Abstract
Background
Although mutations in the promoter region of the telomerase reverse transcriptase (TERTp) gene are the most common alterations in glioblastoma (GBM), their clinical significance remains unclear. Therefore, we investigated the impact of TERTp status on patient outcome and clinicopathological features in patients with GBM over a long period of follow-up.
Methods
We retrospectively analyzed 153 cases of GBM. Six patients with isocitrate dehydrogenase 1 (IDH1) or H3F3A gene mutations were excluded from this study. Among the 147 cases of IDH wild-type GBM, 92 (62.6%) had the TERTp mutation. Clinical, immunohistochemical, and genetic factors (BRAF, TP53 gene mutation, CD133, ATRX expression, O6-methylguanine-DNA methyltransferase [MGMT] promoter methylation) and copy number alterations (CNAs) were investigated.
Results
GBM patients with the TERTp mutation were older at first diagnosis versus those with TERTp wild type (66.0 vs. 60.0 years, respectively, P = .034), and had shorter progression-free survival (7 vs. 10 months, respectively, P = .015) and overall survival (16 vs. 24 months, respectively, P = .017). Notably, magnetic resonance imaging performed showed that TERTp-mutant GBM was strongly associated with multifocal/distant lesions (P = .004). According to the CNA analysis, TERTp mutations were positively correlated with EGFR amp/gain, CDKN2A deletion, and PTEN deletion; however, these mutations were negatively correlated with PDGFR amp/gain, CDK4 gain, and TP53 deletion.
Conclusions
TERTp mutations were strongly correlated with multifocal/distant lesions and poor prognosis in patients with IDH wild-type GBM. Less aggressive GBM with TERTp wild type may be a distinct clinical and molecular subtype of IDH wild-type GBM.
Keywords: distant, glioblastoma, IDH wild type, multifocal, TERT promoter
Key Points.
TERTp mutations strongly correlated with multifocal/distant lesions and poor prognosis in patients with IDH wild-type GBM.
The IDH wild-type GBM with and without TERTp mutations may be a distinct clinical and molecular subtype.
Importance of the Study.
Mutations in the promoter region of the telomerase reverse transcriptase (TERTp) gene are the most common mutations in isocitrate dehydrogenase (IDH) wild-type glioblastoma (GBM). While TERTp mutations are correlated with poor prognosis, aggressive clinicopathological characteristics, and metastasis in other cancers, their clinical significance in GBM remains unclear. Here, we analyzed GBMs to determine whether the TERTp status is associated with other clinical and molecular factors. Particularly, this study focused on whether multifocal/distant lesions were observed during the clinical course. In this study, we demonstrated that TERTp-mutant GBMs are strongly associated with the prognosis and multifocal/distant lesions during a long follow-up period. In addition, TERTp mutation was positively correlated with EGFR amp/gain, CDKN2A deletion, and PTEN deletion; however, it negatively correlated with PDGFR amp/gain, CDK4 gain, and TP53 deletion. Less aggressive GBM with TERTp wild type could be distinct clinical and molecular subtype of IDH wild-type GBM.
Glioblastoma (GBM) is the most common primary malignant tumor affecting the central nervous system in adults.1 Despite of radical surgery combined with concomitant chemoradiation therapy based on temozolomide, the median survival of patients is approximately 18 months.2
According to the World Health Organization revised neuropathological criteria, these tumors are divided into 2 categories, namely isocitrate dehydrogenase (IDH) wild-type and IDH-mutant GBMs. In addition, recent reports indicated that 70%–80% of GBM genomes harbor either C228T or C250T mutations in the promoter region of the telomerase reverse transcriptase (TERTp) gene.3,4 These mutations are associated with enhanced telomere maintenance.5–7 Although several studies reported the prognostic significance of TERTp mutation in patients with GBM, its clinical and pathological roles remain unclear.3–6
Recently, GBM patients with unmethylated O6-methylguanine-DNA methyltransferase (MGMT) and TERTp mutation have a worse prognosis than those with TERTp wild type.3,8 However, the mechanism of interaction of TERTp mutation and MGMT promoter methylation is not well established.
Regarding imaging analysis, necrosis detected through magnetic resonance imaging (MRI) has been reported to indicate the presence of TERTp mutation.9 However, predicting the TERTp status by preoperative imaging study alone remains difficult.
A recent systematic review and meta-analyses stated that the incidence of solitary GBM is 83%.10 Other previous studies showed that 20% of patients with GBM had multiple lesions and their prognosis was worse than that recorded in patients with a single lesion.11
In this study, we analyzed GBMs to determine whether the TERTp status was associated with other clinical and molecular factors. Particularly, this study utilized MRI to determine the development of multifocal/distant lesions during the clinical course.
Materials and Methods
Patients and Samples
This retrospective study was conducted with the approval of the Ethics Committees of the Tohoku University School of Medicine and Yamagata University School of Medicine. Written informed consent was provided by all patients prior to their participation in the study.
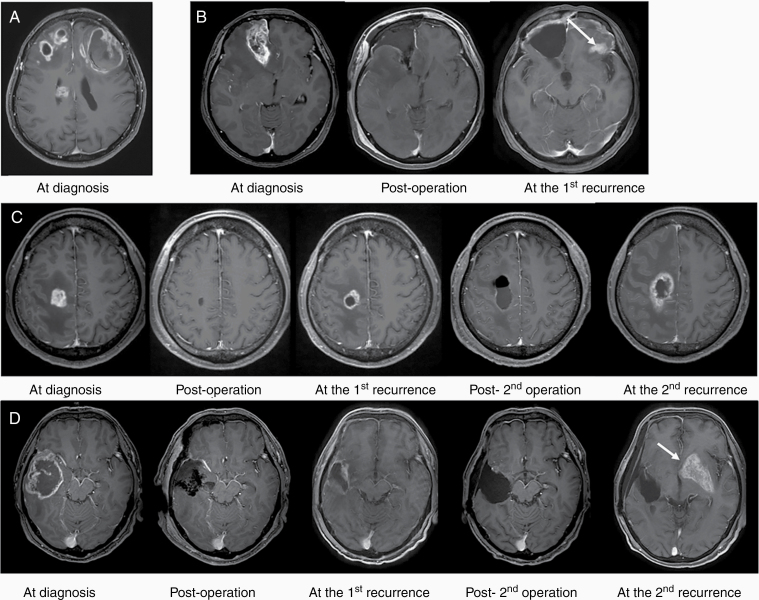
Between January 2009 and October 2019, a total of 153 patients (89 treated at Yamagata University Hospital [Yamagata cohort] and 64 treated at Tohoku University Hospital [Tohoku cohort]) were analyzed. All patients met the following inclusion criteria: (1) diagnosis of GBM, World Health Organization grade IV; (2) no history of lower-grade tumors; (3) availability of genomic DNA; and (4) availability of information regarding events, such as recurrence or death during the follow-up period, or absence of such events for ≥12 months of follow-up. Patients who had previously undergone biopsies were excluded from the study. Tumor specimens were obtained from a lesion that exhibited enhancement on gadolinium-enhanced MRI and immediately stored at −80°C until DNA extraction (Figure 1).
Figure 1.
Definition of multifocal lesion. Representative gadolinium-enhanced MRI scans of patients treated at Yamagata University Hospital. The scans were obtained at diagnosis and after surgery, and at the first and second recurrence. (A) Multifocal lesions at diagnosis. (B) Multifocal/distant lesion at first recurrence. Eight months after surgery, an enhanced lesion was observed at a location distant from the initial lesion (arrow). (C) Local recurrence during the entire follow-up period. (D) Multifocal/distant lesion at second recurrence. Seven months after surgery, local recurrence was observed adjacent to the resection cavity. Ten months later, an enhanced lesion was detected at a distant location (arrow).
Classification of GBM According to Preoperative MRI
MRI sequences were acquired on a 1.5-T or 3.0-T scanner and typically included axial T1-weighted, T2-weighted fast spin-echo, and fluid-attenuated inversion-recovery sequences as well as a postcontrast 3-dimensional spoiled gradient-recalled acquisition in the steady state T1-weighted sequence. Contrast-enhanced lesions (CELs) were assessed to clarify whether they were in contact with the subventricular zone, as previously described.12
Definition of Multifocal/Distant Lesions
One or more enhancing noncontiguous lesions >1 cm distant from the original tumor on preoperative MRI were defined as multifocal/distant lesions at diagnosis.13
In addition, as previously reported, “multifocal/distant lesions at recurrence” were defined as distant or multifocal recurrence. Recurrence was characterized by the development of new CEL centered >3 cm distant from the primary resection cavity or at the margins of the primary residual tumor, or at more than 1 site, with each lesion having a well-defined border and the patient exhibiting normal brain signals.14,15
Clinical Parameters
The clinical profiles of patients were obtained from their medical records. The majority of patients underwent radical surgery followed by chemotherapy (nimustine hydrochloride [ACNU] or temozolomide) and radiotherapy. Total surgical resection was defined as the disappearance of CEL according to pre- and postoperative gadolinium-enhanced MRI studies. In cases in which the primary tumor recurred, patients underwent salvage surgery, second-line chemotherapy, radiotherapy, or palliative therapy. The Ki-67 labeling index was determined by immunohistochemical staining of resected specimens with the Ki-67 antigen (Dako, Agilent Technologies). We also analyzed the expression of CD133 (Miltenyi Biotec), p53 (Dako, Agilent Technologies), and ATRX (Abcam) by immunohistochemical staining. The expression of CD133 in 144 patients among the Yamagata and Tohoku cohorts was previously reported.16,17
Prognosis
Progression-free survival (PFS) was defined as the interval between the day of first surgery and the day of recurrence detection on MRI scans. Overall survival (OS) was defined as the time between the day of the first operation and the day of death or final follow-up.
Molecular Analysis
Genomic DNA was extracted with the QIAamp DNA mini kit (Qiagen), according to the instructions provided by the manufacturer. The isocitrate dehydrogenase1/2 (IDH1/2), H3F3A, HIST1H3B, TP53, BRAF, and TERTp genes were amplified via polymerase chain reaction (PCR), and sequencing was conducted as previously described.18,19 In the MGMT promoter methylation analysis, we performed methylation-specific PCR or quantitative methylation-specific PCR following the bisulfite modification of tumor DNA.19 To assess copy number alterations (CNAs), we performed Multiplex Ligation-dependent Probe Amplification (MLPA) using the SALSA MLPA KIT P105 (version D2), in accordance with the manufacturer’s protocol (MRC Holland).20 The P105 kit is designed to detect CNAs typically found in gliomas and includes probes against the PDGFRA, EGFR, CDKN2A, PTEN, TP53, CDK4, MDM2, and NFKBIA genes. Based on the previous publications, the CNA categories were classified according to the following thresholds: homozygous deletion (x ≤ 0.4), hemizygous deletion (0.4 < x ≤ 0.7), gain (1.3 ≤ x < 2.0), and amplification (x ≥ 2.0).20,21 We used OncoPrinter, a tool provided by the cBioPortal for Cancer Genomics (cbioportal.org/oncoprinter), to visualize and analyze our data with some modifications.22,23
Statistical Analysis
Statistical analyses were performed using the SPSS (IBM Japan) software. The relationship between 2 variables was evaluated using the Mann–Whitney U test and Fisher’s exact test. Estimates of PFS and OS were calculated with the Kaplan–Meier method, and the Log-rank (Mantel–Cox) test was used to evaluate differences between the groups. Cox regression was used for the multivariate analysis. The significance level was set at P < .05.
Results
Population and Tumor Characteristics on MRI
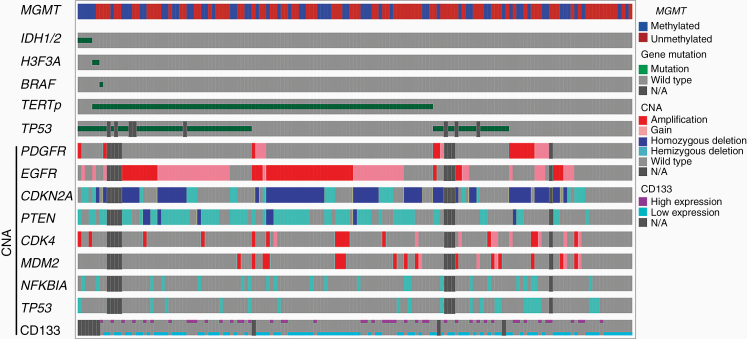
A total of 153 patients, including 82 males and 71 females with a median age of 63 years (range: 27–86 years) and median preoperative Karnofsky Performance Status of 80 (range: 30–100), were included in the present study. Patients in the Yamagata cohort were older than those in the Tohoku cohort (P < .001) (Supplementary Table 1). Genomic DNA and paraffin-embedded samples were obtained from all patients. The median duration of the follow-up period was 17 months (range: 1–152 months), and 119 patients (77.8%) expired. Total surgical resection was achieved in 96 patients (62.7%). In this group, IDH1, H3F3A, and BRAF gene mutations were detected in 4 (2.6%), 2 (1.3%), and 1 patient (0.65%), respectively; however, neither IDH2 nor HIST1H3B gene mutations were detected. TERTp gene mutations were detected in 92 patients (60.1%), including 65 (42.5%) and 27 (17.6%) with C228T and C250T mutations, respectively. Although the frequency of TERTp gene mutations in the Yamagata cohort was higher than that in the Tohoku cohort (P = .019, Supplementary Table 1), there was no significant difference in the mutation frequency in older patients (age ≥60) between the 2 cohorts (P = .348) (data not shown). MGMT gene promoter methylation was found in 62 patients (40.5%). Postoperative treatments consisted of radiation alone for 6 patients, while the remaining 147 patients received combined radiation and chemotherapy with temozolomide (n = 123), ACNU (n = 14), or other agents (n = 10). Bevacizumab was administered as first- and second-line therapy in 1 and 50 patients, respectively. There were no significant differences observed in PFS and OS between patients treated with ACNU and temozolomide (data not shown). TP53 gene mutations and/or strong immunoreactivity of p53 were found in 63 patients (43.4%) (Figure 2). Eleven of 88 patients (12.5%) displayed the loss of ATRX expression. The major CNAs frequently observed in 139 GBMs included EGFR amp/gain (66.2%), CDKN2A deletion (60.4%), and PTEN deletion (51.8%) (Figure 2 and Table 1).
Figure 2.
Genetic distribution in 153 GBMs. Mutations, CNAs, and methylation were generated and visualized by OncoPrinter via the cBioPortal for Cancer Genomics (cbioportal.org/oncoprinter) with some modifications.22,23 The diagram shows the landscape of the molecular characteristics of GBMs, which are sorted by IDH, H3F3A, and TERTp mutations. N/A, not available.
Table 1.
Relationships Between TERTp Status and Other Prognostic Factors
| Total (n = 147) | TERTp wild type (n = 55) | TERTp mutant (n = 92) | P | |||
|---|---|---|---|---|---|---|
| Sex, female, n (%) | 66 (44.8) | 27 (49.1) | 39 (42.4) | .494b | ||
| Age, y, median (range) | 64 (27–86) | 60 (27–82) | 66 (32–86) | .034 a | ||
| Preoperative KPS ≥80, n (%) | 84 (60.0) | 27 (51.9) | 57 (64.8) | .155b | ||
| Gross total resection, n (%) | 93 (63.2) | 33 (60.0) | 60 (65.2) | .597b | ||
| CD133 expression, mean (%) | 12.7 ± 12.9 | 12.1 ± 11.1 | 12.9 ± 13.8 | .729a | ||
| Ki-67 labeling index, mean (%) | 33.8 ± 17.9 | 34.8 ± 17.9 | 33.2 ± 18.0 | .477a | ||
| Multifocal/distant lesions | ||||||
| At diagnosis, n (%) | 21 (14.3) | 4 (7.3) | 17 (18.5) | .087b | ||
| At recurrence, n (%) | 45 (30.6) | 12 (21.8) | 33 (35.9) | .096b | ||
| At the first recurrence, n (%) | 30 (20.4) | 8 (14.5) | 22 (23.9) | .208b | ||
| At the second recurrence, n (%) | 15 (10.2) | 4 (7.3) | 11 (12.0) | .415b | ||
| Total, n (%) | 66 (44.9) | 16 (29.1) | 50 (54.3) | .004 b | ||
| MGMT gene promoter methylation, n (%) | 57 (38.8) | 19 (34.5) | 38 (41.3) | .485b | ||
| TP53 gene mutation, n (%) | 57 (40.7) | 19 (37.3) | 38 (42.7) | .477b | ||
| Loss of ATRX expression, n (%) | 11 (12.5) | 6 (22.2) | 5 (8.2) | .085b | ||
| CNA | PDGFR | Amp, n (%) | 11 (7.9) | 9 (17.6) | 2 (2.3) | .002 b |
| Gain, n (%) | 8 (5.8) | 5 (9.8) | 3 (3.4) | .143b | ||
| Amp/gain, n (%) | 19 (13.7) | 14 (27.5) | 5 (5.7) | .001 b | ||
| EGFR | Amp, n (%) | 46 (33.1) | 8 (15.7) | 38 (43.2) | .001 b | |
| Gain, n (%) | 46 (33.1) | 7 (13.7) | 39 (44.3) | <.0001 b | ||
| Amp/gain, n (%) | 92 (66.2) | 15 (29.4) | 77 (87.5) | <.0001 b | ||
| CDKN2A | Homo, n (%) | 61 (43.9) | 20 (39.2) | 41 (46.6) | .479b | |
| Hemi, n (%) | 23 (16.5) | 5 (9.8) | 18 (20.5) | .154b | ||
| Deletion, n (%) | 84 (60.4) | 25 (49.0) | 59 (67.0) | .048 b | ||
| PTEN | Homo, n (%) | 8 (5.8) | 1 (2.0) | 7 (8.0) | .258b | |
| Hemi, n (%) | 64 (46.0) | 8 (15.7) | 56 (63.6) | <.0001 b | ||
| Deletion, n (%) | 72 (51.8) | 9 (17.6) | 63 (71.6) | <.0001 b | ||
| CDK4 | Amp, n (%) | 13 (9.4) | 4 (7.8) | 9 (10.2) | .768b | |
| Gain, n (%) | 9 (6.5) | 7 (13.7) | 2 (2.3) | .012 b | ||
| Amp/gain, n (%) | 22 (15.8) | 11 (21.6) | 11 (12.5) | .227b | ||
| MDM2 | Amp, n (%) | 16 (11.5) | 6 (11.8) | 10 (11.4) | 1.000b | |
| Gain, n (%) | 5 (3.6) | 4 (7.8) | 1 (1.1) | .061b | ||
| Amp/gain, n (%) | 21 (15.1) | 10 (19.6) | 11 (12.5) | .327b | ||
| NFKBIA | Hemi, n (%) | 20 (14.4) | 8 (15.7) | 12 (13.6) | .804b | |
| TP53 | Hemi, n (%) | 23 (16.5) | 16 (31.4) | 7 (8.1) | .001 b | |
| Mut/Hemi, n (%) | 12 (8.6) | 7 (13.7) | 5 (5.7) | .124b | ||
| SVZ-positive, n (%) | 66 (44.9) | 25 (45.5) | 41 (44.6) | 1.000b |
Amp, amplification; Hemi, hemizygous deletion; Homo, homozygous deletion; KPS, Karnofsky Performance Status; Mut, mutation; SVZ, subventricular zone. P values <0.05 are in bold.
aMann–Whitney test.
bFisher’s exact test.
Correlation Analyses Between the TERTp Status and Other Prognostic Factors
Six patients with IDH1 or H3F3A mutations were excluded from this study. Therefore, we analyzed 147 GBM patients with IDH wild type to determine the factors correlated with the TERTp mutation. The median age was higher in GBM patients with TERTp mutation than those with TERTp wild type (P = .034) (Table 1).
In terms of MRI characteristics, 21 of the 147 patients (14.3%) had multifocal/distant lesions at diagnosis (Table 1). During the follow-up, 129 patients (87.7%) experienced the first recurrence, which included local recurrence and multifocal/distant recurrence in 99 (67.3%) and 30 (20.4%) patients, respectively. Among the patients with a well-controlled first recurrent lesion, 15 patients (10.2%) had new multifocal/distant lesions at second recurrence. Neither local nor distal recurrence was observed at the time of the last observation in the remaining 18 patients (12.3%).
Although multifocal/distant lesions at diagnosis or recurrence were weakly correlated with TERTp mutations (P = .087 and P = .096, respectively), these lesions were significantly more common in patients with TERTp-mutant GBM than in patients with TERTp wild-type GBM during the entire follow-up period (P = .004, Table 1).
The loss of ATRX expression occurred more frequently in TERTp wild-type GBM; however, this difference was not significant (P = .085, Table 1). EGFR amp/gain, CDKN2A deletion, and PTEN deletion were significantly associated with TERTp mutations (P < .0001, P = .048, and P < .0001, respectively, Figure 2 and Table 1). Conversely, PDGFR amp/gain, CDK4 gain, and TP53 hemizygous deletion were more frequently observed in TERTp wild-type GBM (P = .001, P = .012, and P = .001, respectively, Figure 2 and Table 1).
Univariate Analysis for the Prediction of PFS and OS
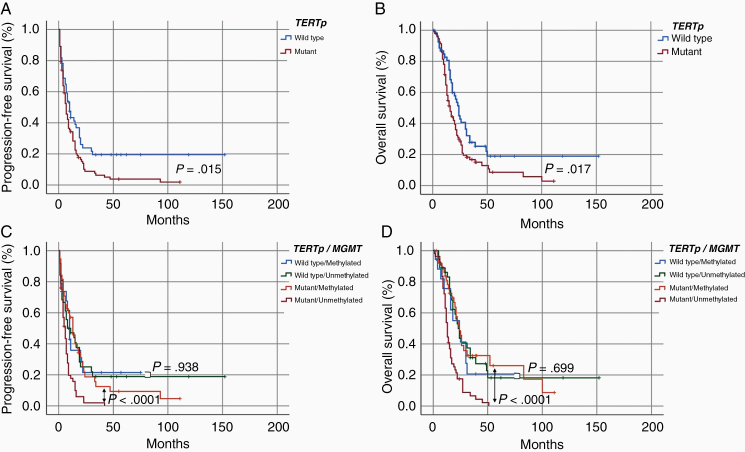
The median PFS and OS for the patients with IDH wild-type GBM were 8 and 18 months, respectively (Table 2). Based on the Kaplan–Meier analysis, longer PFS and OS were correlated with TERTp wild type (P = .015 and P = .017, respectively) (Figure 3A and B; Table 2), gross total resection (P < .001 and P <.001, respectively) (Table 2), MGMT gene promoter methylation (P = .037 and P = .015, respectively) (Table 2), CDK4 amp/gain (P = .015 and P = .042, respectively), and local lesions (P = .006 and P = .001, respectively) (Table 2). The female sex was associated with longer PFS (P = .047) (Table 2).
Table 2.
Clinical and Genetic Parameters Affecting PFS and OS in Primary GBM
| Parameters | No. of patients (n = 147) | PFS | OS | ||
|---|---|---|---|---|---|
| Median (months) | P* | Median (months) | P* | ||
| 147 | 8 | 18 | |||
| TERTp status | |||||
| Mutated | 92 | 7 | 16 | ||
| Wild type | 55 | 10 | .015 | 24 | .017 |
| Sex | |||||
| Female | 66 | 9 | 22 | ||
| Male | 81 | 7 | .047 | 16 | .055 |
| Age at diagnosis | |||||
| <60 years | 52 | 8 | 18 | ||
| >60 years | 95 | 7 | .172 | 19 | .115 |
| Preoperative KPS | |||||
| >80 | 84 | 8 | 21 | ||
| <80 | 56 | 7 | .725 | 15 | .294 |
| Surgery | |||||
| Gross toral resection | 93 | 11 | 23 | ||
| Absence of gross total resection | 54 | 4 | <.001 | 11 | <.001 |
| Ki-67 labeling index | |||||
| Low (<30%) | 55 | 8 | 20 | ||
| High (>30%) | 68 | 7 | .212 | 16 | .061 |
| CD133 expression | |||||
| Low (<15%) | 97 | 8 | 21 | ||
| High (>15%) | 47 | 7 | .480 | 17 | .146 |
| MGMT | |||||
| Methylated | 57 | 13 | 24 | ||
| Unmethylated | 90 | 7 | .037 | 16 | .015 |
| PDGFR | |||||
| Amp/gain | 19 | 10 | 17 | ||
| Retain | 120 | 8 | .916 | 20 | .669 |
| EGFR | |||||
| Amp/gain | 92 | 7 | 17 | ||
| Retain | 47 | 10 | .060 | 24 | .142 |
| CDKN2A | |||||
| Deletion | 84 | 9 | 17 | ||
| Retain | 55 | 8 | .522 | 21 | .350 |
| PTEN | |||||
| Deletion | 72 | 9 | 19 | ||
| Retain | 67 | 8 | .281 | 19 | .497 |
| CDK4 | |||||
| Amp/gain | 22 | 19 | 34 | ||
| Retain | 117 | 7 | .015 | 18 | .042 |
| MDM2 | |||||
| Amp/gain | 21 | 10 | 24 | ||
| Retain | 118 | 8 | .795 | 18 | .368 |
| NFKBIA | |||||
| Deletion | 20 | 13 | 21 | ||
| Retain | 119 | 8 | .802 | 18 | .662 |
| TP53 | |||||
| Mut/deletion | 68 | 10 | 19 | ||
| Wild type | 76 | 7 | .054 | 19 | .580 |
| SVZ | |||||
| Positive | 66 | 7 | 16 | ||
| Negative | 74 | 8 | .952 | 21 | .267 |
| Numbers of lesion | |||||
| Multifocal/distant lesions | 66 | 7 | 16 | ||
| Local lesion | 81 | 9 | .006 | 23 | .001 |
| Cohort site | |||||
| Yamagata | 88 | 7 | 17 | ||
| Tohoku | 59 | 9 | .137 | 22 | .107 |
SVZ, subventricular zone. P values <0.05 are in bold.
*Log-rank test.
Figure 3.
(A and B) Kaplan–Meier curves based on the TERTp mutation in patients with IDH wild-type GBM. (A) PFS. (B) OS. (C and D) Kaplan–Meier curves based on the combination of TERTp mutation and MGMT promoter methylation in patients with IDH wild-type GBM. (C) PFS. (D) OS.
To determine whether the TERTp mutation was negatively correlated with PFS and OS in the non-multifocal/distant group, we analyzed the survival of the 81 patients in the non-multifocal/distant group. The median PFS and OS were 9 and 23 months, respectively, with no significant correlation of PFS and OS with the TERTp mutation (P = .129 and P = .148, respectively) (data not shown).
We also investigated the prognostic value of TERTp mutation in combination with MGMT promoter methylation. Among patients with TERTp mutation, unmethylated MGMT was significantly associated with poor PFS and OS (P < .0001 and P < .0001, respectively) (Figure 3C and D). However, among patients with TERTp wild type, there was no significant difference of PFS and OS between patients with and without MGMT promotor methylation (P = .938 and P = .699, respectively) (Figure 3C and D).
Factors Associated With Multifocal/Distant Lesions
We investigated several factors to determine whether they correlated with multifocal/distant lesions. As shown in Supplementary Table 2, TERTp mutations, the expression of CD133, and PTEN deletion were significantly associated with multifocal lesions (P = .004, P = .004, and P = .004, respectively).
Multivariate Analysis of Prognostic Factors
The factors included in the multivariate analysis for PFS and OS were TERTp status, sex, age, extent of resection, Ki-67 labeling index, MGMT gene promoter methylation, CDK4 amp/gain, number of lesions, and cohort site. We found that TERTp mutation, absence of gross total resection, and MGMT gene promoter unmethylation were independent unfavorable prognostic factors for PFS (hazard ratio [HR]: 2.0, 95% confidence interval [CI]: 1.2–3.3, P = .006; HR: 2.2, 95% CI: 1.3–3.5, P = .002; and HR: 2.0, 95% CI: 1.3–3.0, P = .002, respectively) (Table 3). TERTp mutations (HR: 2.0, 95% CI: 1.2–3.3, P = .010), absence of total resection (HR: 2.9, 95% CI: 1.7–4.8, P < .001), and MGMT gene promoter unmethylation (HR: 2.2, 95% CI: 1.4–3.5, P = .001) were independent unfavorable prognostic factors for OS.
Table 3.
Multivariate Analysis of Independent Prognostic Factors Associated With PFS and OS
| Parameters | PFS | OS | ||||
|---|---|---|---|---|---|---|
| HR | 95% CI | P* | HR | 95% CI | P | |
| TERTp status | ||||||
| Mutant vs. wild type | 2.0 | 1.2–3.3 | .006 | 2.0 | 1.2–3.3 | .010 |
| Sex | ||||||
| Male vs. female | 1.3 | 0.9–2.0 | .218 | 1.4 | 0.9–2.2 | .157 |
| Age | ||||||
| ≥60 vs. <60 | 1.2 | 0.8–2.0 | .364 | 1.1 | 0.7–1.9 | .600 |
| Gross total resection | ||||||
| No vs. Yes | 2.2 | 1.3–3.5 | .002 | 2.9 | 1.7–4.8 | <.001 |
| Ki-67 labeling index | ||||||
| ≥30 vs. <30 | 1.4 | 0.9–2.1 | .129 | 1.5 | 1.0–2.4 | .069 |
| MGMT | ||||||
| Unmethylated vs. methylated | 2.0 | 1.3–3.0 | .002 | 2.2 | 1.4–3.5 | .001 |
| CDK4 | ||||||
| Amp/gain vs. retain | 1.5 | 0.8–2.8 | .261 | 1.5 | 0.7–2.9 | .284 |
| Number of lesions | ||||||
| Multifocal/distant vs. local | 1.3 | 0.8–2.0 | .327 | 1.3 | 0.8–2.2 | .241 |
| Cohort site | ||||||
| Yamagata vs. Tohoku | 1.1 | 0.7–1.7 | .656 | 1.1 | 0.7–1.8 | .589 |
P values <.05 are in bold.
Discussion
TERTp mutation is the most common alteration in GBM; however, the clinical impact of TERTp mutations in GBM remains unclear. To understand the poor prognosis of GBM with TERTp mutations, we hypothesized that malignant clinical features exist in this group. Long-term follow-up revealed that the cumulative incidence of multiple/distant lesions was significantly higher in GBM with TERTp mutations than in patients with TERTp wild-type GBM. Conversely, the non-multifocal/distant group did not show any differences in PFS and OS based on TERTp status. Therefore, we, for the first time, demonstrated that GBM with TERTp mutations has a poor prognosis because of its clinically aggressive behavior. In accordance with this finding, several studies regarding other cancers demonstrated that these mutations were correlated with a poor prognosis, aggressive clinicopathological characteristics, and metastasis.24–28 Xing et al. found that TERTp mutation strongly correlated with vascular invasion in patients with papillary thyroid cancer.25 Yuan et al. reported that thyroid cancer patients with the TERTp mutation have a 4-fold higher risk of distant metastasis than those with TERTp wild type.27
The frequency of TERTp mutations in our study was 62.6%, which is lower than that of previous reports from North America and European countries, which reported mutation frequencies of 73%–75% in IDH wild-type GBMs.3–5 Other reports from Japan also showed relatively low frequencies of TERTp mutations among IDH wild-type GBM, ranging from 50% to 70%.6,9,29 Thus, racial differences in the frequency of TERTp mutations may exist. One possible explanation for the low frequency of TERTp mutations in the Japanese cohort is that other mechanisms involved with replicative immortality in TERTp wild-type GBM. One such mechanism is TERTp hypermethylation, and the other is ATRX or SMARCAL1 gene mutation. TERTp hypermethylation can aberrantly activate telomerase in cancer,30 and the ATRX or SMARCAL1 gene mutations are strongly associated with the maintenance of telomere length, referred to as alternative lengthening of telomeres.31 Indeed, our results indicated the frequent loss of ATRX expression in TERTp wild-type GBM. The other explanation is potential inclusion of other IDH wild-type high grade gliomas such as anaplastic astrocytoma with piloid features.32 Although our cases were histologically confirmed as GBM, further molecular testing may be required to classify into novel entities.
The prognostic significance of the TERTp mutation remains controversial in patients with GBM.3,33–35 In the present study, univariate and multivariate analyses showed that the TERTp mutation was significantly associated with both PFS and OS. In accordance with previous reports, we also found that unmethylated GBM with TERTp mutations presented a poor prognosis.3,8 However, among patients with TERTp wild type, there was no significant difference of PFS and OS between patients with and without MGMT promotor methylation. The reason may be that GBM tumors with the TERTp mutation form multifocal/distant lesions by invading various directions. Nevertheless, those with methylated MGMT were sensitive to treatment with alkylating agents, such as temozolomide. Therefore, TERTp mutated GBM patients with methylated MGMT may survive longer than those with unmethylated MGMT.
Recently, GBMs were divided into 2 groups according to the IDH mutation status. Although IDH mutation is frequently found in lower-grade diffuse glioma, only 5%–10% of patients with GBM had this mutation.36,37 In addition, GBM patients with the IDH mutation are usually young and diagnosed with progression from a lower grade of diffuse astrocytoma. Thus, TERTp mutation, frequently found in GBM is more useful for predicting survival and clinical behavior, such as the pattern of invasion.
Our data showed that TERTp mutations were significantly associated with EGFR amp/gain, CDKN2A deletion, and PTEN deletion and were typically found in IDH wild-type GBM; conversely, the TERTp wild type was associated with PDGFR amp/gain, CDK4 gain, and TP53 deletion. Recently, Williams et al. reported TERTp wild-type GBMs showed frequent PI3K pathway and BAF complex gene family (ATRX, SMARCA4, SMARCB1, and ARID1A) mutations.38 Our data also suggest that TERTp wild-type GBMs are genetically distinct from TERTp-mutant GBMs.
The present study had some limitations. First, since this was a retrospective study, patients were not treated in the same manner. Although we performed a multivariate analysis, differences in treatment may have affected the pattern of recurrence. Second, we demonstrated the malignant features of GBM with the TERTp mutation based on clinicopathological characteristics, but patients with oligodendroglioma (the most benign diffuse glioma) also had the TERTp mutation.6 Third, it has been reported that PTEN, PI3K3A mutation and the expression of CD133 are associated with distant recurrence in patients with GBM.16,17,39,40 In the present study, there was no significant association between CD133 expression and the TERTp mutation, but PTEN deletion was significantly correlated with TERTp mutations and multifocal/distant lesions. The mechanism of invasiveness based on the TERTp mutation warrants further investigation.
Conclusion
We retrospectively investigated whether the TERTp mutation was associated with multifocal/distant lesions in GBM. The results suggested that the TERTp mutations strongly correlated with the multifocal phenotype and poor prognosis in patients with IDH wild-type GBM. We further demonstrated that TERTp mutations were significantly associated with EGFR amp/gain, CDKN2A deletion, and PTEN deletion, whereas the TERTp wild type was correlated with PDGFR amp/gain, CDK4 gain, and TP53 deletion. The IDH wild-type GBM with and without TERTp mutations may be a distinct clinical and molecular subtype.
Supplementary Material
Acknowledgments
We thank Yanagida S. and Sugawara M. for their assistance in extracting genomic DNA and obtaining magnetic resonance images. The authors thank enago (https://www.enago.jp/) for the English language review.
Funding
This work was supported by JSPS KAKENHI [grant numbers JP17K10856, JP20K09363].
Conflict of interest statement. None declared.
Authorship Statement: Experimental design (Z.K., I.S., Y.K., T.T., and Y.S.); implementation (Z.K., I.S., T.Y., E.Y., T.S., Y.K., R.O., K.M., and Y.S.); analysis and interpretation of data (Z.K., I.S., T.Y., E.Y., T.S., R.O., K.M., R.S., M.K., Y.K., T.K., and Y.S.); writing of the manuscript (Z.K., I.S., T.Y., E.Y., T.S., R.O., K.M., R.S., M.K., Y.K., T.K., T.T., and Y.S.).
References
- 1. Louis DN, Brat DJ, Ohgaki H, et al. Glioblatoma, IDH-wildtype. In: Louis DN, Ohgaki H, Wiestler OD, et al. eds. WHO Classification of Tumors of the Central Nervous System. Revised 4th ed. Lyon: IARC Press; 2016. [Google Scholar]
- 2. Stupp R, Mason WP, van den Bent MJ, et al. ; European Organisation for Research and Treatment of Cancer Brain Tumor and Radiotherapy Groups ; National Cancer Institute of Canada Clinical Trials Group. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352(10):987–996. [DOI] [PubMed] [Google Scholar]
- 3. Nguyen HN, Lie A, Li T, et al. Human TERT promoter mutations enables survival advantage from MGMT promoter methylation in IDH1 wild-type primary glioblastoma treated by standard chemoradiotherapy. Neuro Oncol. 2017;19(3):394–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Nencha U, Rahimian A, Giry M, et al. TERT promoter mutations and rs2853669 polymorphism: prognostic impact and interactions with common alterations in glioblastomas. J Neurooncol. 2016;126(3):441–446. [DOI] [PubMed] [Google Scholar]
- 5. Spiegl-Kreinecker S, Lötsch D, Ghanim B, et al. Prognostic quality of activating TERT promoter mutations in glioblastoma: interaction with the rs2853669 polymorphism and patient age at diagnosis. Neuro Oncol. 2015;17(9):1231–1240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Arita H, Narita Y, Fukushima S, et al. Upregulating mutations in the TERT promoter commonly occur in adult malignant gliomas and are strongly associated with total 1p19q loss. Acta Neuropathol. 2013;126(2):267–276. [DOI] [PubMed] [Google Scholar]
- 7. Park CK, Lee SH, Kim JY, et al. Expression level of hTERT is regulated by somatic mutation and common single nucleotide polymorphism at promoter region in glioblastoma. Oncotarget. 2014;5(10):3399–3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Arita H, Yamasaki K, Matsushita Y, et al. A combination of TERT promoter mutation and MGMT methylation status predicts clinically relevant subgroups of newly diagnosed glioblastomas. Acta Neuropathol Commun. 2016;4(1):79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yamashita K, Hatae R, Hiwatashi A, et al. Predicting TERT promoter mutation using MR images in patients with wild-type IDH1 glioblastoma. Diagn Interv Imaging. 2019;100(7-8):411–419. [DOI] [PubMed] [Google Scholar]
- 10. Haque W, Thong Y, Verma V, Rostomily R, Brian Butler E, Teh BS. Patterns of management and outcomes of unifocal versus multifocal glioblastoma. J Clin Neurosci. 2020;74:155–159. doi: 10.1016/j.jocn.2020.01.086. [DOI] [PubMed] [Google Scholar]
- 11. Patil CG, Yi A, Elramsisy A, et al. Prognosis of patients with multifocal glioblastoma: a case-control study. J Neurosurg. 2012;117(4):705–711. [DOI] [PubMed] [Google Scholar]
- 12. Lim DA, Cha S, Mayo MC, et al. Relationship of glioblastoma multiforme to neural stem cell regions predicts invasive and multifocal tumor phenotype. Neuro Oncol. 2007;9(4):424–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Syed M, Liermann J, Verma V, et al. Survival and recurrence patterns of multifocal glioblastoma after radiation therapy. Cancer Manag Res. 2018;10:4229–4235. doi: 10.2147/CMAR.S165956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pope WB, Xia Q, Paton VE, et al. Patterns of progression in patients with recurrent glioblastoma treated with bevacizumab. Neurology. 2011;76(5):432–437. [DOI] [PubMed] [Google Scholar]
- 15. Cachia D, Elshafeey NA, Kamiya-Matsuoka C, et al. Radiographic patterns of progression with associated outcomes after bevacizumab therapy in glioblastoma patients. J Neurooncol. 2017;135(1):75–81. [DOI] [PubMed] [Google Scholar]
- 16. Shibahara I, Sonoda Y, Saito R, et al. The expression status of CD133 is associated with the pattern and timing of primary glioblastoma recurrence. Neuro Oncol. 2013;15(9):1151–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yamaki T, Shibahra I, Matsuda KI, et al. Relationships between recurrence patterns and subventricular zone involvement or CD133 expression in glioblastoma. J Neurooncol. 2020;146(3):489–499. [DOI] [PubMed] [Google Scholar]
- 18. Sonoda Y, Kumabe T, Nakamura T, et al. Analysis of IDH1 and IDH2 mutations in Japanese glioma patients. Cancer Sci. 2009;100(10):1996–1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sasaki T, Fukai J, Kodama Y, et al. Characteristics and outcomes of elderly patients with diffuse gliomas: a multi-institutional cohort study by Kansai Molecular Diagnosis Network for CNS Tumors. J Neurooncol. 2018;140(2):329–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Umehara T, Arita H, Yoshioka E, et al. Distribution differences in prognostic copy number alteration profiles in IDH-wild-type glioblastoma cause survival discrepancies across cohorts. Acta Neuropathol Commun. 2019;7(1):99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jeuken J, Sijben A, Alenda C, et al. Robust detection of EGFR copy number changes and EGFR variant III: technical aspects and relevance for glioma diagnostics. Brain Pathol. 2009;19(4):661–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6(269):pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2(5):401–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wang K, Liu T, Ge N, et al. TERT promoter mutations are associated with distant metastases in upper tract urothelial carcinomas and serve as urinary biomarkers detected by a sensitive castPCR. Oncotarget. 2014;5(23):12428–12439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Xing M, Liu R, Liu X, et al. BRAF V600E and TERT promoter mutations cooperatively identify the most aggressive papillary thyroid cancer with highest recurrence. J Clin Oncol. 2014;32(25):2718–2726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Macerola E, Loggini B, Giannini R, et al. Coexistence of TERT promoter and BRAF mutations in cutaneous melanoma is associated with more clinicopathological features of aggressiveness. Virchows Arch. 2015;467(2):177–184. [DOI] [PubMed] [Google Scholar]
- 27. Yuan P, Cao JL, Abuduwufuer A, et al. Clinical characteristics and prognostic significance of TERT promoter mutations in cancer: a cohort study and a meta-analysis. PLoS One. 2016;11(1):e0146803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Campos MA, Macedo S, Fernandes M, et al. TERT promoter mutations are associated with poor prognosis in cutaneous squamous cell carcinoma. J Am Acad Dermatol. 2019;80(3):660–669.e6. [DOI] [PubMed] [Google Scholar]
- 29. Fukai J, Arita H, Umehara T, et al. Molecular characteristics and clinical outcomes of elderly patients with IDH-wildtype glioblastomas: comparative study of older and younger cases in Kansai Network cohort. Brain Tumor Pathol. 2020;37(2):50–59. [DOI] [PubMed] [Google Scholar]
- 30. Lee DD, Leão R, Komosa M, et al. DNA hypermethylation within TERT promoter upregulates TERT expression in cancer. J Clin Invest. 2019;129(1):223–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Diplas BH, He X, Brosnan-Cashman JA, et al. The genomic landscape of TERT promoter wildtype-IDH wildtype glioblastoma. Nat Commun. 2018;9(1):2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Reinhardt A, Stichel D, Schrimpf D, et al. Anaplastic astrocytoma with piloid features, a novel molecular class of IDH wildtype glioma with recurrent MAPK pathway, CDKN2A/B and ATRX alterations. Acta Neuropathol. 2018;136(2):273–291. [DOI] [PubMed] [Google Scholar]
- 33. Simon M, Hosen I, Gousias K, et al. TERT promoter mutations: a novel independent prognostic factor in primary glioblastomas. Neuro Oncol. 2015;17(1):45–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Nonoguchi N, Ohta T, Oh JE, Kim YH, Kleihues P, Ohgaki H. TERT promoter mutations in primary and secondary glioblastomas. Acta Neuropathol. 2013;126(6):931–937. [DOI] [PubMed] [Google Scholar]
- 35. Shu C, Wang Q, Yan X, Wang J. The TERT promoter mutation status and MGMT promoter methylation status, combined with dichotomized MRI-derived and clinical features, predict adult primary glioblastoma survival. Cancer Med. 2018;7(8):3704–3712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Aldape K, Zadeh G, Mansouri S, Reifenberger G, von Deimling A. Glioblastoma: pathology, molecular mechanisms and markers. Acta Neuropathol. 2015;129(6):829–848. [DOI] [PubMed] [Google Scholar]
- 37. Louis DN, Perry A, Reifenberger G, et al. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary. Acta Neuropathol. 2016;131(6):803–820. [DOI] [PubMed] [Google Scholar]
- 38. Williams EA, Miller JJ, Tummala SS, et al. TERT promoter wild-type glioblastomas show distinct clinical features and frequent PI3K pathway mutations. Acta Neuropathol Commun. 2018;6(1):106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kato H, Fujimura M, Kumabe T, Ishioka C, Kanamaru R, Yoshimoto T. PTEN gene mutation and high MIB-1 labeling index may contribute to dissemination in patients with glioblastoma. J Clin Neurosci. 2004;11(1):37–41. [DOI] [PubMed] [Google Scholar]
- 40. Tanaka S, Batchelor TT, Iafrate AJ, et al. PIK3CA activating mutations are associated with more disseminated disease at presentation and earlier recurrence in glioblastoma. Acta Neuropathol Commun. 2019;7(1):66. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.