Finally, our findings describe for the first time that frataxin is present in MAMs where it interacts with proteins implicated in ER-mitochondria communication highlighting a new role for frataxin in the stability and integrity of these structures. Our results indicate that FRDA pathology goes beyond pure mitochondrial defects and could open new therapeutic avenues for the treatment of the disease.
2. Materials and methods
2.1. Cell culture and production of stable SH-SY5Y cell lines
The human SH-SY5Y neuroblastoma cell line was grown in DMEM-12 (Thermo Fisher Scientific, Waltham, Massachusetts, U.S.) supplemented with 10% fetal bovine serum and 1% l-glutamine and 1% penicillin/streptomycin. Cells were maintained at 37 °C in an atmosphere of 5% CO2 in air. For the generation of stable cell lines with the gene silencing of FXN, SH-SY5Y cells were transfected with pLKO.1 vector (MISSION ® shRNA plasmid DNA, Sigma-Aldrich, St. Louis, Missouri, U.S.) containing a hairpin sequence of FXN (TRCN0000006138) [18]. Transfection were performed using SuperFect Transfection (Qiagen, Venlo, Netherlands) according to the manufacturer's instructions. The stably transfected cells were selected and maintained in medium with 2 μg/ml puromycin.
2.2. Immunofluorescence staining
Cells were seeded on a 13 mm glass at a density of 30,000 cells. After fixation with 4% paraformaldehyde for 10 min, cells were washed with PBS and permeabilized with 0.5% PBS-Triton-X for 10 min. Cells were washed with 0.05% PBS-Triton and blocked with 10% FBS in 0.1% PBS-triton for 1 h. Primary antibodies were incubated overnight at 4 °C: Frataxin (Thermo Fisher Scientific, Waltham, Massachusetts, U.S., MA3-085), VDAC1 (Abcam, Cambridge, United Kingdom, ab14734), GRP75 (Santa Cruz Biotechnology, Dallas, Texas, U.S., sc13967), IP3R (Abcam, Cambridge, United Kingdom, ab125076). Cells were washed with PBS and incubated with secondary antibodies (either with Texas Red™ or Alexa Fluor® 488, Life Technologies) for 1 h at room temperature. Nuclei were detected with DAPI Fluoromount-G® (Southern Biotech, Birmingham, Alabama, U.S.). All the washes were performed three times for 5 min. Images were obtained using a Leica DMi8 with DC9000GT camera and a 63x oil immersion objective.
Cells were seeded in a μ-Dish 35 mm, high Glass Bottom (Ibidi, Gräfelfind, Germany) at a density of 100,000 cells/dish. After fixation with 4% paraformaldehyde for 10 min, cells were washed with PBS and permeabilized with 0.5% PBS-Triton-X for 10 min. Cells were washed with 0.05% PBS-Tween 20 and blocked with Duolink® Blocking solution (Sigma-Aldrich, St. Louis, Missouri, U.S.) for 1 h at 37 °C. After incubation, blocking solution was removed and a combination of two of the following specific antibodies were incubated overnight at 4 °C: Frataxin (Thermo Fisher Scientific, Waltham, Massachusetts, U.S., MA3-085), VDAC1 (Abcam, Cambridge, United Kingdom, ab14734), GRP75 (Santa Cruz Biotechnology, Dallas, Texas, U.S., sc13967), IP3R (Abcam, Cambridge, United Kingdom, ab125076). Cells were incubated at 37 °C for 1 h with Duolink® in situ PLA® Anti-rabbit PLUS and Anti-mouse MINUS (Sigma-Aldrich, St. Louis, Missouri, U.S.) and later washed with 0.01% PBS-tween. Ligation and amplification steps were performed using Duolink® in situ Detection reagents Red (Sigma-Aldrich, St. Louis, Missouri, U.S.). Ligase was incubated for 30 min at 37 °C and washed with 0.01% PBS-tween. Polymerase was incubated for 100 min and washed with 1x and 0.01x SSC buffer respectively for 5 min each. Nuclei were detected with DAPI Fluoromount-G® (Southern Biotech, Birmingham, Alabama, U.S.). All the washes were performed three times for 5 min, at room temperature with agitation. All the incubations at 37 °C were performed in a humidity chamber. Images were obtained using a Leica DMi8 with DC9000GT camera and a 63x oil immersion objective. Interactions were counted manually and expressed as number of puncta per number of nuclei in each field. Puncta per cell of each condition is expressed as percentage over the control condition.
2.3. Calcium imaging
Cells were seeded at a density of 25,000 cells/well in a 6-well plate. 16–20 h after, cells were transfected with pcDNA-4mtD3cpv plasmid using Lipofectamine® 3000 Transfection kit (Thermo Fisher). 24 h after transfection, cells were trypsinized and seeded in a μ-Slide 8 Well chamber (Ibidi, Gräfelfind, Germany, 80,826), suitable for life imaging. Measurements were carried out 48 h after transfection. For all the experiments with carbachol (St. Louis, Missouri, U.S., 212,385), medium was removed prior the beginning of the recordings and replaced with HBSS without Ca2+ (Thermo Fisher Scientific, Waltham, Massachusetts, U.S., 14175095). D3cpv was excited at 435 nm and fluorescence measured at 450–480 nm (CFP) and 520–540 nm (YFP). Images were recorded every 5 s using a DC9000GT camera (Leica, Germany) and a W-view Gemini (Hamamatsu, Hamamatsu city, Japan) for optic splitting imaging. 25 μM, 250 μM, 500 μM or 900 μM carbachol was added at 60 s from the beginning of the experiment. FRET ratio (YFP/CFP) was calculated by measuring fluorescence intensity of the two emission wavelengths, where CFP channel represents fluorophore unbound to Ca2 and YFP, Ca2+-bond. A range of 15–40 cells were analyzed in each condition per replicate.
2.4. Lipid peroxidation measurements
In order to measure lipid peroxidation, cells were loaded with 5 μM C11 BODIPY (581/591) (Thermo Fisher Scientific, Waltham, Massachusetts, U.S.) for 30 min and 4 μg/μL DAPI for 5 min prior to the beginning of the experiments. Fluorescence intensity was detected by flow cytometry with a BD FACSAria™ III cytometer equipped with 5 lasers, 355, 405, 488, 561, 640 nm (BD, DIVA 6.0 software). Parameters FS-A, FS-H, SS-A were used for cell morphology, lipid peroxidation was measured with C11 BODIPY fluorochrome by selecting the RATIO parameter (586/530 nm) the discrimination of dead cells was performed with DAPI (excitation 405, emission 450/50). 5000 single events (FS-A vs FS-H) were acquired at medium speed. The mean fluorescence of the RATIO parameter of living cells was determined.
2.5. Purification and isolation of mitochondria, MAMs and ER from SH-SY5Y cells
The following protocol is based in the one described in Ref. [36] with some modifications. Cell cultures at 80–90% confluence were harvested with trypsin and the pellet washed with 1 ml 1x PBS and centrifuged at 700 g for 3 min. The supernatant was removed, and the pellet resuspended in 500 μl of hypotonic buffer (250 mM Sucrose, 20 mM HEPES pH 7.45, 10 mM KCl, 1.5 mM MgCl2, 1 mM EDTA, 1 mM EGTA, Protease inhibitors) and kept on ice for 30 min. Cells were then disrupted by passing them through a 30G needle for 20 times. The solution was centrifuged at 700 g for 10 min at 4 °C and the supernatant transferred to a 2 ml clean tube. The pellet containing nuclei and intact cells was discarded. The collected solution was centrifuged at 10,000 g for 20 min at 4 °C. The supernatant containing the ER was transferred to a clean 2 ml tube and the pellet containing the crude mitochondria was resuspended in 2 ml isolation medium (250 mM mannitol, 5 mM HEPES pH 7.45, 0.5 mM EGTA, 0.1% BSA). 150 μl of the last suspension was kept as the crude mitochondria (c-mito) fraction. The tube containing the ER was filled with hypotonic buffer, centrifuged at 17,000 g for 35 min at 4 °C, and the new supernatant transferred to an ultracentrifuge tube. The tube was filled with hypotonic buffer and centrifuged at 100,000 g for 1 h at 4 °C. The supernatant was discarded, and the pellet resuspended in 150 μl of hypotonic buffer to obtain the ER fraction. The rest of the solution of isolation medium containing crude mitochondria was transferred to an ultracentrifuge tube filled with 30% Percoll with gradient buffer (30% Percoll, 225 mM mannitol, 25 mM HEPES pH 7.45, 1 mM EGTA, 0.1% BSA) and centrifuged at 95,000 g for 30 min at 4 °C. Two bands or fractions are observed at this point, an upper band or light fraction containing the MAMs and a lower band or heavy fraction containing the pure mitochondria. The lower band was collected with a glass Pasteur pipette, diluted in a clean tube with isolation medium and centrifuged at 6300 g for 10 min at 4 °C. The supernatant was discarded, and the pellet resuspended in 150 μl of isolation medium to obtain the pure mitochondria (p-mito) fraction. The previous upper band containing the MAMs was collected with a glass Pasteur pipette, diluted in isolation medium and centrifuged at 6300 g for 10 min at 4 °C. The supernatant was transferred to an ultracentrifuge tube filled with isolation medium and centrifuged at 100,000 g for 1 h at 4 °C. The supernatant was discarded, and the pellet was resuspended in 150 μl of isolation medium to obtain the MAM fraction.
2.6. Western blot analysis
The isolated ER, c-mito, p-mito and MAM fractions were resuspended in lysis buffer, separated on SDS-polyacrylamide gel, and blotted onto PVDF membranes. Western blots were probed with antibodies against specific protein markers to verify the purity of the fractions and the presence of frataxin. The primary antibodies used were anti-ATP5A (Abcam, Cambridge, United Kingdom, ab110273), anti-Sigma1R (Sigma-Aldrich, St. Louis, Missouri, U.S., HPA018002), anti-GRP75 (Santa Cruz Biotechnology, Dallas, Texas, U.S., sc-13967), anti-VDAC1 (Abcam, Cambridge, United Kingdom, ab14734) and anti-FXN (Abcam, Cambridge, United Kingdom, ab110328). Secondary antibody (anti-mouse or anti-rabbit IgG) coupled to peroxidase was used for detection of the reaction with Amersham ECL Prime (GE Healthcare, Chicago, Illinois, U.S.). Imaging was performed with an ImageQuant LAS4000 (GE Healthcare, Chicago, Illinois, U.S.).
2.7. Drosophila maintenance and stocks
Fly stocks were maintained at 25 °C on standard cornmeal-agar medium (water: 36 l; yeast: 720 g; cornmeal: 3200 g; soy flour: 400 g; light malt extract: 3200 g; sugar beet syrup: 880 g; agar: 320 g, Nipagin 120 g). The crosses between the GAL4 drivers and the UAS responder lines were carried out at 25 °C. UAS constructs were used in heterozygous configurations for all experiments. Genetic interactions were carried out by mating virgin females from the stocks fhRNAi-1/CyO; Repo-GAL4/TM6B tub-GAL80 or fhRNAi-2/CyO; actin-GAL4/TM6B tub-GAL80 with males of the corresponding UAS line. The ability of these two RNAi strains to silence frataxin expression in several tissues by means of the UAS-GAL4 system has been recently reviewed [37]. The stocks used in this work are described in Table S1.
2.8. Hyperoxia and negative geotaxis assays
In all experiments (normal conditions and food supplementation), flies were raised at 25 °C under a 12hr:12hr light/dark cycle and male individuals were collected within 24–48 h post-eclosion, placed at a density of 25 per vial and transferred to vials with fresh food every 2–3 days. Lifespan experiments were conducted in standard cornmeal agar medium and 100–150 males per genotype were used in each experiment. Hyperoxia treatment started one day post-eclosion and was performed by exposing flies in a glass container with a constant flux of 99.5% oxygen under a low positive pressure at 25 °C [31]. Flies were transferred every day to new vials. Locomotor assays were performed as previously described [31]. 15–20 flies per genotype were assessed and each fly was recorded 3 times and the mean value of each fly was used for the subsequent analysis.
2.9. Semiquantitative real time PCR from Drosophila samples
Total RNA was extracted from 15 whole flies, 15 thoraces or 50 heads using peqGold TriFast reagent (PEQLAB Biotechnologie GMBH, Erlangen, Germany) following manufacturer's instructions. 500 ng mRNA were converted into cDNA using QuantiTect Rev. Transcription Kit (Qiagen GmbH, Hilden, Germany) and then used for qPCR with ORA qPCR Green ROX L Mix (HighQu, Kralchtal, Germany) on a CFX connect™ Real-Time PCR Detection System (Bio-rad, Hercules, California, U.S.). The ribosomal protein 49 (rp49) was used as internal control. The results from at least four independent biological replicates were analyzed with the Bio-Rad CFX manager 3.1 software. Gene expression levels were referred to the internal control, the relative quantification was carried out by means of the ΔΔCt method and the results were plotted as relative mRNA expression. Each experiment consisted of 3–5 independent biological replicates. The genes and primers used for the analysis are: MCU - Fw: 5′-CGTCCTGCACCATCGAAAG-3’; Rv: 5′-GTTTGGGAGGATTCACATCCAAT-3′ RP49 - Fw: 5′-CCAAGCACTTCATCCGCCACC-3’; Rv: 5′-GCGGGTGCGCTTGTTCGATCC-3’.
2.10. Analysis of mitochondrial DNA (mtDNA) levels
For DNA extraction, 15 male flies were homogenized in 400 ml Buffer A (100 mM Tris, pH7.5, 100 mM EDTA, 100 mM NaCl, 0.5% SDS). After incubation at 65 °C for 60 min with continuous shaking, 800 ml of LiCl/KAc (5 M KAc and 6 M LiCl in a relation 1:2.5) solution was added, followed by 10 min incubation on ice. After centrifugation (15 min, 13,000 rpm, RT), 1 ml supernatant was transferred in a new tube. The DNA was precipitated by adding 600 ml of isopropanol and vortexing. After centrifugation (15 min, 13,000 rpm, RT) the resulting pellet was washed with 70% ethanol and resuspended in 200 μl H2O. Isolated DNA was quantified by measuring the absorbance at 260 nm with a NanoDrop spectrophotometer. In order to estimate the amount of mtDNA, the levels of mitochondrial cytochrome oxidase I (Fw 50-AACTGTTTACCCACCTTTATCTGCTB-30 and Rv 50-CCCGCTAAGTGTAAAGAAAAAATAGC-30) were referred to the levels of the housekeeping gene GAPDH (Fw 50-GACGAAATCAAGGCTAAGGTCG-30 and Rv 50-AATGGGTGTCGCTGAAGAAGTC-30), using 10 ng of Template DNA per reaction. Reactions were performed with qPCR Green ROX L Mix (HighQu, Kralchtal, Germany) on a CFX connectTM Real-Time PCR Detection System (Bio-rad, Hercules, CA, USA). The relative quantification was carried out by means of the ΔΔCt method and the results were plotted as Relative mtDNA content. Each experiment consisted of 2–3 independent biological replicates.
2.11. Analysis of Drosophila brain integrity and brain lipid homeostasis
Paraffin sections were performed from 35-day-old adult flies. Flies were fixed with carnoy (ethanol:chloroform:acetic acid at a proportion 6:3:1), dehydrated in ethanol, and embedded in paraffin. Paraffin sections (7 μm) from 10 flies of each genotype were analyzed under a fluorescence microscope. Brain vacuolization was quantified from 7 brains per genotype using the Image J 1.48v software. The degeneration was analyzed calculating the affected area that was referred as % of total brain area that includes the lamina (La), the outer chiasm (OC) and the medulla (Me). This analysis was complemented by quantifying the number of vacuoles per genotype in the region of interest (ROI). For examination of accumulation of lipid droplets, flies of appropriate age and genotype were fixed in 4% PFA for 2 h. Brains were then dissected, washed with PBST and incubated 24 h at 4 °C with the Bodipy™ 493/503 (D3922, Thermo Fischer Scientific, MA, USA) at a dilution 1:500 (from a stock solution 1 mg/ml in EtOH). Samples were embedded in Vectashield mounting medium (Vector Laboratories, Burlingame, CA, U.S.). At least 5–10 flies per genotype were scanned using the Confocal Laser Scanning Platform Leica SP8 (Leica, Germany). Samples were excited with an argon laser at 488 nm (20% tube current). Signals were detected using a HC PL APO 40x/1.30 Oil CS2 objective at 490–540 nm. Images were generated at a resolution of 1024 × 1024 pixels. Brains were scanned in z-stacks (1 μm) with 30–35 images per brain. All images were acquired using the same exposure, light intensity and filter settings. Confocal images were further processed with the image processing software Fiji 2.0.0 [38]. In detail, background was subtracted in Fiji via the Rolling Ball method (Radius = 50 pixels). Maximum projections of 30 slices were made and the resulting image was again subjected to background subtraction. Finally, contrast of each image was adjusted to improve quality of signal. In the experiment expressing the UAS-ChiMERA construct, lipids droplets were studied with light microscopy because the ChiMERA protein is coupled to GFP [39] and this prevents the correct observation of Bodipy™ 493/503 signals. In this case, semithin epon plastic sections of 5 flies from 35-day-old adult fly brains were prepared and lipids were identified as characteristic dark droplet-like structures and quantified manually.
2.12. Quantification of ATP amount
ATP was determined using ATP Bioluminescence Assay Kit HS II (Roche, Mannheim, Germany) according to manufacturer's instructions with some modifications. Briefly, five flies were homogenized in 200 μl pre-heated ATP assay buffer (100 mM Tris, 4 mM EDTA, pH 7.75), boiled for 2 min and centrifuged for 1 min at 1000 g. The supernatant was diluted 1:5 in ATP assay buffer and 50 μl of the dilution were used to measure ATP levels. Luminescence was detected with a Tecan Spark™ 10 M plate reader (Tecan Trading AG, Switzerland). Each experiment consisted of 3–5 independent biological replicates. ATP quantities were finally referred to protein amount using the Bradford assay (Coomassie Plus™ Protein Assay Reagent, Thermo Scientific, Schwerte, Germany).
2.13. Statistical analysis
Survival data were analyzed using the Log-rank (Mantel-Cox) and Gehan-Breslow-Wilcoxon tests. In all further experiments, data is represented as mean ± s.e.m of three to five replicates. When comparing 2 samples, equal variances were confirmed by an F test. When comparing multiples samples, equal variances were confirmed by Bartlett and Brown-Forsythe tests. Normality of data was assessed in all cases with Shapiro & Wilk test and parametric or non-parametric tests were used accordingly. For data that passed normality test, significance was determined by two-tailed T-test or by One-way ANOVA with post hoc Dunnett or Tukey Multiple Comparison Test (***, P < 0.001; **, P < 0.01 and *, P < 0.05). Samples that failed normality test were analyzed using non-parametric tests such as Mann-Whitney test or Kluskal-Wallis with post hoc Dunn's Test (***, P < 0.001; **, P < 0.01 and *, P < 0.05). Statistical analysis was carried out using Prism version 8.03 for Windows (GraphPad Software, La Jolla, California, USA).
3. Results
3.1. Frataxin deficiency alters MAMs architecture
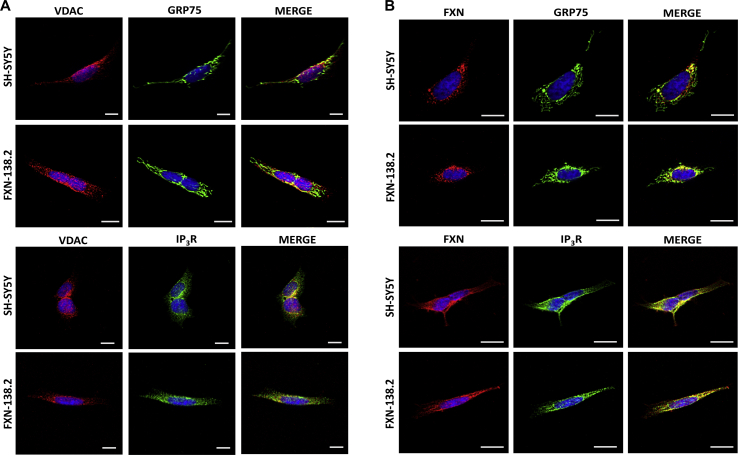
MAMs encompass a wide variety of proteins, from membrane channels to lipid and glucose metabolism enzymes [40]. One of the best characterized tethers that mediates the exchange of Ca2+ between both organelles consists of the voltage-dependent anionic channel 1 (VDAC1), located in the mitochondrial outer membrane, the inositol-1,4,5-trisphosphate receptor (IP3R) in the ER membrane, and the chaperone glucose regulated protein 75 (GRP75), which links the former two.
To better understand if frataxin levels are able to modify MAMs’ stability or expression, we assessed the connections between the ER and mitochondria in frataxin-depleted cells by performing a proximity ligation assay (PLA) (Fig. 1A–C) using a neuroblastoma cell line as a model study. Neuroblastoma is a developmental tumor originated from neural crest, like DRG neurons. FXN-138.1 and FXN-138.2 lines were generated through the silencing of the FXN gene using two different hairpins, obtaining 82 and 78% of frataxin reduction, respectively [18]. First, antibodies were validated by immunofluorescence to determine the correct working dilutions before performing the PLA experiments. This way, we ensured the specificity of the PLA technique (Fig. S1).
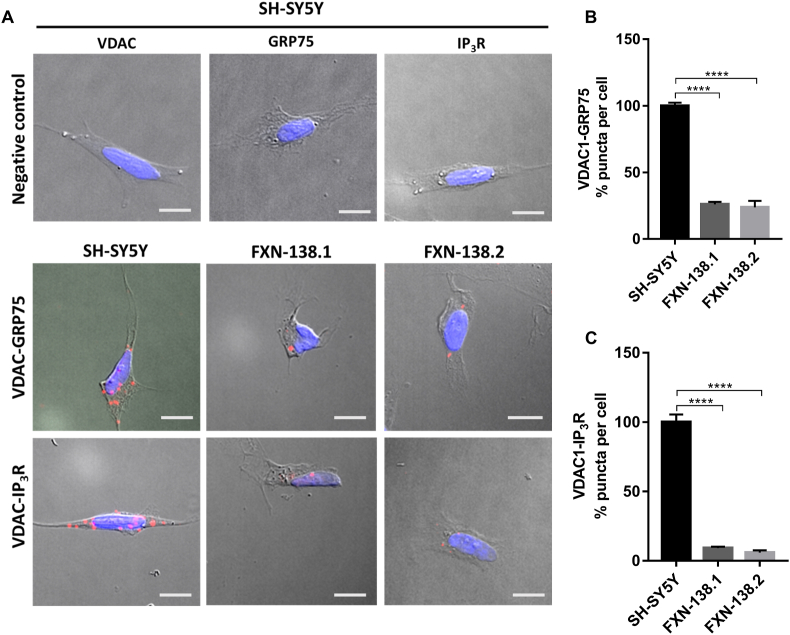
Fig. 1.
MAMs interactions are decreased in frataxin-deficiency cells. (A) Representative microscopy images of the PLA evaluating the interactions between VDAC1-GRP75 and VDAC1-IP3R in baseline conditions. Red dots represent the physical interactions of the two proteins and nucleus is marked in blue with DAPI. VDAC1, GRP75 and IP3R antibodies were used as negative controls in SH-SY5Y cell line (Upper panel). Scale bars: 12 μm (B) Normalized levels of interactions between VDAC1-GRP75 (puncta) per cell in each frataxin-silenced clone compared to SH-SY5Y control. (C) Normalized levels of interactions between VDAC1- IP3R (puncta) per cell in each frataxin-silenced clone compared to SH-SY5Y control. Results (n ≥ 4) are represented as mean ± SEM. A minimum of 500 cells were analyzed in each independent experiment. ANOVA significance: ****p ≤ 0.0001 over SH-SY5Y. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Upon frataxin downregulation, we observed a dramatic decrease in the number of interactions between VDAC1 and GRP75 compared to control cells (SH-SY5Y, 100 ± 2%; FXN-138.1, 26 ± 1.91%; FXN-138.2, 23.78 ± 4.9%) (Fig. 1B). Accordingly, interactions between VDAC1 and IP3R were also reduced (SH-SY5Y, 100 ± 5.48%; FXN-138.1, 9.14 ± 1.07%; FXN-138.2, 5.99 ± 1.5%) in cells with deficiency in frataxin (Fig. 1C). Our results indicate that the mitochondrial-endoplasmic reticulum interaction is disturbed in frataxin deficient cells and such impairment could contribute to the mitochondrial Ca2+ dysregulation described in frataxin deficiency models, including our previous results [18].
3.2. Trolox recovers MAMs interactions and Ca2+ transfer into mitochondria
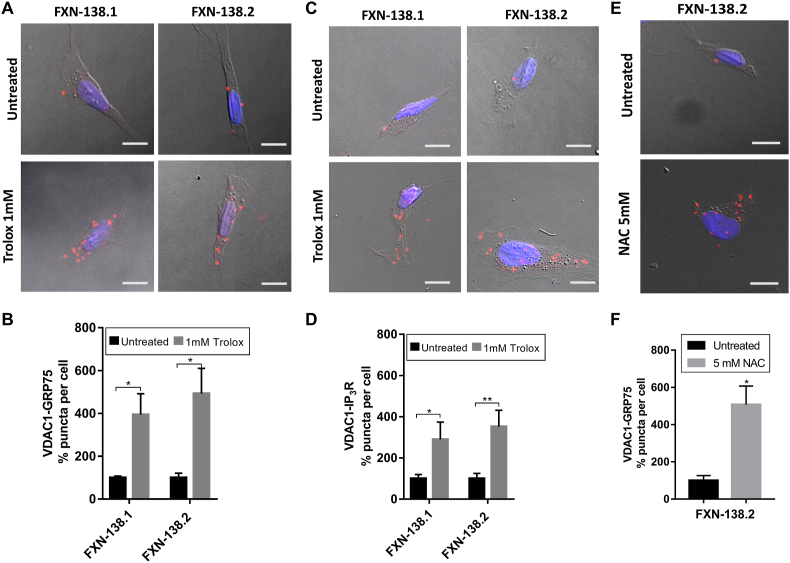
In order to elucidate the likely relationship between MAMs and Ca2+ dyshomeostasis in FRDA, we decided to treat our FRDA cells with the purpose of improving Ca2+ management. Trolox is a water-soluble analogue of Vitamin E that acts as lipid peroxidation scavenger and therefore contributes to the stabilization of cellular membranes [41]. Interestingly, Abeti and coworkers have recently shown that vitamin E recovers mitochondrial Ca2+ uptake in frataxin-depleted cerebellar granule neurons and cardiomyocytes [42]. Thus, we have analyzed the impact of Trolox in the integrity of the ER-mitochondrial contacts. We incubated FXN-138.1 and FXN-138.2 clones with 1 mM Trolox for 24 h. Importantly, we observed a marked increase (FXN-138.1: untreated 100 ± 19.8%, treated 290.1 ± 83.79%; FXN-138.2: untreated 100 ± 25%, treated 352,2 ± 79.4%) in the number of interactions between VDAC1-GRP75 (Fig. 2A–B), which was also confirmed with the VDAC1-IP3R interactions analysis (Fig. 2C–D). Our results indicate that Trolox restores communication between these two compartments, probably by reducing oxidative stress environment in mitochondrial and ER membranes.
Fig. 2.
Antioxidants rescue MAMs connections in frataxin-deficiency cells. (A) Representative microscopy images of the PLA evaluating the interactions between VDAC1 and GRP75 in FXN-138.1 and FXN-138.2 cells after 1 mM Trolox treatment compared to untreated conditions. (B) Normalized number of VDAC1-GRP75 interactions (puncta) per cell in each frataxin-silenced clone treated with Trolox compared to untreated conditions. (C) Representative microscopy images of the PLA evaluating the interactions between VDAC1 and IP3R in FXN-138.1 and FXN-138.2 cells after 1 mM Trolox treatment compared to untreated conditions. (D) Normalized number of VDAC1-IP3R interactions (puncta) per cell in each frataxin-silenced clone treated with Trolox compared to untreated conditions. (E) Representative microscopy images of the PLA evaluating the interactions between VDAC1 and GRP75 in FXN-138.2 cells after 5 mM NAC treatment compared to untreated conditions. (F) Normalized number of VDAC1-GRP75 interactions (puncta) per cell in FXN-138.2 cells treated with NAC compared to untreated conditions. Results (n ≥ 4) are represented as mean ± SEM. A minimum of 500 cells were analyzed in each independent experiment. Student's t-test significance: *p ≤ 0.05; **p ≤ 0.01 over untreated cells.
Furthermore, we also treated FXN-138.2 cells with 5 mM N-acetylcysteine (NAC). NAC is a versatile thiol molecule related to three main antioxidant mechanisms in the cell. It has both direct and indirect antioxidant effects over oxidative species, including the restorage of thiol groups and cysteine donor in Glutathione synthesis, regulating the redox state [43]. In agreement with the Trolox results, we observed a recovery in the number of VDAC1-GRP75 interactions (FXN-138.2: untreated 100 ± 26%, treated 506.95 ± 100.46%) (Fig. 2E–F).
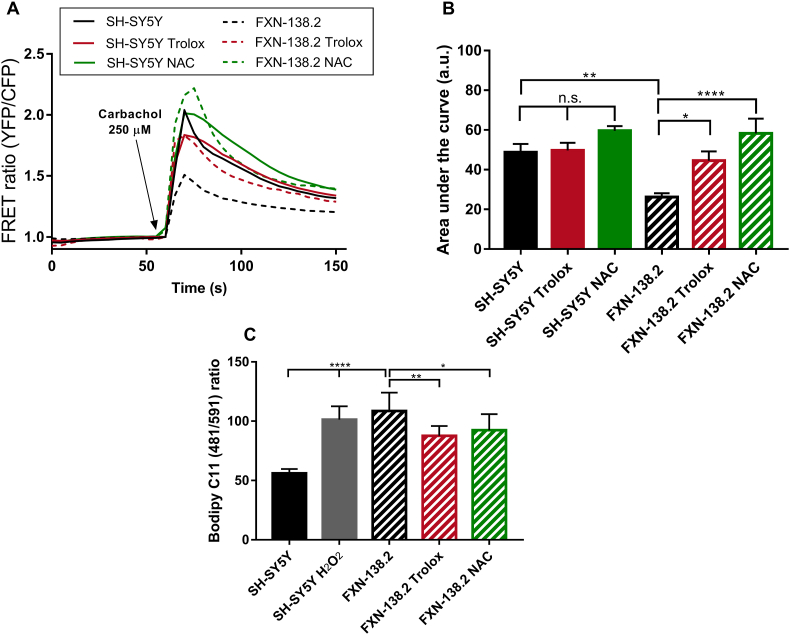
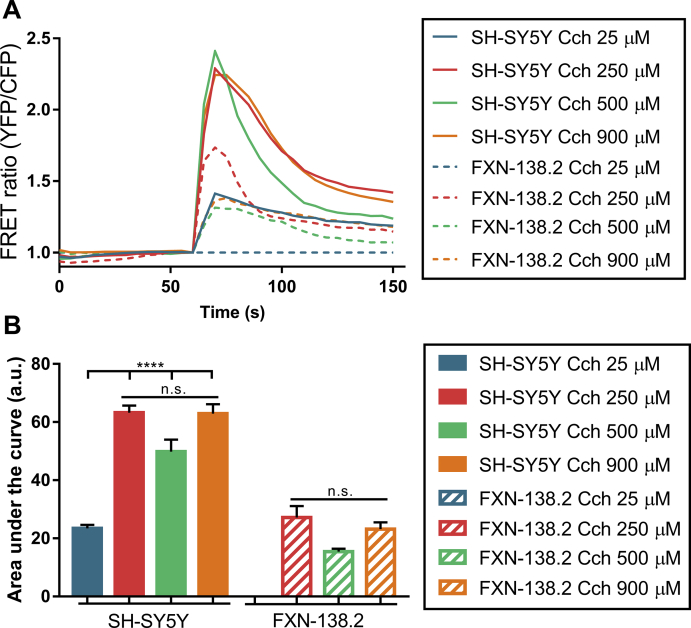
These results prompted us to analyze in our model the mitochondrial capability of Ca2+ uptake after antioxidant treatments. FXN-138.2 and control cells were transfected with a mitochondrially targeted chameleon (4mtD3cv) and challenged with carbachol in order to release ER Ca2+ through IP3R. Concentration curves of carbachol indicated 250 μM was the most suitable concentration that gave the maximum response (Fig. S2). Our results clearly show that cells with frataxin deficiency displayed a significant lesser response to carbachol compared to control cells (Fig. 3A). Additionally, SH-SH5Y cells do not show significant changes after Trolox or NAC treatment in the area under curve analysis (SH-SY5Y: Untreated 48.79 ± 4.14, Trolox 49.71 ± 3.87, NAC 59.6 ± 2.37). Importantly, the analysis of the mitochondrial response to carbachol showed that both compounds successfully boosted Ca2+ uptake by mitochondria in cells with frataxin deficiency (Fig. 3B, FXN-138.2: Untreated 26.17 ± 1.97, Trolox 44.54 ± 4.64, NAC 58.32 ± 7.38). These results support the hypothesis that Trolox and NAC recover the network connecting mitochondria and ER and this is sufficient to improve Ca2+ transfer between both compartments in frataxin-silenced cells.
Fig. 3.
Antioxidants recover mitochondrial calcium uptake and alleviate lipid peroxidation in FXN-138.2 cells. (A) FRET analysis of mitochondrial calcium levels measured through 4mtD3cpv chameleon. 250 μM Carbachol was used to stimulate Ca2+ release from the ER through IP3R channels. Cells were then treated either with 1 mM Trolox or 5 mM NAC for 24 h. Results (n ≥ 3) are represented as normalized levels of fluorescence intensity over carbachol loading value. A range of 5–34 cells were analyzed in each condition per replicate. (B) Area Under curve (AUC) analysis of the mitochondrial response to 250 μM carbachol. (C) Flow cytometry detection of lipid peroxidation. SH-SY5Y cells were treated with 500 μM H2O2 as positive control, and FXN-138.2 were treated either with 1 mM Trolox or 5 mM NAC for 24 h. Cells were stained with 5 μM Bodipy C11 (581/591) for 30 min. Results (n ≥ 4) are represented as 481/591 ratio of the fluorescence intensity of the dye (mean ± SEM). ANOVA significance: *p ≤ 0.05; **p ≤ 0.01; ****p ≤ 0.0001.
After confirming that antioxidants improve ER-mitochondria communication, in both structural and functional ways, we analyzed whether its effects in reducing lipid peroxidation might be underpinning the observed rescue. Quantification of lipid peroxidation by means of Bodipy C11 (481/491) fluorescence intensity and flow cytometry (Fig. 3C) confirmed a significant increase of these oxidative species in our frataxin-deficient cells. Moreover, we also detected that both Trolox and NAC, were able to alleviate lipid peroxidation in FXN-138.2 cells, suggesting that oxidative insults affect MAMs integrity in FRDA cells. These results reinforce the idea of an antioxidant-dependent recovery of MAMs and suggest oxidative stress has an important role in Ca2+ homeostasis impairment in our FRDA model.
3.3. Frataxin is a member of the protein network of MAMs
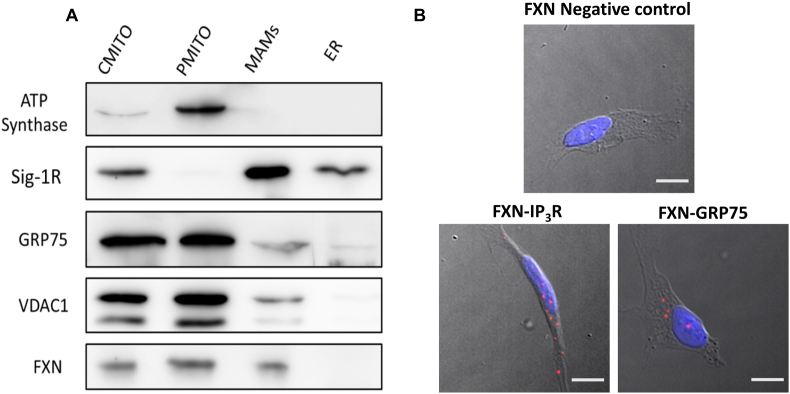
Frataxin has been extensively considered as a mitochondrial protein [44], but interestingly some authors have also described an additional extramitochondrial location [45,46]. Thus, given the reduced levels of MAMs interactions present in our frataxin silenced models, we wondered whether frataxin might be also present at the core of this structures. To elucidate this question, we isolated in SH-SY5Y cell line the subcellular fractions implicated in the architecture of MAMs (mitochondria, MAMs, ER) and performed a Western blot analysis in order to assess the presence of frataxin in the different fractions. Interestingly, we observed that frataxin is clearly located in MAMs, in addition to the characteristic mitochondrial presence (Fig. 4A).
Fig. 4.
Frataxin is located in MAMs and interacts with two of its main proteins. (A) Representative Western blot analysis of frataxin after the isolation of the subcellular fractions implicated in MAMs. ATP Synthase was used as mitochondrial marker. Sig-1R was used as MAMs marker. ATP-synthase, Sig-1R, GRP75 and VDAC1 were used as subcellular fraction markers. Results are representative of at least three independent experiments. CMITO: crude mitochondria fraction (PMITO + MAMs), PMITO: pure enriched mitochondria fraction, MAMs: Mitochondria-ER Associated membranes fraction, ER: endoplasmic reticulum fraction. (B) Representative microscopy images of the PLA evaluating the interactions between frataxin-IP3R and frataxin-GRP75 in baseline conditions. Red dots represent the physical interactions of the two proteins (either frataxin-IP3R or frataxin-GRP75). DNA is marked in blue with DAPI. Frataxin antibody was used as negative control. Scale bars: 12 μm. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
In order to further investigate the possibility of a direct implication of frataxin in the structure of MAMs, we performed a PLA by analyzing the interaction in SH-SY5Y cells of frataxin with GRP75 and IP3R, two of the main proteins associated to the mitochondria-ER connections. Importantly, we observed a clear and robust direct relationship of frataxin with both proteins (Fig. 4B), which suggests a pivotal role of frataxin in the regulation and maintenance of this protein network. Interestingly, the interaction between frataxin and GRP75 has already been described by Dong and collaborators [47]. So, our data do not only confirm the interaction between the proteins, but also represent a positive control of our PLA-based approach. Altogether, these results indicate that frataxin could have a direct role in the regulation of the contact sites set by mitochondria and the ER.
3.4. The induction of ER-mitochondrial contacts recovers brain degeneration in frataxin deficient flies
Next, we decided to evaluate whether the observations obtained in our cellular model might be also relevant in a multicellular organism. We chose a Drosophila model in which frataxin silencing was targeted to glia cells because in this model, the interactions between ER and mitochondria have already been shown to participate in the development and progression of the pathology [21]. The loss of frataxin in Drosophila glia cells using the Repo-GAL4 line (Repo-G4) and a strong RNAi line (fhRNAi-1) induces 3 main defects, a locomotor dysfunction, a strong brain vacuolization and the accumulation of lipids within the fly brain [31].
In our first approach, we decided to express the ChiMERA construct in glia cells. The ChiMERA protein is the fusion of the mitochondrial protein Tom70 and the ER protein Ubc6. This combination of proteins bridges the mitochondria and the ER membranes. Remarkably, this has been shown to successfully increase functional mitochondria-ER contacts in yeast and, more importantly, also in flies [39,48]. Thus, we assessed whether promoting the formation of bridges between ER and mitochondria was sufficient to recover loss of frataxin in the fly glial cells. Fig. 5A shows the ROI as well as the anatomic areas analyzed (La, Me, and OC). Our experiments showed that compared to age-matched control flies (Fig. 5A), glial downregulation of frataxin triggered a vacuolization of the lamina, medulla and the outer chiasm (asterisk in Fig. 5B and quantification in Fig. 5D). Interestingly, expression of ChiMERA strongly reduced brain degeneration in terms of number of vacuoles and degenerated area (Fig. 5C and quantification in Fig. 5D). However, no improvement on the lipid accumulation (arrows in Fig. 5B–C and quantification in Fig. 5E) or on the locomotor performance of the flies (Fig. 5F) was observed. Therefore, increasing mitochondrial-ER contacts seems to have a positive albeit minor impact on FRDA phenotypes in glia.
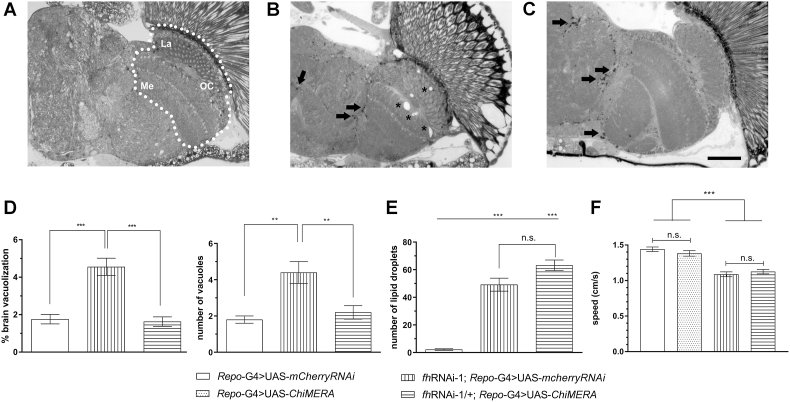
Fig. 5.
Effects of promoting ER-mitochondrial contacts in frataxin-deficient glia. (A–C) Representative light microscopy images of semithin plastic sections from controls (A) or brains from frataxin-deficient flies co-expressing either a control UAS (B) or the ChiMERA construct (C). Asterisk indicate the presence of the vacuolization and arrows denote the presence of lipid deposits. (D) Quantification of area occupied by vacuoles as well as their number. Compared to controls (Repo-G4>UAS-mCherryRNAi, white column), frataxin deficient flies (fhRNAi-1; Repo-G4>UAS- mCherryRNAi, vertical stripes) display a higher vacuolization levels in terms of affected area and number of vacuoles that were reverted to control values by co-expression of ChiMERA (fhRNAi-1;Repo-G4>UAS-ChiMERA, horizontal stripes). (E–F) Compared to controls (Repo-G4>UAS-mCherryRNAi, white column), co-expression of ChiMERA (fhRNAi-1;Repo-G4>UAS-ChiMERA, horizontal stripes) failed to restore lipid accumulation (E) and impaired locomotion (F) of frataxin deficient flies (fhRNAi-1;Repo-G4> UAS-mCherryRNAi, vertical stripes). Graphs represent means ± SEM. In D, E and F, data was analyzed by One-way ANOVA with post-hoc Tukey Multiple Comparison test. **, P < 0.01; ***, P < 0.001, Scale bar: 50 μm. La, Me and OC stand for lamina, medulla and outer chiasm, respectively and n.s. stands for not significant.
3.5. Promotion of Ca2+ import into the mitochondria recovers frataxin-deficient phenotypes in Drosophila
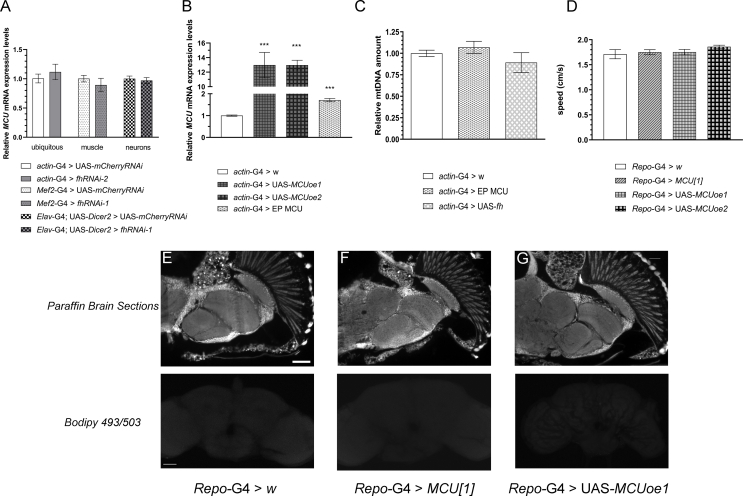
The observations that reactivation of MAMs successfully increased mitochondrial Ca2+ uptake in our cell culture model, encouraged us to manipulate Ca2+ transport into the mitochondria in our in vivo Drosophila model. Although we did not observe altered expression levels of the mitochondrial Ca2+ uniporter (MCU) in frataxin deficient flies (Fig. S3A), we decided to genetically manipulate its expression since misexpression of MCU (or other components of the MCU complex) has been proven to alter the mitochondrial Ca2+content [[49], [50], [51], [52]]. First, we speculated that MCU downregulation should worsen the phenotypes associated to frataxin silencing. In agreement, MCU silencing using a RNAi line triggered preadult lethality when co-expressed with the fhRNAi-1 line, whereas such lethality was not observed in the control cross. To bypass this problem, we decided to use a null mutant allele of MCU (MCU[1], described in Ref. [52]) in a heterozygous configuration in order to achieve 50% loss of MCU expression. Combination of frataxin-deficiency in glia cells and this level of reduction of MCU expression failed to exacerbate locomotor deficit of FRDA flies (Fig. S4A) and the brain vacuolization (Fig. S4B and C–E upper panel). In order to detect the accumulation of lipids in vivo, we have used a probe based on a Bodipy fluorophore that allows detection of neutral lipids (see material and methods). As described by Kis and collaborators [53] no lipids were detected in control flies (Fig. S4C lower panel). In agreement with our previous results [31] loss of frataxin in glia cells triggered a strong accumulation of lipids (Fig. S4D lower panel). Reduction of MCU expression also did not influence the presence of such lipid deposits (Fig. S4E lower panel). No defect or dysfunction in any of the three parameters analyzed (negative geotaxis, brain integrity and brain lipid levels) was observed in the heterozygous MCU[1] flies when compared to controls (Fig. S3D-F).
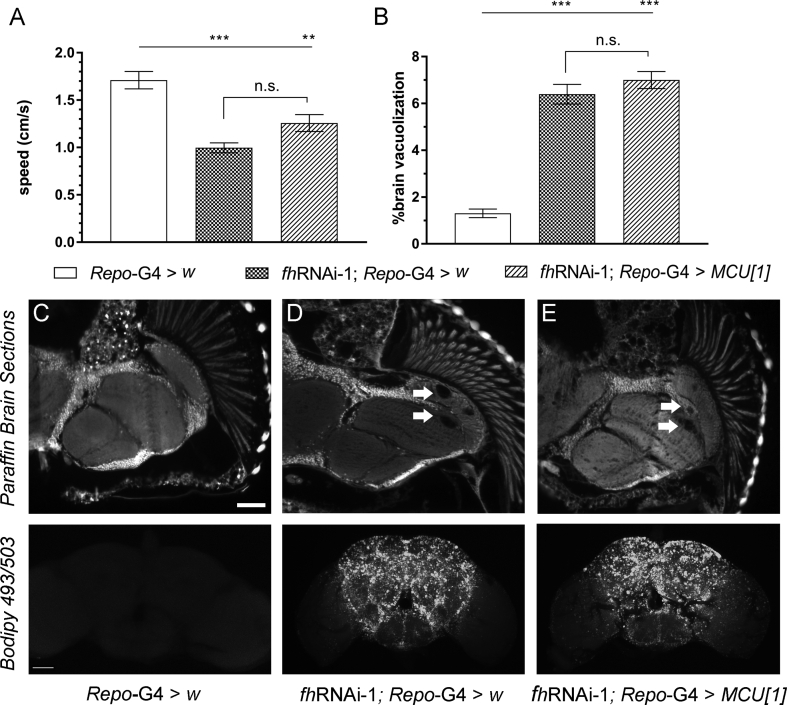
Next, we decided to increase MCU expression by means of two independent UAS lines [52]. Both lines trigger a 10-fold increase of MCU expression (Fig. S3B). Interestingly, overexpression of MCU in frataxin deficient glia completely suppressed the locomotion impairment (Fig. 6A) as well as the percentage of area affected and the number of vacuoles (quantification in Fig. 6B and representative pictures in Fig. 6C–E upper panels). Visualization of neutral lipid accumulation by means of a Bodipy dye (Fig. 6C–E lower panels) showed that MCU overexpression (Fig. 6E) was sufficient to completely suppress the accumulation of lipids observed in frataxin-deficient flies (Fig. 6D). Since both UAS lines display similar MCU mRNA levels (Fig. S3B) and the same effects in a frataxin-deficient background (as observed in Fig. 6A–B), only histological findings from line 1 (UAS-MCUoe1) are shown and only UAS-MCUoe1 was used in the rest of the experiments. Reduced longevity under oxidative stress conditions (hyperoxia atmosphere) is another phenotype that is displayed by frataxin deficient flies in glia [31]. We confirmed such a defect and observed that, remarkably, overexpression of MCU almost duplicated the median and maximum longevity of FRDA flies under this stressor (Fig. 6F). Finally, we wanted to assess whether such rescues of FRDA phenotypes in the Drosophila glia were directly linked to an improvement of the mitochondrial function. In this case, we used a second frataxin RNAi line because fhRNAi-2 is compatible with a normal development compared to the lethality induced by fhRNAi-1 (reviewed in Ref. [37]) when the silencing of frataxin is ubiquitous (actin-G4). Unexpectedly, the strong ubiquitous overexpression of MCU achieved with the UAS-MCUoe lines led to some deleterious effects even in control flies. Therefore, we decided to take advantage of a Drosophila Stock containing an EP transposable element in the 5′ untranslated region of the MCU promoter. The EP elements also contain UAS sequences in their structure and therefore they can be used to increase the expression of a nearby gene [54]. The EP MCU line only triggers a mild overexpression of MCU (Fig. S3B and [55]). As it can be seen in Fig. 6G, ubiquitous frataxin knockdown (KD) with fhRNAi-2 reduced ATP production around 65% in 20 day-old-flies and MCU overexpression was sufficient to restore ATP production in frataxin deficient mitochondria and to trigger a 70% increase of ATP amounts in a control scenario. It is important to consider that mammalian MCU has been also related to mitochondrial biogenesis [56] and thus an increase in mitochondrial mass would also explain the rescue of ATP amounts in frataxin deficient flies. In order to assess this possibility, we have quantified the amount of mtDNA as an indirect measure of mitochondrial mass using an approach that we previously validated in our laboratory [21]. Our results showed that a mild overexpression of MCU did not altered the amount of mtDNA in the flies (Fig. S3C). Similarly, frataxin overexpression also failed to increase the generation of mitochondria (Fig. S3C). All these results might exclude a compensatory effect of MCU in mitochondrial biogenesis as the main cause underlying the rescue observed in the fly models of FRDA and suggest that lack of mitochondrial Ca2+ as a core element of the FRDA pathology.
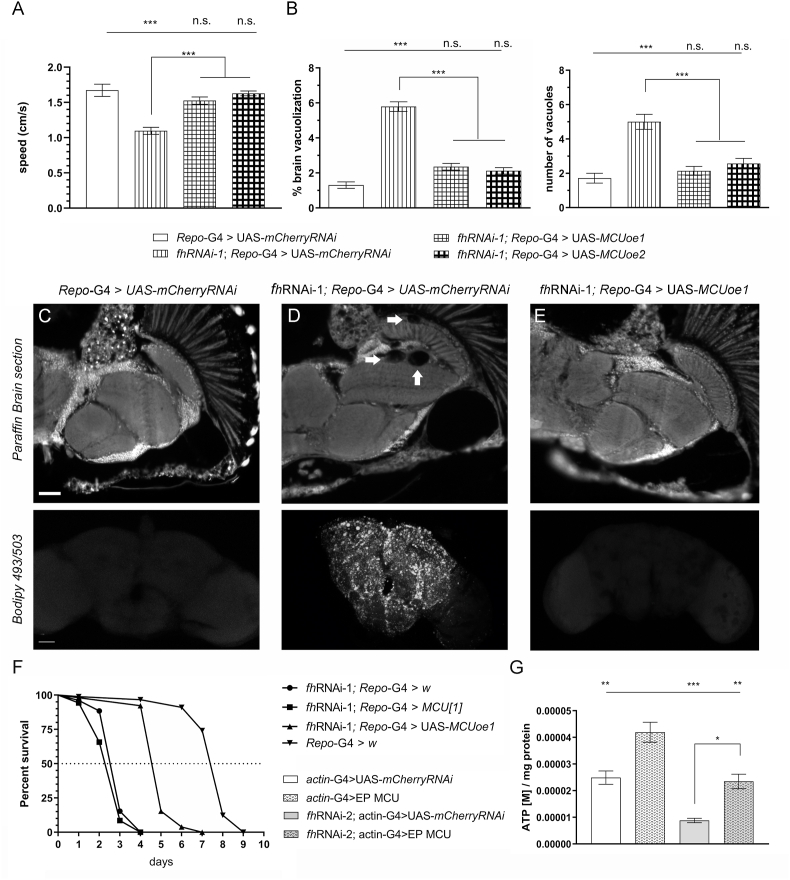
Fig. 6.
Overexpression of Mitochondrial Calcium Uniporter improves several FRDA-like conditions in flies. (A–B) Compared to control flies (Repo-G4>UAS-mCherryRNAi, white column), overexpression of MCU along with frataxin silencing (fhRNAi-1;Repo-G4>UAS-MCUoe1 and fhRNAi-1;Repo-G4>UAS-MCUoe2) successfully restored the locomotor performance (A) as well as the levels of brain degeneration and the number of vacuoles (B) of FRDA flies (fhRNAi-1;Repo-G4>UAS-mCherryRNAi) to normal values. (C–E) Representative paraffin brain sections and Bodipy stainings of controls (C), FRDA flies (D) or overexpression of MCU along with frataxin silencing flies (E). Arrows indicate the presence of the vacuolization in frataxin-deficient brains with normal levels of MCU (D). Importantly, increased expression of MCU in glia was sufficient to reverse the degenerative phenotypes (E, and quantification in B). Complete absence of lipid deposits in controls (C), a strong accumulation in FRDA flies (D) and normal lipid levels upon overexpression of MCU along with frataxin silencing (E). (F) In agreement to our previous results, frataxin silencing in glia shortened longevity under oxidative stress. Importantly, MCU overexpression strongly improved this defect. (G) Similarly, overexpression of MCU (EP-MCU) is sufficient to increase ATP values in controls (actin-G4>EP-MCU) and FRDA flies (fhRNAi-2;actin-G4>EP-MCU). Graphs represent means ± SEM. In A, B and G data was analyzed by One-way ANOVA with post-hoc Tukey Multiple Comparison test. In F, data was analyzed by Log-rank (Mantel-Cox) and Gehan-Breslow-Wilcoxon tests. *, P < 0.05; **, P < 0.01; ***, P < 0.001. Scale bars represent 50 μm.
4. Discussion
Communication between the mitochondria and the ER is especially important in order to maintain a proper Ca2+ transfer required to regulate processes such as energy supply and cell survival. Specially in the nervous system, there are important physiological functions closely related to mitochondrial Ca2+ homeostasis such as synapse assembly, generation of action potentials and synaptic transmission [57]. Variations in the number and structure of MAMs are present in the neurobiology of several neurodegenerative disorders such as Alzheimer's disease (AD), Parkinson's disease (PD), Amyotrophic Lateral Sclerosis, hereditary spastic paraplegia, and peripheral neuropathies. Interestingly, many proteins involved in these diseases have been found in the ER-mitochondria interface and exhibit an implication in the regulation of these structures. For instance, mutations in AD related proteins presenilin 1 and 2 increase ER-mitochondria contacts, upregulating mitochondrial Ca2+ uptake. Also, huntingtin, implicated in Huntington's disease (HD), is essential for ER morphology, so mutated huntingtin activates ER stress and alters Ca2+ transfer in MAMs domain [26,58]. However, little is known about the role of MAMs in the physiopathology of FRDA. In order to address this question, in this work we have performed the first comprehensive analysis of the functionality and integrity of ER-mitochondria contacts also known as MAMs under frataxin-loss conditions.
In the last years we described both in neuroblastoma cells [18] as well as in frataxin deficient sensory neurons of the YG8R mouse model [17], a clear loss of buffering Ca2+ capacity in the mitochondria leading to increased levels of Ca2+ in the cytosol. In agreement, Ca2+ chelators were able to improve FRDA conditions [17]. More recently, Abeti and co-workers also observed that primary cultures of cerebellar granule neurons and cardiomyocytes from the YG8R mouse model presented a reduced mitochondrial Ca2+ uptake that was concomitantly accompanied by lower Ca2+ amount in the mitochondria and in the ER [59]. However, the mechanisms underlying an improper mitochondrial Ca2+ buffering under frataxin loss conditions have not been studied in detail. It is known that Ca2+ is transferred from ER to mitochondria throughout a scaffold formed by IP3R, VDAC in the outer mitochondrial membrane (OMM) and the MCU in the inner mitochondrial membrane (IMM). Therefore, alterations in MAMs in frataxin deficient cells would induce a clear Ca2+ deregulation. Using a novel experimental approach named proximity ligation assay that analyses interactions between proteins, we determined that the number of MAMs was strongly reduced in a neuroblastoma model of FRDA, which correlates with an impairment of Ca2+ buffering by the mitochondria.
In order to analyze the impact of MAMs’ dysfunction in the frataxin-deficiency conditions over Ca2+ dyshomeostasis, we improved the mitochondrial Ca2+ uptake with antioxidant treatments to associate it with a recovery of MAMs interactions. In cardiomyocytes from the YG8R mouse model treated with vitamin E, the mitochondrial capability of Ca2+ buffering was restored [42]. Accordingly, treatment with the vitamin E mimic Trolox also improved Ca2+ buffering capability in our neuroblastoma model. And more importantly, this was possible by increasing the ER-mitochondria contacts. Thus, the recovery of the network connecting mitochondria and ER is sufficient to improve mitochondrial Ca2+ uptake.
To test this hypothesis in an in vivo model, we decided to take advantage of the outstanding tools for genetic studies offered by the fruit fly (Drosophila melanogaster). Drosophila FRDA models have provided along the last decade several evidences about FRDA-dependent mechanisms beyond the state of the art [37]. Our results with the overexpression of MCU are in favor of the hypothesis that increasing the Ca2+ uptake by the mitochondria is neuroprotective in FRDA flies. Using different sensors to visualize and quantify mitochondrial Ca2+, overexpression of MCU has been shown to increase basal concentration of mitochondrial Ca2+ as well as to enhance Ca2+ uptake in drosophila cultured embryonic cells [60] and in living flies [50]. Mitochondrial biogenesis has not been observed in the fly since overexpression of MCU or other components of the MCU complex such as MICU1 and MICU3 did not alter mitochondrial density [60] or mitochondrial porin levels [52]. These observations agree with our results in which MCU overexpression did not increase the amount of mtDNA and subsequently of the mitochondrial mass. Therefore, in our experimental scenario, overexpression of MCU would increase the mitochondrial Ca2+ content and this would be sufficient to recover several loss-of frataxin phenotypes in the fly. It is known that the cellular bioenergetics are sustained by the transfer of Ca2+ from ER to mitochondria via MAMs [61]. Ca2+ seems to influence the activity of some enzymes such as mitochondrial ATP synthase pyruvate dehydrogenase complex (PDC), isocitrate dehydrogenase and α-ketoglutarate dehydrogenase [62]. All these evidences are validated in our in vivo model where we recovered ATP levels due to the reintroduction of Ca2+ in the mitochondria.
There are several lines of evidence highlighting the key role of antioxidants in FRDA [63]. In 2008, Cooper et al. found promising improvements with a combined Coenzyme Q10-Vitamin E therapy in FRDA patients and NAC, a direct donor of cysteines, has recently been used as a Nuclear factor E2-related factor 2 (NRF2) modulator in FRDA fibroblasts, exhibiting neuroprotective features and increasing frataxin expression [64]. We have recovered function and structure of MAMs with NAC or Trolox treatment, demonstrating a direct relationship between Ca2+ dyshomeostasis and oxidative stress. Several authors have described oxidative stress together with Ca2+ dysregulation in different FRDA models [19,42,65,66]. It is interesting to emphasize that lipid peroxidation has been described in FRDA patients [67] as well as in flies and different tissues from FRDA mice models [29,31,68,69]. Here we show that NAC and Trolox treatment decreases membrane lipid peroxidation in our frataxin deficient cell model recovering Ca2+ homeostasis. The impact of lipid peroxidation in vivo was already tested a decade ago [31] in the fly model used in this work. We observed along with the lipid accumulation, a concomitant increase of lipid peroxides. Interestingly, we also found that co-expression of a lipid peroxides’ scavenger was able to improve some frataxin-deficient conditions in the fly [31]. These results underscore the importance of a reduced cytosolic microenvironment, surrounding the MAMs to preserve its physiological architecture. Thus, in our model, although the decrease in peroxidation levels after antioxidant treatments are minor, they are sufficient to restore ER-mitochondrial contacts.
Although a structural role of frataxin in the MAMs still needs to be elucidated, bringing together these results, we hypothesize that frataxin could participate in MAMs in two different but not necessarily exclusive ways. Firstly, our findings describe for the first time frataxin as a member of the protein network of MAMs, indicating a pivotal role in the processes regulated by these structures, including a proper redox environment. Secondly, the fact that frataxin has a direct interaction with GRP75 and IP3R, two of the main proteins implicated in mitochondrial Ca2+ transfer from the ER, suggesting its implication in the VDAC-GRP75-IP3R protein bridge stabilization. These results contribute to understand the mechanisms of Ca2+ dysregulation.
Furthermore, our findings agree with previous results obtained after artificially increasing the amount of mitochondria-ER connections in other fly models of neurodegenerative diseases. Genetic manipulation of mitochondrial-ER bridges has been shown to be a successful counteracting strategy in Drosophila models of AD [70], where the promotion of contacts was beneficial. Here we observed that expression of ChiMERA in glia cells was only able to revert the neurodegeneration, whereas locomotion and lipid homeostasis were not improved. Such a limited effect might further support the role of frataxin in the stability of mitochondria-ER contacts, and more concretely in the mitochondrial calcium uptake.
In conclusion, our findings indicate that frataxin deficiency causes an impairment in both the ER-mitochondria communication and in the dysregulation of Ca2+ homeostasis (Graphical abstract), which provides a new approach regarding frataxin functionality and mitochondrial imbalance in FRDA. These results raise the question whether reduction of the interactions between these two organelles is the critical event in the progression of the pathology, offering a new field of investigation regarding MAMs as therapeutic targets.
Funding information
This work was supported by grants from the Ministerio de Economía y Competitividad de España [Grant no. SAF2015-66625-R] within the framework of the National R + D + I Plan and co-funded by the Instituto de Salud Carlos III (ISCIII)-Subdirección General de Evaluación y Fomento de la Investigación and FEDER funds; Fundación Ramón Areces (CIVP18A3899); the Generalitat Valenciana (PROMETEO/2018/135); Conselleria de Sanitat Universal i Salut Pública de la Generalitat Valenciana (Plan GenT CDEI-04/20-C); CIBERER (ACCI-2018-22); Joint Research Funding Project from the French National Research Agency (ANR) and the German Research Foundation (DFG) to S.S. (DFG-SCHN 558/9-1). CIBERER is an initiative developed by the Instituto de Salud Carlos III in cooperative and translational research on rare diseases.
Author contributions
LRR conducted and designed experiments, analyzed the results and wrote the manuscript. PCQ and TLL performed experiments. FVP and SS interpreted data and wrote the manuscript. JAN and PG-C designed the study, supervised the experiments, analyzed data and wrote the manuscript. All authors read and approved the final manuscript.
Declaration of competing interest
The authors declare there is no conflict of interest.
Acknowledgements
We would like to thank Gudrun Karch and Ursula Roth for technical assistance as well as Alex Whitworth for the generous gift of MCU fly lines. We also thank the Bloomington Drosophila Stock Center (NIH P40OD018537) and the Transgenic RNAi Project (TRiP) at Harvard Medical School (NIH/NIGMS R01-GM084947) for providing transgenic fly stocks used in this study. We thank Dr. Jorgina Satrústegui (Centro de Biología molecular Severo Ochoa, Spain) for providing the pcDNA-4mtD3cpv plasmid.
Footnotes
Table S1.
| Name | Genotype | Kindly Provided |
|---|---|---|
| W1118 | w [1118] | Bloomington Stock 3605 |
| actin-GAL4 | y [1]w [*]; P{w [+mC] = Act5C-GAL4}17bFO1/TM6B, Tb [1] | Bloomington Stock 3954 |
| Repo-GAL4 | w [1118]; P{w [+m*] = GAL4}repo/TM3, Sb [1] | Bloomington Stock 7415 |
| Mef2-GAL4 | y [1] w [*]; P{w [+mC] = GAL4-Mef2.R}3 | Bloomington Stock 27390 |
| Elav-GAL4, UAS-Dicer 2 | P{w [+mW.hs] = GawB}elav [C155] w [1118]; P{w [+mC] = UAS-Dcr-2.D}2 | Bloomington Stock 25750 |
| UAS-mCherryRNAi | w [1118]; P{w [+mC] = UAS-fhRNAi-1} | Bloomington Stock 35787 |
| fhRNAi-1 | w [1118]; P{w [+mC] = UAS-fhRNAi-1} | Prof. John P. Phillips |
| fhRNAi-2 | y [1] w [*]; P{w [+mC] = UAS-fhRNAi-2}2 | Prof. Maria D. Moltó |
| UAS-fh | y [1]w [*]; P{w [+mC] = UAS- fh}2 | Prof. Maria D. Moltó |
| UAS-mitoGFP | P{w [+mC] = UAS-mito-HA-GFP.AP}/CyO | Bloomington Stock 8442 |
| UAS-MCURNAi (TRiP) | y1 sc* v1 sev 21; P{TRiP.HMS05618}attP40 | Bloomington Stock 67857 |
| MCU[1] | w [1118]; MCU[1]/TM6B, P{w [+mW.hs] = Ubi-GFP.S65T}PAD2, Tb [1] | Prof. Alex Whitworth |
| UAS-MCUoe1 | y [1]w [*]; P{w [+mC] = UAS-MCU-flag}attP40 | Prof. Alex Whitworth |
| UAS-MCUoe2 | y [1]w [*]; P{w [+mC] = UAS-MCU-flag}attP2 | Prof. Alex Whitworth |
| EP MCU | y [1] w [67c23]; P{w [+mC] y [+mDint2] = EPgy2} MCU [EY08610] | Bloomington Stock 19933 |
| UAS-ChiMERA | y [1] M{vas-int.Dm}ZH-2A w [*]; P{w [+mC] = UAS-ChiMERA}VK00037 | Prof. Patrik Verstrecken |
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Fig. S1.
Immunostainings of the proteins used in PLA assay. The immunofluorescence of VDAC1, IP3R, GRP75 and FXN were performed as control for PLA technique specificity in the SH-SY5Y and FXN-138.2 cells. (A) Representative images of VDAC either with GRP75 or IP3R. (B) Representative images of FXN either with GRP75 or IP3R.
Fig. S2.
Dose response to carbachol in SH-SY5Y and FXN-138.2 cells. (A) FRET analysis of mitochondrial calcium levels measured through 4mtD3cpv chameleon. 25 μM, 250 μM, 500 μM and 900 μM carbachol (Cch) were used to stimulate Ca2+ release from the ER through IP3R channels. FXN-138.2 cell line did not change in response to 25 μM carbachol. 250 μM carbachol was chosen as the most suitable concentration that gave the maximum response. Results (n ≥ 3) are represented as normalized levels of fluorescence intensity over carbachol loading value. A range of 11–31 cells were analyzed in each condition per replicate. (B) Area Under curve (AUC) analysis of the mitochondrial response to the different concentrations of carbachol.
Fig. S3.
Impact of MCU manipulation in glia cell on fly behavior, brain integrity and lipid metabolism. (A) MCU levels are not modified upon frataxin downregulation ubiquitously in the whole fly or specifically in fly muscles or neurons. (B) Compared to control flies, UAS MCU lines are able to upregulate MCU expression by ten times, whereas EP MCU line only upregulates it by 2-fold. (C) Quantification of mtDNA as indirect measurement of mitochondrial mass showed that neither mild upregulation of MCU expression nor frataxin overexpression seemed to promote the biogenesis of mitochondria. (D) Nor MCU downregulation neither targeted overexpression of MCU in glia impair locomotor activity in Drosophila. (E–G) Representative paraffin brain sections and Bodipy stainings showing no vacuolization and absence of lipid deposits in controls in 35-day-old controls (E, Repo-G4>w), MCU-deficient flies (F,Repo-G4>MCU[1]) and flies with a targeted glial expression of MCU (G,Repo-G4>UAS-MCUoe1). Graphs represent means ± SEM. In A, data was analyzed by two-tailed T-test, whereas in B, C and D, data was analyzed by One-way ANOVA with post-hoc Tukey Multiple Comparison test. ***, P < 0.001. Scale bars represent 50 μm.
Fig. S4.
Moderate loss of Mitochondrial Calcium Uniporter (MCU) does not worsen loss-of-frataxin conditions. (A–B) Compared to control flies (Repo-G4>w), 50% loss of MCU levels (fhRNAi-1;Repo-G4>MCU[1]) did not modified the negative geotaxis ability (A) as well as the levels of brain degeneration (B) of FRDA flies (fhRNAi-1;Repo-G4>w). (C–E) Representative paraffin brain sections and Bodipy staining of controls (C, Repo-G4>w), frataxin-deficient flies (D,fhRNAi-1;Repo-G4>w) or combination of frataxin deficiency and one MCU mutant allele (E,fhRNAi-1;Repo-G4>MCU[1]). Arrows indicated the presence of the vacuolization in frataxin-deficient brains with normal levels of MCU (D) and with 50% reduction of MCU expression (E). Bodipy staining of whole fly brains showed complete absence of lipid deposits in controls (C) and a strong accumulation in frataxin deficient flies (D) that was not modified by 50% inactivation of MCU (E). Graphs represent means ± SEM In A and B data was analyzed by One-way ANOVA with post-hoc Tukey Multiple Comparison test. **, P < 0.01 ***, P < 0.001, Scale bars: 50 μm.
References
- 1.Campuzano V., Montermini L., Molto M.D., Pianese L., Cossee M., Cavalcanti F., Monros E., Rodius F., Duclos F., Monticelli A., Zara F., Canizares J., Koutnikova H., Bidichandani S.I., Gellera C., Brice A., Trouillas P., De Michele G., Filla A., De Frutos R., Palau F., Patel P.I., Di Donato S., Mandel J.-L., Cocozza S., Koenig M., Pandolfo M. Friedreich's ataxia: autosomal recessive disease caused by an intronic GAA triplet repeat expansion. Science. 1996;271(80):1423–1427. doi: 10.1126/science.271.5254.1423. [DOI] [PubMed] [Google Scholar]
- 2.Parkinson M.H., Boesch S., Nachbauer W., Mariotti C., Giunti P. Clinical features of Friedreich's ataxia: classical and atypical phenotypes. J. Neurochem. 2013;126(Suppl):103–117. doi: 10.1111/jnc.12317. [DOI] [PubMed] [Google Scholar]
- 3.Koeppen A.H., Mazurkiewicz J.E. Friedreich ataxia: neuropathology revised. J. Neuropathol. Exp. Neurol. 2013;72:78–90. doi: 10.1097/NEN.0b013e31827e5762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Llorens J.V., Soriano S., Calap-Quintana P., Gonzalez-Cabo P., Moltó M.D. The role of iron in friedreich's ataxia: insights from studies in human tissues and cellular and animal models. Front. Neurosci. 2019;13:75. doi: 10.3389/fnins.2019.00075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gerber J., Mühlenhoff U., Lill R. An interaction between frataxin and Isu1/Nfs1 that is crucial for Fe/S cluster synthesis on Isu1. EMBO Rep. 2003;4:906–911. doi: 10.1038/sj.embor.embor918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Muhlenhoff U., Gerber J., Richhardt N., Lill R. Components involved in assembly and dislocation of iron-sulfur clusters on the scaffold protein Isu1p. EMBO J. 2003;22:4815–4825. doi: 10.1093/emboj/cdg446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stehling O., Elsässer H.-P., Brückel B., Mühlenhoff U., Lill R. Iron-sulfur protein maturation in human cells: evidence for a function of frataxin. Hum. Mol. Genet. 2004;13:3007–3015. doi: 10.1093/hmg/ddh324. [DOI] [PubMed] [Google Scholar]
- 8.Napier I., Ponka P., Richardson D.R. Iron trafficking in the mitochondrion: novel pathways revealed by disease. Blood. 2005;105:1867–1874. doi: 10.1182/blood-2004-10-3856. [DOI] [PubMed] [Google Scholar]
- 9.Yoon T., Dizin E., Cowan J.A. N-terminal iron-mediated self-cleavage of human frataxin: regulation of iron binding and complex formation with target proteins. JBIC J. Biol. Inorg. Chem. 2007;12:535–542. doi: 10.1007/s00775-007-0205-2. [DOI] [PubMed] [Google Scholar]
- 10.Yoon T., Cowan J.A. Frataxin-mediated iron delivery to ferrochelatase in the final step of heme biosynthesis. J. Biol. Chem. 2004;279:25943–25946. doi: 10.1074/jbc.C400107200. [DOI] [PubMed] [Google Scholar]
- 11.Schoenfeld R.A., Napoli E., Wong A., Zhan S., Reutenauer L., Morin D., Buckpitt A.R., Taroni F., Lonnerdal B., Ristow M., Puccio H., Cortopassi G.A. Frataxin deficiency alters heme pathway transcripts and decreases mitochondrial heme metabolites in mammalian cells. Hum. Mol. Genet. 2005;14:3787–3799. doi: 10.1093/hmg/ddi393. [DOI] [PubMed] [Google Scholar]
- 12.Ristow M., Pfister M.F., Yee A.J., Schubert M., Michael L., Zhang C.-Y., Ueki K., Michael M.D., Lowell B.B., Kahn C.R. Frataxin activates mitochondrial energy conversion and oxidative phosphorylation. Proc. Natl. Acad. Sci. Unit. States Am. 2000;97:12239–12243. doi: 10.1073/pnas.220403797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.González-Cabo P., Vázquez-Manrique R.P., García-Gimeno M.A., Sanz P., Palau F. Frataxin interacts functionally with mitochondrial electron transport chain proteins. Hum. Mol. Genet. 2005;14:2091–2098. doi: 10.1093/hmg/ddi214. [DOI] [PubMed] [Google Scholar]
- 14.Napoli E., Taroni F., Cortopassi G.A. Frataxin, iron–sulfur clusters, heme, ROS, and aging. Antioxidants Redox Signal. 2006;8:506–516. doi: 10.1089/ars.2006.8.506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Santos R., Lefevre S., Sliwa D., Seguin A., Camadro J.-M., Lesuisse E. Friedreich ataxia: molecular mechanisms, redox considerations, and therapeutic opportunities. Antioxidants Redox Signal. 2010;13:651–690. doi: 10.1089/ars.2009.3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tamarit J., Obis È., Ros J. Oxidative stress and altered lipid metabolism in Friedreich ataxia. Free Radic. Biol. Med. 2016;100:138–146. doi: 10.1016/j.freeradbiomed.2016.06.007. [DOI] [PubMed] [Google Scholar]
- 17.Mollá B., Muñoz-Lasso D.C., Riveiro F., Bolinches-Amorós A., Pallardó F.V., Fernandez-Vilata A., de la Iglesia-Vaya M., Palau F., Gonzalez-Cabo P. Reversible axonal dystrophy by calcium modulation in frataxin-deficient sensory neurons of YG8R mice. Front. Mol. Neurosci. 2017;10 doi: 10.3389/fnmol.2017.00264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bolinches-Amorós A., Mollá B., Pla-Martí-n D., Palau F., González-Cabo P. Mitochondrial dysfunction induced by frataxin deficiency is associated with cellular senescence and abnormal calcium metabolism. Front. Cell. Neurosci. 2014;8:124. doi: 10.3389/fncel.2014.00124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chiang S., Kalinowski D.S., Jansson P.J., Richardson D.R., Huang M.L.-H. Mitochondrial dysfunction in the neuro-degenerative and cardio-degenerative disease. Friedreich’s ataxia, Neurochem. Int. 2018;117:35–48. doi: 10.1016/j.neuint.2017.08.002. [DOI] [PubMed] [Google Scholar]
- 20.González-Cabo P., Palau F. Mitochondrial pathophysiology in Friedreich's ataxia. J. Neurochem. 2013;126(Suppl 1):53–64. doi: 10.1111/jnc.12303. [DOI] [PubMed] [Google Scholar]
- 21.Edenharter O., Schneuwly S., Navarro J.A. Mitofusin-dependent ER stress triggers glial dysfunction and nervous system degeneration in a Drosophila model of friedreich's ataxia. Front. Mol. Neurosci. 2018;11 doi: 10.3389/fnmol.2018.00038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pinton P. Mitochondria-associated membranes (MAMs) and pathologies. Cell Death Dis. 2018;9:413. doi: 10.1038/s41419-018-0424-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van Vliet A.R., Verfaillie T., Agostinis P. New functions of mitochondria associated membranes in cellular signaling. Biochim. Biophys. Acta Mol. Cell Res. 2014;1843:2253–2262. doi: 10.1016/J.BBAMCR.2014.03.009. [DOI] [PubMed] [Google Scholar]
- 24.Hamasaki M., Furuta N., Matsuda A., Nezu A., Yamamoto A., Fujita N., Oomori H., Noda T., Haraguchi T., Hiraoka Y., Amano A., Yoshimori T. Autophagosomes form at ER-mitochondria contact sites. Nature. 2013;495:389–393. doi: 10.1038/nature11910. [DOI] [PubMed] [Google Scholar]
- 25.Marchi S., Patergnani S., Missiroli S., Morciano G., Rimessi A., Wieckowski M.R., Giorgi C., Pinton P. Mitochondrial and endoplasmic reticulum calcium homeostasis and cell death. Cell Calcium. 2018;69:62–72. doi: 10.1016/j.ceca.2017.05.003. [DOI] [PubMed] [Google Scholar]
- 26.Rodríguez-Arribas M., Yakhine-Diop S.M.S., Pedro J.M.B.S., Gómez-Suaga P., Gómez-Sánchez R., Martínez-Chacón G., Fuentes J.M., González-Polo R.A., Niso-Santano M. Mitochondria-associated membranes (MAMs): overview and its role in Parkinson's disease. Mol. Neurobiol. 2017;54:6287–6303. doi: 10.1007/s12035-016-0140-8. [DOI] [PubMed] [Google Scholar]
- 27.Prause J., Goswami A., Katona I., Roos A., Schnizler M., Bushuven E., Dreier A., Buchkremer S., Johann S., Beyer C., Deschauer M., Troost D., Weis J. Altered localization, abnormal modification and loss of function of Sigma receptor-1 in amyotrophic lateral sclerosis. Hum. Mol. Genet. 2013;22:1581–1600. doi: 10.1093/hmg/ddt008. [DOI] [PubMed] [Google Scholar]
- 28.Schon E.A., Area-Gomez E. Mitochondria-associated ER membranes in Alzheimer disease. Mol. Cell. Neurosci. 2013;55:26–36. doi: 10.1016/j.mcn.2012.07.011. [DOI] [PubMed] [Google Scholar]
- 29.Abeti R., Parkinson M.H., Hargreaves I.P., Angelova P.R., Sandi C., Pook M.A., Giunti P., Abramov A.Y. Mitochondrial energy imbalance and lipid peroxidation cause cell death in friedreich's ataxia. Cell Death Dis. 2016;7:e2237. doi: 10.1038/cddis.2016.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Simon D. Friedreich ataxia mouse models with progressive cerebellar and sensory ataxia reveal autophagic neurodegeneration in dorsal root ganglia. J. Neurosci. 2004;24 doi: 10.1523/JNEUROSCI.4549-03.2004. 1987–1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Navarro J.A., Ohmann E., Sanchez D., Botella J.A., Liebisch G., Moltó M.D., Ganfornina M.D., Schmitz G., Schneuwly S. Altered lipid metabolism in a Drosophila model of Friedreich's ataxia. Hum. Mol. Genet. 2010;19:2828–2840. doi: 10.1093/hmg/ddq183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Giorgi C., Missiroli S., Patergnani S., Duszynski J., Wieckowski M.R., Pinton P. Mitochondria-associated membranes: composition, molecular mechanisms, and physiopathological implications. Antioxidants Redox Signal. 2015;22:995–1019. doi: 10.1089/ars.2014.6223. [DOI] [PubMed] [Google Scholar]
- 33.Kerkhofs M., Bittremieux M., Morciano G., Giorgi C., Pinton P., Parys J.B., Bultynck G. Emerging molecular mechanisms in chemotherapy: Ca 2+ signaling at the mitochondria-associated endoplasmic reticulum membranes. Cell Death Dis. 2018;9 doi: 10.1038/s41419-017-0179-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Britti E., Delaspre F., Tamarit J., Ros J. Mitochondrial calcium signalling and neurodegenerative diseases. Neuronal Signal. 2018;2 doi: 10.1042/ns20180061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Berridge M.J., Bootman M.D., Roderick H.L. Calcium signalling: dynamics, homeostasis and remodelling. Nat. Rev. Mol. Cell Biol. 2003;4:517–529. doi: 10.1038/nrm1155. [DOI] [PubMed] [Google Scholar]
- 36.Annunziata I., Patterson A., d'Azzo A. Isolation of mitochondria-associated ER membranes (MAMs) and glycosphingolipid-enriched microdomains (GEMs) from brain tissues and neuronal cells. Methods Mol. Biol. 2015;1264:25–33. doi: 10.1007/978-1-4939-2257-4_3. [DOI] [PubMed] [Google Scholar]
- 37.Monnier V., Llorens J.V., Navarro J.A. Impact of drosophila models in the study and treatment of friedreich's ataxia. Int. J. Mol. Sci. 2018;19 doi: 10.3390/ijms19071989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., Tinevez J.-Y., White D.J., Hartenstein V., Eliceiri K., Tomancak P., Cardona A. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Valadas J.S., Esposito G., Vandekerkhove D., Miskiewicz K., Deaulmerie L., Raitano S., Seibler P., Klein C., Verstreken P. ER lipid defects in neuropeptidergic neurons impair sleep patterns in Parkinson's disease. Neuron. 2018;98:1155–1169. doi: 10.1016/j.neuron.2018.05.022. e6. [DOI] [PubMed] [Google Scholar]
- 40.Patergnani S., Suski J.M., Agnoletto C., Bononi A., Bonora M., De Marchi E., Giorgi C., Marchi S., Missiroli S., Poletti F., Rimessi A., Duszynski J., Wieckowski M.R., Pinton P. Calcium signaling around mitochondria associated membranes (MAMs) Cell Commun. Signal. 2011;9:19. doi: 10.1186/1478-811X-9-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Çelik E.E., Rubio J.M.A., Gökmen V. Behaviour of Trolox with macromolecule-bound antioxidants in aqueous medium: inhibition of auto-regeneration mechanism. Food Chem. 2018;243:428–434. doi: 10.1016/j.foodchem.2017.10.009. [DOI] [PubMed] [Google Scholar]
- 42.Abeti R., Brown A.F., Maiolino M., Patel S., Giunti P. Calcium deregulation: novel insights to understand friedreich's ataxia pathophysiology. Front. Cell. Neurosci. 2018;12:1–13. doi: 10.3389/fncel.2018.00264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Aldini G., Altomare A., Baron G., Vistoli G., Carini M., Borsani L., Sergio F. N-Acetylcysteine as an antioxidant and disulphide breaking agent: the reasons why. Free Radic. Res. 2018;52:751–762. doi: 10.1080/10715762.2018.1468564. [DOI] [PubMed] [Google Scholar]
- 44.Schmucker S., Argentini M., Carelle-Calmels N., Martelli A., Puccio H. The in vivo mitochondrial two-step maturation of human frataxin. Hum. Mol. Genet. 2008;17:3521–3531. doi: 10.1093/hmg/ddn244. [DOI] [PubMed] [Google Scholar]
- 45.Acquaviva F., De Biase I., Nezi L., Ruggiero G., Tatangelo F., Pisano C., Monticelli A., Garbi C., Acquaviva A.M., Cocozza S. Extra-mitochondrial localisation of frataxin and its association with IscU1 during enterocyte-like differentiation of the human colon adenocarcinoma cell line Caco-2. J. Cell Sci. 2005;118:3917–3924. doi: 10.1242/jcs.02516. [DOI] [PubMed] [Google Scholar]
- 46.Condò I., Malisan F., Guccini I., Serio D., Rufini A., Testi R. Molecular control of the cytosolic aconitase/IRP1 switch by extramitochondrial frataxin. Hum. Mol. Genet. 2010;19:1221–1229. doi: 10.1093/hmg/ddp592. [DOI] [PubMed] [Google Scholar]
- 47.Dong Y.N., McMillan E., Clark E.M., Lin H., Lynch D.R. GRP75 overexpression rescues frataxin deficiency and mitochondrial phenotypes in Friedreich ataxia cellular models. Hum. Mol. Genet. 2019;28:1594–1607. doi: 10.1093/hmg/ddy448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kornmann B., Currie E., Collins S.R., Schuldiner M., Nunnari J., Weissman J.S., Walter P. An ER-mitochondria tethering complex revealed by a synthetic biology screen. Science. 2009;325(80):477–481. doi: 10.1126/science.1175088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Drago I., Davis R.L. Inhibiting the mitochondrial calcium uniporter during development impairs memory in adult Drosophila. Cell Rep. 2016;16:2763–2776. doi: 10.1016/j.celrep.2016.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Choi S., Quan X., Bang S., Yoo H., Kim J., Park J., Park K.-S., Chung J. Mitochondrial calcium uniporter in Drosophila transfers calcium between the endoplasmic reticulum and mitochondria in oxidative stress-induced cell death. J. Biol. Chem. 2017;292:14473–14485. doi: 10.1074/jbc.M116.765578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee K.-S., Huh S., Lee S., Wu Z., Kim A.-K., Kang H.-Y., Lu B. Altered ER–mitochondria contact impacts mitochondria calcium homeostasis and contributes to neurodegeneration in vivo in disease models. Proc. Natl. Acad. Sci. Unit. States Am. 2018;115:E8844–E8853. doi: 10.1073/pnas.1721136115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tufi R., Gleeson T.P., von Stockum S., Hewitt V.L., Lee J.J., Terriente-Felix A., Sanchez-Martinez A., Ziviani E., Whitworth A.J. Comprehensive genetic characterization of mitochondrial Ca2+ uniporter components reveals their different physiological requirements in vivo. Cell Rep. 2019;27:1541–1550. doi: 10.1016/j.celrep.2019.04.033. e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kis V., Barti B., Lippai M., Sass M. Specialized cortex glial cells accumulate lipid droplets in Drosophila melanogaster. PloS One. 2015;10 doi: 10.1371/journal.pone.0131250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bellen H.J., Levis R.W., Liao G., He Y., Carlson J.W., Tsang G., Evans-Holm M., Hiesinger P.R., Schulze K.L., Rubin G.M., Hoskins R.A., Spradling A.C. The BDGP gene disruption Project. Genetics. 2004;167:761–781. doi: 10.1534/genetics.104.026427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lee S., Lee K.-S., Huh S., Liu S., Lee D.-Y., Hong S.H., Yu K., Lu B. Polo kinase phosphorylates miro to control ER-mitochondria contact sites and mitochondrial Ca 2+ homeostasis in neural stem cell development. Dev. Cell. 2016;37:174–189. doi: 10.1016/j.devcel.2016.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu Y., Jin M., Wang Y., Zhu J., Tan R., Zhao J., Ji X., Jin C., Jia Y., Ren T., Xing J. MCU-induced mitochondrial calcium uptake promotes mitochondrial biogenesis and colorectal cancer growth. Signal Transduct. Target. Ther. 2020;5:59. doi: 10.1038/s41392-020-0155-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Islam M.T. Oxidative stress and mitochondrial dysfunction-linked neurodegenerative disorders. Neurol. Res. 2017;39:73–82. doi: 10.1080/01616412.2016.1251711. [DOI] [PubMed] [Google Scholar]
- 58.Paillusson S., Stoica R., Gomez-Suaga P., Lau D.H.W., Mueller S., Miller T., Miller C.C.J. There's something wrong with my MAM; the ER-mitochondria Axis and neurodegenerative diseases. Trends Neurosci. 2016;39:146–157. doi: 10.1016/j.tins.2016.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Abeti R., Baccaro A., Esteras N., Giunti P. Novel Nrf2-inducer prevents mitochondrial defects and oxidative stress in friedreich's ataxia models. Front. Cell. Neurosci. 2018;12:188. doi: 10.3389/fncel.2018.00188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chaudhuri D., Artiga D.J., Abiria S.A., Clapham D.E. Mitochondrial calcium uniporter regulator 1 (MCUR1) regulates the calcium threshold for the mitochondrial permeability transition. Proc. Natl. Acad. Sci. Unit. States Am. 2016;113:E1872–E1880. doi: 10.1073/pnas.1602264113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cárdenas C., Miller R.A., Smith I., Bui T., Molgó J., Müller M., Vais H., Cheung K.-H., Yang J., Parker I., Thompson C.B., Birnbaum M.J., Hallows K.R., Foskett J.K. Essential regulation of cell bioenergetics by constitutive InsP3 receptor Ca2+ transfer to mitochondria. Cell. 2010;142:270–283. doi: 10.1016/j.cell.2010.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tarasov A.I., Griffiths E.J., Rutter G.A. Regulation of ATP production by mitochondrial Ca2+ Cell Calcium. 2012;52:28–35. doi: 10.1016/j.ceca.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.V Aranca T., Jones T.M., Shaw J.D., Staffetti J.S., Ashizawa T., Kuo S.-H., Fogel B.L., Wilmot G.R., Perlman S.L., Onyike C.U., Ying S.H., Zesiewicz T.A. Emerging therapies in Friedreich's ataxia. Neurodegener. Dis. Manag. 2016;6:49–65. doi: 10.2217/nmt.15.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rodríguez L.R., Lapeña T., Calap‐quintana P., Moltó M.D., Gonzalez‐cabo P., Langa J.A.N. Antioxidant therapies and oxidative stress in friedreich's ataxia: the right path or just a diversion? Antioxidants. 2020;9:1–23. doi: 10.3390/antiox9080664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wong A., Yang J., Cavadini P., Gellera C., Lonnerdal B., Taroni F., Cortopassi G. The Friedreich's ataxia mutation confers cellular sensitivity to oxidant stress which is rescued by chelators of iron and calcium and inhibitors of apoptosis. Hum. Mol. Genet. 1999;8:425–430. doi: 10.1093/hmg/8.3.425. [DOI] [PubMed] [Google Scholar]
- 66.Mollá B., Muñoz-Lasso D.C., Calap P., Fernandez-Vilata A., de la Iglesia-Vaya M., Pallardó F.V., Moltó M.D., Palau F., Gonzalez-Cabo P. Phosphodiesterase inhibitors revert axonal dystrophy in friedreich's ataxia mouse model. Neurotherapeutics. 2019;16:432–449. doi: 10.1007/s13311-018-00706-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Emond M., Lepage G., Vanasse M., Pandolfo M. Increased levels of plasma malondialdehyde in Friedreich ataxia. Neurology. 2000;55:1752–1753. doi: 10.1212/wnl.55.11.1752. [DOI] [PubMed] [Google Scholar]
- 68.Al-Mahdawi S., Pinto R.M., Varshney D., Lawrence L., Lowrie M.B., Hughes S., Webster Z., Blake J., Cooper J.M., King R., Pook M.A. GAA repeat expansion mutation mouse models of Friedreich ataxia exhibit oxidative stress leading to progressive neuronal and cardiac pathology. Genomics. 2006;88:580–590. doi: 10.1016/j.ygeno.2006.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Abeti R., Uzun E., Renganathan I., Honda T., Pook M.A., Giunti P. Targeting lipid peroxidation and mitochondrial imbalance in Friedreich's ataxia. Pharmacol. Res. 2015;99:344–350. doi: 10.1016/j.phrs.2015.05.015. [DOI] [PubMed] [Google Scholar]
- 70.Garrido-Maraver J., Loh S.H.Y., Martins L.M. Forcing contacts between mitochondria and the endoplasmic reticulum extends lifespan in a Drosophila model of Alzheimer's disease. Biol. Open. 2020;9 doi: 10.1242/bio.047530. [DOI] [PMC free article] [PubMed] [Google Scholar]