Highlights
-
•
Subpopulations of cisplatin-resistant cells are cross-resistant to anti-microtubule drugs.
-
•
Resistance mechanisms in cisplatin-resistant ovarian cancer cells are diverse.
-
•
Increased TNF and NFκB signaling found in cisplatin-resistant subpopulations treated with anti-microtubule drugs.
Keywords: Ovarian cancer, Cisplatin, Drug resistance, Anti-microtubule drugs, TNF, NFκB
Abbreviations: GTP, guanosine triphosphate; STR, short tandem repeat; MEM, minimum essential medium; FBS, fetal bovine serum; PBS, phosphate-buffered saline; IMEM, improved minimal essential medium; SEM, standard error of mean; GSEA, gene set enrichment analysis; TNF, tumor necrosis factor; EMT, epithelial-to-mesenchymal transition; FTC, fumitremorgin C; COL, colchicine; PTX, paclitaxel; VIN, vincristine
Abstract
Although the first line of therapy for epithelial ovarian cancer typically consists of taxane-platinum combination therapy, many patients develop a platinum-resistant tumor within a year. Several previous studies have looked at this cross-resistance between cisplatin and anti-microtubule drugs, but their findings have been somewhat conflicting. Here, we developed cisplatin-resistant cell lines that are resistant to low and high levels of cisplatin and explored the effects of three anti-microtubule drugs (paclitaxel, vincristine, and colchicine) on the parental and cisplatin-resistant cells. We found that cells resistant to lower levels of cisplatin were no more resistant to anti-microtubule drugs than parental cells, while cells that were resistant to higher levels of cisplatin had a subpopulation of cells that were cross-resistant to anti-microtubule drugs, clarifying discrepancies within the field. We then isolated this subpopulation by applying selective pressure with anti-microtubule drugs and performed RNA sequencing and gene set enrichment analysis to identify resistance mechanisms. This subpopulation was found to express increased levels of pro-survival TNF/NFκB signaling, among other enriched pathways, suggesting that cross-resistance was due to more general survival mechanisms found in the cisplatin-selected cells.
Introduction
Cisplatin is a platinum-based chemotherapeutic drug that acts by cross-linking with DNA, resulting in DNA damage and subsequent induction of apoptosis [1,2]. While cisplatin is initially effective against ovarian cancer cells, a significant portion of those cells will develop resistance. Mechanisms of resistance include increased tolerance to DNA damage [3], decreased accumulation of cisplatin [4], and increased uptake of thiol-containing species such as glutathione, which facilitates cisplatin export from the cell by forming glutathione-bound cisplatin conjugates [5]. More recently, carboplatin is used as a treatment for ovarian cancer. Because cisplatin is thoroughly studied and we wanted to have a reference point to test the validity of our study, we decided to utilize cisplatin instead of carboplatin for our study. Although some studies have suggested that microtubules play a role in cisplatin resistance [6,7], this is an area of research that needs to be further explored. Several reports have suggested that cisplatin-resistant cells are also cross-resistant to anti-microtubule drugs, but the mechanism for this has not been clearly defined [8–10]. In contrast, it has also been reported that there is no cross-resistance between cisplatin-resistant cells and anti-microtubule drugs [11]. Irrespective of the potential role of microtubules in cisplatin resistance, paclitaxel, a microtubule-stabilizing agent, is an effective treatment for ovarian cancer and is often used prior to or in conjunction with cisplatin. Thus, we investigated whether selection for cisplatin resistance confers resistance to anti-microtubule drugs to clarify whether there is a relationship between these two types of resistance.
There are many anti-microtubule (polymerizing and depolymerizing) drugs. One of these, paclitaxel, functions by binding to a microtubule polymer and augmenting the polymerization of tubulin. Typically, guanosine triphosphate (GTP) is a necessary component for the polymerization of microtubules; however, paclitaxel promotes polymerization without GTP by binding to the β-tubulin subunit of microtubules [12]. In contrast, vincristine binds to β-tubulin and prevents proteins from polymerizing into microtubules, resulting in metaphase arrest and eventual apoptosis [13]. Similarly, colchicine blocks mitotic cells in metaphase by binding to soluble tubulin, which then attaches to the ends of microtubules, preventing further elongation of the polymer [14]. Supplementary Fig. 1 displays the structure of an αβ-tubulin heterodimer with the various binding sites of these three anti-microtubule drugs (Supplementary Fig. 1A) [15], [16], [17]–18] and their chemical structures (Supplementary Fig. 1B) [19].
The objective of this study is to explore whether selection for cisplatin resistance confers resistance to anti-microtubule drugs. We hypothesized that resistance to increasing concentrations of cisplatin results in cross-resistance to anti-microtubule drugs. We tested this by generating the cisplatin (CP)-resistant cell lines OVCAR8-CP1 and OVCAR8-CP5 from the parental cell line, OVCAR8, by gradually exposing the cells to 1 μM and 5 μM cisplatin, respectively. We found that the less resistant cell line, OVCAR8-CP1, displayed no greater resistance to the three anti-microtubule drugs than did the parental cell line. However, we found that the more resistant cell line, OVCAR8-CP5, included a subpopulation of cells that showed cross-resistance to the same three drugs. Through RNA sequencing and gene set enrichment analysis, we identified increased levels of pro-survival TNF/NFκB signaling within this specific subpopulation that demonstrated cross-resistance to both cisplatin and anti-microtubule drugs. Our data suggest that even though both of these cell populations are resistant to cisplatin, the differing levels of cisplatin resistance in the two populations may contribute to differing mechanisms of resistance that could potentially explain the conflicting data in the literature.
Materials and methods
Visualization of tubulin structure
The structure of the αβ-tubulin dimer was downloaded from The Protein Data Bank (PDB ID: 6E7B; www.rcsb.org) and presented using PyMOL Molecular Graphics System, Version 2.0 Schrödinger, LLC (www.pymol.org).
Cell culture
The OVCAR8 cell line is an NCI-60 cell line obtained from the National Cancer Institute Division of Cancer Treatment and Diagnosis Tumor Repository (NCI DCTD; Frederick, MD). OVCAR8 cell line was utilized in our study because based on its functional characteristics, it closely resembled high grade serous ovarian cancer [20]. The parental ovarian cancer cell line, OVCAR8 (here, termed OVCAR8-CP0, was grown in 0 μM cisplatin (CP)), in Roswell Park Memorial Institute medium 1640 (RPMI; Gibco; Cat#11,875-093) supplemented with 10% fetal bovine serum (FBS; Atlanta Biologicals; Cat# S11150) and 1% penicillin and 1% streptomycin (Gibco; Cat# 15,140-148) in a 37 °C incubator with 5% CO2. OVCAR8-CP1 and OVCAR8-CP5 cells were grown in the same media supplemented with 1 μM and 5 μM cis-Diamineplatinum (II) dichloride (cisplatin; Sigma-Aldrich; Cat# P4394) dissolved in phosphate-buffered saline (PBS; KD Medical; Cat# RGE-3190), respectively. Both resistant cell lines, OVCAR8-CP1 and OVCAR8-CP5, were established from their parental cell line, the OVCAR8 ovarian adenocarcinoma cell. All cell lines were authenticated by short tandem repeat (STR) analysis and tested for mycoplasma using a MycoAlert Plus Mycoplasma Detection Kit (Lonza; Cat# LT07-710). Positive control cell lines used for efflux assays included MDR19 (ABCB1 positive) and R5 (ABCG2 positive) [21]. These cell lines were cultured in Minimum Essential Medium (MEM; Corning; Cat# 10-010-CM) supplemented with 10% FBS (Gibco; Cat# 26,140-079), 1% penicillin and 1% streptomycin, and 2 mg/mL G418 selecting agent (Corning; Cat# 61–234-RG).
Generation of resistant cell lines
OVCAR8-CP0 was used to generate cisplatin-resistant cells. The OVCAR8-CP0 resistant cell line variants, OVCAR8-CP1 and OVCAR8-CP5, were established by continual exposure to gradually increasing concentrations of cisplatin with final concentrations of 1 μM and 5 μM cisplatin, respectively. The OVCAR8-CP5 cells were derived from OVCAR8-CP1 cells. Cells were passaged to reach an equivalent passage number to avoid variability caused by passage number. The resistant cells retained their resistance for several days (we have not tested for months).
Cell viability assay
All ovarian cancer cell lines (OVCAR8-CP0, OVCAR8-CP1, and OVCAR8-CP5) were plated in 96-well plates with 2–5 × 103 cells per well and treated with a serial dilution of cisplatin, paclitaxel (Sigma-Aldrich; Cat# T7402), vincristine (Sigma-Aldrich; Cat# V8879), or colchicine (Sigma-Aldrich; Cat# C9754). Cisplatin was dissolved in PBS while paclitaxel, vincristine, and colchicine were dissolved in dimethyl sulfoxide (DMSO; Sigma-Aldrich; Cat# D2660). Specific drug concentrations are indicated either in the Fig. or in the Fig. legends and were optimized to develop a full killing curve. Cellular proliferation was measured using a CellTiter-Glo Luminescent Cell Viability Assay (Promega; Cat# G7573) at 72 h and luminescence values were measured at an integration time of 100 ms on a SpectraMax ID3 (Molecular Devices).
Cell growth assay
OVCAR8-CP0, OVCAR8-CP1, and OVCAR8-CP5 cells were plated on 60 × 15 mm polystyrene plates at a density of approximately 1×105 cells per plate. Plates were washed with PBS eight days after plating, fixed with 4% paraformaldehyde (Electron Microscopy Sciences; Cat# 15710) for 12 min, and stained with crystal violet (Sigma-Aldrich; Cat# C3886). Plates were then imaged using a BioTek Lionheart FX machine at an optical density of 595 nm. The object sum area obtained from the BioTek Lionheart FX machine was used to calculate the rate of cell proliferation.
Cell cycle flow cytometry analysis
All cell lines were plated at a density of 2.5 × 105 per well in a 6-well plate and treated with a vehicle control (PBS) or cisplatin after 24 h. Specific drug concentrations are indicated either in the figure or in the figure legends. Cells were then harvested by trypsin 24 h post-treatment and incubated with 50 μg/mL propidium iodide (Sigma-Aldrich; Cat# P4170), 0.1% Triton X-100 (Sigma-Aldrich; Cat# 648462), and 200 U/mL RNase A (USB Corporation; Cat# 27033002) for 15 min. Samples were then read using a FACSCanto II Flow Cytometer (BD Biosciences; Cat# 338960) and FlowJo software was used to determine the percentage of cells in cell cycle phases.
Efflux flow cytometry analysis
To examine ABCB1- or ABCG2-mediated transport in the cisplatin-resistant progression cell lines, we conducted an efflux assay via flow cytometry analysis as described previously [22]. Briefly, all cell lines were harvested by trypsin and resuspended in phenol red-free improved minimal essential medium (IMEM; Corning; Cat# 10–026-CV) supplemented with 10% FBS (Gibco; Cat# 26140-079), and 1% penicillin and 1% streptomycin. In order to detect ABCB1 mediated transport, cells were then incubated with 0.5 μg rhodamine/mL (ABCB1 fluorescent substrate; Sigma-Aldrich; Cat# 83702) in the absence or presence of 3 μM valspodar (ABCB1 inhibitor; MedChemExpress; Cat# HY-17384) for 30 min in a 37 °C incubator with 5% CO2. Cells were subsequently incubated in substrate-free medium in the absence or presence of valspodar for two hours. Samples were then washed with cold PBS and intracellular substrate fluorescence was measured by flow cytometry on a FACSCanto II Flow Cytometer. To detect ABCG2-mediated transport, cells were incubated with 5 μM pheophorbide A (PHA; ABCG2 fluorescent substrate; Frontier Scientific; Cat# Pha-592) in the absence or presence of 10 μM fumitremorgin C (FTC; ABCG2 inhibitor; isolated by Thomas McCloud, NCI Developmental Therapeutics Program, Natural Products Extraction Laboratory) for 30 min in a 37 °C incubator with 5% CO2. Cells were then incubated in substrate-free medium in the absence or presence of FTC for two hours. Samples were subsequently washed with cold PBS and intracellular substrate fluorescence was measured by flow cytometry on a FACSCanto II Flow Cytometer. FlowJo software was later used to determine the mean fluorescence in the presence of inhibitor and the mean fluorescence in the absence of inhibitor for both ABCB1- and ABCG2-mediated transport.
Annexin V flow cytometry analysis
All cell lines were plated at a density of 5 × 105 cells per well in a 6-well plate and treated with DMSO, cisplatin, paclitaxel, vincristine, or colchicine after 24 h. Specific drug concentrations are indicated either in the figure or in the figure legends. Cells were harvested six days post-treatment and incubated with allophycocyanin-conjugated annexin V (APC Annexin V; BioLegend; Cat# 640941) and SYTOX green (Invitrogen; Cat# S7020) for 20 min. Samples were read using a FACSCanto II Flow Cytometer and FlowJo software was used to determine the percentage of annexin-positive cells.
Immunoblot analysis
OVCAR8-CP0, OVCAR8-CP1, and OVCAR8-CP5 cell lines were plated in a 100 × 20 mm polystyrene plate at a density of 1 × 106 cells per plate and treated with indicated treatments after 24 h. Cells were harvested 24 h post-treatment and re-suspended in lysis buffer (50 mM Tris-HCl pH 7.4, 150 mM NaCl, 1% NP-40, 1% protease inhibitor cocktail (Bimake; Cat# B14001), sonicated, and centrifuged to remove cell debris. Supernatant was reserved and protein was loaded into a 4–12% Bis-Tris NuPAGE gel (ThermoFisher; Cat# NP0321PK2), subjected to electrophoresis and transferred to a 0.2 μm pore nitrocellulose membrane (VitaScientific; Cat# DBOC80004). The resulting membrane was blocked in Odyssey PBS Blocking Buffer (LI-COR; Cat# 927-40000) for 1 h at room temperature and subsequently incubated with the following primary antibodies overnight: anti-cleaved Caspase-3 (1:1000; Cell Signaling Technology; Cat# 9661S), anti-ABCB1 (C219) (1:1000 dilution; Fujirebio Diagnostics, Inc., Malvern, PA), anti-ABCG2 (also called as breast cancer resistance protein) (1:1000; Enzo Life Sciences; Cat# ALX-801-029-C250); anti-NFkB p65 (1:1000; Cell Signaling; Cat# 6956); anti-phospho-NFkB p65 (1:1000; Cell Signaling; Cat# 3033), anti-detyrosinated alpha tubulin antibody (1:1000; Abcam; Cat# ab48389), anti-alpha tubulin antibody (DM1A) (1:5000; Abcam, Cat# 7291), and anti-GAPDH (1:1000; American Research Products, Inc; Cat# 05-50118). GAPDH was used as a reference control in our study. ABCB1 and ABCG2 overexpressing cells were used as positive controls. These cells were generated as described previously [22]. Membranes were then washed with 0.5X TBS (KD Medical) containing 0.5% TBS-Tween-20 (Boston BioProducts; Cat# IBB-181) three times for 5 min prior to and after the addition of an IRDye Goat anti-Rabbit (LI-COR; Cat# 926-32211) or anti-Mouse (LI-COR; Cat# 926-68070) secondary antibody. Proteins were visualized using an Odyssey CLx imaging system (LI-COR). Relative expression of cleaved Caspase-3 was quantified using Image Studio Lite Quantification Software (LI-COR).
Confocal imaging
OVCAR8-CP0 and OVCAR8-CP5 cell lines were plated in 4-well chamber slides (Thermo Fisher Scientific; Cat# 154526) at a density of 1 × 104 cells per well. After 24 h of plating, the cells were fixed with 4% paraformaldehyde for 15 min. Cells were rinsed with PBS and were blocked with blocking buffer (PBS containing 0.3% Triton X-100 with 1% BSA). Cells were treated with anti-detyrosinated alpha tubulin antibody (1:100; Abcam; Cat# ab48389) in blocking buffer for 3 h at room temperature. Cells were then washed with PBS at least 5 times. Cells were treated with secondary antibody (Alexa Fluor 488) for 1 h followed with by hoechst stain (1:10,000; Thermo Fisher Scientific; Cat# H3569) for 5 min. Cells were washed and imaged to determine their fluorescence levels using the Zeiss LSM780 confocal microscope.
RNA sequencing
OVCAR8-CP5 cells were treated with DMSO, 0.1 μM paclitaxel, 1 μM vincristine, or 0.2 μM colchicine for 72 h to isolate the subpopulation of OVCAR8-CP5 cells resistant to these anti-microtubule drugs. Libraries constructed from CP-resistant OVCAR8-CP5 cell lines and pooled subpopulations of OVCAR8-CP5 cells conferring cross-resistance to paclitaxel, vincristine, and colchicine were sequenced on a NextSeq 500 using Illumina's TruSeq Stranded mRNA Library Prep Kit. The sequencing quality of 14–25 million reads per sample was assessed using FastQC (version 0.11.5), Preseq (version 2.0.3) [23] Picard tools (version 1.119), and RSeQC (version 2.6.4). Reads were trimmed using Cutadapt (version 1.18) [24] to remove adapter sequences, prior to mapping to the human reference genome, hg38, using STAR (version 2.7.0f) in two-pass mode [25]. Across all samples, the average percentage of mapped reads was 93.2%. Expression levels were quantified using RSEM version 1.3.0 [26] with GENCODE annotation version 30 [27]. Low count genes were removed prior to differential expression analysis. Genes with counts per million > 0.5 in at least two samples were considered for downstream analysis. The RSEM counts were then normalized using the voom algorithm [28] from the Limma R package (version 3.40.6) [29]. These normalized counts were used for clustering and data visualization. The R package EnhancedVolcano (version 1.2.0) was used to distinguish genes into four separate groups using the following thresholds: p-value < 10e-6 and abs|log2(fold-change)| > 1.0. The autoplot function within the R package ggplot2 (version 3.2.1) was used to generate a PCA plot and pheatmap (version 1.0.12) was used to visualize expression of ABC transporters. The R package ggpubr (version 0.2.3) was used to generate the box plot of gene expression for ABCB1 and ABCG2. The Limma package was used to test for differential gene expression between experimental conditions. Significant differentially expressed genes were identified with a false-discovery rate ≤ 0.05. Pathway enrichment was performed using pre-ranked Gene Set Enrichment Analysis (GSEA) [30] with the KEGG genesets [31] from the Molecular Signatures Database [32].
Statistical analyses
Unless noted otherwise, all statistical analyses including standard error of mean (SEM) and statistical significance were determined using GraphPad Prism 8. Data in this study was considered as statistically significant when p < 0.05 using a two-tailed Student's t-test.
Results
Establishment of cisplatin-resistant cell lines
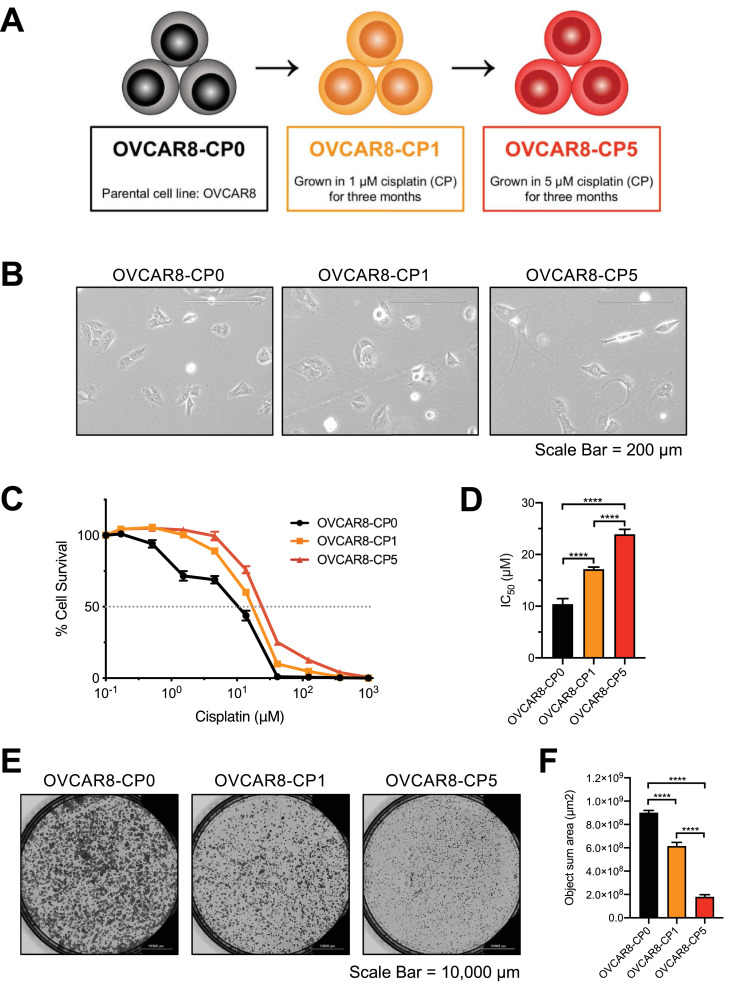
In order to develop cisplatin-resistant cell lines, OVCAR8 cells were first grown in the presence of 1 μM cisplatin for three months to select for the less resistant cell line, OVCAR8-CP1. Beginning with this resistant population, the cisplatin concentration was gradually increased to 5 μM over a time period of three more months to establish the more resistant OVCAR8-CP5 cell line. Both OVCAR8-CP1 and OVCAR8-CP5 were grown in the presence of cisplatin for four additional months prior to beginning experiments and continued to grow in cisplatin for the remainder of the study (Fig. 1A). The OVCAR8-CP0 and OVCAR8-CP1 cell populations were also passaged to reach an equivalent passage number as OVCAR8-CP5. We observed changes in cell morphology between the parental and resistant cell lines (Fig. 1B). Cell elongation in cisplatin-resistant cells have been observed previously [33]. It is possible that the cell elongation could be due to microtubule involvement. To test that hypothesis, we analyzed levels of polymerized microtubules in the three cell lines and found increased microtubule polymerization in both cisplatin-resistant cell lines, with more polymerization observed in the more resistant OVCAR8-CP5 cells (Supplementary Fig. 2A and B). Therefore, it is clear that cells become more elongated as they become more resistant to cisplatin. A cell viability assay was performed to confirm cisplatin resistance (Fig. 1C). The IC50 values of OVCAR8-CP0, OVCAR8-CP1, and OVCAR8-CP5 were 9.56 ± 0.913, 16.51± 0.59, and 26.1 ± 1.49 μM, respectively (Fig. 1D). We also observed that the cisplatin-resistant cells grew more slowly than the parental cells (Fig. 1E and 1F). The object sum area for OVCAR8-CP0, OVCAR8-CP1, and OVCAR8-CP5 was 9.00 × 108 ± 2.04 × 107, 6.14 × 108 ± 3.27 × 107, and 1.80 × 108 ± 1.92 × 107 μm2, respectively. The OVCAR8-CP1 cells showed a 31.78% decrease in rate of growth and the OVCAR8-CP5 displayed an 80.04% decrease in rate of growth. These data are consistent with reports suggesting that cisplatin resistance mechanisms require additional resources for proliferation, which leads to a deficiency in the cells’ fitness [34].
Fig. 1.
Generation and confirmation of cisplatin resistance in OVCAR8-CP1 and OVCAR8-CP5 cell lines. (A) Schematic depicting the development of the cisplatin-resistant cell lines. (B) Bright-field microscopic images of OVCAR8-CP0, OVCAR8-CP1, and OVCAR8-CP5. Scale bar = 200 μm. (C) All three cell lines were incubated with various concentrations of cisplatin, with a final concentration of 1000 μM cisplatin, for 72 h. Cell viability was determined by the CellTiter-Glo assay system. Values shown represent the mean of eight biological replicates, with the standard error of mean indicated by error bars. (D) IC50 levels are displayed as the mean of eight biological replicates at which% cell survival is at y = 50%. n = 8, **** p < 0.0001. (E) Bright-field microscopic images of OVCAR8-CP0, OVCAR8-CP1, and OVCAR8-CP5 cells 8 days after plating. Scale bar = 10,000 μm (F) Object sum area (μm2) of all plates was determined using BioTek Lionheart FX and is shown as the mean of six biological replicates with the standard error of mean indicated by error bars. n = 6, **** p < 0.0001.
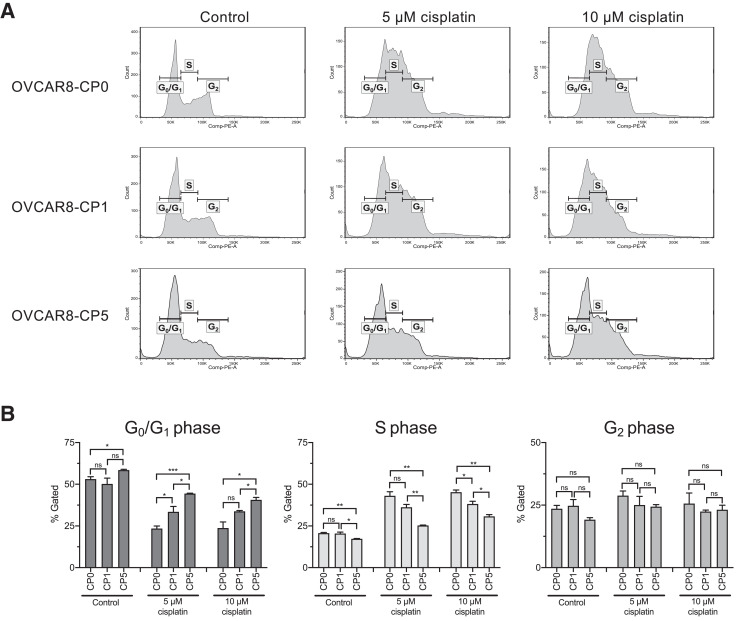
Increased cell cycle arrest upon cisplatin treatment is seen in the parental cell line as compared to cisplatin-resistant cells
We then determined the distribution of cell cycle phases of our cell lines in the presence or absence of cisplatin (Fig. 2A and B). After treatment with cisplatin, cells with increased resistance to cisplatin exhibited less arrest in the S phase. With no drug treatment, the percentage of cells in the S phase for OVCAR8-CP0, OVCAR8-CP1, and OVCAR8-CP5 was similar (20.67% ± 0.41, 20.43% ± 0.89, 17.27% ± 0.20, respectively). With 5 μM and 10 μM cisplatin treatment, the percentage of OVCAR8-CP0 cells in the S phase increased to 43.07% and 45.20%, respectively. In the case of the less resistant line, OVCAR8-CP1, the percentage of cells in S phase was 36.17% ± 1.73 and 38.17% ± 1.62, respectively. Consistent with the extent of resistance, the more resistant line, OVCAR8-CP5, showed the least amount of change with the addition of cisplatin (25.27% ± 0.27 and 30.70% ± 1.12 upon 5 μM and 10 μM treatment of cisplatin).
Fig. 2.
Comparison of cell cycle analyses of parental cell line and cisplatin-resistant cells. (A) OVCAR8-CP0, OVCAR8-CP1, and OVCAR8-CP5 cells were treated for 24 h with cisplatin as indicated, stained with propidium iodide, and analyzed by flow cytometry. Cell cycle histograms of one biological replicate of all three cell lines depicting populations of various cell cycle phases is shown. (B) Bar graph displaying the quantitative analysis of distribution of cells in G0/G1, S, and G2 phases of the cell cycle represented as the mean of three biological replicates with the standard error of mean indicated by error bars. n = 3.
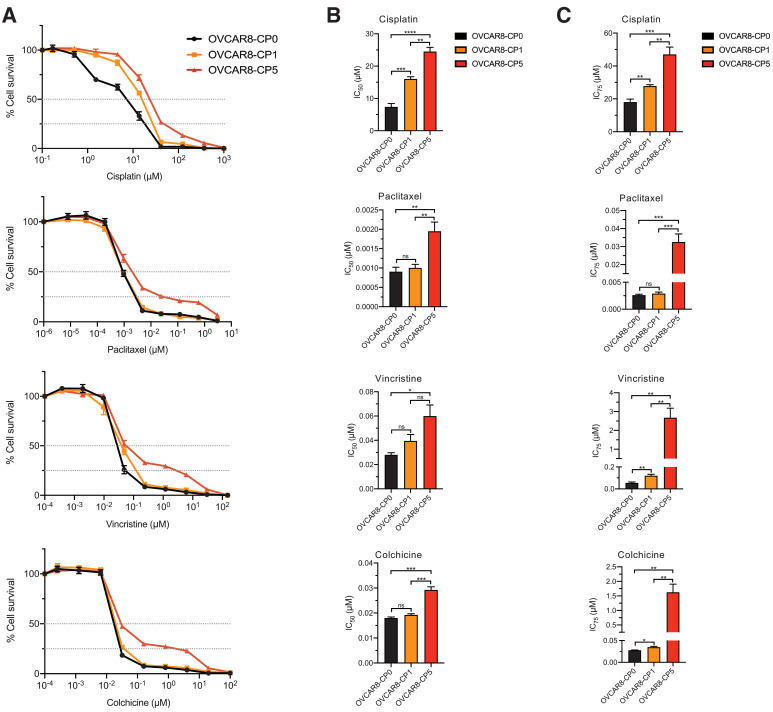
A subpopulation of cisplatin-resistant cells is cross-resistant to anti-microtubule drugs
Since ovarian cancer patients are often treated with paclitaxel in conjunction with cisplatin or carboplatin, we wanted to examine how anti-microtubule drugs (paclitaxel, vincristine, and colchicine) in combination with cisplatin affect the viability of cisplatin-resistant cells. We found that upon treatment with anti-microtubule drugs, the more resistant cell line, OVCAR8-CP5, does exhibit cross-resistance, while the less resistant cell line, OVCAR8-CP1, does not. By conducting cell viability assays, we detected the IC50 values for each treatment (Fig. 3A and B) as well as the IC75 values (Fig. 3C). We provide both the IC50 and IC75 values to illustrate the level of resistance of the different cell lines to anti-microtubule drugs. The parental and less cisplatin-resistant cells, OVCAR8-CP0 and OVCAR8-CP1, showed similar levels of sensitivity to paclitaxel, vincristine, and colchicine, consistent with previous studies that suggest there is no cross-resistance between cisplatin-resistant cells and anti-microtubule drugs [11]. On the other hand, it is evident that OVCAR8-CP5 cells have significantly higher resistance to anti-microtubule drugs compared to the OVCAR8-CP1. At a very high drug concentration, OVCAR8-CP5 cells plateau at approximately 25% cell survival, as indicated by the dotted line. This suggests that a subset of these cells was able to survive high concentrations of anti-microtubule drug. We wanted to investigate the transcriptional profile of these cell lines to extrapolate their resistance mechanism.
Fig. 3.
Cell viability of parental cell line and cisplatin-resistant cells when treated with anti-microtubule drugs. (A) OVCAR8-CP0, OVCAR8-CP1, and OVCAR8-CP5 cells were incubated with various concentrations of cisplatin, paclitaxel, vincristine, or colchicine as indicated for 72 h and cell viability was determined by the CellTiter-Glo assay system. Values shown represent the mean of four biological replicates with the standard error of mean indicated by error bars. (B) IC50 levels are displayed as the mean of four biological replicates at which% cell survival is at y = 50x%. n = 4, *** p < 0.001, ** p < 0.01, * p < 0.05, ns > 0.05. (C) IC75 levels are displayed as the mean of four biological replicates at which% cell survival is at y = 25%. n = 4, *** p 〈 0.001, ** p < 0.01, * p < 0.05, ns 〉 0.05.
Cisplatin-resistant cell lines do not show increased activity of either ABCB1 or ABCG2
In an effort to characterize the subpopulation of OVCAR8-CP5 cells that survived these higher concentrations, we looked at the activity of two common multidrug resistance proteins, ATP-binding cassette subfamily B member 1 (ABCB1) and ATP-binding cassette subfamily G member 2 (ABCG2). We conducted efflux assays via flow cytometry analyses to determine if any of the cell lines contained ABCB1 or ABCG2, but we did not detect significant expression or activity of either protein in the cell lines (Supplementary Fig. 3A–C), suggesting other resistance mechanisms were responsible for the cross-resistance to anti-microtubule drugs.
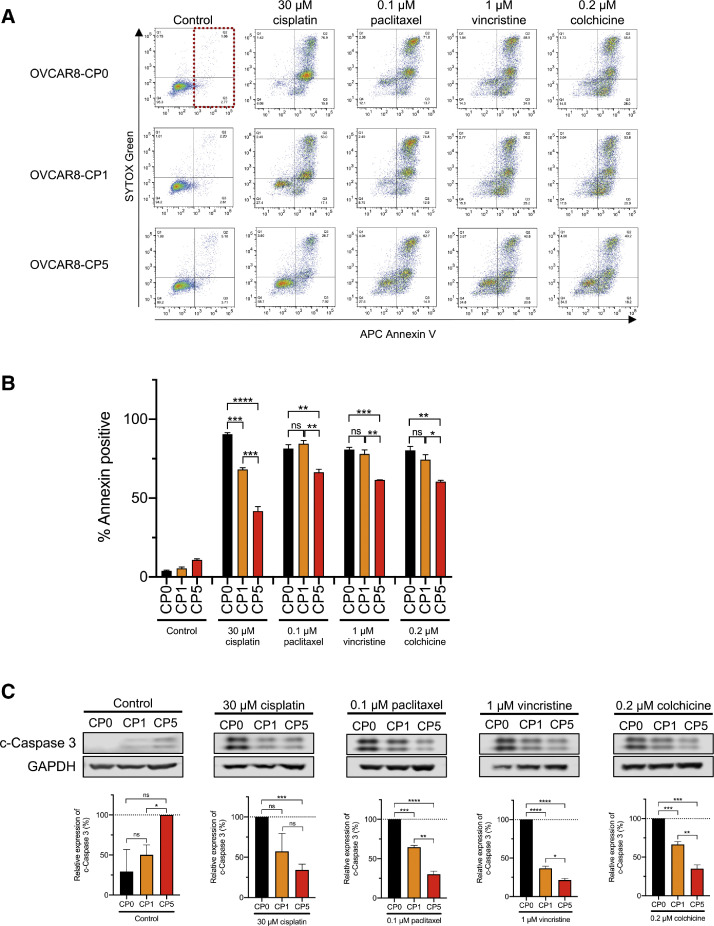
Cisplatin-resistant cells exhibit less apoptosis than parental cells when treated with anti-microtubule drugs
To better understand how the higher concentrations of various anti-microtubule drugs may be affecting apoptosis in the parental and cisplatin-resistant cell lines, we performed an annexin V assay by flow cytometry (Fig. 4A). We found that after treatment with anti-microtubule drugs, OVCAR8-CP5 displayed the least amount of apoptosis among the three cell lines. Although there was no significant difference in the level of annexin-positive cells between OVCAR8-CP0 and OVCAR8-CP1 when treated with each of the anti-microtubule drugs, there was a significant decrease in the percent of annexin-positive cells in OVCAR8-CP5 (Fig. 4B). This decreased apoptosis was further verified by examining caspase cleavage in all three cell lines after treatment with cisplatin, paclitaxel, vincristine, and colchicine (Fig. 4C). A low basal level of cleaved caspase 3 was observed in the untreated control OVCAR8-CP5 cells. Despite this, OVCAR8-CP0 displayed the most cleaved caspase 3 when compared to OVCAR8-CP1 and OVCAR8-CP5 after treatment with anti-microtubule drugs. Furthermore, OVCAR8-CP1 exhibited less cleaved caspase 3 than OVCAR8-CP0, but OVCAR8-CP5 had even less cleaved caspase 3.
Fig. 4.
Effect of anti-microtubule drugs on apoptosis of parental and cisplatin-resistant cell lines. (A) OVCAR8-CP0, OVCAR8-CP1, and OVCAR8-CP5 cells were treated for 72 h with drugs as indicated, stained with Annexin V and SYTOX green, and analyzed by flow cytometry. The two right-hand quadrants of each graph (see dotted box as an example) represent annexin-positive cells. (B) Quantitative analysis of annexin-positive cells represented as the mean of three biological replicates with the standard error of mean indicated by error bars. n = 3, **** p < 0.0001, *** p < 0.001, ** p < 0.01, * p < 0.05, ns 〉 0.05. (C) Immunoblot images of OVCAR8-CP0, OVCAR8-CP1, and OVCAR8-CP5 cells treated with drug as indicated. GAPDH was used as a loading control. Representative immunoblots are shown from at least 3 independent experiments, with quantitative data shown below each image. n = 3, **** p 〈 0.0001, *** p < 0.001, ** p < 0.01, * p < 0.05, ns 〉 0.05. The untreated control group was normalized to OVCAR8-CP5 instead of OVCAR8-CP0 because no expression was observed in OVCAR8-CP0.
Gene set enrichment analysis (GSEA) reveals enrichment for survival pathways within a subpopulation of OVCAR8-CP5 cells
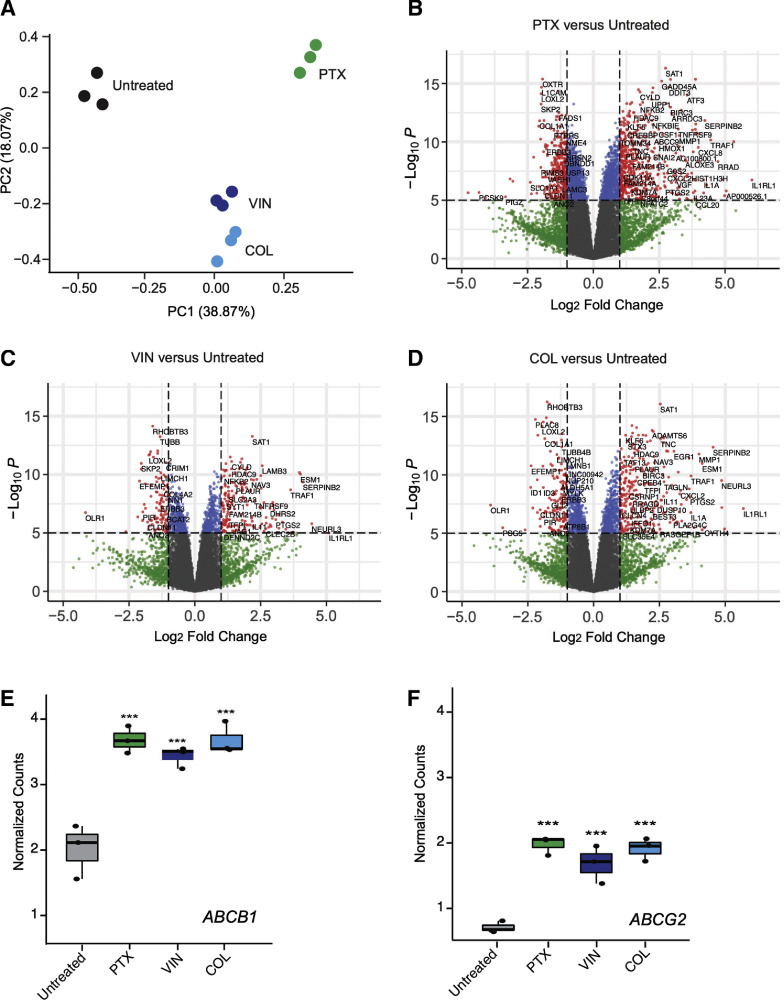
We further characterized the subpopulation of OVCAR8-CP5 cells that survived higher concentrations of paclitaxel (0.1 μM), vincristine (1 μM), and colchicine (0.2 μM). Subpopulations from each treatment group were pooled for RNA sequencing to identify the most predominant resistance mechanism. We then performed a differential gene expression analysis of the subpopulations that were resistant to paclitaxel, vincristine, and colchicine in comparison to the untreated OVCAR8-CP5 cells. A principal component analysis (PCA) was implemented to evaluate the reproducibility of the biological replicates within the RNA sequencing data and also to compare how different the cells treated with anti-microtubule drugs were to the untreated cells or to other treatment groups (Fig. 5A). We found that OVCAR8-CP5 cells treated with anti-microtubule drugs clustered closer together; variance was further reduced between replicates of cells treated with a microtubule-destabilizing agent (vincristine or colchicine) versus a microtubule-stabilizing agent (paclitaxel). We looked at differential gene expression of each treatment group against the untreated control to observe which genes were upregulated and downregulated when the cells were treated with an anti-microtubule drug for 72 h (Fig. 5Band D). In addition, we found this subpopulation of cells expressed higher levels of ABCB1 (Fig. 5E) and ABCG2 (Fig. 5F) than the general population of cisplatin-resistant OVCAR8-CP5 cells (Supplementary Fig. 3A–C). Upon further observation, we found altered gene expression in several ABC transporters that survived anti-microtubule drug treatment in comparison to the untreated population (Supplementary Fig. 4A). ABCB1 and ABCG2 efflux assays demonstrated that despite upregulation of these gene transcripts, ABCB1 transporter activity only slightly increases in the isolated subpopulation of OVCAR8-CP5, while hardly any changes are seen in ABCG2-mediated transport (Supplementary Fig. 4B and C).
Fig. 5.
RNA sequencing analysis identifies increased levels of ABCB1 and ABCG2 in highly resistant OVCAR8-CP5 subpopulation. OVCAR8-CP5 cells were treated with 0.1 μM paclitaxel, 1 μM vincristine, or 0.2 μM colchicine for 72 h to isolate the subpopulation of OVCAR8-CP5 cells resistant to these anti-microtubule drugs. RNA sequencing was performed to analyze for differential gene expression among various treatment groups. The subpopulation is named by the cells they were derived from and the treatment (paclitaxel = PTX, vincristine = VIN, colchicine = COL) that was applied to isolate them from the parental OVCAR8-CP5. (A) Principal component analysis comparing untreated OVCAR8-CP5 cells (black) and OVCAR8-CP5 cells treated with paclitaxel (green), vincristine (navy), or colchicine (blue). Differential gene expression of RNA sequencing analysis for (B) paclitaxel versus untreated group, (C) vincristine versus untreated group, and (D) colchicine versus untreated group were displayed as –log10(p-value) against the log2(fold-change). Genes with a p-value > 1e-5 and log2(fold-change) < 1 are displayed as gray; genes with a p-value > 1e-5 and log2(fold-change) > 1 are displayed as green; genes with a p-value < 1e-5 and log2(fold-change) < 1 are displayed as blue; and genes with a p-value < 1e-5 and log2(fold-change) > 1 are displayed as red. Box plot corresponding to the normalized counts of the (E) ABCB1 and (F) ABCG2 mRNA transcripts in all sample groups. Significance for these values was calculated by moderated T test through the Limma R package. *** p < 0.001.
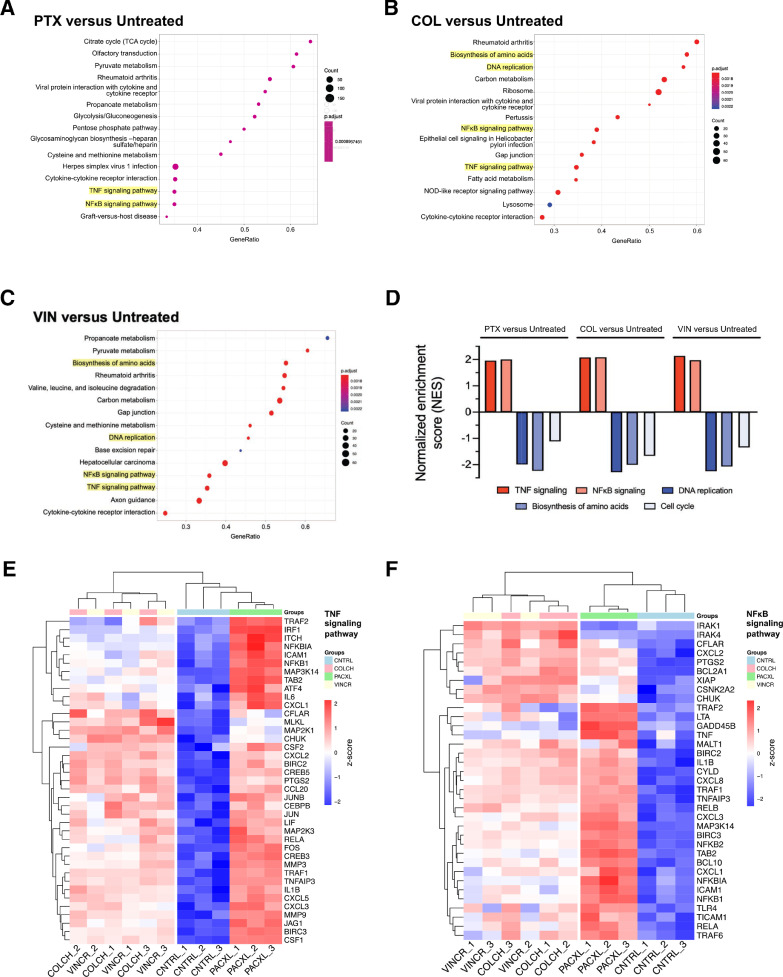
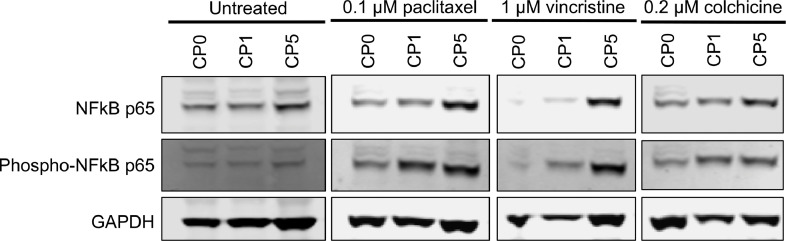
In order to broaden our understanding of affected pathways, we performed gene set enrichment analysis (GSEA) and identified significantly affected pathways in OVCAR8-CP5 cells treated with paclitaxel, vincristine, and colchicine (Fig. 6A–C). As expected, of the 15 most significant pathways, 7 of the pathways were shared between OVCAR8-CP5 cells treated with the destabilizing anti-microtubule drugs vincristine or colchicine. We found multiple cell survival-associated pathways that were common to all three groups (Fig. 6A–C, Supplementary Fig. 5A–C). Upregulated pathways included the pro-survival pathways of nuclear factor kappa-light-chain-enhancer of activated B cells (NFκB) and tumor necrosis factor (TNF), both of which have been implicated in cisplatin resistance [35], [36]–37]. Downregulated pathways included DNA replication and the biosynthesis of amino acids, correlating with our results (Fig. 1D and E) showing that resistant cells proliferate more slowly. Interestingly, components of the cell cycle were downregulated, but to a lesser degree than DNA replication and biosynthesis of amino acids, suggesting that the upregulated pro-survival pathways enabled the cells to progress through the cell cycle without succumbing to mitotic arrest (Fig. 6D–F). We further verified our RNA-seq finding that aside from the upregulation of NFKB1 mRNA transcripts in OVCAR8-CP5 cells upon treatment with anti-microtubule drugs, the protein level was also upregulated (Fig. 7). In addition, unlike the untreated cells, OVCAR8-CP5 cells had increased NFκB activity upon treatment. More interestingly, NFκB expression and activity were highest in OVCAR-CP5 cells treated with anti-microtubule drugs compared to OVCAR8-CP1, suggesting a different response mechanism. Taken together, the results of our study indicate that although both OVCAR8-CP1 and OVCAR8-CP5 cells are resistant to cisplatin, the differing levels of resistance in the two populations may contribute to differing mechanisms of resistance that could potentially explain the conflicting data in the literature.
Fig. 6.
RNA sequencing analysis identifies cellular growth and survival pathways as being significantly impacted in isolated OVCAR8-CP5 subpopulation. Comparison of the most significant KEGG pathways enriched in (A) OVCAR8-CP5-PTX, (B) OVCAR8-CP5-COL, and (C) OVCAR8-CP5-VIN treatment groups. The GeneRatio of a cluster represents the ratio of the number of genes within one cluster to the number of differentially expressed genes. (D) Normalized enrichment scores (NES) for all five enriched pathways that are focused on in the three treatment sets. Hierarchical clustering analysis was used to generate a heatmap of z-scores displaying expression levels of gene transcripts related to (E) TNF and (F) NFκB signaling pathways.
Fig. 7.
Upregulation of NFκB expression and activity in OVCAR8-CP5 subpopulation treated with anti-microtubule drugs. Immunoblot images of OVCAR8-CP0, OVCAR8-CP1, and OVCAR8-CP5 cells treated with anti-microtubule drugs that were stained with anti-NFκB, anti-phospho- NFκB, and anti-GAPDH antibodies. GAPDH was used as a loading control.
Discussion
Several studies have suggested that microtubules play a role in cisplatin resistance, and ovarian cancer patients are often treated with paclitaxel, an anti-microtubule drug, in conjunction with cisplatin. However, previous findings have also suggested that there is no cross-resistance between cisplatin or carboplatin-resistant cells and anti-microtubule drugs. In this study, we observed the effects of the anti-microtubule drugs paclitaxel, vincristine, and colchicine on the parental cell line and on two different cisplatin-resistant ovarian cancer cell lines to better understand the reason for discrepancies in the literature.
As mentioned earlier, since various reports have characterized cisplatin-resistant cells as both cross-resistant and not cross-resistant to anti-microtubule drugs, we wanted to investigate the reason for these inconsistencies. Previous studies have typically reported IC50 values for a wide range of drugs, using those values to draw conclusions regarding cross-resistance to other drugs. In our analysis, we focus on a deeper understanding of three specific anti-microtubule drugs: paclitaxel, vincristine, and colchicine. For that purpose, we chose to focus our studies on one cell line to extensively study how cisplatin resistance may contribute to anti-microtubule drug cross-resistance. We determined that less-resistant cells (OVCAR8-CP1) acted in a manner similar to OVCAR8-CP0 in all of our studies using anti-microtubule drugs; both of these cell lines showed similar cross-resistance to paclitaxel, vincristine, or colchicine. In contrast, more-resistant cells (OVCAR8-CP5) included a subpopulation of cells that were simultaneously resistant to the same three anti-microtubule drugs. Since anti-microtubule drugs preferentially target rapidly proliferating cells [38] it is possible that the OVCAR8-CP5 cells are evading anti-microtubule-mediated killing by proliferating slowly.
While previous studies have reported that cisplatin-resistant cells are also cross-resistant to anti-microtubule drugs, the mechanism for this has not been clearly defined [8–10]. One potential useful study would be to generate resistant cells with a 2-drug combination (cisplatin and paclitaxel). However, we decided to not add cisplatin and paclitaxel at the same time because their half-lives are not the same. The combined treatment would select for cells for cisplatin (longer half-life) longer than paclitaxel and it would be very difficult to regulate and extrapolate what the underlying cause of resistance. We decided that it would be best to first better have a clear understanding of cisplatin resistant cells’ response to anti-microtubule drugs. For a future study, it would be useful to generate paclitaxel-resistant cells and then expose them to cisplatin to understand the cross-resistance of the drugs. In this study, in an attempt to understand why a subpopulation of OVCAR8-CP5 was able to survive higher concentrations of anti-microtubule drugs, we conducted efflux assays via flow cytometry analysis to determine if these cells expressed two common multidrug resistance proteins (ABCB1 or ABCG2). All three anti-microtubule drugs, paclitaxel, vincristine, and colchicine, used in this study are ABCB1 substrates [39]. It would be unusual, but not impossible, for cisplatin-resistant cells to display increased levels of ABCB1 [10] since cisplatin is not a substrate for ABCB1 transport. In addition, even though paclitaxel, vincristine, and colchicine are not ABCG2 substrates [40], some studies have suggested that ABCG2 plays a role in cross-resistance in cisplatin-resistant cells [9]. Despite these inferences, the efflux assays we conducted showed little to no increase in activity of ABCB1 (Supplementary Fig. 3A, Supplementary Fig. 4B) or ABCG2 (Supplementary Fig. 3B, Supplementary Fig. 4C) with increasing resistance to cisplatin. Previous reports have suggested ABCB1 or ABCG2 are involved in conferring cisplatin and anti-microtubule drug cross-resistance. Our studies indicate that although these transporters may account for some of the resistance, there are other underlying mechanisms that need further investigation.
Our RNA-sequencing data on OVCAR8-CP5 cells treated with anti-microtubule drugs show that cells treated with a microtubule-destabilizing agent (vincristine or colchicine) have a distinct transcriptional profile compared to cells treated with a microtubule-stabilizing agent (paclitaxel) (Fig. 5A). While vincristine and colchicine have similar mechanisms of action, treatment with these drugs leads to observable transcriptional differences that may be due to differences in their ability to bind to tubulins [41]. While colchicine takes about 4 h to reach maximum binding capacity to tubulin, vincristine takes less than 5 min. This may potentially affect downstream pathways. Furthermore, we found that NFκB, a master transcriptional regulator, is more highly activated in OVCAR8-CP5 cells. This also affects what pathways are activated, and may explain why gene enrichment analysis showing similar hits would also exhibit differences in enriched pathways, suggesting different response/resistance mechanisms in the OVCAR8-CP5 and OVCAR8-CP1 cells (Fig. 6A–C).
Our study first generated cisplatin resistant cells before exposing the cells to anti-microtubule drugs. It is very likely that the acquired resistance mechanisms from cisplatin selection is also involved in anti-microtubule drug resistance. As mentioned earlier, through gene set enrichment analysis of our RNA-sequencing data, we found multiple pathways related to cellular survival, such as the TNF signaling pathway and its downstream NFκB signaling pathway to be significantly enhanced upon treatment with all three of our anti-microtubule drugs. Several cisplatin resistance mechanisms have been described previously such as alteration of glutathione levels, altered transport of cisplatin, involvement of ATP7A and ATP7B, and activation of DNA repair pathways [2]. However, when cisplatin resistant cells were treated with anti-microtubule drugs, these resistance mechanisms were not the main drivers that we found to be involved in resistant mechanisms to anti-microtubule drugs (Fig. 6). In our models, we found genes associated with TNF/NFκB signaling to be enriched in cells that were also resistant to anti-microtubule drugs (Figs. 6 and 7).
The TNF cytokines and NFκB transcription factors have been implicated in several pathways including the regulation of inflammation, cellular survival, and proliferation. As the name suggests, TNF is largely known for its cytotoxic abilities; however, it is also an important inducer of NFκB, a transcription factor that, when activated, mediates the transcription of proteins involved in cell survival and proliferation, which could possibly mask the death-inducing capabilities of TNF [42].
Although not much has been published concerning the involvement of the cytoskeleton in NFκB regulation, it has been shown that short exposures to extremely high concentrations of anti-microtubule drugs can affect TNF induction of NFκB [43]. Jackman and colleagues showed that paclitaxel, a microtubule-stabilizer, in the absence of other inducers including TNF, can induce NFκB in a TNF-independent manner. Importantly, paclitaxel plus TNF does not show an additive effect, indicating that cells activate NFκB either by TNF or paclitaxel, but not both. Moreover, colchicine, a microtubule-destabilizing agent, inhibited TNF activation of NFκB. Since our data shows TNF and NFκB signaling is upregulated in cells treated with either a microtubule-stabilizing or -destabilizing agent, this suggests that the upregulation is a consequence of the cisplatin resistance, and not the 72 h anti-microtubule drug treatment.
Furthermore, the cell cycle pathway seems to be relatively unaffected in cells that survive anti-microtubule drug treatment, despite decreases in DNA replication and biosynthesis of amino acids (Fig. 6D–F). As anti-microtubule drugs are known to induce apoptosis in cells by inducing mitotic arrest [44], average enrichment scores for the cell cycle pathway indicate that although the cells were proliferating more slowly, as signified by decreases in DNA replication in the biosynthesis of amino acids, the anti-microtubule drugs were unable to exert their anti-mitotic effects on these cells. Although there are many potential mechanisms of resistance against anti-microtubule drugs, it is possible that cisplatin-induced NFκB activation through TNF signaling is contributing to the cross-resistance in these cells, as NFκB has been found to promote survival during mitotic cell cycle arrest [45]. Future studies including investigation of the mechanism of resistance to cisplatin may help define the role of cell survival mechanisms and whether these alterations directly affect anti-microtubule drug resistance.
CRediT authorship contribution statement
Ruchi P. Patel: Conceptualization, Investigation, Validation, Methodology, Writing - original draft, Writing - review & editing, Funding acquisition, Visualization, Formal analysis. Skyler Kuhn: Visualization, Writing - original draft, Formal analysis. Da Yin: Visualization, Formal analysis. Jordan Hotz: Investigation, Validation, Methodology. Frances A. Maher: Investigation, Methodology. Robert W. Robey: Investigation, Methodology, Formal analysis. Michael M. Gottesman: Conceptualization, Resources, Funding acquisition, Writing - review & editing. Sachi Horibata: Conceptualization, Investigation, Validation, Methodology, Writing - original draft, Writing - review & editing, Funding acquisition, Visualization, Formal analysis, Supervision, Project administration.
Declaration of Competing Interest
The authors of this study do not report any conflicts of interest.
Acknowledgments
Acknowledgments
We thank D. Kambach for her technical assistance in using the BioTek Lionheart FX machine, X. Bai and S. Lusvarghi for their technical assistance for visualization of the tubulin structure, L.M. Huff for technical advice, and G. Leiman for editorial assistance. We also thank T. McCloud for providing fumitremorgin C (FTC) and C. Annunziata and Y. Shibuya for their technical advice and NFκB antibody. We also appreciate the Center for Cancer Research (CCR) Genomics Core (S. Shema and Q. Wei) in Bethesda, Maryland for their help with assessing RNA integrity, the CCR Sequencing Facility (B. Tran, J. Shetty, and Y. Zhao) in Frederick, Maryland for performing RNA-sequencing on our samples, the CCR Collaborative Bioinformatics Resource (M. Cam and P. Jailwala) in Bethesda, Maryland for technical assistance, and the Confocal Core (L. Langston) in Bethesda, Maryland for technical assistance..
Funding
This work was supported by the Intramural Research Program at the National Cancer Institute (NCI) of the National Institutes of Health (ZIA BC 005598 and ZIA BC 010830), the Japan Society for the Promotion of Science (JSPS) (JSPS Research Fellowship for Japanese Biomedical and Behavioral Researcher at NIH to S.H.), and the joint Johns Hopkins University and NCI Molecular Target and Drug Discovery Fellowship Program (Fellowship to R.P.P.).
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.tranon.2020.100917.
Contributor Information
Ruchi P. Patel, Email: ruchi124@gmail.com.
Skyler Kuhn, Email: skyler.kuhn@nih.gov.
Da Yin, Email: da.yin@nih.gov.
Jordan Hotz, Email: jordan.hotz@nih.gov.
Frances A. Maher, Email: frances.t.maher@gmail.com.
Robert W. Robey, Email: robeyr@mail.nih.gov.
Michael M. Gottesman, Email: mgottesman@nih.gov.
Sachi Horibata, Email: horibatas@nih.gov.
Appendix. Supplementary materials
References
- 1.Fichtinger-Schepman A.M., van Oosterom A.T., Lohman P.H., Berends F. cis-Diamminedichloroplatinum(II)-induced DNA adducts in peripheral leukocytes from seven cancer patients: quantitative immunochemical detection of the adduct induction and removal after a single dose of cis-diamminedichloroplatinum(II) Cancer Res. 1987;47:3000–3004. [PubMed] [Google Scholar]
- 2.Shen D.W., Pouliot L.M., Hall M.D., Gottesman M.M. Cisplatin resistance: a cellular self-defense mechanism resulting from multiple epigenetic and genetic changes. Pharmacol. Rev. 2012;64:706–721. doi: 10.1124/pr.111.005637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fink D., Nebel S., Aebi S., Zheng H., Cenni B., Nehme A., Christen R.D., Howell S.B. The role of DNA mismatch repair in platinum drug resistance. Cancer Res. 1996;56:4881–4886. [PubMed] [Google Scholar]
- 4.Kelland L.R., Mistry P., Abel G., Loh S.Y., O'Neill C.F., Murrer B.A., Harrap K.R. Mechanism-related circumvention of acquired cis-diamminedichloroplatinum(II) resistance using two pairs of human ovarian carcinoma cell lines by ammine/amine platinum(IV) dicarboxylates. Cancer Res. 1992;52:3857–3864. [PubMed] [Google Scholar]
- 5.Mistry P., Kelland L.R., Abel G., Sidhar S., Harrap K.R. The relationships between glutathione, glutathione-S-transferase and cytotoxicity of platinum drugs and melphalan in eight human ovarian carcinoma cell lines. Br. J. Cancer. 1991;64:215–220. doi: 10.1038/bjc.1991.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hubaux R., Thu K.L., Vucic E.A., Pikor L.A., Kung S.H., Martinez V.D., Mosslemi M., Becker-Santos D.D., Gazdar A.F., Lam S., Lam W.L. Microtubule affinity-regulating kinase 2 is associated with DNA damage response and cisplatin resistance in non-small cell lung cancer. Int. J. Cancer. 2015;137:2072–2082. doi: 10.1002/ijc.29577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li L., Huang X., Huang R., Gou S., Wang Z., Wang H. Pt(IV) prodrugs containing microtubule inhibitors displayed potent antitumor activity and ability to overcome cisplatin resistance. Eur. J. Med. Chem. 2018;156:666–679. doi: 10.1016/j.ejmech.2018.07.016. [DOI] [PubMed] [Google Scholar]
- 8.Hamaguchi K., Godwin A.K., Yakushiji M., O'Dwyer P.J., Ozols R.F., Hamilton T.C. Cross-resistance to diverse drugs is associated with primary cisplatin resistance in ovarian cancer cell lines. Cancer Res. 1993;53:5225–5232. [PubMed] [Google Scholar]
- 9.Iwasaki I., Sugiyama H., Kanazawa S., Hemmi H. Establishment of cisplatin-resistant variants of human neuroblastoma cell lines, TGW and GOTO, and their drug cross-resistance profiles. Cancer Chemother. Pharmacol. 2002;49:438–444. doi: 10.1007/s00280-002-0452-4. [DOI] [PubMed] [Google Scholar]
- 10.Stordal B., Hamon M., McEneaney V., Roche S., Gillet J.P., O'Leary J.J., Gottesman M., Clynes M. Resistance to paclitaxel in a cisplatin-resistant ovarian cancer cell line is mediated by P-glycoprotein. PLoS One. 2012;7:e40717. doi: 10.1371/journal.pone.0040717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Januchowski R., Wojtowicz K., Sujka-Kordowska P., Andrzejewska M., Zabel M. MDR gene expression analysis of six drug-resistant ovarian cancer cell lines. Biomed. Res. Int. 2013;2013 doi: 10.1155/2013/241763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Parness J., Horwitz S.B. Taxol binds to polymerized tubulin in vitro. J. Cell Biol. 1981;91:479–487. doi: 10.1083/jcb.91.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brunton L.L., Hilal-Dandan R., Knollman B.C. Pharmacotherapy of neoplastic disease. In: Shanahan J.F., Lebowitz H., editors. Goodman & Gilman's: The Pharmacological Basis of Therapetuics. McGraw-Hill Education; 2018. Eds. [Google Scholar]
- 14.Leung Y.Y., Yao Hui L.L., Kraus V.B. Colchicine–Update on mechanisms of action and therapeutic uses. Semin. Arthritis Rheum. 2015;45:341–350. doi: 10.1016/j.semarthrit.2015.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chi S., Xie W., Zhang J., Xu S. Theoretical insight into the structural mechanism for the binding of vinblastine with tubulin. J. Biomol. Struct. Dyn. 2015;33:2234–2254. doi: 10.1080/07391102.2014.999256. [DOI] [PubMed] [Google Scholar]
- 16.Massarotti A., Coluccia A., Silvestri R., Sorba G., Brancale A. The tubulin colchicine domain: a molecular modeling perspective. ChemMedChem. 2012;7:33–42. doi: 10.1002/cmdc.201100361. [DOI] [PubMed] [Google Scholar]
- 17.Nogales E., Wolf S.G., Downing K.H. Structure of the alpha beta tubulin dimer by electron crystallography. Nature. 1998;391:199–203. doi: 10.1038/34465. [DOI] [PubMed] [Google Scholar]
- 18.Rao S., He L., Chakravarty S., Ojima I., Orr G.A., Horwitz S.B. Characterization of the Taxol binding site on the microtubule. Identification of Arg(282) in beta-tubulin as the site of photoincorporation of a 7-benzophenone analogue of Taxol. J. Biol. Chem. 1999;274:37990–37994. doi: 10.1074/jbc.274.53.37990. [DOI] [PubMed] [Google Scholar]
- 19.Kim S., Chen J., Cheng T., Gindulyte A., He J., He S., Li Q., Shoemaker B.A., Thiessen P.A., Yu B., Zaslavsky L., Zhang J., Bolton E.E., update PubChem 2019. improved access to chemical data. Nucleic Acids Res. 2019;47:D1102–D1109. doi: 10.1093/nar/gky1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haley J., Tomar S., Pulliam N., Xiong S., Perkins S.M., Karpf A.R., Mitra S., Nephew K.P., Mitra A.K. Functional characterization of a panel of high-grade serous ovarian cancer cell lines as representative experimental models of the disease. Oncotarget. 2016;7:32810–32820. doi: 10.18632/oncotarget.9053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Robey R.W., Lin B., Qiu J., Chan L.L., Bates S.E. Rapid detection of ABC transporter interaction: potential utility in pharmacology. J. Pharmacol. Toxicol. Methods. 2011;63:217–222. doi: 10.1016/j.vascn.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Robey R.W., Shukla S., Finley E.M., Oldham R.K., Barnett D., Ambudkar S.V., Fojo T., Bates S.E. Inhibition of P-glycoprotein (ABCB1)- and multidrug resistance-associated protein 1 (ABCC1)-mediated transport by the orally administered inhibitor, CBT-1((R)) Biochem. Pharmacol. 2008;75:1302–1312. doi: 10.1016/j.bcp.2007.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Daley T., Smith A.D. Predicting the molecular complexity of sequencing libraries. Nat. Methods. 2013;10:325–327. doi: 10.1038/nmeth.2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal. 2011;17:10–12. [Google Scholar]
- 25.Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li B., Dewey C.N. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harrow J., Frankish A., Gonzalez J.M., Tapanari E., Diekhans M., Kokocinski F., Aken B.L., Barrell D., Zadissa A., Searle S., Barnes I., Bignell A., Boychenko V., Hunt T., Kay M., Mukherjee G., Rajan J., Despacio-Reyes G., Saunders G., Steward C., Harte R., Lin M., Howald C., Tanzer A., Derrien T., Chrast J., Walters N., Balasubramanian S., Pei B., Tress M., Rodriguez J.M., Ezkurdia I., van Baren J., Brent M., Haussler D., Kellis M., Valencia A., Reymond A., Gerstein M., Guigo R., Hubbard T.J. GENCODE: the reference human genome annotation for the ENCODE project. Genome Res. 2012;22:1760–1774. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Law C.W., Chen Y., Shi W., Smyth G.K. voom: precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 2014;15:R29. doi: 10.1186/gb-2014-15-2-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smyth G.K. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 2004;3 doi: 10.2202/1544-6115.1027. Article 3. [DOI] [PubMed] [Google Scholar]
- 30.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., Mesirov J.P. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fabregat A., Jupe S., Matthews L., Sidiropoulos K., Gillespie M., Garapati P., Haw R., Jassal B., Korninger F., May B., Milacic M., Roca C.D., Rothfels K., Sevilla C., Shamovsky V., Shorser S., Varusai T., Viteri G., Weiser J., Wu G., Stein L., Hermjakob H., D'Eustachio P. The reactome pathway knowledgebase. Nucleic Acids Res. 2018;46:D649–D655. doi: 10.1093/nar/gkx1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liberzon A., Subramanian A., Pinchback R., Thorvaldsdottir H., Tamayo P., Mesirov J.P. Molecular signatures database (MSigDB) 3.0. Bioinformatics. 2011;27:1739–1740. doi: 10.1093/bioinformatics/btr260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Michalak M., Lach M.S., Antoszczak M., Huczynski A., Suchorska W.M. Overcoming resistance to platinum-based drugs in ovarian cancer by salinomycin and its derivatives – an in vitro study. Molecules. 2020;25:537. doi: 10.3390/molecules25030537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Duan G., Tang Q., Yan H., Xie L., Wang Y., Zheng X.E., Zhuge Y., Shen S., Zhang B., Zhang X., Wang J., Wang W., Zou X. A strategy to delay the development of cisplatin resistance by maintaining a certain amount of cisplatin-sensitive cells. Sci. Rep. 2017;7:432. doi: 10.1038/s41598-017-00422-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Almeida L.O., Abrahao A.C., Rosselli-Murai L.K., Giudice F.S., Zagni C., Leopoldino A.M., Squarize C.H., Castilho R.M. NFkappaB mediates cisplatin resistance through histone modifications in head and neck squamous cell carcinoma (HNSCC) FEBS Open Bio. 2014;4:96–104. doi: 10.1016/j.fob.2013.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lagunas V.M., Melendez-Zajgla J. Nuclear factor-kappa b as a resistance factor to platinum-based antineoplasic drugs. Met. Based Drugs. 2008;2008 doi: 10.1155/2008/576104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zuo J., Zhao M., Liu B., Han X., Li Y., Wang W., Zhang Q., Lv P., Xing L., Shen H., Zhang X. TNFalphamediated upregulation of SOD2 contributes to cell proliferation and cisplatin resistance in esophageal squamous cell carcinoma. Oncol. Rep. 2019;42:1497–1506. doi: 10.3892/or.2019.7252. [DOI] [PubMed] [Google Scholar]
- 38.Mukhtar E., Adhami V.M., Mukhtar H. Targeting microtubules by natural agents for cancer therapy. Mol. Cancer Ther. 2014;13:275–284. doi: 10.1158/1535-7163.MCT-13-0791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Robey R.W., Pluchino K.M., Hall M.D., Fojo A.T., Bates S.E., Gottesman M.M. Revisiting the role of ABC transporters in multidrug-resistant cancer. Nat. Rev. Cancer. 2018;18:452–464. doi: 10.1038/s41568-018-0005-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stacy A.E., Jansson P.J., Richardson D.R. Molecular pharmacology of ABCG2 and its role in chemoresistance. Mol. Pharmacol. 2013;84:655–669. doi: 10.1124/mol.113.088609. [DOI] [PubMed] [Google Scholar]
- 41.Owellen R.J., Owens A.H., Jr., Donigian D.W. The binding of vincristine, vinblastine and colchicine to tubulin. Biochem. Biophys. Res. Commun. 1972;47:685–691. doi: 10.1016/0006-291x(72)90546-3. [DOI] [PubMed] [Google Scholar]
- 42.Wajant H., Pfizenmaier K., Scheurich P. Tumor necrosis factor signaling. Cell Death Differ. 2003;10:45–65. doi: 10.1038/sj.cdd.4401189. [DOI] [PubMed] [Google Scholar]
- 43.Jackman R.W., Rhoads M.G., Cornwell E., Kandarian S.C. Microtubule-mediated NF-kappaB activation in the TNF-alpha signaling pathway. Exp. Cell Res. 2009;315:3242–3249. doi: 10.1016/j.yexcr.2009.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van Vuuren R.J., Visagie M.H., Theron A.E., Joubert A.M. Antimitotic drugs in the treatment of cancer. Cancer Chemother. Pharmacol. 2015;76:1101–1112. doi: 10.1007/s00280-015-2903-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mistry P., Deacon K., Mistry S., Blank J., Patel R. NF-kappaB promotes survival during mitotic cell cycle arrest. J. Biol. Chem. 2004;279:1482–1490. doi: 10.1074/jbc.M310413200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.