Abstract
The raw datasets of oxysterol quantifications from whole cell and mitochondrial fractions of THP-1 monocytes and macrophages, neuronal-like SH-SH5Y cells and human peripheral blood mononuclear cells are presented. Oxysterols were quantified using a new liquid chromatography-mass spectrometry (LC-MS) and multiple reaction monitoring analysis published in the article “A quantitative LC-MS/MS method for analysis of mitochondrial-specific oxysterol metabolism” in Redox Biology [1]. This method showed improved extraction efficiency and recovery of mono and dihydroxycholesterols from cellular matrix. The datasets derived from the three cell lines are included in the appendix. These datasets provide new information about the oxysterol distribution in THP-1 monocytes and macrophages, SH-SY5Y cells and peripheral blood mononuclear cells. These datasets can be used as a guide for oxysterol distribution in the three cell lines for future studies, and can used for future method optimization, and for comparison of oxysterol recovery with other analytical techniques.
Keywords: Oxysterol, Cholesterol, Metabolism, Mitochondria, Liquid chromatography-mass spectrometry, Peripheral blood mononuclear cell
Specifications Table
| Subject | Biochemistry, Genetics and Molecular Biology, Analytical chemistry |
| Specific subject area | Development of new mass spectrometry method for analysis of mitochondrial-specific oxysterol metabolism |
| Type of data | Figure Raw and analysed datasets are included in appendix as Data File S1 and supplementary |
| How data were acquired | Liquid chromatography-mass spectrometry (targeted analysis) Multiple reaction monitoring method Instrument: Waters Xevo TQ-S Triple Quadrupole Mass Spectrometer; Waters ACQUITY Ultra performance liquid chromatography (UPLC) quaternary system; NUCLEOSIL C18 column (100–5 125/2) |
| Data format | .raw files Analyzed data sets in excel spreadsheet |
| Parameters for data collection | Samples extracted at room temperature. Samples in LC were kept at 10° celcius during LC-MS analysis. Raw data were used for quantification and normalized to the external standard data for each sample |
| Description of data collection | Mass spectrometry data was collected with the Waters LC-MS instrument. Peak Detection and integration was performed with ApexTrack integration feature built in MassLynx. Peak areas were extracted for the quantifier MRM transition for each analyte. Graphs and statistical analysis including two-tailed t-tests, ANOVA analysis and regression analysis were performed using Graphpad prism 8.0 and in Microsoft excel. The full details of the method is published in Borah et al., 2020 [1] |
| Data source location | Institution: University of Surrey City/Town/Region: Guildford Country: United Kingdom |
| Data accessibility | Analysed datasets are included in appendix as Data File S1. Raw mass spectra files are available in the Mendeley data repository (http://dx.doi.org/10.17632/npcc7yrfjk.1) and also in supplementary. |
| Related research article | K. Borah, O.J. Rickman, N. Voutsina, I. Ampong, D. Gao, E.L. Baple, I.H.Dias, A.H. Crosby, H.R. Griffiths, A quantitative LC-MS/MS method for analysis of mitochondrial -specificoxysterol metabolism, Redox Biology (2020). Doi:https://doi.org/10.1016/j.redox.2020.101595 |
Value of the Data
-
•
The datasets include oxysterols distributions in whole cell and mitochondrial fractions extracted from THP-1, SH-SY5Y and peripheral blood mononuclear cells (PBMC) and they provide new information on the oxysterol distribution in these cell lines.
-
•
Researchers can use these oxysterol datasets as a reference for comparison with that of genetically modified or drug treated THP-1, SH-SY5Y and PBMCs.
-
•
Researchers can use the LC-MS/MS method to extract and quantify oxysterols in various cell lines and also for future method development and optimisations.
1. Data Description
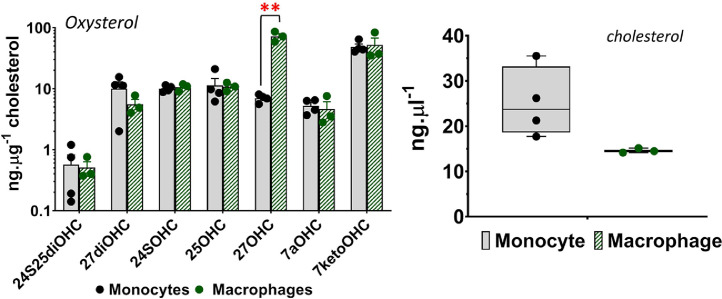
The analysed datasets showing mono and dihydroxycholesterol distribution in THP-1, SH-SY5Y and PBMCs are included in appendix as Data file S1. The mass spectrometry data generated in the study are available as raw mass spectra files deposited in the Mendeley data repository (http://dx.doi.org/10.17632/npcc7yrfjk.1) [2], and as supplementary material included in this manuscript. We also measured oxysterol and cholesterol distribution in THP-1 macrophages and compared it to monocytes (Fig. 1). Oxysterols were extracted using Triton and dimethyl sulfoxide cell lysis, followed by solid phase extraction (SPE) of oxysterols. We quantified oxysterols using our newly developed LC-MS/MS method [1]. The raw and analysed data are included in Data File S1. We measured significantly higher 27-hydroxycholesterol (27OHC) levels in macrophages, demonstrating the sensitivity of our LC-MS/MS method to detect cell-specific oxysterol metabolism.
Fig. 1.
Oxysterol (A) and cholesterol (B) quantification in THP-1 monocytes and macrophages. Values are mean ± Standard deviation (n = 3–4 biological replicates). Statistical significance was determined using Holm-Sidak test with α=0.05; **, P<0.005.
The datasets for whole cell and mitochondrial oxysterols in THP-1 monocytes, SH-SY5Y and PBMCs were used to support results and conclusions in Borah et al., 2020 [1]. We quantified oxysterols in the mitochondria and whole cells and demonstrated distinctive mitochondrial-specific oxysterol metabolism in the three cell lines [1]. For oxysterol analysis in PBMCs, blood samples were collected from healthy individuals and were stored for 1 h at room temperature, prior to PBMC and mitochondrial isolation. 7ketocholesterol (7ketoOHC) was the predominant oxysterol in both mitochondrial and whole cell fractions (see Data File S1). In PBMC mitochondrial fractions, 24S-hydroxycholesterol (24SOHC) and 24S-dihydroxycholesterol + 25-dihydroxycholesterol (24S25diOHC) were the least abundant. In PBMC whole cells, 7α-hydroxycholesterol was the least abundant. We measured mitochondrial-specific oxysterol distribution in both THP-1 monocytes and SH-SY5Y cells (see Data File S1). In THP-1 monocytes, 7ketoOHC was the predominant oxysterol in both mitochondrial and whole cell fractions. In SH-SY5Y cells, 27OHC concentrations were higher than the other enzymatically derived mono- and di-hydroxycholesterols in mitochondria, while 7ketoOHC was the dominant oxysterol in whole cell fractions. 24SOHC was the least abundant oxysterol in mitochondria, while 24S25diOHC was the least abundant in whole cells.
2. Experimental Design, Materials and Methods
2.1. Cell culture and oxysterol extraction
Blood was collected from healthy individuals with approval from ethics committee. PBMC isolation and cultivation of THP-1 human monocytic cell line and SH-SY5Y cells in their respective growth medium were performed as described in the related article [1]. THP-1 monocytes were treated with 100 ng.mL−1 phorbol myristate acetate for three days to differentiate them into macrophages prior to cell harvest and oxysterol analysis.
The detailed methodology for cell lysis and oxysterol extraction is described in the related article [1]. 22SOHC-D7 was used as the external standard in preparation of standards and samples. Briefly, whole cells and mitochondrial fractions from different cell types were sonicated in the lysis buffer (Triton, DMSO and butylated hydroxytoluene). Polar and non-polar metabolites were separated using biphasic solvent extraction with methanol, dichloromethane and water. Samples were spun and lower non-polar phase containing lipids, cholesterol and oxysterols were dried under N2 gas, followed by solid phase extraction (SPE) to isolate oxysterols [1].
2.2. LC-MS/MS analysis and data processing
Oxysterols were analysed using LC-MS/MS multiple reaction monitoring technique set up for nine oxysterols including 24(S) hydroxycholesterol, 25-hydroxycholesterol, 27-hydroxycholesterol, 7α-hydroxycholesterol, 7 ketocholesterol, 7α,27-dihydroxycholesterol, 7α,24 (R/S)-dihydroxycholesterol, 7α,25-dihydroxycholesterol and using Xevo TQ-S Triple Quadrupole mass spectrometer. The details of the LC method, mass spectrometer parameters and MS/MS transitions for the nine oxysterols are available in the related article [1]. 7α,24 (R/S)-dihydroxycholesterol (24SdiOHC) and 7α,25-dihydroxycholesterol (25diOHC) were analysed together as 24S25diOHC for detection and quantification due to their identical elution and MS/MS transitions.
Mass Lynx was used for extraction and processing of mass spectra. Oxysterols were identified based on the peak retention times and their MS/MS transitions. Standard curves were generated using the peak areas and linear regression analysis using authentic oxysterol standards analysed in a range of concentrations (0, 0.005, 0.010, 0.025, 0.050, 0.075, 0.1, 0.25, 0.5, 0.75 and 1 ng.µl−1), and raw data are included in Data File S1. The raw mass spectra are available in the data repository (http://dx.doi.org/10.17632/npcc7yrfjk.1) [2] and in the supplementary material. These data can be visualised and processed using freely available tools such as Proteowizard and Msconvert.
2.3. Statistical analysis
Standard curves were generated using linear regression analysis. Holm Sidak test was used to compare oxysterols in THP-1 monocytes and macrophages with α=0.05. These analyses were performed in Prism 8.2.
Ethical statement for blood samples
Blood samples were collected from healthy volunteers with ethics committee approval and with informed consent.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships which have, or could be perceived to have, influenced the work reported in this article.
Acknowledgement
K Borah and HR Griffiths acknowledge INClusilver funded by the European Union, grant number H2020‐INNOSUP‐2017‐2017 731349; NeutroCure funded by the European Union, grant number H2020-FETOPEN-01-2018-2019-2020 861878 and Faculty Research Support Fund (FRSF) fund from the University of Surrey 2019–2020. IHKD acknowledges funding from Alzheimer's research UK midlands network grant 2019. AH Crosby and EL Baple acknowledge support from the Hereditary Spastic Paraplegia Support Group and The Diamond Jubilee Doctoral Scholarship Fund.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.dib.2020.106382.
Appendix
The analysed data sets are included in appendix as excel file (Data File S1). The supplementary material includes mass spectra (.raw files) generated in this study and in the related article [1].
Appendix B. Supplementary Materials
References
- 1.Borah K., Rickman O.J., Voutsina N., Ampong I., Gao D., Baple E.L., I.H.Dias A.H.Crosby, Griffiths H.R. A quantitative LC-MS/MS method for analysis of mitochondrial–specific oxysterol metabolism. Redox Biol. 2020 doi: 10.1016/j.redox.2020.101595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Borah K., Griffiths H.R. Oxysterol_LCMS/MS raw data_whole cell/mitochondria. Mendeley Data. 2020 doi: 10.17632/npcc7yrfjk.1. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.