Abstract
Background:
In recent years, accumulating studies have found that circular RNA (circRNA) exerts a great effect on tumor progression. Circ_0000215, a novel circRNA, remains largely unknown in terms of its effect and mechanism in glioma.
Method:
Quantitative real-time polymerase chain reaction (qRT-PCR) was carried out to detect the expressions of circ_0000215, miR-495-3p and CXCR2 in human glial cell line HEB and glioma cell lines (A172, U251, U87, SHG-44, LN-18), human glioma tissues and adjacent healthy tissues. Gain- and loss-assays of circ_0000215 were conducted. Cell proliferation ability was detected via the CCK8 assay, and cell invasion ability was examined by Transwell assay. CXCR2 expression was evaluated via RT-PCR and Western blot. Moreover, bioinformatics was applied to analyze the targeting molecules of circ_0000215 and CXCR2. Verification of the relationship between these molecules were supported through the dual-luciferase reporter gene and RNA immunocoprecipitation (RIP) assay.
Results:
Circ_0000215 and CXCR2 were remarkably upregulated in glioma tissues and cells. Overexpression of circ_0000215 notably promoted the proliferation, invasion and epithelial-mesenchymal transition (EMT) but inhibited apoptosis of glioma cells, while knocking down circ_0000215 had the opposite effects. Additionally, miR-495-3p, a sponge RNA of circ_0000215, inhibited the growth, invasion and EMT of glioma cells. Mechanistically, miR-495-3p targeted CXCR2 and negatively regulated CXCR2/PI3K/Akt pathway. However, the effects of miR-495-3p were all dampened by overexpression of circ_0000215.
Conclusion:
These data demonstrated that circ_0000215 functions as a competitive endogenous RNA by sponging miR-495-3p, thus accelerating glioma progression through CXCR2 axis.
Keywords: glioma, proliferation, invasion, circ_0000215, miR-495-3p, CXCR2
Introduction
Glioma is a prevailing fatal malignancy in human central nervous system.1 The median survival time of glioma patients only lasts 12 ∼ 14 months due to its invasive growth mode, the unclear boundary with normal brain tissue, incomplete surgical resection, high postoperative recurrence rate and insensitivity to radiotherapy and chemotherapy.2 Therefore, it is particularly important to explore the pathogenesis of glioma and to find new methods for its diagnosis and treatment.
As a non-coding RNA, circularRNA (circRNA) has s closed circular structure that exists widely in mammalian cells.3,4 It is mainly formed by trans-splicing to connect the 3’-end and 5’- end with covalent bonds.5 With the development of new bioinformatics methods and deep sequencing technologies, a large number of circRNAs have been found to be involved in gene regulation post-transcriptionally.6,7 Studies have stated that circRNAs act as a microRNA sponge to affect the expression of related genes, and then affect the proliferation, migration and invasion of tumor cells.8-10 Taking circ_0005075 as an example, it is abnormally highly expressed in liver cancer tissues and functions as ceRNA to sponge miR-23b-5p.11 Circ-ITCH is low expressed in esophageal cancer tissues and affects on inhibiting tumor progression. Functionally, circ-ITCH not only sponges multiple miRNA molecules such as miR-7, miR-17 and miR-214, but also regulates the Wnt/β-catenin pathway and participates in the occurrence and development of esophageal cancer.12 Circ_0000215 is a circRNA newly discovered in recent years, but its role in glioma has not been reported.
MicroRNAs (miRNAs) are non-coding RNAs composed of 21-24 nucleotides. Through binding to the targeted gene at 3’UTR region, microRNAs regulate gene expression and participate in tumor cell proliferation, differentiation and apoptosis.13,14 MiRNAs are abnormally expressed in various tumors: such as breast, colon, lung, liver, and pancreatic cancers.15-17 For example, miR-124 reduces cell proliferation and inhibits cell migration and invasion by targeting multiple protein expressions, thus inhibiting tumor growth.18 MiR-21 also functions as an oncogene in the occurrence and development of glioma, and its expression in glioblastoma multiforme (GBM) is considerably upregulated, which accelerates tumor formation by inhibiting insulin-like growth factor-binding protein 3 (IGFBP3) expression.19 Studies have reported that miR-495-3p inhibits the progression of colorectal cancer by regulating CDK6 and inhibiting the apoptotic cycle in colorectal cancer.20 However, there is little knowledge about the effect of miR-495-3p on gliomas.
CXC chemokine receptor 2 (CXCR2) exists in the G protein-coupled receptor group and is located on chromosome 2q33-q36.21 CXCR2 is mainly expressed on the surface of T cells, monocytes, melanoma cells, neutrophils, synovial fibroblasts, HL-60, THP-1 myeloid progenitor cells and other cells, playing a prominent role in the body’s anti-infection and anti-virus immunity.22 In recent decades, it has been reported that CXCR2 promotes the proliferation, invasion and migration of different tumor cells and makes a great difference in tumor biology.23-25 For example, in lung cancer cell lines and mouse-derived K-ras/p53 mutant lung adenocarcinoma animal models, CXCR2 knockdown results in decreased tumor invasion and metastatic abilities.26 In breast cancer, down-regulation or knockout of CXCR2 inhibits lung metastasis, and CXCR2 neutralizing antibodies or small molecule antagonists (SB265610) inhibits the migration effect of bone-derived stromal stem cell-conditioned medium on breast tumor model PyMT cells.27,28 However, the molecular mechanism of CXCR2 in glioma needs further investigation.
It has been reported that circRNA was highly expressed in both oligodendroglioma and glioblastoma by bioinformatics analysis.29 In this study, we firstly found that miR-495-3p was downregulated in glioma and inhibited the progression of glioma. Thus, we conducted bioinformatics analysis to predict the upstream target circRNA and downstream target mRNA of miR-495-3p. Interestingly, circ_0000215 and CXCR2 were the candidates which were also overexpressed in glioma tissues. Therefore, we further conducted in vitro experiments to explore the underlying network of circ_0000215/miR-495-3p/CXCR2 axis in glioma progression.
Materials and Methods
Clinical Sample Collection
Thirty-six primary glioma tissues from patients and the corresponding paracancerous healthy tissues were collected from Second Affiliated Hospital of Xinjiang Medical University during June 2016 and June 2018. All patients had not previously went through chemotherapy or radiation therapy. Histological features of the tissue were diagnosed independently by 2 pathologists. The Second Affiliated Hospital of Xinjiang Medical University ethics committee approved this study for human research (approve number: 20181207-04). All of the patients agreed to be involved in this study and signed paper consent.
Cell Culture and Transfection
Human Glioma cell lines (A172, U251, U87, SHG-44, LN-18) and healthy glial cell line HEB were obtained from American Type Culture Collection (ATCC). All of the cells were cultured in RPMI 1640 medium (HyClone, South Logan, UT, USA) containing 10% fetal bovine serum (FBS) at 37°C in 5% CO2 environment. The medium was supplemented with 10% heat-inactivated fetal bovine serum (Invitrogen, Grand Island, NY, USA), 100 U/ml penicillin and 100 μg/ml streptomycin.
The siRNA specifically targeting circ_0000215, the overexpression circ_0000215 plasmids, and their negative controls were designed and synthetized by GenePharma (Shanghai, China). The miR-495-3p mimics, miR-495-3p inhibitors and miRNA negative controls were purchased from RiboBio (Guangzhou, China). Finally, we transfected glioma cells with the expression vectors by lipofectamine 3000 (Thermo Fisher Scientific, IL, USA) in accordance with the manufacturer’s instructions.
qRT-PCR
The expressions of circ_0000215, miR-495-3p and CXCR2 were detected by fluorescent qRT-PCR. The tissues and cells were collected and mechanically dissociated with TRIzol lysates (Thermo Fisher Scientific, Shanghai, China). According to the instructions of the reverse transcription kit (Thermo Fisher Scientific, Shanghai, China), 2 µg RNA samples were taken to prepare 20 reverse transcription system, which was catalyzed by reverse transcriptase to reverse transcription into cDNA. 1 µL of cDNA sample in each group was taken, and the quantitative PCR reaction system (50 μL) was prepared, which contains SYBRGreen fluorescence and forward primers and reverse primers primers. PCR was performed on Step-One plus real-time PCR system (Applied Biosystems). GAPDH was used as the endogenous control of circ_0000215 and CXCR2 and the endogenous control of miR-495-3p is U6. The relative expression is calculated using the 2- ΔΔCT method. The primers used in this study are as follows: circ_0000215, forward primer, 5’-AAACTGCAGAGGAGGAAGCT-3’, reverse primer, 5’-GGTTCCGAGGGTCTCTTTGA-3’; CXCR2, forward primer, 5’-GCATCAGTGTGGACCGTTAC-3’, reverse primer, 5’-GGCTGGGCTAACATTGGATG-3’; miR-495-3p, forward primer, 5’-AACACGCAAACAAACATGGTGC-3’, reverse primer, 5’-CAGTGCAGGGTCCGAGGT-3’.
CCK8 Assay
Human glioma cell lines A172 and LN-18 were inoculated on 96-well plates with 5 × 103 cells per well at different time points (12 h, 24 h, 48 h, 72 h). The previous medium was exchanged by fresh compete medium and then 10 µL CCK-8 medium mixture was added into each well. After incubation in a standard incubator for 1 h, the absorbance value at 490 nm was measured with a Microplate Reader (Thermo, Multiskan GO).
Transwell Assay
Cell invasion ability was detected by Transwell assay. A number of 5 × 104 A172/LN-18 cells were inoculated in the upper chamber (Corning, Beijing, China). The chamber was with 8 µm pore size and coated with Matrigel. 400 µL RPMI-1640 medium containing 10% FBS were placed in the lower chamber. After incubation at 37°C for 24 h, the cells that failed to invade into the lower chamber were removed from the upper chamber. Next, the cells were secured with 4% paraformaldehyde for 10 min and then stained by 0.5% crystal violet. Finally, the invasive cells were counted under an inverted microscope. The experiment was performed for 3 times with triplicated wells.
Western Blot
The cells were collected and split with RIPA lysate (Roche, Shanghai, China). 5 µg of total protein was separated by SDS-PAGE electrophoresis at 100 V for 2 h. After that, the proteins were electrically transferred to polyvinylidene fluoride (PVDF) membranes. After 1-hour block with 5% skimmed milk powder at room temperature, the membranes were washed with TBST for 3 times and 10 min each time. Next, the membranes were incubated with primary antibodies CXCR2 (Abcam, ab225732, 1: 1000), Bax (Abcam, ab32503, 1:1000), Bcl2(Abcam, ab32124, 1:1000), Caspase3(Abcam, ab13847, 1:1000), E-cadherin (Abcam, ab40772, 1:1000), N-cadherin(Abcam, ab18203, 1:1000), Vimentin(Abcam, ab92547, 1:1000), p-PI3 K (Abcam, ab182651, 1:1000), PI3 K (Abcam, ab191606, 1:1000), p-Akt (Abcam, ab38449, 1:1000), Akt(Abcam, ab8805, 1:1000) overnight at 4°C. Afterward, we washed the membranes with TBST and incubated them with horseradish peroxidase (HRP) labeled anti-rabbit secondary antibody (concentration 1: 3000) at room temperature for 1 h. Then, TBST was used for washing the membranes for 3 times (10 min each time). Finally, the membranes were exposed using an ECL kit, and gray value which marked the relative expression of proteins was analyzed through Image J.
Dual-Luciferase Reporter Gene Assay
Luciferase reporter assay was performed in accordance with the dual-luciferase reporter assay system (Promega, Madison, WI, USA). We constructed and integrated wild-type and mutant-type target fragments into the pGL3 vector (Promega, Madison, WI, USA) to build pGL3-circ_0000215-wild type (circ_0000215-WT), pGL3-circ_0000215-mutant (circ_0000215-MUT), pGL3-CXCR2-wild type (CXCR2-WT), pGL3-CXCR2-mutant (CXCR2-MUT) reporter gene vector. Circ_0000215-WT, circ_0000215-MUT, CXCR2-WT, CXCR2-MUT were co-transfected with miR-495-3p mimics or negative controls to A172 or LN-18 cells. Forty-eight hours after transfection, luciferase viability was detected following the manufacturer’s instructions.
RNA Binding Protein Immunoprecipitation (RIP)
The EZMagna RIP kit (Millipore, Billerica, MA, USA) was employed to detect the binding relationships between circ_0000215 and miR-495-3p, miR-495-3p and CXCR2 following the manufacturer’s protocol. Human anti-Ago2 antibody (Millipore) was used to enrich circ_0000215, miR-495-3p and CXCR2, and healthy mouse immunoglobulin G (IgG; Millipore) was used as a negative control. The purified RNA was extracted from immunoprecipitation of cell lysates and qRT-PCR were conducted to test enriched circ_0000215, miR-495-3p and CXCR2.
Flow Cytometry
The apoptosis of A172 and LN-18 cells was detected by flow cytometry using an Annexin V-FITC apoptosis detection kit (eBioscience, Shanghai, China). Tumor cells were cultured in 6-well plates at 4 × 105 cells/well. Stable transfected A172 and LN-18 cells were washed with 1 × PBS and resuspended in 500 µL binding buffer. Then the cells were supplemented with 5 µL V-FITC and 5 µL propidium iodine (PI), and incubated in the darkness at room temperature for 10 min. Finally, apoptosis was detected by fluorescence-activated cell sorting (FACS).
Statistical Analysis
Analyses of significance were performed using SPSS17.0 statistical software (SPSS Inc., Chicago, IL, USA). Data are presented means±s.d. A standard 2-tailed unpaired Student’s t-test was used for 2 groups. One-way ANOVA was carried out to compare multiple groups. P value <0.05 was considered statistically significant. All of the experiments were repeated at least for 3 times.
Results
Circ_0000215 Expression and Clinical Significance in Glioma Tissues and Cells
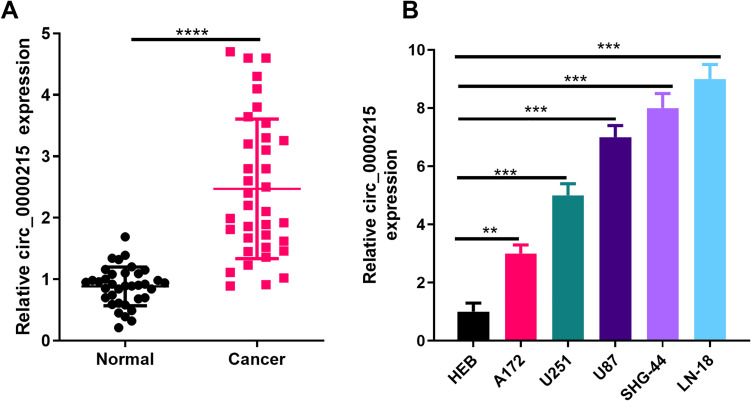
In order to verify the level of circ_0000215 in glioma tissue, we performed RT-PCR experiments. The results illustrated that compared with healthy tissues adjacent to the cancer, the expression level of circ_0000215 in glioma tissues was remarkably increased (P < 0.05, Figure 1A). Further analysis revealed that circ_0000215 overexpression was significantly associated with tumor size (Table 1). Besides, we also detected the expression of circ_0000215 in different glioma cells by RT-PCR. The results showed that circ_0000215 expression in glioma cell lines (A172, U251, U87, SHG-44, LN-18) was remarkably upregulated compared with human healthy glial cell line HEB (P < 0.05, Figure 1B).
Figure 1.
The expression of circ _0000215 in glioma tissues and cell lines. A: Circ_0000215 expression in the tissues of glioma patients and normal tissues adjacent to the cancer was measured via qRT-PCR. ***represents P < 0.0001. B. Circ_0000215 expression in normal glioma cell line HEB and glioma cell lines (A172, U251, U87, SHG-44, LN-18) was determined by qRT-PCR. *** P < 0.0001.
Table 1.
Correlation Between circ_0000215, miR-495-3p and CXCR2 Levels and Clinical Features in Glioma Patients.
| Pathological parameters | n | Circ_0000215 expression | p-Value | miR-495-3p expression | p-Value | CXCR2 expression | p-Value | |||
|---|---|---|---|---|---|---|---|---|---|---|
| Low | High | Low | High | Low | High | |||||
| Gender | 0.4941 | 0.4941 | 0.9684 | |||||||
| Male | 22 | 12 | 10 | 10 | 12 | 8 | 14 | |||
| Female | 14 | 6 | 8 | 8 | 6 | 5 | 9 | |||
| Age (years) | 0.7384 | 0.3165 | 0.7384 | |||||||
| <40 | 19 | 10 | 9 | 8 | 11 | 9 | 10 | |||
| ≥40 | 17 | 8 | 9 | 10 | 7 | 9 | 8 | |||
| Tumor grade | 0.0951 | 0.0194 | 0.3165 | |||||||
| I- II | 17 | 6 | 11 | 12 | 5 | 7 | 10 | |||
| III-IV | 19 | 12 | 7 | 6 | 13 | 11 | 8 | |||
| Tumor size | 0.0023 | 0.0179 | 0.091 | |||||||
| <5 cm | 15 | 12 | 3 | 4 | 11 | 10 | 5 | |||
| >5 cm | 21 | 6 | 15 | 14 | 7 | 8 | 13 | |||
Effects of Circ_0000215 on Glioma Cell Proliferation, Apoptosis and Metastasis
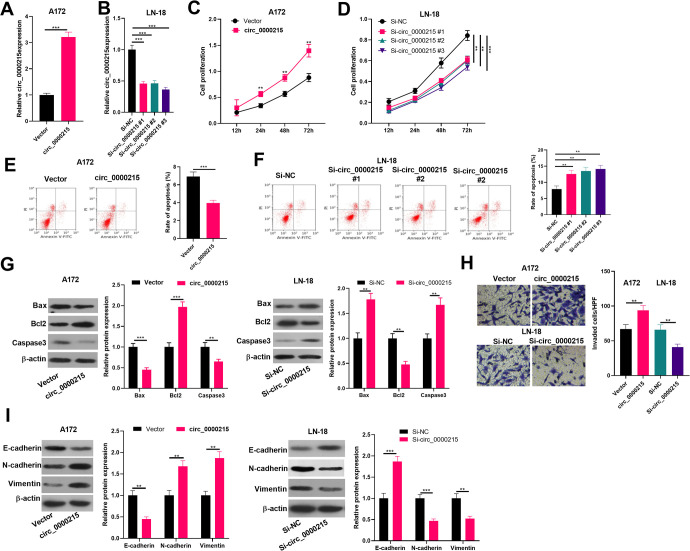
To investigate circ_0000215 function in gliomas, we successfully constructed circ_0000215 overexpression and knockdown cell lines in glioma cell lines A-172 and LN-18 (Figure 2A and B). CCK8 assay was conducted to detect cell proliferation. It was found that compared with the control group, glioma cell proliferation level was notably increased after overexpressing circ_0000215 (Figure 2C and D). Next, cell apoptosis of A-172 and LN-18 cells were detected by flow cytometry and western blot. The results showed that the apoptosis rate and apoptosis related proteins of Bax and Caspase3 were all downregulated in circ_0000215 overexpressed cells, while Bcl2 was upregulated. On the contrary, downregulation of circ_0000215 significantly enhanced apoptosis (Figure 2E-G). Moreover, Transwell assay showed that circ_0000215 upregulation significantly enhanced cell invasion ability and EMT, while circ_0000215 downregulation had the opposite effects (P < 0.05, Figure 2H and I). Collectively, the statistics suggested that circ_0000215 promoted the proliferation, invasion and EMT, but inhibited apoptosis of glioma cells.
Figure 2.
The role of circ _0000215 in glioma cell proliferation, apoptosis and metastasis. A-B. Circ_0000215 overexpression and downexpression model were conducted on A172 (A) and LN-18 (B) cells, and the relative expression of circ_0000215 was determined by qRT-PCR. C-D: The effect of circ_0000215 on cell proliferation was determined via CCK-8. E and F. Cell apoptosis of A-172 and LN-18 cells were detected by flow cytometry. G. Apoptosis related proteins including Bax, Bcl2 and Caspase3 was detected by western blot. H: The effect of circ_0000215 on the invasion ability of glioma cells was determined by Transwell assay. I. Western blot was used to detect EMT markers (E-cadherin, N-cadherin and Vimentin). *,**,*** represent P < 0.05, P < 0.01 and P < 0.001 compared with vector or si-NC group respectively.
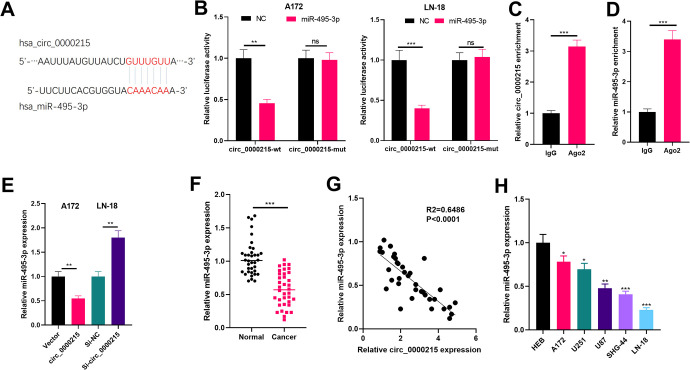
Circ_0000215 Targeted miR-495-3p
Aiming on the exploration of circ_0000215 downstream molecular mechanism, we browsed the bioinformatics database Circular RNA Interactome (https://circinteractome.nia.nih.gov/) to find potential target genes for circ_0000215 and found that miR-495-3p is its crucial target (Figure 3A). Next, we conducted dual-luciferase reporter gene assay and RIP assay to verify the binding relationship between circ_0000215 and miR-495-3p. The results demonstrated that miR-495-3p was a promising target of circ_0000215 (Figure 3B-D). Besides, miR-495-3p in the circ_0000215 overexpression group was considerably reduced while the si-circ_0000215 group had converse effect (Figure 3E). What’s more, we measured the expression of miR-495-3p in different glioma tissues and cells via qRT-PCR. The results suggested that miR-495-3p was both downregulated in glioma tissues (compared with normal adjacent tissues) and glioma cell lines (A172, U251, U87, SHG-44, LN-18) (compared with human normal glial cell line HEB) (P < 0.05, Figure 3F and H). Further experiments demonstrated that the expression levels of circ_0000215 and miR-495-3p were negatively correlated in glioma tissues (Figure 3G). In summary, these data proved that circ_0000215 functioned as a competitive endogenous RNA targeting miR-495-3p and inhibited its expression.
Figure 3.
Circ _0000215 targeted miR-495-3p in glioma cells. A: Circ_0000215 has a potential binding site for miR-495-3p (predicted by Circular RNA Interactome (https://circinteractome.nia.nih.gov/)). B: The binding relationship between miR-495-3p and circ_0000215 was verified through the luciferase reporter assay. C-D: RIP assay was used to further confirm the targeted relationship of circ_0000215 and miR-495-3p, the enrichment of them in immunoprecipitation of cell lysates were detected by qRT-PCR. E. miR-495-3p expression after transfection with circ_0000215 or si-circ_0000215 was determined via qRT-PCR. F. The expression of miR-495-3p in glioma tissues and normal tissues adjacent to the cancer was measured by qRT-PCR. NS, **,*** represents P > 0.05, P < 0.01 and P < 0.001. G. Linear regression analysis was used to analyze the relationship between circ_0000215 and miR-495-3p expression in glioma tissues. H. miR-495-3p expression in normal glioma cell line HEB and glioma cell lines was determined by qRT-PCR (A172, U251, U87, SHG-44, LN-18), *,**,*** represents P < 0.05, P < 0.01 and P < 0.001 compared with HEB group.
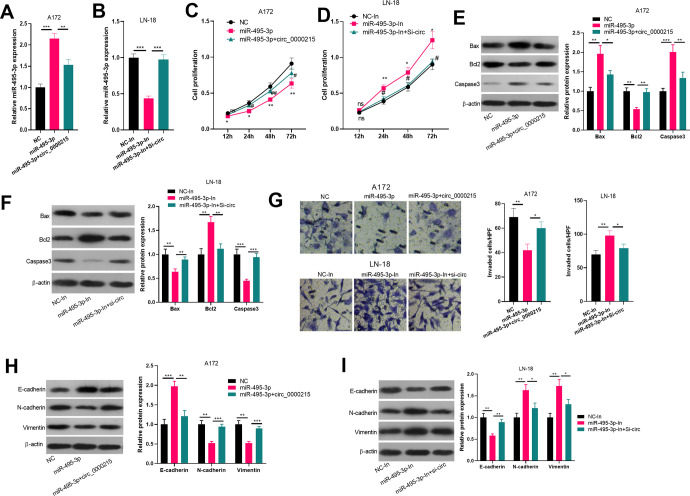
Circ_0000215/miR-495-3p Regulated Glioma Cell Proliferation and Metastasis
To verify the role of the circ_0000215/miR-495-3p axis in modulating the progression of glioma, gain- and loss- assays of circ_0000215/miR-495-3p were conducted. The results revealed that circ_0000215 overexpression markedly reduced miR-495-3p expression while inhibiting circ_0000215 through si-circ_0000215 increased miR-495-3p expression (Figure 4A and B). Next, tumor cell proliferation, apoptosis and metastasis were also examined. The data suggested that miR-495-3p upregulation inhibited glioma cell proliferation, invasion capability, EMT and promoted apoptosis, while knocking down miR-495-3p had adverse effects (Figure 4C-I), indicating that miR-495-3p plays an anti-tumor role in glioma progression. However, overexpressing circ_0000215 crippled the effects of miR-495-3p upregulation. Thus, circ_000015 functioned as an oncogene by sponging miR-495-3p.
Figure 4.
Circ_0000215/miR-495-3p regulated glioma cell proliferation and metastasis. A-B. A172(A) and LN-18 (B) cells were transfected miR-495-3p mimics or circ_0000215 overexpression plasmids and miR-495-3p inhibitor or si-circ_0000215 vector. The expression of miR-495-3p in the cells was detected via RT-PCR. ** P < 0.01, *** P < 0.001. C and D. Cell proliferation was determined via CCK-8, *,**represent P < 0.05 and P < 0.01 compared with . E-F. Apoptosis related proteins including Bax, Bcl2 and Caspase3 was detected by western blot. G: The effect of circ_0000215/miR-495-3p on the invasion ability of glioma cells was determined by Transwell assay. H-I. Western blot was used to detect EMT markers (E-cadherin, N-cadherin and Vimentin). *,**,*** represents P < 0.05, P < 0.01 and P < 0.001 compared with vector or si-NC group respectively.
Circ_0000215 Upregulated CXCR2 Expression Through miR-495-3p
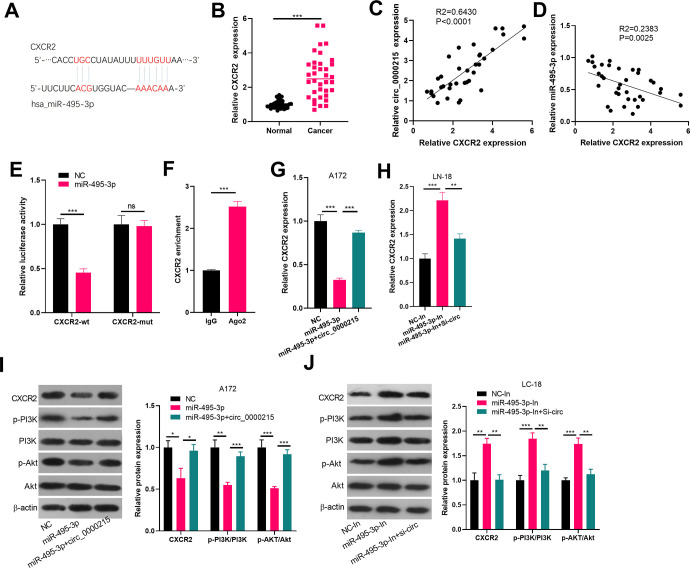
Evidence indicated that miRNAs regulate mRNA expression by targeting its 3′UTR end. For exploration of miR-495-3p functional targets, we selected miR-495-3p candidate targets in TargetScan (http://www.targetscan.org/vert_72/). The results showed that CXCR2 mRNA 3′UTR sites had a binding region of miR-495-3p, suggesting that the gene CXCR2 is likely to be a target gene for miR-495-3p (Figure 5A). Next, we detected the mRNA level of CXCR2 in glioma tissues and analyzed its level with circ-0000215 and miR-495-3p. interestingly, CXCR2 was also upregulated in glioma tissues (compared normal tissues) (Figure 5B). besides, the CXCR2 level had a positive correlation with circ_0000215 (Figure 5C). but a negative correlation with miR-495-3p (Figure 5D). Next, through luciferase activity and RIP experiments, we further confirmed that CXCR2 was an effective target for miR-495-3p (Figure 5E and F). Besides, CXCR2 expression was detected via Real-Time PCR and Western blot. It was found that the protein expression and mRNA of CXCR2 was significantly reduced after miR-495-3p overexpression while increased after miR-495-3p downexpression (Figure 5G-J). Moreover, CXCR2 expression was notably upregulated after further overexpression of circ_0000215 (compared with miR-495-3p group (Figure 5G-J). More importantly, the detection of PI3K/Akt pathway by western blot illustrated that miR-495-3p inhibited the phosphorylated of both PI3 K and Akt, while upregulating circ_0000215 dampened this effect. Allover, these results revealed that circ_0000215 regulates CXCR2 expression through miR-495-3p and circ_0000215/miR-495-3p/CXCR2 might mediate glioma progression via PI3K/Akt axis.
Figure 5.
CXCR2 is a functional target of miR-495-3p in glioma cells. A: miR-495-3p contains a potential binding site for CXCR2 as predicted by TargetScan (http://www.targetscan.org/vert_72/). B. RT-PCR was used to detect CXCR2 mRNA level in glioma and normal tissues. C and D. Pearson repression analysis was used to compare the relationships of circ_0000215 and CXCR2 (C), miR-495-3p and CXCR2 (D). E. Dual-luciferase reporter gene assay was used to verify the binding relationship between miR-495-3p and CXCR2. F. Enrichment of CXCR2 in immunoprecipitation of cell lysates were detected by qRT-PCR in RIP assay. G and H: The expression of CXCR2 was detected via qRT-PCR. I and J: The protein level of CXCR2, PI3 K, Akt were detected by western blot. NS, *,**,*** represents P > 0.05, P < 0.05, P < 0.01 and P < 0.001.
Discussion
Glioma is a common life-threatening malignancy of the central nervous system (CNS).1 Hence, exploring new biomarkers involved in glioma progression and prognosis makes much sense for the diagnosis and treatment of glioma. Here, circ_0000215 expression was found significantly upregulated in glioma tissues and cells. Further experiments confirmed that circ_000021 accelerated glioma progression through miR-495-3p/CXCR2/PI3K/Akt.
CircRNA is a newly discovered endogenous non-coding RNA. In recent years, the role of circRNA in tumors has been gradually revealed. Aberrant expression of circRNA can be used as a molecular marker for the diagnosis of malignant tumors, and significantly regulates the progress of malignant tumors.30 Taking circLMTK2 as an example, its expression is upregulated in gastric cancer tissues and is closely connected with worse overall gastric cancer patient survival. In the meantime, circLMTK2 remarkably accelerates gastric cancer cell proliferation, tumor formation, migration and invasion.31 In multiple myeloma, Circ-AMARCA5 expression is downregulated. Overexpression of Circ-AMARCA5 can inhibit myeloma cell proliferation and accelerate apoptosis.32 In the progress of gliomas, a variety of circRNAs exert vital effects. For example, circPTN,33 circ_000173034 are both notably overexpressed in glioma tissues and promote glioma cell proliferation and metastasis. Circ_0000215 was first identified in 2013.35 Our results initially demonstrated that circ_0000215 was significantly overexpressed in glioma tissues, as well as cell lines. Further gain- and loss- of functions verified that circ_0000215 significantly promoted glioma cell proliferation and metastasis, indicating that circ_0000215 was a well candidate for the diagnosis and treatment of glioma.
Mounting studies have shed light on the differential expression of miRNAs in tumors, which directly participate in and affect the development of tumors. In gliomas, miRNAs, as a tumor suppressor or oncogenes, are involved in the EMT, growth, apoptosis, migration, invasion and new angiogenesis of glioma via modulating various biological processes.36 For instance, miR-128 promotes intercellular adhesion by reducing EPH receptor B2 (Eph receptor B2, EphB2), thereby inhibiting glioma cell migration and invasion.37 Through this study, we proved that miR-495-3p was not only downregulated in glioma tissues and cells, and also significantly inhibited the growth, EMT and invasion of these glioma cells. It can be speculated that miR-495-3p works as a tumor suppressor gene in Glioma.
Accumulating studies have found that circRNA, as a competitive endogenous RNA, plays a vital role in regulating tumor development and progression in gene expression.38 For example, circTCF25 enhances the proliferation of bladder cancer cells by regulating miR-103a-3p and promoting the expression of CDK6.39 Besides, Circ-TTBK2 is overexpressed in glioma tissues and cell lines, and it acts as a miR-217 sponge in a sequence-specific manner to mediate the malignant progression of gliomas.40 In the present research, circ_0000215 was predicted and verified to be a ceRNA sponge of miR-495-3p in glioma by luciferase reporter gene experiment and RIP assay. Moreover, overexpression of circ_0000215 significantly inhibited miR-495-3p level in glioma cells and weakened the anti-cancer effects of miR-495-3p. This manifested that circ_0000215 affects glioma cell proliferation, apoptosis, EMT and invasion by targeting miR-495-3p.
In recent years, CXCR2 has been involved in the tumorigenesis, development and treatment of a variety of tumors, and the effect of the CXCR2 axis on the tumor microenvironment has received more and more attention.41 For instance, CXCR2 activated by IL-8 promotes ovarian cancer EMT via Wnt/β-catenin pathway.42 In addition, CXCR2 is also found to be regulated by miRNA. By blocking CXCL12β/CXCR2/4 axis, miR-141 modulates trophoblast apoptosis, invasion, and vascularization in pre-eclampsia.43 MiR-940 downregulates the expression of CXCR2, thus inhibiting the migration and invasion of hepatocellular carcinoma.44 Through this study, CXCR2 was found to be an effective target of miR-495-3p, and indirectly upregulated by circ_0000215, suggesting that circ_0000215/miR-495-3p promotes glioma development through CXCR2. On the other hand, PI3K/AKT signaling, a classical pathway participated in tumor growth, metastasis, apoptosis and EMT, also plays a vital role in glioma development.45,46 Interestingly, CXCR2 also affects tumor development by mediating PI3K/AKT signaling.47 Here, our data revealed that miR-495-3p inhibited phosphorylated level of PI3K/Akt, while overexpressing circ_0000215 promoted the PI3K/Akt activation, which was consistent with CXCR2 expression, suggesting that circ_0000215/miR-495-3p could promote glioma progression via activating PI3K/Akt through CXCR2.
Conclusion
Overall, we confirmed that overexpressed circ_0000215 can promote glioma progression by targeting the miR-495-3p/CXCR2/PI3K/Akt axis. This study is helpful for the early diagnosis and treatment of glioma and provides necessary help for the stage and prognosis of glioma.
Footnotes
Authors’ Note: Conceived and designed the experiments: Xinping Luan, Nurehemaiti Mutalifu; Performed the experiments: Nurehemaiti Mutalifu, Jingjing Zhan, Halik Akbar; Statistical analysis: Baofeng Yan, Sulaiman Alimu, Lingxiao Tong; Wrote the paper: Nurehemaiti Mutalifu, Peng Du. All authors read and approved the final manuscript. The data sets used and analyzed during the current study are available from the corresponding author on reasonable request. Our study was approved by the Ethics Committee of the Second Affiliated Hospital of Xinjiang Medical University (No. 20181207-04).
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by the Xinjiang Uygur Autonomous Region Science and technology innovations (talents, base) construction project (Natural Science Foundation plan fund project of Xinjiang) (No. 2018d01c227).
ORCID iD: Xinping Luan  https://orcid.org/0000-0002-7836-2744
https://orcid.org/0000-0002-7836-2744
References
- 1. Frosina G. Development of therapeutics for high grade gliomas using orthotopic rodent models. Curr Med Chem. 2013;20(26):3272–3299. [DOI] [PubMed] [Google Scholar]
- 2. Hansen TB, Wiklund ED, Bramsen JB, et al. miRNA dependent gene silencing involving A go2-mediated cleavage of a circular autisense RNA. EMBO J. 2011;30(21):4414–4422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. SMzman J, Gawad C, Wang PL, Lacayo N, Brown PO. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One. 2012;7(2):e30733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hentze MW, Preiss T. Circular RNAs: splicing’s enigma variations. EMBO J. 2013;32(7):923–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Dang Y, Yan L, Hu B, et al. Tracing the expression of circular RNAs in human pre-implantation embryos. Genome Biol. 2016;17(1):130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zhang XO, Wang HB, Zhang Y, Lu X, Chen LL, Yang L. Complementary sequence-mediated exon circularization. Cell. 2014;159(1):134–147. [DOI] [PubMed] [Google Scholar]
- 7. Danan M, Schwartz S, Edelheit S, Sorek R. Transcriptome-wide discovery of circular RNAs in Archaea. Nucleic Acids Res. 2011;40(7):3131–3142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Salzman J, Gawad C, Wang PL, Lacayo N, Brown PO. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PloS One. 2012;7(2):e30733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhang Y, Zhang X O, Chen T, et al. Circular intronic long noncoding RNAs. Molecular Cell. 2013;51(6):792–806. [DOI] [PubMed] [Google Scholar]
- 10. Shang X, Li G, Liu H, et al. Comprehensive circular RNA profiling reveals that hsa-circ-0005075, a new circular rna biomarker, is involved in hepatocellular carcinoma development. Medicine (Baltimore). 2016;95(22):e38l1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Li F, Zhang L, Li W, et al. Circular RNA ITCH has inhibitory effect on ESCC by suppressing the Wnt/β-catenin pathway. Oncotarget. 2015;6(8):6001–6013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sand M, Beehara FG, Gambichler T, et al. Circular RNA expression in cutaneous squamous cell carcinoma. Dermmol Sci. 2016;83(3):210–218. [DOI] [PubMed] [Google Scholar]
- 13. Li P, Chen S, Chen H, et al. Using circular RNA as a novel type of biomarker in the screening of gastric cancer. Clin Chim Acta. 2015;444:132–136. [DOI] [PubMed] [Google Scholar]
- 14. Shenoy A, Blelloch RH. Regulation of microRNA function in somatic stem cell proliferation and differentiation. Nat Rev Mol Cell Biol. 2014;15(9):565–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010;11(9):597–610. [DOI] [PubMed] [Google Scholar]
- 16. Schetter AJ, Leung SY, Sohn JJ, et al. MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenoearcinoma. JAMA. 2008;299(4):425–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Varnholt H, Drebber U, Schulze F, et al. MicroRNA gene expression profile of hepatitis C virus associated hepatocellular carcinoma. Hepatology. 2008;47(4):1223–1232. [DOI] [PubMed] [Google Scholar]
- 18. Yu SL, Chen HY, Chang GC, et al. MicroRNA signature predicts survival and relapse in lung cancer. Cancer Cell. 2008;13(1):48–57. [DOI] [PubMed] [Google Scholar]
- 19. Zhao WH, Wu SQ, Zhang YD. Downregulation of miR-124 promotes the growth and invasiveness of glioblastoma cells involving upregulation of PPP1R13L. Int J Mol Med. 2013;32(1):101–107. [DOI] [PubMed] [Google Scholar]
- 20. He Z, Dang J, Song A, Cui X, Ma Z, Zhang Z. NEAT1 promotes colon cancer progression through sponging miR-495-3p and activating CDK6 in vitro and in vivo. J Cell Physiol. 2019;234(11):19582–19591. [DOI] [PubMed] [Google Scholar]
- 21. Ahuja S K, Ozcelik T, Milatovitch A, Francke U, Murphy PM. Molecular evolution of the human interleukin-8 receptor gene cluster. Nat Genet. 1992;2(1):31–36. [DOI] [PubMed] [Google Scholar]
- 22. Chuntharapai A, Lee J, Hebert CA, Kim KJ. Monoclonal antibodies detect different distribution patterns of IL-8 receptor A and IL-8 receptor B on human peripheral blood leukocytes. J Immunol. 1994;153(12):5682–5688. [PubMed] [Google Scholar]
- 23. Kuo PL, Chen YH, Chen TC, Shen KH, Hsu YL. CXCL5/ENA78 increased cell migration and epithelial-to-mesenchymal transition of hormone-independent prostate cancer by early growth response-1/snail signaling pathway. J Cell Physiol. 2011;226(5):1224–1231. [DOI] [PubMed] [Google Scholar]
- 24. Miyazaki H, Patel V, Wang H, Edmunds RK, Gutkind JS, Yeudall WA. Down-regulation of CXCL5 inhibits squamous carcinogenesis. Cancer Res. 2006,66(8):4279–4284. [DOI] [PubMed] [Google Scholar]
- 25. Kawamura M, Toiyama Y, Tanaka K, et al. CXCL5, a promoter of cell proliferation, migration and invasion, is a novel serum prognostic marker in patients with colorectal cancer. Eur J Cancer. 2012;48(14):2244–2251. [DOI] [PubMed] [Google Scholar]
- 26. Saintigny P, Massarelli E, Lin S, et al. CXCR2 expression in tumor cells is a poor prognostic factor and promotes invasion and metastasis in lung adenocarcinoma. Cancer Res. 2013;73(2):571–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Nannuru KC, Sharma B, Varney ML, Singh RK. Role of chemokine receptor CXCR2 expression in mammary tumor growth, angiogenesis and metastasis. J Carcinog. 2011;10:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Halpern JL, Kilbarger A, Lynch CC. Mesenchymal stem cells promote mammary cancer cell migration in vitro via the CXCR2 receptor. Cancer Lett. 2011;308(1):91–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Del Río R, Ochoa C, Alarcon A, Arnáez J, Blanco D, García-Alix A. Amplitude integrated electroencephalogram as a prognostic tool in neonates with hypoxic- ischemic encephalopathy: a systematic review. PLoS One. 2016;11(11):e0165744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Meng S, Zhou H, Feng Z, et al. CircRNA: functions and properties of a novel potential biomarker for cancer. Mol Cancer. 2017;16(1):94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wang S, Tang D, Wang W, et al. circLMTK2 acts as a sponge of miR-150-5p and promotes proliferation and metastasis in gastric cancer. Mol Cancer. 2019;18(1):162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Liu H, Wu Y, Wang S, et al. Circ-SMARCA5 suppresses progression of multiple myeloma by targeting miR-767-5p. BMC Cancer. 2019;19(1):937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chen J, Chen T, Zhu Y, et al. circPTN sponges miR-145-5p/miR-330-5p to promote proliferation and stemness in glioma. J Exp Clin Cancer Res. 2019;38(1):398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lu Y, Deng X, Xiao G, Zheng X, Ma L, Huang W. circ_0001730 promotes proliferation and invasion via the miR-326/Wnt7B axis in glioma cells. Epigenomics. 2019;11(11):1335–1352. [DOI] [PubMed] [Google Scholar]
- 35. Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333–338. [DOI] [PubMed] [Google Scholar]
- 36. Homstein E., Mansfield JH, Yekta S, et al. The microRNA miR-196 acts upstream of Hoxb8 and Shh in limb development. Nature. 2005;438(7068):671–674. [DOI] [PubMed] [Google Scholar]
- 37. Lin L, Chen X, Peng X, et al. MicroRNA-128 promotes cell-cell adhesion in U87 glioma cells via regulation of EphB2. Oncol Rep. 2013;30(3):1239–1248. [DOI] [PubMed] [Google Scholar]
- 38. Zheng Q, Bao C, Guo W, et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun. 2016;7:11215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhong Z, Lv M, Chen J. Screening differential circular RNA expression profiles reveals the regulatory role of circTCF25-miR-103a-3p/miR-107-CDK6 pathway in bladder carcinoma. Sci Rep. 2016;6:30919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zheng J, Liu X, Xue Y, et al. TTBK2 circular RNA promotes glioma malignancy by regulating miR-217/HNF1β/Derlin-1 pathway. J Hematol Oncol. 2017;10(1):52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Acker G, Zollfrank J, Jelgersma C, et al. The CXCR2/CXCL2 signalling pathway—an alternative therapeutic approach in high-grade glioma. Eur J Cancer. 2020;126:106–115. [DOI] [PubMed] [Google Scholar]
- 42. Wen J, Zhao Z, Huang L, Wang L, Miao Y, Wu J. IL-8 promotes cell migration through regulating EMT by activating the Wnt/β-catenin pathway in ovarian cancer. J Cell Mol Med. 2020;24(2):1588–1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wu D, Chen X, Wang L, Chen F, Cen H, Shi L. Hypoxia-induced microRNA-141 regulates trophoblast apoptosis, invasion, and vascularization by blocking CXCL12β/CXCR2/4 signal transduction. Biomed Pharmacother. 2019;116:108836. [DOI] [PubMed] [Google Scholar]
- 44. Ding D, Zhang Y, Yang R, et al. miR-940 suppresses tumor cell invasion and migration via regulation of cxcr2 in hepatocellular carcinoma. Biomed Res Int. 2016;2016:7618342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zhong C, Li X, Tao B, et al. LIM and SH3 protein 1 induces glioma growth and invasion through PI3K/AKT signaling and epithelial-mesenchymal transition. Biomed Pharmacother. 2019;116:109013. [DOI] [PubMed] [Google Scholar]
- 46. Nanta R, Shrivastava A, Sharma J, Shankar S, Srivastava RK. Inhibition of sonic hedgehog and PI3K/Akt/mTOR pathways cooperate in suppressing survival, self-renewal and tumorigenic potential of glioblastoma-initiating cells. Mol Cell Biochem. 2019;454(1-2):11–23. [DOI] [PubMed] [Google Scholar]
- 47. Sun F, Wang J, Sun Q, et al. Interleukin-8 promotes integrin β3 upregulation and cell invasion through PI3K/Akt pathway in hepatocellular carcinoma. J Exp Clin Cancer Res. 2019;38(1):449. [DOI] [PMC free article] [PubMed] [Google Scholar]