Abstract
Ciliary neurotrophic factor (CNTF) is a member of interleukin-6 type cytokine family. The CNTF receptor complex is a heterodimer including gp130 and CNTF receptor α (CNTFRɑ) proteins triggering the activation of multiple intracellular signaling pathways including AKT/PI3K, MAPK/ERK and Jak/STAT pathways. At present no data are available on the localization of CNTF and CNTFRɑ in prostate as well as on the role of CNTF in this organ. In this study we have analyzed the localization of CNTF and CNTFRɑ by immunohistochemistry and we have used PWR-1E cell line as a model for normal glandular cell to investigate the role of this cytokine. Our results show that CNTF and CNTFRɑ are expressed in the staminal compart of the prostate and that CNTF selectively inhibits ERK pathway. In conclusion, we suggest that CNTF could be considered as key molecule to maintain the epithelium homeostasis via pERK downregulation by an autocrine mechanism. Further CNTF studies in prostate cancer could be useful to verify the potential role of this cytokine in carcinogenesis.
Key words: PWR-1E cell line, immunohistochemistry, Western blotting, cytokine
Introduction
Ciliary neurotrophic factor (CNTF) was originally discovered in intraocular tissue of embryonic chick, having a role in promoting the survival of ciliary ganglion neurons during embryonic development.1 It was subsequently identified also in mammals neuronal and non-neuronal cell types, showing trophic actions.2 CNTF is a member of interleukin (IL)-6 type cytokine family including leukemia inhibitory factor (LIF), interleukin-6 (IL-6), IL-11, cardiotrophin 1 (CT-1), cardiotrophin-like cytokine (CLC), interleukin-27 (IL-27), and oncostatin M (OSM).3 All IL-6 type family cytokines, need additional membrane-bound non-signaling receptors to activate signal transduction via gp130,4-7 in particular CNTF acts by using the CNTF receptor α (CNTFRɑ).6,8,9 The formation of the CNTF receptor complex triggers the activation of multiple intracellular signaling pathways such as MAPK/ERK10, AKT/PI3K11 and Jak/STAT12 pathways, which mediate various biological effects, such as survival and/or differentiation in different cell types.13-25 Most of the studies involving CNTF has been carried out on nervous system and adipose tissues concerning food intake and the association among obesity, hyperglycemia, hyperinsulinemia, and hyperlipidemia.26-29 Recently, Chen et al.30 provided new insights into the role of CNTF on the migration of corneal epithelial stem/progenitor cells and on AKT signaling pathway. At the best of our knowledge, less is known about the role of CNTF and its receptor CNTFRɑ in epithelial prostate tissues. The normal prostatic glandular epithelium consists of three cell types, i.e., basal, luminal-secretory and neuroendocrine cells expressing specific markers and organized in a double layer surrounded by stromal tissue.31-36
The most widely accepted explanation for the balanced relationship between basal and secretory cells is due to the presence of non-altered stem cells in the basal cell compartment. The alteration of these normal stem cells leads to tumorigenesis.37-40 At present, molecular and biological basis as well as the pathogenesis of prostate cancer are still a matter of debate although prostate cancer, i.e., adenocarcinoma, is the second most common malignancy in males and the fifth leading cause of cancer mortality.41-43 Generally, prostate adenocarcinoma is clinically localized and not aggressive. For patients with advanced prostate adenocarcinoma, androgen deprivation therapy is usually used, however, a part of these patients can have poor clinical outcomes for onset of castration resistant prostate cancer.44 Thus, it is crucial to investigate the molecules and factors playing a pivotal role in processes such as cellular proliferation and migration. So, given the remarkable effects of CNTF on stem cell30 and its potential on cell migration, we performed a series of experiments with the aim: i) to identify the presence of CNTF and its receptor CNTFRɑ in human prostate tissues; ii) to characterize the signaling systems modulated by CNTF treatments using cultured prostate cells. We have investigated ERK, AKT and STAT3 pathways because they have an important role in triggering prostate cancer45-48 and it has been proven that these three pathways can be involved in CNTF signaling through its receptor CNTFRɑ.10-12,30 In addition, these pathways mediate processes such as cellular proliferation, survival, differentiation, migration that play a pivotal role in tissue homeostasis and in the cancer onset.13-25
Materials and Methods
Tissue collection
In this study we analyzed a total of 15 normal human prostate samples: 5 from benign prostatic hyperplasia (BPH), 5 normallooking samples from radical prostatectomy (NL-RP) and 5 from cystoprostatectomy (CYP). A pathologist (RMa) reviewed the samples stained with hematoxylin and eosin for selecting the samples used in this study.
All the samples were obtained from pathological files from the Pathology Services of the Polytechnic University of the Marche Region-United Hospitals. The procedures followed for the collection of samples were in accordance with the Helsinki Declaration of 1975, as revised in 2013.
Immunohistochemistry
All prostate samples were fixed in 10% neutral buffered formalin and routinely processed for paraffin embedding. Immunohistochemical staining was performed as previously described.49 Briefly, after dewaxing, paraffin sections were rinsed in phosphate buffered saline (PBS), incubated with 3% hydrogen peroxide for 40 min to block endogenous peroxidase. Pre-treatment by heat in 10 mM citrate buffer, pH 6.0 for 5 min was used for CNTF and p63 while pre-treatment by 100 ng/mL Proteinase K (Sigma-Aldrich, St. Louis, MO, USA) 5 min at 37°C was used for CNTFRɑ. After pre-treatment, sections were rinsed with PBS and incubated with normal horse serum (Vector Laboratories, Burlingame, CA, USA) diluted 1:75 in PBS for 1 h at room temperature (RT). Sections were then incubated with anti-CNTFRɑ, anti-CNTF and anti-p63 (Table 1) primary antibodies diluted in PBS, overnight at 4°C. After a thorough rinse in PBS, sections were incubated with the appropriate biotinylated secondary antibody (Vector Laboratories) diluted 1:200 v/v solution for 30 min at RT. Vectastain ABC Kit (Vector Laboratories) for 1h at RT and 3′,3′- diaminobenzidine hydrochloride (Sigma-Aldrich) were used to develop the immunohistochemistry reaction. Sections were counterstained with Mayer’s hematoxylin, dehydrated and mounted using Eukitt solution (Kindler GmbH and Co., Freiburg, Germany). Negative controls were performed by omitting the first or secondary antibody for all the immunohistochemical reactions performed in this study. p63 was used as nuclear marker of basal prostate cells. Nervous tissues (fibers and ganglions) are used as positive internal control for CNTF and CNTFRɑ.50-52
Table 1.
Primary antibodies used in this study.
| Antibody | IHC | WB | IF | Company |
|---|---|---|---|---|
| pAb Rabbit anti-human CNTF (#ab190985) | 1:500 | // | 1:100 | Abcam, Cambridge, UK |
| pAb Rabbit anti-human CNTFRɑ (#PA5-45053) | // | 1:400 | // | Thermo Fisher Scientific, Waltham, USA |
| mAb Mouse anti-human CNTFRɑ (#ab89333) | 1:150 | // | 1:100 | Abcam, Cambridge, UK |
| mAb Rabbit anti-human pAKT (#4060) | // | 1: 1000 | // | Cell Signaling Technology, Danvers, USA |
| pAb Rabbit anti-human AKT (#9272) | // | 1: 1000 | // | Cell Signaling Technology, Danvers, USA |
| mAb Rabbit anti-human pERK1/2 (#4377) | // | 1:800 | // | Cell Signaling Technology, Danvers, USA |
| mAb Rabbit anti-human ERK1/2 (#4695) | // | 1:1000 | // | Cell Signaling Technology, Danvers, USA |
| mAb Mouse anti-human pSTAT3 (#4113) | // | 1: 800 | // | Cell Signaling Technology, Danvers, USA |
| mAb Rabbit anti-human STAT3 (#4904) | // | 1: 1000 | // | Cell Signaling Technology, Danvers, USA |
| mAb Mouse anti-human p63 (#M7317) | 1:50 | // | // | DAKO, Glostrup, Denmark |
mAb, monoclonal antibody; pAb, polyclonal antibody; IHC, immunohistochemistry; WB, Western blotting; IF, immunofluorescence.
Cell culture
Normal human prostate epithelial cells PWR-1E (ATCC/LGC Standards, Manassas, VA, USA) were cultured in serum-free keratinocyte cell culture media (K-SFM) supplemented with human EGF (5 ng/mL), bovine pituitary extract (25 mg/mL), 100 U/mL penicillin and streptomycin (Gibco, Thermo Fisher Scientific, MA, USA) at 37°C, 95% humidity and 5% CO2. The medium was changed 3 times a week and cells were split 1:4 every 3/4 days.
Immunofluorescence
PWR-1E cells were washed in Dulbecco’s PBS (Lifetechnology, Monza, Italy), fixed in 4% paraformaldehyde in PBS for 10 min at RT, and permeabilized in PBS 0.1 M added with 0.1% Triton X-100 (Sigma, Milan, Italy) for 5 min. After washing in PBS at RT, cells were blocked with 10% Normal Donkey Serum (Jackson ImmunoResearch, West Grove, PA, USA) in PBS 0.1 M and incubated overnight at 4° C with the anti-human CNTF (1:100) and CNTFRɑ (1:150) antibodies (Table 1). Cells were then washed three times in PBS and incubated with the FITC-conjugated donkey anti-rabbit (for CNTF) and TRITC-conjugated anti-mouse (for CNTFRɑ) IgG secondary antibodies (both from Jackson ImmunoResearch) for 30 min at RT. TOTO3 probe was used for nuclear staining. Finally, the slides were cover-slipped with propyl gallate and evaluated with a Leica TCS-SL spectral confocal microscope.
CNTFRɑ detection in cell lines by Western blotting
Once PWR-1E cells reached 80% confluence, cells were lysed by using the following lysis buffer: 0.1M PBS, 0.1% (w/v) SDS, 1% (w/w) NONIDET-P40, 1mM (w/v) Na orthovanadate, 1mM (w/w) PMSF (phenyl methane sulfonyl fluoride), 12 mM (w/v) Na deoxycholate, 1.7 μg/mL Aprotinin, pH 7.5. Cell lysates were centrifuged at 20,000 g for 20 min at 4°C and the supernatants were aliquoted and stored at −80°C. Viable counts using the Trypan blue dye exclusion test were routinely performed. All experiments were performed in duplicate and were repeated at least three times. The proteins concentrations were determined by a Bradford protein assay (Bio-Rad Laboratories, Milan, Italy). All protein samples were analyzed by Western blotting technique. They were fractionated on 10% SDS-polyacrylamide gels (SDS-PAGE) and electrophoretically transferred (Trans-Blot® Turbo™ Transfer System; Bio-Rad Laboratories Inc., Richmond, CA, USA) to nitrocellulose membranes, and subjected to Western blot analysis. Non-specific protein binding was blocked with 5% (w/v) non-fat-dried milk (Bio-Rad Laboratories) in Tris-buffered saline (TBS/0.05% Tween 20 (TBS-T) for 1 h. Blots were incubated with 1:400 anti- CNTFRɑ (Thermo Fisher Scientific, Waltham, MA, USA) primary antibody overnight at 4°C. After washing, blots were incubated with anti-rabbit secondary antibody conjugated with horseradish peroxidase (Amersham Italia Srl, Milan, Italy) diluted 1:5000 in TBS-T. Detection of bound antibodies was performed with the Clarity Western ECL Substrate (Bio-Rad Laboratories) and images were acquired with Chemidoc (Bio-Rad Laboratories). Bands were analyzed using the ImageJ software (https://imagej.nih.gov/ij/ download.html) for quantification, and normalization was completed using β-actin band intensities.
Figure 1.
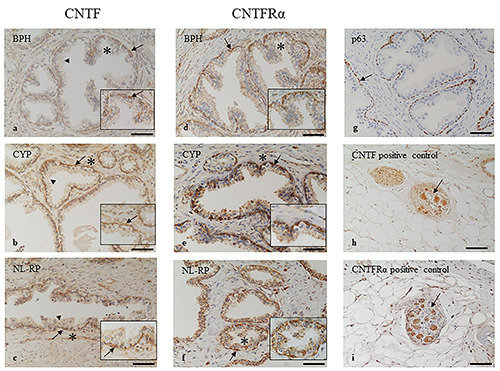
Immunohistochemistry localization of CNTF and CNTFRa in prostate samples. CNTF is highly expressed in basal layer (arrows) of BPH (a), CYP (b) and NL-RP (c) while the secretory layer (arrowheads) is mainly negative. The stromal tissues are weakly stained for CNTF in all samples analysed. CNTFRa is highly expressed in basal layer (arrows) of BPH (d), CYP (e) and NL-RP (f) while the other tissues are mainly negative. The basal layer of glandular epithelium is identified by p63 marker (arrow, g). Pictures in h) and i) show a ganglion (arrow) positive for CNTF (h) and for CNTFR (i) used as positive internal controls. The insets show higher magnification of the area indicated by asterisk, scale bars: 30 μm. Scale bars: a,b,c,d,f,g) 100 μm; e,h,i) 200 μm.
CNTF signaling pathway in PWR-1E cell line
After verified the presence of the CNTFRɑ in PWR-1E cells, a dose/responsive curve was performed to test the best CNTF concentration showing a significant response for cellular treatments. These cells were treated with 0, 2, 10 and 20 ng/ml by recombinant human CNTF (rhCNTF) for 15 min, to detect which of the following signaling pathway was trigged: pERK/ERK, pAKT/AKT and pSTAT3/STAT3. These signaling pathways were analyzed by Western blotting as above described using the primary antibodies shown in Table 1.
Results were calculated in arbitrary units (AU) and reported as bars of a histogram. pERK1/2, pAKT and pSTAT3 quantities were normalized using total ERK1/2, AKT and STAT3 respectively.
Statistical analysis
Data represent the mean ± SD, and were analyzed for statistical significance (p<0.05) using Student’s t-test by Graphpad Prism ver. 8 program.
Results
CNTF and CNTFRɑ localization in human prostate tissue by immunohistochemistry
CNTF and CNTFRɑ were localized mainly in the basal layer of the prostate epithelium in BPH (Figure 1 a,d), CYP (Figure 1 b,e) and NL-RP (Figure 1 c,f), while the secretory luminal layer was mainly negative or very weakly positive for the two molecules in all samples analyzed (Figure 1 a-f). Stromal components of prostate tissue were weakly positive for CNTF (Figure 1 a-c) and mainly negative for CNTFRɑ (Figure 1 d-f).
Figure 2.
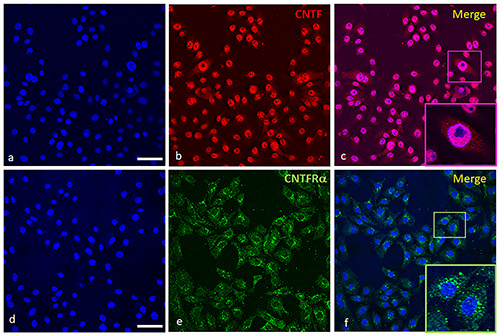
Immunofluorescence of CNTF and CNTFRɑ in PWR-1E cell line. In (a) and (d) are stained the nuclei in blue. CNTF (b, red staining) is localized mainly in nuclei while the cytoplasm is weakly stained for CNTF (b,c). In (c), the nucleoli are negative as depicted in the inset (c, Merge). CNTFRɑ (e, green staining) is localized in the cytoplasm of the cells as shown in (f ) and it is especially intense in the perinuclear region (see inset in f, Merge). Scale bars: a,b,c,d,e,f ) 50 μm; Insets in c,f ) 17 μm.
CNTF and CNTFRɑ localization in PWR-1E cell line by immunofluorescence
In Figure 2 the immunopositivity for CNTF is mainly nuclear (on chromatin, the nucleoli are negative; inset in panel c), while the cytoplasm is weakly stained (panels b and c). The green signal for CNTFRɑ is cytoplasmic (panels e and f) and especially intense in the perinuclear region (inset in panel f).
CNTF signaling pathways analyzed by Western blotting
rhCNTF induced pERK de-phosphorylation at 10 ng/mL and 20 ng/mL, whereas 2 ng/mL of rhCNTF was insufficient to induce de-phosphorylation of pERK (Figure 3a, see the representative Western blotting image). The expression level of pERK was very weakly or negative detected performing Western blotting analyses. The quantitative analysis of pERK expression levels normalized on ERK expression levels was represented by the histogram on the right side of the Western blotting (Figure 3a). The statistical analysis showed a significant pERK decrease after rhCNTF treatments at 10 ng/mL (p=0.008) and 20 ng/mL (p=0.006) (Figure 3a). RhcNTF did not have any effect on phosphorylation/de-phosphorylation of pAKT/AKT (Figure 3b) and pSTAT3/STAT3 (Figure 3c). The histograms representing the quantitative analysis of pAKT expression levels normalized by AKT (Figure 3b) and pSTAT3 expression levels normalized by STAT3 (Figure 3c) did not show any statistical differences (p>0.05) among treatments at the different rhCNTF concentrations (see pAKT/AKT and pSTAT3/STAT3 histograms in Figure 3b c, respectively).
Discussion
This is the first study that describes the localization of CNTF and its receptor CNTFRɑ in human prostate tissues, and investigates the activation pathways of CNTF in human normal prostate cell line.
In order to characterize the location of CNTF and its receptor CNTFRɑ we have used prostate tissues obtained from different zones of the prostate, i.e. transitional and peripherical zones. In particular, we analyzed benign prostatic hyperplasia (BPH) from transitional zone and normal-looking samples, from both radical prostatectomy (NL-RP) and cystoprostatectomy (CYP), from peripheral zone where at least 75% of cancers originate.53 The normal glandular epithelium of prostate is highly organized containing, basal cells, which account for ~40% of total epithelial cell numbers, and a layer of luminal-secretory cells that make up the rest of the epithelium.54 We have demonstrated by immunohistochemistry the presence of CNTF and CNTFRɑ in the basal cell layer in the prostatic normal glandular epithelium, in which reside stem cells.55 Under normal conditions the basal cells through differentiation replace the terminally differentiated luminal cells that regularly shed in the lumen of the gland, while an impaired differentiation of the normal epithelial lineage is present in prostate cancer. 55,56 It has been supposed that the genesis of prostate cancers takes into account both genetic and epigenetic changes, such as DNA methylation, chromatin remodeling and transcriptional regulation, involving a single cell or a cancerous progenitor cell which sources a dysregulated differentiation program to form the tumor.54,57,58 Although the prostate cancer presents a dominant luminal phenotype, recently, a progenitor cell on the basal layer seems to be an increasingly supported alternative for the origin of prostate cancer and basal to luminal cellular differentiation appears a critical event.54,57
Multiple signaling pathways normally involved in prostate differentiation can be also linked to prostate cancer development, including the MAPK/ERK, AKT/PI3K, and Jak/STAT pathways.45-48 In addition, it has been reported that alteration of the axis PTEN/AKT/PI3K together with activation of RAS/MAPK/ERK signaling are correlated with prostate cancer progression and metastasis and that RAS/MAPK/ERK pathway alone is significantly elevated in both primary and metastatic lesion.59,60 Moreover, it is known that the activation of Jak/ STAT3 pathway by IL-6 is crucial for maintenance of tumor progenitor cell phenotype and that CNTF binding to its receptor CNTFRɑ can active the three pathways described above.10-12,29,30
Figure 3.
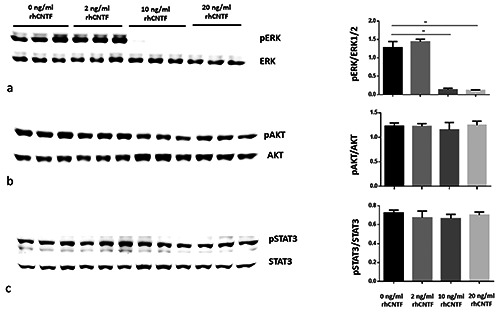
Dose-dependent response of PWR-1E normal prostate cell line to rhCNTF. Densitometrical analysis of the bands show a significant downregulation of pERK1/2 (a) while pAKT (b) and pSTAT3 (c) do not show any significant modulation. Results were calculated in arbitrary units (AU) and reported in the histograms. Note the significant decrease (**) of pERK1/2 in PWR-1E cell lines treated with 10 and 20 ng/mL rhCNTF compared to the untreated control (mean ± SD; **p<0.01).
Our in vitro results showed that AKT and STAT3 phosphorylation were not modified under rhCNTF treatments while rhCNTF selectively inhibits ERK pathway, in fact we have detected a decrease of ERK activation, i.e. the decrease of pERK under rhCNTF stimulation, in normal prostate cellular model. Our data are in line with that of Nickols and colleagues61 showing a correlation of prostate cancer recurrence with the increase of pERK expression levels. Moreover, the same authors demonstrated that the activation of ERK is characteristic of castration resistant prostate cancer while others found a correlation between increased ERK phosphorylation with both stage T and Gleason grade of prostate cancer.61,62 So, we can hypothesize that inhibition of ERK phosphorylation can decrease migration and invasion processes downstream of this pathway. As a result, we can speculate that CNTF promotes epithelium homeostasis via pERK downregulation by an autocrine mechanism in normal prostate glandular basal compartment and that dysregulation of this mechanism could contribute to the onset of prostate cancer. All these findings suggest that further CNTF studies in prostate cancer are needed to verify the potential role of this cytokine in carcinogenesis.
Acknowledgments
Funding for this work was provided by Scientific Research Grant from Università Politecnica delle Marche (2018-2019) to DM, RMa and by FFABR 2017 to RMa. GT is a recipient of a fellowship Starting Grant 2018 of the Italian Ministry of Health.
References
- 1.Adler R, Landa KB, Manthorpe M, Varon S. Cholinergic neuronotrophic factors: intraocular distribution of trophic activity for ciliary neurons. Science 1979;204:1434-6. doi: 10.1126/science.451576 [DOI] [PubMed] [Google Scholar]
- 2.Sendtner M, Carrol P, Holtmann B, Hughes RA, Thoenen H. Ciliary neurotrophic factor. J Neurobiol 1994;25:1436-53. doi: 10.1002/neu.480251110 [DOI] [PubMed] [Google Scholar]
- 3.Lin LF, Mismer D, Lile JD, Armes LG, Butler ET 3rd, Vannice JL, et al. Purification, cloning and expression of ciliary neurotrophic factor (CNTF). Science 1989;246:1023-5. doi: 10.1126/science.2587985 [DOI] [PubMed] [Google Scholar]
- 4.Garbers C, Hermanns HM, Schaper F, Müller-Newen G, Grötzinger J, Rose-John S, et al. Plasticity and cross-talk of interleukin 6-type cytokines. Cytokine Growth Factor Rev 2012;23:85-97. doi: 10.1016/j.cytogfr.2012.04.001 [DOI] [PubMed] [Google Scholar]
- 5.Hirano T, Matsuda T, Nakajima K. Signal transduction through gp130 that is shared among the receptors for the interleukin 6 related cytokine subfamily. Stem Cells 1994;12:262-77. doi: 10.1002/stem.5530120303 [DOI] [PubMed] [Google Scholar]
- 6.Ip NY, Nye SH, Boulton TG, Davis S, Taga T, Li Y, Birren SJ, et al. CNTF and LIF act on neuronal cells via shared signaling pathways that involve the IL-6 signal transducing receptor component gp130. Cell 1992;69:1121-32. doi: 10.1016/0092-8674(92)90634-o [DOI] [PubMed] [Google Scholar]
- 7.Jones SA, Scheller J, Rose-John S. Therapeutic strategies for the clinical blockade of IL-6/gp130 signaling. J Clin Invest 2011;121:3375-83. doi: 10.1172/JCI57158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rose-John S. Interleukin-6 family cytokines. Schreiber Cold Spring Harb Perspect Biol 2018;10:a028415. doi: 10.1101/cshperspect. a028415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wagener EM, Aurich M, Aparicio-Siegmund S, Floss DM, Garbers C, Breusing K, et al. The amino acid exchange R28E in ciliary neurotrophic factor (CNTF) abrogates interleukin-6 receptor-dependent but retains CNTF receptor-dependent signaling via glycoprotein 130 (gp130)/leukemia inhibitory factor receptor (LIFR). J Biol Chem 2014;289:18442-50. doi: 10.1074/jbc.M114.568857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boulton TG, Stahl N, Yancopoulos GD. Ciliary neurotrophic factor/ leukemia inhibitory factor/interleukin 6/oncostatin M family of cytokines induces tyrosine phosphorylation of a common set of proteins overlapping those induced by other cytokines and growth factors. J Biol Chem 1994;269:11648-55. [PubMed] [Google Scholar]
- 11.Oh H, Fujio Y, Kunisada K, Hirota H, Matsui H, Kishimoto T, et al. Activation of phosphatidylinositol 3-kinase through glycoprotein 130 induces protein kinase B and p70 S6 kinase phosphorylation in cardiac myocytes. J Biol Chem 1998;273:9703-10. doi: 10.1074/jbc.273.16.9703 [DOI] [PubMed] [Google Scholar]
- 12.Bonni A, Frank DA, Schindler C, Greenberg ME. Characterization of a pathway for ciliary neurotrophic factor signaling to the nucleus. Science 1993;262:1575-9. doi: 10.1126/science.7504325 [DOI] [PubMed] [Google Scholar]
- 13.Arakawa Y, Sendtner M, Thoenen H. Survival effect of ciliary neurotrophic factor (CNTF) on chick embryonic motoneurons in culture: comparison with other neurotrophic factors and cytokines. J Neurosci 1990;10:3507-15. doi: 10.1523/JNEUROSCI.10-11-03507.1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Conover JC, Ip NY, Poueymirou WT, Bates B, Goldfarb MP, DeChiara TM, et al. Ciliary neurotrophic factor maintains the pluripotentiality of embryonic stem cells. Development 1993;119:559-65. [DOI] [PubMed] [Google Scholar]
- 15.Davis S, Aldrich TH, Ip NY, Stahl N, Scherer S, Farruggella T, et al. Released form of CNTF receptor alpha component as a soluble mediator of CNTF responses. Science 1993;259:1736-9. doi: 10.1126/science.7681218 [DOI] [PubMed] [Google Scholar]
- 16.Di Marco A, Gloaguen I, Demartis A, Saggio I, Graziani R, Paonessa G, et al. Agonistic and antagonistic variants of ciliary neurotrophic factor (CNTF) reveal functional differences between membrane-bound and soluble CNTF alpha-receptor. J Biol Chem 1997;272:23069-75. doi: 10.1074/jbc.272.37.230 69 [DOI] [PubMed] [Google Scholar]
- 17.Gimble JM, Wanker F, Wang CS, Bass H, Wu X, Kelly K, et al. Regulation of bone marrow stromal cell differentiation by cytokines whose receptors share the gp130 protein. J Cell Biochem 2012;54:122-33. doi: /10.1002/jcb.240540113 [DOI] [PubMed] [Google Scholar]
- 18.Hughes SM, Lillien LE, Raff MC, Rohrer H, Sendtner M. Ciliary neurotrophic factor induces type-2 astrocyte differentiation in culture. Nature 1988;335:70-3. doi: 10.1038/335070a0 [DOI] [PubMed] [Google Scholar]
- 19.Ip NY, Yancopoulos GD. The neurotrophins and CNTF: two families of collaborative neurotrophic factors. Annu Rev Neurosci 1996;19:491-515. doi: 10.1146/annurev.ne.19.030 196.002423 [DOI] [PubMed] [Google Scholar]
- 20.Johnson DE, O'Keefe RA, Grandis JR. Targeting the IL- 6/JAK/STAT3 signalling axis in cancer. Nat Rev Clin Oncol 2018;15:234-48. doi: 10.1038/nrclinonc.2018.8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Louis JC, Magal E, Takayama S, Varon S. CNTF protection of oligodendrocytes against natural and tumor necrosis factorinduced death. Science 1993;259:689-92. doi: 10.1126/science. 8430320 [DOI] [PubMed] [Google Scholar]
- 22.Manthorpe M, Louis JC, Hagg T, Varon S. Ciliary neurotrophic factor. Loughlin SE, Fallon JH, editors. Neurotrophic factors. San Diego: Academic Press; 1993. p. 443-73. [Google Scholar]
- 23.Schooltink H, Stoyan T, Roeb E, Heinrich PC, Rose-John S. Ciliary neurotrophic factor induces acute-phase protein expression in hepatocytes. FEBS Lett 1992;314:280-4. doi: 10.1016/0014-5793(92)81489-9 [DOI] [PubMed] [Google Scholar]
- 24.Yu H, Lee H, Herrmann A, Buettner R, Jove R. Revisiting STAT3 signalling in cancer: new and unexpected biological functions. Nat Rev Cancer 2014;14:736-46. doi: 10.1038/nrc3818 [DOI] [PubMed] [Google Scholar]
- 25.Zhang XG, Gu JJ, Lu ZY, Yasukawa K, Yancopoulos GD, Turner K, et al. Ciliary neurotropic factor, interleukin 11, leukemia inhibitory factor, and oncostatin M are growth factors for human myeloma cell lines using the interleukin 6 signal transducer gp130. J Exp Med 1994;179:1337-42. doi: 10.1084/jem.179.4.1337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Anderson DJ, Stahl S, Yancopoulos GD. CNTF and LIF act on neuronal cells via shared signaling pathways that involve the IL-6 signal transducing receptor component gp130. Cell 1992; 69:1121-32. 10.1016/0092-8674(92)90634-o [DOI] [PubMed] [Google Scholar]
- 27.Janoschek R, Plum L, Koch L, Munzberg H, Diano S, Shanabrough M, et al. Gp130 signaling in proopiomelanocortin neurons mediates the acute anorectic response to centrally applied ciliary neurotrophic factor. Proc Natl Acad Sci USA 2006;103:10707-12. doi: 10.1073/pnas.0600425103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lambert PD, Anderson KD, Sleeman MW, Wong V, Tan J, Hijarunguru A, et al. Ciliary neurotrophic factor activates leptinlike pathways and reduces body fat, without cachexia or rebound weight gain, even inleptin-resistant obesity. Proc Natl Acad Sci USA 2001;98:4652-7. doi: 10.1073/pnas.061034298 7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rhee KD, Nusinowitz S, Chao K, Yu F, Bok D, Yang XJ. CNTF-mediated protection of photoreceptors requires initial activation of the cytokine receptor gp130 in Müller glial cells. Proc Natl Acad Sci USA 2013;110:E4520-9. doi: 10.1073/pnas.1303604110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen J, Chen P, Backman LJ, Zhou Q, Danielson P. Ciliary neurotrophic factor promotes the migration of corneal epithelial stem/progenitor cells by up-regulation of MMPs through the phosphorylation of Akt. Sci Rep 2016;6:25870. doi: 10.1038/srep25870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bonkhoff H, Remberger K. Differentiation pathways and histogenetic aspects of normal and abnormal prostatic growth: a stem cell model. Prostate 1996;28:98-106. doi: 10.1002/(SICI)1097-0045(199602)28:2<98::AIDPROS4>3.0.CO;2-J [DOI] [PubMed] [Google Scholar]
- 32.Hudson DL, Guy AT, Fry P, O’Hare MJ, Watt FM, Masters JR. Epithelial cell differentiation pathways in the human prostate: identification of intermediate phenotypes by keratin expression. J Histochem Cytochem 2001;49:271-8. doi: 10.1177/002215540104900214 [DOI] [PubMed] [Google Scholar]
- 33.Mayer MJ, Klotz LH, Venkateswaran V. Metformin and prostate cancer stem cells: a novel therapeutic target. Prostate Cancer Prostatic Dis 2015;18:303-9 doi: 10.1038/pcan.2015.35 [DOI] [PubMed] [Google Scholar]
- 34.Peehl DM, Leung GK, Wong ST. Keratin expression: a measure of phenotypic modulation of human prostatic epithelial cells by growth inhibitory factors. Cell Tissue Res 1994;277:11-8. doi: 10.1007/bf00303075 [DOI] [PubMed] [Google Scholar]
- 35.Shibata M, Shen MM. Stem cells in genetically-engineered mouse models of prostate cancer. Endocr Relat Cancer 2015; 22:T199-208. doi: 10.1530/ERC-15-0367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Signoretti S, Loda M. Prostate stem cells: from development to cancer. Semin Cancer Biol 2007;17:219-24. doi: 10.1016/j.semcancer.2006.04.004 [DOI] [PubMed] [Google Scholar]
- 37.De Marzo AM, Nelson WG, Meeker AK, Coffey DS. Stem cell features of benign and malignant prostate epithelial cells. J Urol 1998;160:2381-92. doi: 10.1097/00005392-199812020-00004 [DOI] [PubMed] [Google Scholar]
- 38.Frank SB, Miranti CK. Disruption of prostate epithelial differentiation pathways and prostate cancer development. Front Oncol 2013;3:273. doi: 10.3389/fonc.2013.00273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Huang CK, Luo J, Lee SO, Chang C. Concise review: androgen receptor differential roles in stem/progenitor cells including prostate, embryonic, stromal, and hematopoietic lineages. Stem Cells 2014;32:2299-308. doi: 10.1002/stem.1722 [DOI] [PubMed] [Google Scholar]
- 40.Leão R, Domingos C, Figueiredo A, Hamilton R, Tabori U, Castelo-Branco P. Cancer stem cells in prostate cancer: implications for targeted therapy. Urol Int 2017;99:125-36. doi: 10.1159/000455160. [DOI] [PubMed] [Google Scholar]
- 41.Humphrey PA, Amin MB, Berney DM, Billis A, Cao D, Cheng L, et al. Acinar adenocarcinoma. Moch H, Humphrey PA, Ulbright TM, Reuter VE, editors. WHO Classification of tumours of the urinary system and male genital organs. IARC: Lyon; 2016. p. 138-61. [Google Scholar]
- 42.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. Cancer J Clin 2013;63:11-30. doi: 10.3322/caac.21166 [DOI] [PubMed] [Google Scholar]
- 43.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, et al. Incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136 E359-86. doi: 10.1002/ijc.29210 [DOI] [PubMed] [Google Scholar]
- 44.Boukovala M, Spetsieris N, Justin A, Weldon JA, Tsikkinis A, Hoang A. A candidate androgen signalling signature predictive of response to abiraterone acetate in men with metastatic castration- resistant prostate cancer. Eur J Cancer 2020;127:67-75. doi: 10.1016/j.ejca.2019.12.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lu Z, Ding L, Hong H, Hoggard J, Lu Q, Chen YH. Claudin- 7 inhibits human lung cancer cell migration and invasion through ERK/MAPK signaling pathway. Exp Cell Res 2011;317:1935-46. doi: 10.1016/j.yexcr.2011.05.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McCubrey JA, Steelman LS, Chappell WH, Abrams SL, Wong EW, Chang F, et al. Roles of the Raf/MEK/ERK pathway in cell growth, malignant transformation and drug resistance. Biochim Biophys Acta 2007;1773:1263-84. doi: 10.1016/j.bbamcr.2006.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Thomas MU, Messex JK, Dang T, Abdulkadir SA, Jorcyk CL, Liou GY. Macrophages expedite cell proliferation of prostate intraepithelial neoplasia through their downstream target ERK. FEBS J 2020. doi: 10.1111/febs.15541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xu CJ, Dong LL, Kang XL, Li ZM, Zhang HY. Leptin promotes proliferation and inhibits apoptosis of prostate cancer cells by regulating ERK1/2 signaling pathway. Eur Rev Med Pharmacol Sci 2020;24:8341-8. doi: 10.26355/eurrev_202 008_22630. [DOI] [PubMed] [Google Scholar]
- 49.Avellini C, Licini C, Lazzarini R, Gesuita R, Guerra E, Tossetta G, et al. The trophoblast cell surface antigen 2 and miR-125b axis in urothelial bladder cancer. Oncotarget 2017;8:58642-53. doi: 10.18632/oncotarget.17407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hafidi A, Decourt B, MacLennan AJ. CNTFR→ and CNTF expressions in the auditory brainstem: light and electron microscopy study. Hearing Res 2004;194:14-24. doi: 10.1016/j.heares.2004.04.004 [DOI] [PubMed] [Google Scholar]
- 51.MacLennan AJ, Vinson EN, Marks L, McLaurin DL, Pfeifer M, Lee N. Immunohistochemical localization of ciliary neurotrophic factor receptor CY expression in the rat nervous system. J Neurosci 1996;76:621630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Squinto SP, Aldrich TH, Lindsay RM, Morrissey DM, Panayotatos N, Bianco SM, et al. Identification of functional receptors for ciliary neurotrophic factor on neuronal cell lines and primary neurons. Neuron 1990;5:757-66. [DOI] [PubMed] [Google Scholar]
- 53.McNeal JE, Redwine EA, Freiha FS, Stamey TA. Zonal distribution of prostatic adenocarcinoma. Correlation with histologic pattern and direction of spread. Am J Surg Pathol 1988;12:897-906. doi: 10.1097/00000478-198812000-00001 [DOI] [PubMed] [Google Scholar]
- 54.Packer JR, Maitland NJ. The molecular and cellular origin of human prostate cancer. Biochim Biophys Acta 2016;1863:1238-60. doi: 10.1016/j.bbamcr.2016.02.016 [DOI] [PubMed] [Google Scholar]
- 55.Lang SH, Frame FM, Collins AT. Prostate cancer stem cells. J Pathol 2009;217:299-306. doi: 10.1002/path.2478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang D, Zhao S, Li X, Kirk JS, Dean G., Tang DG. Prostate luminal progenitor cells in development and cancer. Trends Cancer 2018;4:769-83. doi: 10.1016/j.trecan.2018.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Strand DW, Goldstein AS. The many ways to make a luminal cell and a prostate cancer cell. Endocr Relat Cancer 2015;22;187-97. doi: 10.1530/ERC-15-0195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ge R, Wang Z, Montironi R, Jiang Z, Cheng M, Santoni M, et al. Epigenetic modulations and lineage plasticity in advanced prostate cancer. Ann Oncol 2020;31:470-9. doi: 10.1016/j.annonc.2020.02.002 [DOI] [PubMed] [Google Scholar]
- 59.Butler DE, Marlein C, Walker HF, Frame FM, Mann VM, Simms MS, et al. Inhibition of the PI3K/AKT/mTOR pathway activates autophagy and compensatory Ras/Raf/MEK/ERK signalling in prostate cancer. Oncotarget 2017;22;8:56698-713. doi: 10.18632/oncotarget.18082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mulholland DJ, Kobayashi N, Ruscetti M, Zhi A, Tran LM, Huang J, et al. Pten loss and RAS/MAPK activation cooperate to promote EMT and metastasis initiated from prostate cancer stem/progenitor cells. Cancer Res 2012;72:1878-89. doi: 10.1158/0008-5472.CAN-11-3132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nickols NG, Nazarian R, Zhao SG, Tan V, Uzunangelov V, Xia Z, et al. MEK-ERK signaling is a therapeutic target in metastatic castration resistant prostate cancer. Prostate Cancer and Prostatic Dis 201922:531-8. doi: 10.1038/s41391-019-0134-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gioeli D, Mandell JW, Petroni GR, Frierson HF, Weber MJ. Activation of mitogen-activated protein kinase associated with prostate cancer progression. Cancer Res 1999;59:279-84. [PubMed] [Google Scholar]


