Abstract
The 2019 novel coronavirus disease (COVID-19) pandemic caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has affected all aspects of human life. Rapid, accurate, sensitive and user friendly detection method is urgently needed to facilitate early intervention and control the spread of SARS-CoV-2. Here, we propose a one-pot visual SARS-CoV-2 detection system named “opvCRISPR” by integrating reverse transcription loop-mediated isothermal amplification (RT-LAMP) and Cas12a cleavage in a single reaction system. We demonstrate that the collateral activity against single-stranded DNA (ssDNA) reporters of activated Cas12a triggered by RT-LAMP amplicon increases detection sensitivity and makes detection results observable with naked eye. The opvCRISPR enables detection at nearly single molecule level in 45 min. We validate this method with 50 SARS-CoV-2 potentially infected clinical samples. The opvCRISPR diagnostic results provide 100% agreement with the Centers for Disease Control and Prevention (CDC)-approved quantitative RT-PCR assay. The opvCRISPR holds great potential for SARS-CoV-2 detection in next-generation point-of-care molecular diagnostics.
Keywords: SARS-Cov-2, CRISPR, RT-LAMP, One-pot, Visual detection, Molecular diagnosis
Highlights
-
•
We have proposed an ultrasensitive visual SARS-CoV-2 detection method that could be accomplished within 45 min.
-
•
This visual detection method has fundamental differences with colorimetric methods.
-
•
This method integrated RT-LAMP and CRISPR/Cas cleavage in one-pot and totally avoided amplicon contamination.
-
•
The operation is greatly simplified with only a thermoblock and blue light required.
-
•
This method could detect SARS-CoV-2 at nearly single molecule level and demonstrated 100% positive predictive agreement.
1. Introduction
The outbreak of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the causative agent of 2019 novel coronavirus disease (COVID-19), has led to a global pandemic, with 21, 073, 456 reported cases and 757,479 deaths worldwide as of 14, August 2020 (Lu et al., 2020a, Lu et al., 2020b). Person-to-person transmission from infected individuals with no or mild symptoms has been reported (Bai et al., 2020; Rothe et al., 2020). Rapid, accessible and accurate detection of SARS-CoV-2 is urgently needed to facilitate early intervention and reduce disease transmission risk.
Quantitative reverse transcription polymerase chain reaction (qRT-PCR) has been considered as the ‘gold standard’ and is the most commonly used detection method for SARS-CoV-2 (Chu et al., 2020; Corman et al., 2020). Although the efficiency is established, it requires professional personnel, long operation time and expensive equipment for implementation. For these reasons, it is impractical for the point-of-care (POC) diagnostic applications.
In such a backdrop, clustered regularly interspaced short palindromic repeats (CRISPR)-associated (Cas) nuclease based methods with properties of ultrasensitive, cheaper and portable diagnostic tests for the assessment of suspected cases, could help advance the diagnosis of SARS-CoV-2, regardless of the presence of qualified personnel or sophisticated equipment for virus detection (Chen et al., 2018; Gootenberg et al., 2017; Li et al., 2018). The CRISPR-Cas nucleases, including RNA-guided RNases (Cas13a-d) and RNA-guided DNases (Cas12 and Cas14), display collateral cleavage activity (Kellner et al., 2019; Harrington et al., 2018). Upon recognition of their RNA or DNA targets, activated CRISPR-Cas nucleases indiscriminately cleave nearby single-stranded non-targeted nucleic acids (Chen et al., 2018). Combining with nucleic acid amplification, amplified products will trigger CRISPR-Cas nucleases for collateral cleavage of reporters to achieve additional sensitivity (Kellner et al., 2019; Wang et al., 2020a, Wang et al., 2020b). Researchers have developed CRISPR-based tests by integrating PCR or recombinase polymerase amplification (RPA) with CRISPR cleavage (Ding et al., 2020; Guo et al., 2020; Tian et al., 2020; Wang et al., 2019, Wang et al., 2020a, Wang et al., 2020b; Zhang et al., 2020a, Zhang et al., 2020b). But these approaches have limitations, such as two separate reaction steps, long incubation time or weak signals for limited template detection. Loop-mediated isothermal amplification (LAMP) has been coupled with CRISPR for SARS-CoV-2 detection (Broughton et al., 2020; Joung et al., 2020; Li et al., 2019; Qian et al., 2019). LAMP is highly specific and generates a high yield of amplicon in a short time, with reagents more affordable and accessible (Notomi et al., 2000). However, the current LAMP-CRISPR strategies have drawbacks such as uncapped operation, long reaction time and complicated operations.
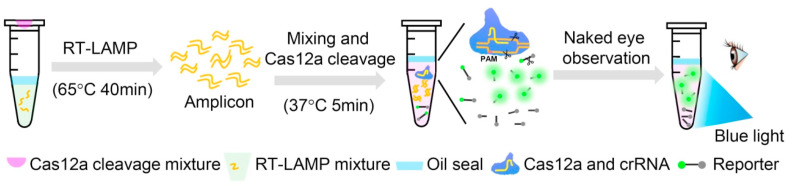
Here, we developed a one-pot visual reverse transcription (RT)-LAMP-CRISPR (opvCRISPR) method for ultrasensitive visual detection of SARS-CoV-2, which simplified the operations and avoided contamination. The RT-LAMP reagents are incubated at the bottom of the tube. The CRISPR/Cas12a reaction reagents are added on the lid. SARS-CoV-2 RNA templates extracted from the respiratory swab are amplified by RT-LAMP, followed by mixing with the Cas12a reagents for cleavage. Once the Cas12a nuclease is activated by recognizing DNA target, it splits the quenched fluorescent single-stranded DNA (ssDNA) reporter (FAM-TTATT-BHQ1) indiscriminately, generating the fluorescence signal visible to the naked eye under blue light (Fig. 1 ). Besides, the minimum equipment required to operate the protocol after RNA extraction includes only pipettes, reagent tubes, a thermo block and a blue light. All the equipment could be integrated into portable suitcase for point-of-care applications (Fig. S1).
Fig. 1.
The scheme of one-pot visual RT-LAMP-CRISPR (opvCRISPR) detection method. For SARS-CoV-2 detection, 20 μL RNA templates extracted from the respiratory swab is added into RT-LAMP mixture. The RT-LAMP mixture is placed at the bottom of the tube and sealed with 25 μL oil. The CRISPR/Cas12a reaction reagents are added inside the lid. After 40 min of RT-LAMP amplification at 65 °C, shake the tube to mix with Cas12a reagents for cleavage. Once the Cas12a nuclease is activated by recognizing DNA target, it splits the quenched fluorescent ssDNA reporter indiscriminately, generating fluorescence signal visible to the naked eye under blue light. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
2. Material and methods
2.1. Materials
All primers, ssDNA probe and CRISPR RNA (crRNA) were synthesized by Sangon Biotech. (Shanghai, China). EngenLba Cas12a, DNase I, HiScribe T7 High YieldRNA Synthesis Kit and WarmStart LAMP Kit were purchased from New EnglandBiolabs (Ipswich, MA, UK). " to "EngenLba Cas12a, DNase I, HiScribe T7 High Yield RNA Synthesis Kit and WarmStart LAMP Kit were purchased or kindly free-provided by New England Biolabs (Ipswich, MA, USA). RNase inhibitor and Taq Hot Start Version were purchased from Takara Bio Inc. (Dalian, China). QIAGEN OneStep RT-PCR kit was purchased from Qiagen (Frederick, MD, USA). RNAXP clean beads were purchased from Beckman Coulter Inc. (Indianapolis, IN, USA). A pUC57 plasmid with SARS-CoV-2 gene sequences was kindly provided by BGI (Beijing, China). Plasmids with three SARS-like coronaviruses (SARS-CoV-2 (Genebank: NC_045512.2), bat SARS-like coronavirus (bat-SL-CoVZC45, Genebank: MG772933.1) and SARS-CoV (Genebank: NC_004718.3)) and one human coronaviruses (HKU1 (Genebank: NC_006577.2) were synthesized by Sangon Biotech. (Shanghai, China). RNA of clinical samples were provided by the First People's Hospital of Yuhang district (Hangzhou, China). The study was approved by the Scientific Research Ethics Review Committee of the First People's Hospital of Yuhang district. Written informed (oral) consent was obtained from all the patients involved. Nasopharyngeal swab samples were collected from all patients at admission.
2.2. In vitro RNA preparation with T7 RNA polymerase
For in vitro transcribed RNA template preparation, 493 nt S gene sequence in plasmid was firstly PCR-amplified with primers containing T7 promotor (Fig. S2). Then, the PCR amplicon were extracted and purified with Gel Extraction Kit to form in vitro transcribed templates. Thereafter, RNA was synthesized from PCR amplicon by incubating at 37 °C for 4 h using High Yield RNA Synthesis Kit. Next, the synthesized RNA was treated by DNase I to remove DNA template. Finally, the obtained RNA was extracted and purified with RNAXP clean beads and stored at −80 °C for further use.
The stock concentration of purified in vitro transcribed RNA was determined with NanoDrop ND-1000 (Thermo Fisher Scientific Inc., Waltham, MA, USA) and calculated to be 5 × 1010 copies/μL. The target site for opvCRISPR detection was firstly PCR-amplified with RT-LAMP outer primers, followed by sequencing and blasting to confirm its accuracy for SARS-CoV-2 detection (Fig. S3).
2.3. Cas12a-mediated cleavage assay
The Cas12a-mediated cleavage assay contained 1 × NEB buffer 2.1, 0.2 μM of EnGenLba Cas12a, 0.6 μM of crRNA, 1 μM of ssDNA reporter, 4 U of RNase inhibitor with 10 ng SARS-CoV-2 plasmid in 20 μL reaction volume. The reaction was performed at 37 °C on a thermo block (Suzhou beaver Biomedical Engineering Co., Ltd, Suzhou, China) for 20 min and photographed immediately under blue light.
2.4. One-step RT-PCR assay
QIAGEN OneStep RT-PCR kit was employed for RT-PCR assay. RT-PCR primers targeting SARS-CoV-2 S gene were designed with primer premier 5.0. OneStep RT-PCR assay contained 1 × QIAGEN OneStep RT-PCR Buffer (Mg2+ plus), 400 μM of each dNTP, 0.6 μM of each primer (F and R), 2 μL of QIAGEN OneStep RT-PCR Enzyme Mix, 1 × EvaGreen Dye, 1 μL of in vitro transcribed RNA template or 20 μL of RNA from clinical samples, and RNase-free water up to 50 μL. Thermal cycling was carried out with 30 min at 50 °C for reverse transcription, 15 min at 95 °C for HotStarTaq DNA Polymerase activation, followed by 45 cycles of denaturation at 94 °C for 30 s, annealing at 60 °C for 30 s and extension at 72 °C for 1 min. After amplification, 2 μL of amplified products were electrophoresed at 3% agarose gel under 120 V for 30 min.
2.5. Real-time RT-LAMP assay
The LAMP primer group contains six primers targeting eight regions of S gene sequence was designed with PrimerExplorer V5 (http://primerexplorer.jp/e/index.html). RT-LAMP was performed in 40 μL reaction mixture containing 1 × WarmStart LAMP Master Mix, 1.6 μM of FIP and BIP, 0.2 μM F3 and B3, 0.4 μM LF and LB, 1 × Fluorescent Dye and 1 μL of in vitro transcribed RNA template. The reaction mixture was incubated at 65 °C for 40 min via LightCycler 480 System (Roche Diagnostics Co., Indianapolis, IN, USA). 2 μL of amplified products were electrophoresed at 3% agarose gel under 120 V for 30 min and imaged in ChemiDoc XRS + System (Bio-Rad Laboratories Inc., Hercules, CA, USA).
2.6. One-pot visual RT-LAMP-CRISPR detection
For opvCRISPR detection, 40 μL of RT-LAMP reaction mixture (Fluorescent Dye-free) including 1 μL of in vitro transcribed RNA template or 20 μL of RNA from clinical samples was added at the bottom of tube and covered with 25 μL mineral oil. 20 μL Cas12a cleavage system containing 0.4 μM of EnGenLba Cas12a, 2 × NEB buffer 2.1, 1.2 μM of crRNA, 2 μM of ssDNA reporter, RNase inhibitor 8 U was added in the lid. Cas12a cleavage system was physically separated from the high-temperature zone at the bottom by a section of air column and oil phase. After 40 min of RT-LAMP amplification, Cas12a cleavage system was mixed with LAMP product by shaking operation. Then, Cas12a cleavage reaction was conducted at 37 °C for 5 min and fluorescent results were observed immediately with naked eye under blue light illuminator (LABGIC Inc., Beijing, China).
3. Results
3.1. SARS-CoV-2 specific crRNA design
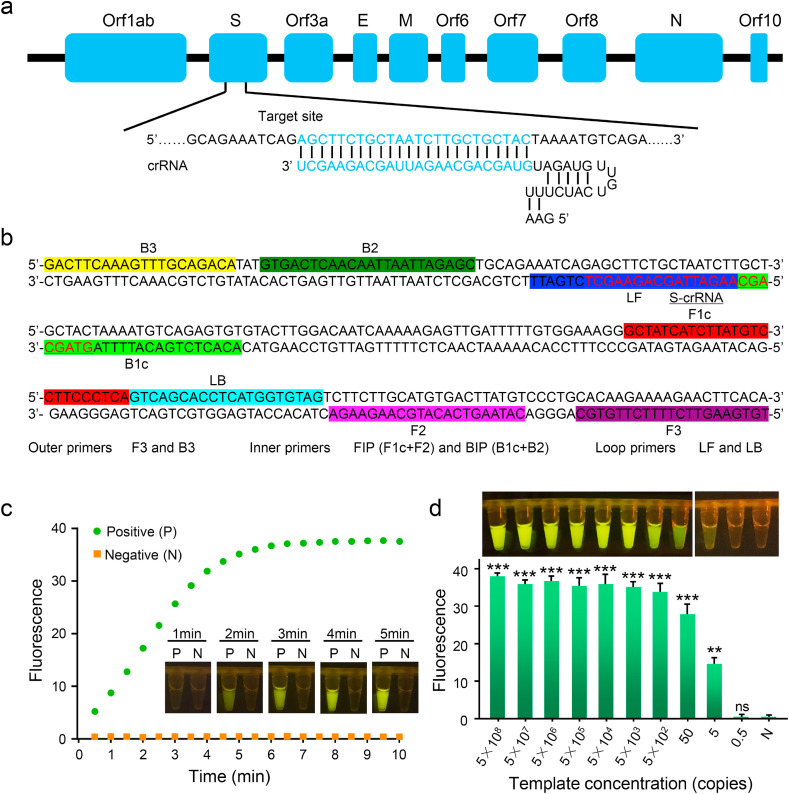
To achieve high detection sensitivity, we firstly optimized the crRNA design. A total of 7 SARS-CoV-2 specific crRNAs targeting different domains of orf1ab, S and N gene were designed. To evaluate the efficiency of crRNAs, the individual crRNA was incubated with Cas12a, synthetic SARS-CoV-2 target DNA and ssDNA reporter. The results revealed that 4 out of 7 crRNAs generated fluorescence and the S-targeted crRNA produced the strongest signal (Fig. S4). In the following experiments, we selected crRNA targeting S gene for SARS-CoV-2 detection (Fig. 2 a).
Fig. 2.
(a) Genome map shows the target site and crRNA sequence on the SARS-CoV-2 S gene. (b) The sequences and locations of RT-LAMP primers and S-crRNA in this assay. (c) Real-time fluorescence records sample signals at different time points during Cas12a digestion. Real-time images of the samples at different time points are shown. For each time point, three repeats of samples are photographed and one represented image is shown for each time point. P: positive; N: No template added control. (d) Fluorescence signals of opvCRISPR with 10-fold gradually diluted templates are quantified by Light Cycler 480 fluorescence detection system (top, n = 3, error bars showed mean ± SEM). The corresponding sample images with one represented tube for one template concentration are shown above. N: no template added control.
3.2. One-pot visual RT-LAMP-CRISPR system built up
Next, we attempted to integrate RT-LAMP and Cas12a cleavage in a single tube. We designed a set of 6 RT-LAMP primers for the target amplification (Fig. 2b). The RT-LAMP mixture, including 1 × WarmStart LAMP Master Mix, 1.6 μM of FIP and BIP, 0.2 μM F3 and B3, 0.4 μM LF and LB and 1 μL of vitro transcribed RNA template, was added to the bottom of the tube. RT-LAMP took place at 65 °C, which may inactivate Cas12a enzyme. To avoid inactivation of Cas12a, mineral oil was coated on the RT-LAMP mixture to prevent heat transfer. The CRISPR/Cas12a reaction reagents, including Cas12a enzyme, crRNAs, and ssDNA reporter were added on the lid. To optimize the ratio of Cas12a mixture and RT-LAMP mixture, 20 μL of Cas12a mixture combined with different volumes of RT-LAMP mixture were employed. For each volume of RT-LAMP mixture, either fixed copy number (5 × 105 copies) or fixed concentration (0.2 × 105 copies per μL) of SARS-CoV-2 RNA was added as positive sample with no template added as negative control. After 40 min of RT-LAMP amplification, the reaction was transferred to a 37 °C heater and Cas12a mixture was shaken into amplified products for digestion for 10 min. Five to 40 μL of RT-LAMP mixture generated significant fluorescence (Figs. S5a–c). Considering the large clinical sample size, 40 μL of RT-LAMP mixture combined with 20 μL of Cas12a mixture was used in the following experiments.
To evaluate the detection specificity, we tested three SARS-like coronaviruses (SARS-CoV-2 (Genebank: NC_045512.2), bat SARS-like coronavirus (bat-SL-CoVZC45, Genebank: MG772933.1) and SARS-CoV (Genebank: NC_004718.3)) and one human coronaviruses (HKU1 (Genebank: NC_006577.2). Results showed that only the RNA from SARS-CoV-2 produced signals, whereas RNAs from other pathogens did not produce any detected signals, demonstrating good specificity for SARS-CoV-2 determination (Fig. S6).
3.3. Optimization of opvCRISPR method for SARS-cov-2 detection
Thereafter, we optimized the Cas12a cleavage time. We prepared a batch of samples containing 500 copies of RNA templates. Pure water was used as blank control and RNA extracted from uninfected people were used as negative control. After RT-LAMP amplification, the products were mixed together with Cas12a and incubated at 37 °C for different time. Cas12a enzyme was heat-inactivated immediately after incubation. The results revealed that the fluorescence signals could be observed after 2 min of incubation and increased over time (Fig. 2c). There was almost no difference between 4 and 5 min of incubation. No fluorescence was observed in blank control or negative control. The real-time fluorescent signals were quantified by the Light Cycler 480 fluorescence detection system, which showed that the fluorescence signals increased over time and reached the plateau in 5 min. After 10-min reaction, there was no fluorescence generated in negative control (Fig. S7). Therefore, 5 min were used as the Cas12a cleavage time in the opvCRISPR detection.
3.4. Sensitivity evaluation of opvCRISPR method
Next, we explored the sensitivity of opvCRISPR by testing eight 10-fold serially diluted RNA templates. The results revealed that the detection limit of opvCRISPR was 5 copies (Fig. 2d). We compared the sensitivity of opvCRISPR with RT-PCR and RT-LAMP. The sensitivity of opvCRISPR was comparable to that of RT-PCR and ten times higher than that of RT-LAMP (Fig. 3 a and b). To confirm that Cas12a cleavage improved sensitivity, RT-LAMP amplification was stopped at different time and the products were digested by Cas12a for visual detection. The results revealed that the amplicons accumulated by RT-LAMP within 20 min were below the fluorescence threshold but could be detected by Cas12a cleavage (Fig. 3c). Therefore, Cas12a cleavage increased RT-LAMP sensitivity, resulting in improved detection sensitivity to nearly single-molecule level.
Fig. 3.
Real-time fluorescence of RT-LAMP (a) and RT-PCR (b) amplification with 10-fold gradually diluted in vitro transcribed RNA as templates. For each template concentration, three repeats are tested simultaneously and one fluorescence curve for each template concentration is shown here. 2 μL of amplified products are gel electrophoresed and imaged shown in the bottom. (c) Visual detection results after RT-LAMP amplified for 5, 10, 15, 20, 25, 30, 35, 40 min, respectively. For each time point, three repeats of samples are photographed at the same time and one represented image is shown here.
3.5. OpvCRISPR method for SARS-cov-2 infected clinical samples determination
To investigate the diagnostic accuracy and reliability of the opvCRISPR, respiratory swab samples from 26 infected clinical cases were tested for SARS-CoV-2. These samples have been previously verified to be SARS-CoV-2 positive by real-time RT-PCR. We also tested respiratory swab samples from 24 uninfected clinical cases. In this study, real-time RT-PCR was conducted as the standard control. The opvCRISPR and RT-PCR were performed by adding 20 μL of the extracted RNA as templates. All infected samples were determined to be SARS-CoV-2 positive while all uninfected samples tested to be negative by both opvCRISPR and RT-PCR (Fig. 4 , Figs. S8 and S9).
Fig. 4.
SARS-CoV-2 detection in 26 SARS-CoV-2 infected clinical samples with proposed opvCRISPR method and real-time RT-PCR. P: positive control with in vitro transcribed RNA template; N: No template added control.
4. Discussion
Although RT-PCR is the gold standard and most widely used for SARS-CoV-2 determination with high detection sensitivity and reliability, it is not suitable for large-scale point-of-care diagnostics due to severe staffing and critical supply shortages, the lack of sophisticated equipment and long reaction time (Zhou et al., 2020; Corman et al., 2020). Our proposed method is affordable, user-friendly, rapid and robust. It integrates RT-LAMP amplification and CRISPR cleavage in one-pot reaction with detection sensitivity just as high as RT-PCR. The minimum equipment required to operate the protocol include only pipettes, reagent tubes, a thermo block and a blue light. All the equipment can be integrated into a 60 × 50 cm2 suitcase (Fig. S1). Thus, the proposed method has great potential to enable point-of-care testing outside of the clinical diagnostic laboratory, such as airports, local emergency departments and clinics and other locations.
Colorimetric methods coupled with RT-LAMP amplification have been mostly reported for point-of-care SARS-CoV-2 detection (Zhang et al., 2020a, Zhang et al., 2020b; Lau et al., 2020; Park et al., 2020; Yang et al., 2020). Our method has advantage over colorimetric methods. First, it is more specific than colorimetric methods. The biggest drawback of colorimetric methods is that most of them are lack of detection specificity because their detection objects are based on by-products of LAMP amplification including H+ induced pH change (Zhang et al., 2020a, Zhang et al., 2020b; Huang et al., 2020; Lu et al., 2020a, Lu et al., 2020b), double stranded DNA secondary structure (Wang et al., 2020a, Wang et al., 2020b; Park et al., 2020) and Magnesium pyrophosphate (Lau et al., 2020; Yang et al., 2020). Our proposed method employed CRISPR, which even could achieve single base mutation (SNP) detection by crRNA specially targeting target sequence, greatly improving detection specificity. Second, our method is more sensitive than colorimetric methods. We demonstrated that CRISPR-Cas12a cleavage of fluorescence quenched reporters can make second round signal amplification triggered by LAMP amplicon, increasing the detection sensitivity by 10-fold. Third, our method is more distinguishable with naked eye than colorimetric methods. Colorimetric methods have high background for naked eye observation because the color always changes from one color to an adjacent one (Wang et al., 2020a, Wang et al., 2020b; Lau et al., 2020; Park et al., 2020). Our proposed method is based on fluorescence detection, which is more distinguishable with naked eye than colorimetric methods.
The opvCRISPR has advantage over existing CRISPR-based SARS-CoV-2 detection. Several two-step CRISPR-based methods for SARS-CoV-2 detection have been developed (Broughton et al., 2020; Ding et al., 2020; Guo et al., 2020; Lucia et al., 2020), but the separate reaction steps increase operation complexity and the likelihood of amplicon cross-contamination. One-step CRISPR-based methods coupled with RPA have been developed (Ding et al., 2020; Li et al., 2019), but they either require long incubation time (120 min) or present high background during visual detection due to multiple enzymes in the RPA system (Ding et al., 2020). In addition, amplicon yield of RPA was lower than that of LAMP, which decreased the sensitivity of CRISPR detection. Therefore, we provided an efficient, accurate and easy-to-implement solution for increasing testing capacity during SARS-CoV-2 determination.
5. Conclusions
We have proposed a one-pot visual method for SARS-CoV-2 detection by integrating RT-LAMP amplification with Cas12a cleavage. The detection sensitivity is at nearly single molecule level and the whole detection process could be accomplished within 45 min. The proposed method only requires minimal equipment, demonstrating great potential in enabling next-generation molecular diagnosis towards point-of-care diagnosis. However, the present method requires additional step to extract RNA. Further efforts need to be made to combine the RNA extraction module with the opvCRISPR to achieve from sampling to result nucleic acid detection.
CRediT authorship contribution statement
Rui Wang: Methodology, Data curation, Validation, Writing - original draft. Chunyan Qian: Resources, Data curation, Validation. Yanan Pang: Formal analysis, Writing - review & editing. Miaomiao Li: Resources. Yu Yang: Resources. Haijing Ma: Resources. Manying Zhao: Resources. Feng Qian: Resources. Hang Yu: Resources. Zhenping Liu: Resources. Ting Ni: Conceptualization, Validation, Writing - review & editing, Supervision, Conceptualization, Validation, Writing - review & editing, Supervision. Yan Zheng: Conceptualization, Validation, Writing - review & editing, Supervision.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (32001786), Special project of China postdoctoral science foundation for prevention and control of novel coronavirus pneumonia (2020T130018ZX), the internal grants from Fudan University and Hangzhou First People's Hospital, the National Natural Science Foundation of China (81870199), Science and Technology Research Program of Shanghai (Grant Number 19DZ2282100). The Research and Development Project for Novel Technology and Products in Zhejiang Provincial Health Commission (2021PY018).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bios.2020.112766.
Appendix B. Supplementary data
The following is the Supplementary data to this article:
References
- Bai Y., Yao L., Wei T., Tian F., Jin D.Y., Chen L., Wang M. Jama. 2020;323(14):1406–1407. doi: 10.1001/jama.2020.2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broughton J.P., Deng X., Yu G., Fasching C.L., Servellita V., Singh J., Miao X., Streithorst J.A., Granados A., Sotomayor-Gonzalez A., Zorn K., Gopez A., Hsu E., Gu W., Miller S., Pan C.Y., Guevara H., Wadford D.A., Chen J.S., Chiu C.Y. Nat. Biotechnol. 2020;38(7):870–874. doi: 10.1038/s41587-020-0513-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J.S., Ma E., Harrington L.B., Da Costa M., Tian X., Palefsky J.M., Doudna J.A. Science. 2018;360(6387):436–439. doi: 10.1126/science.aar6245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu D.K.W., Pan Y., Cheng S.M.S., Hui K.P.Y., Krishnan P., Liu Y., Ng D.Y.M., Wan C.K.C., Yang P., Wang Q., Peiris M., Poon L.L.M. Clin. Chem. 2020;66(4):549–555. doi: 10.1093/clinchem/hvaa029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K., Bleicker T., Brünink S., Schneider J., Schmidt M.L., Mulders D.G., Haagmans B.L., van der Veer B., van den Brink S., Wijsman L., Goderski G., Romette J.L., Ellis J., Zambon M., Peiris M., Goossens H., Reusken C., Koopmans M.P., Drosten C. Euro Surveill. 2020;25(3) [Google Scholar]
- Ding, X., Yin, K., Li, Z., Liu, C., 2020. bioRxiv http://doi.org/10.1101/2020.03.19.998724.
- Gootenberg J.S., Abudayyeh O.O., Lee J.W., Essletzbichler P., Dy A.J., Joung J., Verdine V., Donghia N., Daringer N.M., Freije C.A., Myhrvold C., Bhattacharyya R.P., Livny J., Regev A., Koonin E.V., Hung D.T., Sabeti P.C., Collins J.J., Zhang F. Science. 2017;356(6336):438–442. doi: 10.1126/science.aam9321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L., Sun X., Wang X., Liang C., Jiang H., Gao Q., Dai M., Qu B., Fang S., Mao Y., Chen Y., Feng G., Gu Q., Wang R.R., Zhou Q., Li W. Cell discovery. 2020;6:34. doi: 10.1038/s41421-020-0174-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrington L.B., Burstein D., Chen J.S., Paez-Espino D., Ma E., Witte I.P., Cofsky J.C., Kyrpides N.C., Banfield J.F., Doudna J.A. Science. 2018;362(6416):839–842. doi: 10.1126/science.aav4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W.E., Lim B., Hsu C.C., Xiong D., Wu W., Yu Y., Jia H., Wang Y., Zeng Y., Ji M., Chang H., Zhang X., Wang H., Cui Z. Microbial biotechnology. 2020;13(4):950–961. doi: 10.1111/1751-7915.13586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joung, J., Ladha, A., Saito, M., Segel, M., Bruneau, R., Huang, M.W., Kim, N.G., Yu, X., Li, J., Walker, B.D., Greninger, A.L., Jerome, K.R., Gootenberg, J.S., Abudayyeh, O.O., Zhang, F., 2020. medRxiv http://doi.org/10.1101/2020.05.04.20091231.
- Kellner M.J., Koob J.G., Gootenberg J.S., Abudayyeh O.O., Zhang F. Nat. Protoc. 2019;14(10):2986–3012. doi: 10.1038/s41596-019-0210-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau Y.L., Ismail I., Mustapa N.I., Lai M.Y., Soh T.S.T., Hassan A., Peariasamy K., Lee Y.L., Chong Y.M., Sam I.C., Goh P.P. PeerJ. 2020;8:9278. doi: 10.7717/peerj.9278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Li S., Wu N., Wu J., Wang G., Zhao G., Wang J. ACS Synth. Biol. 2019;8(10):2228–2237. doi: 10.1021/acssynbio.9b00209. [DOI] [PubMed] [Google Scholar]
- Li S.Y., Cheng Q.X., Wang J.M., Li X.Y., Zhang Z.L., Gao S., Cao R.B., Zhao G.P., Wang J. Cell discovery. 2018;4:20. doi: 10.1038/s41421-018-0028-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R., Wu X., Wan Z., Li Y., Jin X., Zhang C. Int. J. Mol. Sci. 2020;21:2826. doi: 10.3390/ijms21082826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R., Zhao X., Li J., Niu P., Yang B., Wu H., Wang W., Song H., Huang B., Zhu N., Bi Y., Ma X., Zhan F., Wang L., Hu T., Zhou H., Hu Z., Zhou W., Zhao L., Chen J., Meng Y., Wang J., Lin Y., Yuan J., Xie Z., Ma J., Liu W.J., Wang D., Xu W., Holmes E.C., Gao G.F., Wu G., Chen W., Shi W., Tan W. Lancet. 2020;395(10224):565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notomi T., Okayama H., Masubuchi H., Yonekawa T., Watanabe K., Amino N., Hase T. Nucleic Acids Res. 2000;28(12):63. doi: 10.1093/nar/28.12.e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park G.S., Ku K., Baek S.H., Kim S.J., Kim S.I., Kim B.T., Maeng J.S. J. Mol. Diagn. 2020;22:6. doi: 10.1016/j.jmoldx.2020.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian C., Wang R., Wu H., Zhang F., Wu J., Wang L. Anal. Chem. 2019;91(17):11362–11366. doi: 10.1021/acs.analchem.9b02554. [DOI] [PubMed] [Google Scholar]
- Rothe C., Schunk M., Sothmann P., Bretzel G., Froeschl G., Wallrauch C., Zimmer T., Thiel V., Janke C., Guggemos W., Seilmaier M., Drosten C., Vollmar P., Zwirglmaier K., Zange S., Wölfel R., Hoelscher M. N. Engl. J. Med. 2020;382(10):970–971. doi: 10.1056/NEJMc2001468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian B., Minero G.A.S., Fock J., Dufva M., Hansen M.F. Nucleic Acids Res. 2020;48(5):30. doi: 10.1093/nar/gkaa017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B., Wang R., Wang D., Wu J., Li J., Wang J., Liu H., Wang Y. Anal. Chem. 2019;91(19):12156–12161. doi: 10.1021/acs.analchem.9b01526. [DOI] [PubMed] [Google Scholar]
- Wang R., Chen R., Qian C., Pang Y., Wu J., Li F. Sensor. Actuator. B Chem. 2020;326 [Google Scholar]
- Wang X., Zhong M., Liu Y., Ma P., Dang L., Meng Q., Wan W., Ma X., Liu J., Yang G., Yang Z., Huang X., Liu M. Sci. Bull. 2020;65(17):1436–1439. doi: 10.1016/j.scib.2020.04.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, W., Dang, X., Wang, Q., Xu, M., Zhao, Q., Zhou, Y., Zhao, H., Wang, L., Xu, Y., Wang, J., Han, S., Wang, M., Pei, F., Wan, Y., 2020. medRxiv https://doi.org/10.1101/2020.03.02.20030130.
- Zhang M., Liu C., Shi Y., Wu J., Chen H. Talanta. 2020;214 doi: 10.1016/j.talanta.2020.120818. [DOI] [PubMed] [Google Scholar]
- Zhang, Y., Odiwuor, N., Xiong, J., Sun, L., Nyaruaba, R.O., Wei, H., Tanner, N.A., 2020b. medRxiv https://doi.org/10.1101/2020.02.26.20028373.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.