Highlights
-
•
Bacterial adhesion on dental surfaces depends on the bacterial strain used.
-
•
The magnification used for observation with a scanning electron microscope depends on the size of the bacteria to be observed.
-
•
The quality of dental tissues is determinant in the process of bacterial adhesion.
Keywords: Bacterial adhesion, Tooth, Microscopy, Electron, Scanning, Streptococcus mutans, Lactobacillus casei, Amelogenesis imperfecta
Abstract
AIM
To describe the in vitro bacterial adhesion protocol of Streptococcus mutans and Lactobacillus casei on dental surfaces for a qualitative approach by Scanning Electron Microscopy (SEM) observations. A control and Amelogenesis Imperfecta (AI) affected teeth were used to validate the protocol.
METHOD DETAILS
Eight teeth were collected and fixed in 10% formalin during 10 days. Crowns were fragmented into 4 parts and kept in the freshly prepared artificial saliva. For the preparation of bacterial suspensions, bacterial strains (S. mutans and L. casei) were incubated in a freshly prepared culture medium. After two successive cultures at 37 °C and 3 rinces, bacterial suspensions were prepared in artificial saliva and adjusted to correspond to 108 CFU ml−1. Bacterial adhesion was carried out by sedimentation. Dental fragments were immersed in bacterial suspensions and rinsed with PBS to remove non adherent bacteria. Adherent bacteria were fixed with glutaraldehyde. Finally, teeth samples were dehydrated, coated, dried and observed using high-resolution SEM (JEOL, JSM-5400).
RESULTS
SEM observations showed adherence of spheric stuctures, identified as S. mutans and bacilic structures identified as L. casei.
CONCLUSION
Adhesion of bacteria could be observed by SEM and depends on the quality of dental mineralized tissues.
Graphical abstract

Specifications table
| Subject area: | Dentistry/ Microscopy/ Microbiology |
| More specific subject area: | Bacterial adhesion |
| Method name: | Bacterial adhesion to dental tissues analysed by scanning electron microscopy |
| Name and reference of original method: | R. Kammoun, T. Zmantar, A. Labidi, I. Abbes, L. Mansour, S. Ghoul, Dental Caries And Hypoplastic Amelogenesis Imperfecta: Clinical, Structural, Biochemical And Molecular Approaches, Microb.Pathog, 135 (2019) 103615 |
| Ressource availability: | https://www.sigmaaldrich.com/catalog/product/sial/g5882?lang=en®ion=TN&gclid=CjwKCAjw5Kv7BRBSEiwAXGDElcUpbBJK8XElroB2FxDc_Da1_QNSpkzSP9qg2GT6HBZhGySs4xgY2BoCPPoQAvD_BwE. |
Introduction
Bacterial adhesion is a situation where bacteria adhere firmly to a surface by complete physicochemical interactions between them [1]. Several steps have been described during the bacterial adhesion as the formation of biofilm and bacterial plaque [2,3]. However, the accurate descriptions of bacterial adhesion were mainly detailed either quantitatively or qualitatively [4].
Quantitatively, the bacteria colonies adhering to a dental surfaces could be evaluated by different approaches such as counting bacteria elaborated manually or using automatic tools [5]. Concerning the qualitative approaches, a number of microscopy techniques have been used for a deeper understanding about bacterial adhesion [6]. Scanning Electron Microscopy (SEM) is a gold standard to observe and to identify the morphology of bacteria and to describe the density of their adhesion to dental surfaces [7].
This technique allows access to a nanometer resolution and characterize the state and shape of bacteria following their adhesion to the surface of teeth [8].
Several studies have described natural and spontaneous bacterial adhesion to dental surfaces [9,10]. In the study proposed by Kammoun et al. (2019) [11], a qualitative in vitro approach describes the bacterial adhesion protocol to dental surfaces.
Tooth surfaces are mineralized dental tissues. Some pathologies such as Amelogenesis Imperfecta (AI) are characterized by an alteration of these mineralized tissues. Among the bacteria which adhere the most to these dental surfaces, Streptococcus mutans and Lactobacillus casei are specially described during the initiation of a carious process. They are also easily identifiable by the SEM technique due to their specific morphology respectively spherical or bacilic [12].
The aim of this study was to describe the in vitro bacterial adhesion protocol of S. mutans and L. casei on dental surfaces for a qualitative approach by Scanning Electron Microscopy observations. A control and AI affected teeth were used to validate the protocol.
Method details
Sample collection
Eight teeth were collected from patients attending the Dental Clinic of Monastir (Tunisia). Four molars free of caries or structural abnormalities, extracted for orthodontic reasons were used as control. Four others molars were collected from Hypoplastic AI patients for prosthetic reasons. All the teeth were fixed in 10% formalin during 10 days.
Dental samples preparation
After cleaning the teeth, only crowns were used in this study. Crowns were sectioned in the mesio-distal and vestibulo-lingual directions using an abrasive disk mounted on a low-speed handpiece. Therefore, each crown was fragmented into 4 parts, obtaining a total of thirty-two dental fragments. Dental fragments were kept in the freshly prepared artificial saliva which composition was (CaCl2, 0.44 g l − 1; NaCl, 6 g l − 1; K2HPO4, 0.35 g l − 1 at pH:7.4).
Bacterial suspensions preparation
Streptococcus mutans and Lactobacillus casei previously isolated from Tunisian children mouth with dental caries [13] and identified by conventional and molecular methods were used in this study. S. mutans were isolated on supplemented agar with 5% sheep blood (Biorad, France). L. casei were isolated on Man Rogosa Sharpe agar (MRS) (Biorad, France).
The bacterial strains were respectively incubated in a freshly prepared culture medium, the Brain Heart Infusion broth (BHI) for S. mutans and the Man Rogosa Sharpe broth (MRS) (Biorad, France) for L. casei. After two successive cultures at 37 °C, cells were rinsed 3 times with a solution of NaCl (8.7 gl−1) by centrifugation for 15 min at 7000 g and 4 °C to remove residues of culture medium. Then, bacterial suspensions were prepared in artificial saliva and adjusted to an optical density ranged between 0.7 and 0.8 at 405 nm corresponding to 108 CFU ml−1.
Bacterial adhesion protocol
Bacterial adhesion was carried out by sedimentation. The steps of this experiment are described as follows (Fig. 1) :
Fig. 1.
Schematic presentation of bacterial adhesion protocol.
A: Dental fragments were put in 24 wells plate
B: Ten milliliters of bacterial suspension in artificial saliva (OD405 = 0.8) were dropped on dental fragments
C: Incubation for 3 h at 37 °C
D: Dental fragments were gently rinsed with PBS three times to remove non adherent bacteria
E: Adherent bacteria were fixed with glutaraldehyde (2.5%) on dental surfaces for 4 h at 4 °C and dental fragments were washed 3 times with PBS
F: The adherent bacteria were dehydrated in gradient ethanol concentration on (70%, 80%, 90% and 100%)
G: Dental samples were conserved in 100% ethanol at obscurity before SEM observation at 4 °C
H: The dental samples were mounted onto an aluminum stub with carbon tape, sputter coated with gold before examination in SEM.
Immersion of dental fragments in bacterial suspensions
For this step, a 24 wells plate was used. In each well, dental fragments were oriented to expose the enamel surface to the outside (Fig. 1-A). Using a pipette, 10 ml of the bacterial suspensions (OD 405 = 0.8) were taken and added on the side of the well (Fig. 1-B). Afterwards, bacterial suspension were homogenized by gentle agitation of the plate. Using aluminum paper, the plate was covered. Finally, an incubation for 3 h at 37 °C at obscurity was performed for bacterial adhesion (Fig. 1-C).
Rinsing with PBS to remove non adherent bacteria
Bacterial suspensions were removed with a pipette. After that, 10 ml of PBS was slowly pour on the side and the plate was gently shaked. Few seconds later, the PBS was aspired. These steps were repeated 3 times (Fig. 1-D).
Fixation with glutaraldehyde
Fixation of the adherent bacteria requires the use of diluted glutaraldehyde (2.5%). One ml of 25% glutaraldehyde solution (Sigma-Aldrich) was mixed with 9 ml of PBS. The mixture was pour into the plate. This volume was devoted to cover the dental fragments with the adherent bacteria. Then, plates were covered with aluminum paper and placed into the refrigerator for 4 h to fix bacteria that adhered to dental surfaces (Fig. 1-E). In a second time, three rinsings with PBS for 5 min were carried out at the same conditions already described. Finally, samples were conserved in the PBS until the next step.
Observation under scanning electron microscopy
Teeth samples were dehydrated in ethanol solutions of increasing gradients (50%, 70%, 80%, 90% and 100%). Each bath required 5 min (Fig. 1-F). Samples were conserved in ethanol 100% at 4 °C untel be used (Fig. 1-G). These samples were sputter-coated with gold using a JEOL (JFC-1100E) coater (Fig. 1-H) and then dried under vaccum and observed using high-resolution SEM (JEOL, JSM-5400) at 5000X and 10000X magnifications.
Results and discussion
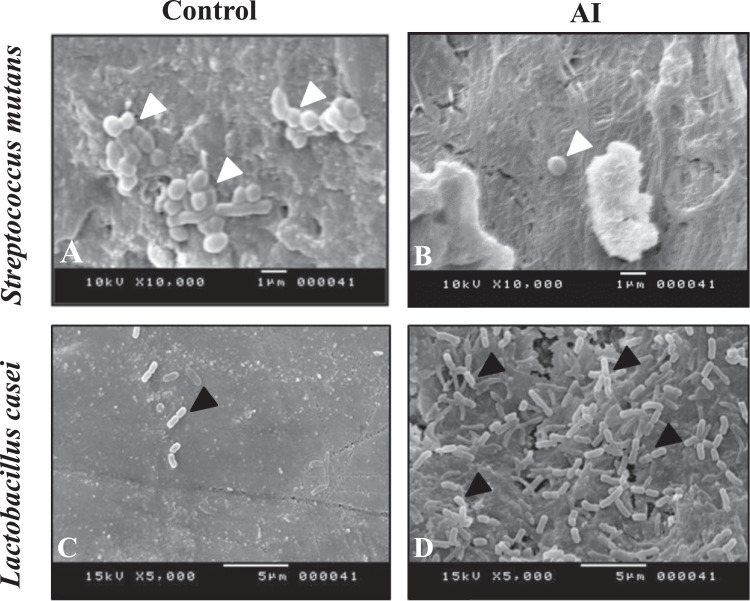
SEM observations showed adherence of spheric stuctures, identified as Streptococcus mutans on the enamel surface of the control teeth (Fig. 2-A). Few S. mutans were comparatively noticed on the studied group of AI teeth (Fig. 2-B). Concerning Lactobacillus casei, few bacilic structures were noticed on the enamel surface of control teeth (Fig. 2-C) compared to that of AI teeth where several adherent L. casei were observed (Fig. 2-D).
Fig. 2.
SEM observations of bacterial adhesion on dental hard tissues.
A: Adhesion of Streptococcus mutans (white arrows) on control dental fragments
B: Adhesion of Streptococcus mutans (white arrow) on AI dental fragments
C: Adhesion of Lactobacillus casei (black arrows) on control dental surfaces
D: Adhesion of Lactobacillus casei (black arrows) on dental surfaces with AI.
Amelogenesis Imperfecta (AI) as a genetic disorder has been shown to affect all the mineralized tissues in the teeth [14,15]. The quality of the mineralized tissues seems to determine the adhesion of the bacteria.
Declaration of Competing Interest
Authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Co-Submission: DOI: 10.1016/j.micpath.2019.103615.
References
- 1.Carniello V., Peterson B.W., van der Mei H.C., Busscher H.J. Physico-chemistry from initial bacterial adhesion to surface-programmed biofilm growth. Adv. Colloid. Interface Sci. 2018;261:1–14. doi: 10.1016/j.cis.2018.10.005. [DOI] [PubMed] [Google Scholar]
- 2.Wessel S.W., Chen Y., Maitra A., van den Heuvel E.R., Slomp A.M., Busscher H.J., van, der Mei H.C. Adhesion forces and composition of planktonic and adhering oral microbiomes. J. Dent. Res. 2014;93:84–88. doi: 10.1177/0022034513511822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Donlan R.M. Bacterial adhesion and biofilms on surfaces. Emerg. Infect. Dis. 2002;8:881–890. doi: 10.3201/eid0809.020063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Katsikogianni M., Missirlis Y.F. Concise review of mechanisms of bacterial adhesion to biomaterials and of techniques used in estimating bacteria-material interactions. Eur. Cell. Mater. 2004;8:37–57. doi: 10.22203/ecm.v008a05. [DOI] [PubMed] [Google Scholar]
- 5.Lee Y.K., Lim C.Y., Teng W.L., Ouwehand A.C., Tuomola E.M., Salminen S. Quantitative approach in the study of adhesion of lactic acid bacteria to intestinal cells and their competition with enterobacteria. Appl. Environ. Microbiol. 2000;66:3692–3697. doi: 10.1128/aem.66.9.3692-3697.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Scarano A., Piattelli M., Caputi S., Favero G.A., Piattelli A. Bacterial adhesion on commercially pure titanium and zirconium oxide disks: an in vivo human study. J. Periodontol. 2004;75:292–296. doi: 10.1902/jop.2004.75.2.292. [DOI] [PubMed] [Google Scholar]
- 7.Kodjikian L., Burillon C., Roques C., Pellon G., Freney J., Renaud F.N. Bacterial adherence of Staphylococcus epidermidis to intraocular lenses: a bioluminescence and scanning electron microscopy study. Invest. Ophthalmol. Vis. Sci. 2003;44:4388–4394. doi: 10.1167/iovs.03-0186. [DOI] [PubMed] [Google Scholar]
- 8.Grin I., Schwarz H., Linke D. Electron microscopy techniques to study bacterial adhesion. Adv. Exp. Med. Biol. 2011;715:257–269. doi: 10.1007/978-94-007-0940-9_16. [DOI] [PubMed] [Google Scholar]
- 9.Stenudd C., Nordlund A., Ryberg M., Johansson I., Källestål C., Strömberg N. The association of bacterial adhesion with dental caries. J. Dent. Res. 2001;80:2005–2010. doi: 10.1177/00220345010800111101. [DOI] [PubMed] [Google Scholar]
- 10.Nam S.H., Ok S.M., Kim G.C. Tooth bleaching with low-temperature plasma lowers surface roughness and Streptococcus mutans adhesion. Int. Endod. 2018;51:479–488. doi: 10.1111/iej.12860. [DOI] [PubMed] [Google Scholar]
- 11.Kammoun R., Zmantar T., Labidi A., Abbes I., Mansour L., Ghoul-Mazgar S. Dental caries and hypoplastic amelogenesis imperfecta: clinical, structural, biochemical and molecular approaches. Microb. Pathog. 2019;135 doi: 10.1016/j.micpath.2019.103615. [DOI] [PubMed] [Google Scholar]
- 12.Amend S., Frankenberger R., Lücker S., Domann E., Krämer N. Secondary caries formation with a two-species biofilm artificial mouth. Dent. Mater. 2018;34:786–796. doi: 10.1016/j.dental.2018.02.002. [DOI] [PubMed] [Google Scholar]
- 13.Kouidhi B., Zmantar T., Jrah H., Souiden Y., Chaieb K., Mahdouani K., Bakhrouf A. Antibacterial and resistance-modifying activities of thymoquinone against oral pathogens. Ann. Clin. Microbiol. Aimicrob. 2011;10:29. doi: 10.1186/1476-0711-10-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Crawford P.J., Aldred M., Bloch-Zupan A. Amelogenesis imperfecta. Orphanet. J. Rare. Dis. 2007;4:17. doi: 10.1186/1750-1172-2-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kammoun R., Behets C., Mansour L., Ghoul-Mazgar S. Mineral features of connective dental hard tissues in hypoplastic amelogenesis imperfecta. Oral. Dis. 2018;24:384–392. doi: 10.1111/odi.12724. [DOI] [PubMed] [Google Scholar]