Abstract
Given the crucial role of microRNAs in the cellular proliferation of various types of cancers, we aimed to analyze the expression and function of a cellular proliferation-associated miR-188-5p in papillary thyroid carcinoma (PTC). Here we demonstrate that miR-188-5p is downregulated in PTC tumor tissues compared with the associated noncancerous tissues. We also validate that the miR-188-5p overexpression suppressed the PTC cancer cell proliferation. In addition, fibroblast growth factor 5 (FGF5) is observed to be downregulated in the PTC tumor tissues compared with the associated noncancerous tissues. Subsequently, FGF5 is identified as the direct functional target of miR-188-5p. Moreover, the silencing of FGF5 was found to inhibit PTC cell proliferation, which is the same pattern as miR-188-5p overexpression. These results suggest that miR-188-5p-associated silencing of FGF5 inhibits tumor cell proliferation in PTC. It also highlights the importance of further evaluating miR-188-5p as a potential biomarker and therapy target in PTC.
Keywords: miR-188-5p, FGF5, cell proliferation
Introduction
Papillary thyroid carcinoma (PTC) remains the most common type of all thyroid carcinomas, wherein diagnostic limitations and lack of accurate prognostic factors are important clinical challenges. The incidence of thyroid cancer has increased dramatically worldwide, especially in Asian countries including China1,2. Although great progress has been made in understanding the molecular pathogenesis of thyroid cancer to develop more-effective treatment strategies, the disease remains incurable. Therefore, it is essential to develop novel therapeutic strategies.
MicroRNAs (miRNAs) consist of a large family of noncoding, single-stranded RNA molecules that are approximately 22 nucleotides long, which are generally involved in posttranscriptional gene regulation and stability3. This regulation is achieved through incomplete and complete sequence matching between miRNAs and the 3′untranslated regions (UTRs) of target mRNA4. Since miRNAs were first identified in 1993, it has been demonstrated that miRNAs contribute to the progression of different types of cancers, such as colorectal, lung, brain, breast, and liver cancers5,6. Numerous studies have revealed that miRNA profiling is critical for the diagnosis and prognosis of patients with cancer, while certain miRNAs possess the potential to be used as diagnostic and prognostic biomarkers or therapeutic targets in cancer. Therefore, it is of value to illuminate whether dysregulation of miRNA-regulatory networks contributes to the progression of PTC. However, the role of miR-188-5p in PTC has not been determined. So the present study aimed to clarify the biological functions and underlying molecular mechanisms of miR-188-5p in PTC.
Fibroblast growth factor-5 (FGF-5) is a member of a group of 23 related fibroblast growth factor genes that are reported to participate in a variety of biological processes such as development, morphogenesis, tissue growth, and repair7. FGF5 has been demonstrated oncogenic activity of FGF5 in astrocytic brain tumors and attribute to both autocrine effects on the tumor cells as well as paracrine effects on endothelial cells8. Zhou et al have demonstrated that the downregulation of FGF5 inhibited cell growth and invasion of human non-small-cell lung cancer cells9. FGF5 expression was found to be significantly upregulated in breast cancer patients relative to that in normal controls (P < 0.0001), and breast cancer patients in the FGF5 low-expression group were correlated with better clinical characteristics10. Despite these findings in and the proposed tumor-promoting role of FGF5 in other malignancies, FGF5 has not been further investigated in PTC so far.
Therefore, we asked whether miR-188-5p/FGF5 may contribute to the progression of PTC. In this study, we showed that miR-188-5p is downregulated in PTC tumor tissues compared with the associated noncancerous tissues. We also found that the upregulation of miR-188-5p suppressed the PTC cancer cell proliferation. In addition, FGF5 is observed to be downregulated in the PTC tumor tissues and identified as the direct functional target of miR-188-5p. Furthermore, the silencing of FGF5 also inhibits PTC cell proliferation, which is the same as miR-188-5p overexpression.
Materials and Methods
Clinical Specimens and Cell Lines
PTC tumor and normal tissues were obtained from patients who were diagnosed with PTC and who had undergone surgery at the Second Hospital of Wuhan and Steel Group between 2017 and 2018. In total, 30 pairs of tissue samples were freshly frozen in liquid nitrogen and stored at −80°C until RNA extraction. The use of tissues for this study was approved by the Ethics Committee of the Second Hospital of Wuhan Iron and Steel Group.
Human PTC cell TPC-1, K1, and normal thyroid epithelial cells Nthy-ori 3-1 cells as a control were obtained from China Center for Type Culture Collection in Wuhan university (Wuhan, Hubei, China). These cells were cultured in Dulbecco’s modified Eagle medium (Sigma-Aldrich, Shanghai, China) supplemented with 10% (v/v) fetal bovine serum (Sigma-Aldrich, 95% air, 5% CO2 at 37°C.
Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR) to Detect the miR-188-5p and FGF5
The miR-188-5p level and FGF5 were detected by qRT-PCR as previously described11. Total RNA was extracted from cells and tissues by the mirVanaTM miRNA isolation kit (Thermo Fisher Scientific, Shanghai, China) as described by the manufacturer.
Then the RNA was reverse transcribed using a TaqManTM advanced miRNA cDNA synthesis kit (Thermo Fisher Scientific). The qRT-PCR for miR-188-5p was performed using TaqMan Universal PCR Master Mix (Thermo Fisher Scientific) by TaqManTM analysis run on a QuantStudio 6 Flex PCR system (Thermo Fisher Scientific). RNA U6 (Qiagen, Shanghai, China) was used as an internal control.
To detect the FGF expression level, RNA was reverse-transcribed into cDNA by the high-capacity RNA-to-cDNA kit (Thermo Fisher Scientific) after removal of the residual DNA by DNA-free DNase (Norgen Biotek, Thorold, ON, Canada). Taqman qRT-PCR was used to detect the FGF gene expression. The relative FGF expression level was normalized to β-actin.
qRT-PCR primers for FGF and miR-188-5p expression were available from Applied Biosystems. All studies were performed in triplicate and the data were presented as mean ± standard error (SE).
Western Blotting Analysis
Protein lysates from tissues and cells were separated on 10% sodium dodecyl sulfate-polyacrylamide gel (Invitrogen, Beijing, China) and electrophoretically transferred onto a nitrocellulose membrane. Membranes were blocked with 5% dried nonfat milk powder in TBST for 1 h and incubated with primary antibodies overnight at 4°C. After being washed with TBST three times, the membranes were incubated with the corresponded secondary antibody (Bio-Rad, Hercules CA, USA) for 2 h at room temperature. The band signals of target proteins were visualized using an enhanced chemiluminescence kit (Abcam, Shanghai, China).
All the experiments were performed in triplicate. In order to avoid possible problems related to incomplete stripping, all the results are from separate blots.
The primary antibodies: anti-FGF5 (1:600, Abcam, Cambridge, MA, USA) and β-Actin (1:1200, Sigma-Aldrich, St Louis, MO, USA).
MiR-188-5p Overexpression
To stably overexpress miR-188-5p, miR-188-5p-expressing vector was constructed as previously described12. Pri-miR-188-5p was amplified and cloned into pcDNA3.1 (Invitrogen). The cells were seeded into the six-well plate at a density of 0.25 M cells/ well and transfected with 400 mg pcDNA3.1-miR-188-5p with Effectene (Qiagen) followed the manufacturer’s instruction. The control vector (miR-NC) contains a nonrelevant sequence as a negative control.
Dual Luciferase Gene Reporter assay
The putative targeting gene (FGF5) of miR-188-5p was predicted by TargetScan (http://www.targetscan.org). The 3′ UTR of FGF5 luciferase reporter was commercially available at ORIGENE (Rockville, MD, USA). The cells (TPC-1 and K1) were seeded in a six-well plate (Thermo Fisher Scientific) at a density of 0.25 M cells/well with the complete media for 24 h. Then, cells were cotransfected with 1 µg FGF5 3′-UTR luciferase reporter construct with 20 nM miR-188-5p mimic or miR-NC using lipofectamine 2000 (Invitrogen) with Opti-MEN (Gibco, Shanghai, China) for 48 h. Luciferase assays were performed using the dual-luciferase reporter assay reagent from GeneCopoeia (Rockville, MD, USA). Data are expressed as the ratio of firefly luciferase activity by Renilla luciferase activity.
FGF-5 Silencing
To silence FGF-5, PTC cells were transduced with FGF-5-targeting shRNA lentiviruses (sh-FGF5, Sigma-Aldrich) following the manufacturer’s instructions. Then, cells were selected with puromycin and expanded. The control virus (sh-Ctrl) was also commercially available from Sigma-Aldrich.
Cell Proliferation
The effect of miR-188-5p on the growth of MPM cells was monitored by Cyquant assay (Thermo Fisher Scientific). Cells were seeded in the 96-well plate (Thermo Fisher Scientific) at a density of 5 × 103 cells/well. Plates were frozen at the indicated time for 24, 48, 72, 96, and 120 h. One hundred microliters of fresh prepared Cyquant solution was added to the wells and incubated in the dark for 45 min at room temperature. Plates were read at excitation at 497 nm and emission at 520 nm. All studies were performed in triplicate.
Soft Agar Assay
The soft agar colony formation assay is a traditional method to determine the anchorage-independent growth in vitro, which is considered the most stringent assay for detecting the malignance of cells. Briefly, we melt 1.4% (v/v) agarose (Sigma-Aldrich) in a microwave and cooled to room temperature. Then, equal volumes of the 1.4% melted agarose were mixed with the complete cell culture medium. After that, 2 ml of 0.7% (v/v) low-melting point agar was added into each well of the six-well plate and set aside to allow agarose to solidify. Cells (5,000/ well) were mixed with 1.4% agarose in complete culture medium, plated on top of the solidified layer to form colonies in 1–3 wk. Cells were fed with complete culture medium every 3 d.
The colony is defined to consist of at least 50 cells13. After the cells have formed sufficiently large clones, we removed the medium above the cells and rinsed carefully with PBS. Then, colonies were fixed with 4% paraformaldehyde (Sigma-Aldrich) for 10 min and stained with crystal violet solution (0.5%, v/v) (Sigma-Aldrich) for at least 30 min. After removal of the crystal violet solution and carefully washing with tap water until excess dye was removed, the plates were left to dry at room temperature. Colonies were counted manually by light microscopy.
Statistics
Results are shown as means ± SE. Student’s t-test for two groups or one-way analysis of variance test, followed by Tukey’s multiple comparison for multiple groups were used to compare the significance of differences between the mean of different groups. SPSS 15.0 (SPSS Inc. Chicago, IL, USA) was used to calculate the significance. The value of P <0.05 indicated the statistical significance.
Results
miR-188-5p was Downregulated in PTC Tumor Tissues
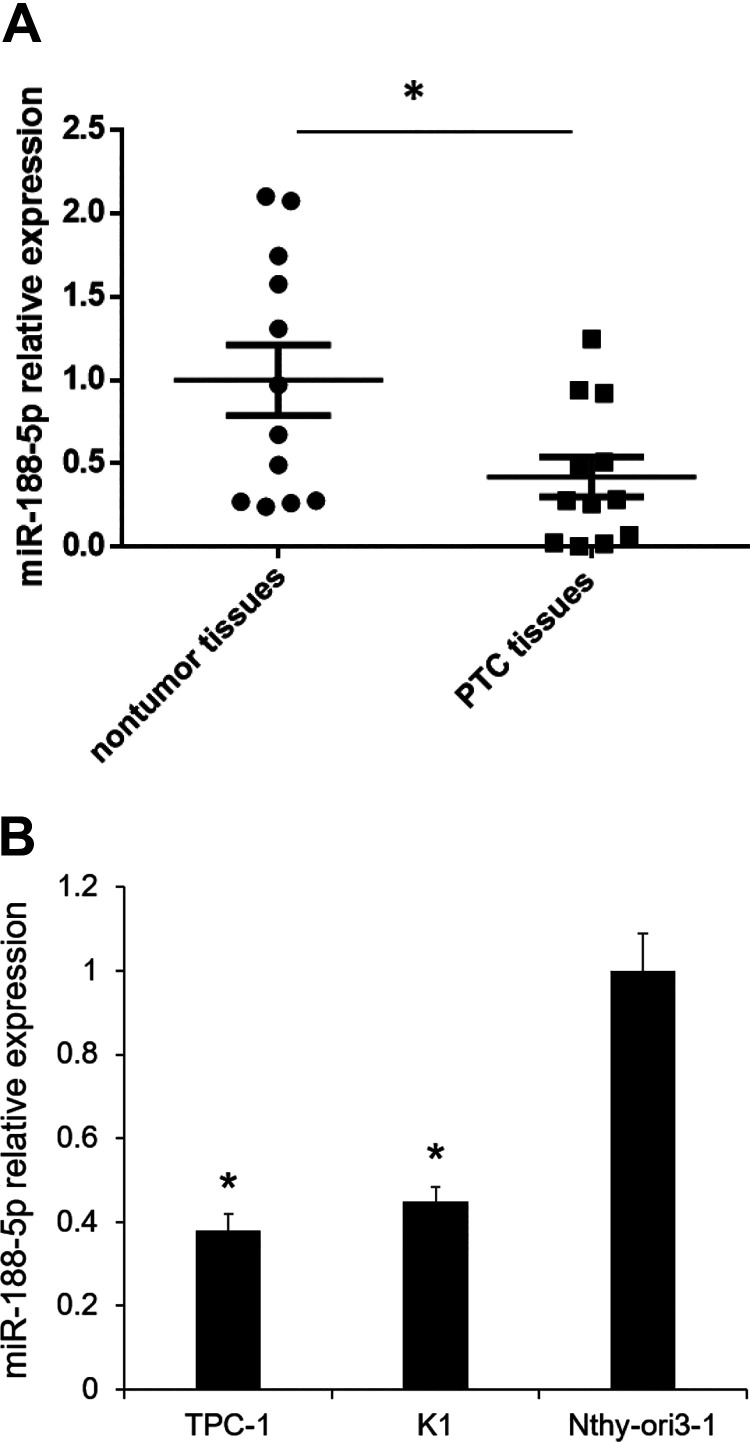
Based on our preliminary miRNA microarray study, we found that miR-188-5p was one of the downregulated miRNAs in PTC tissue specimens compared with the adjacent nontumor tissues. We investigated the expression of miR-188-5p in 12 PTC tissues and their paired adjacent nontumor tissues by qRT-PCR analysis. We found that miR-188-5p was downregulated by 2.3-fold (P < 0.05, Fig. 1A). We also determined the expression of miR-188-5p in TPC-1, K1, and Nthy-ori 3-1. miR-188-5p was revealed to be downregulated in PTC cell lines compared with the Nthy-ori 3-1, human thyroid epithelial cell line (Fig. 1B). These results suggested that low miR-188-5p may be associated with PTC progression.
Fig. 1.
miR-188-5p expression was downregulated in PTC tissues. (A) miR-188-5p expression was determined by qualitative real-time polymerase chain reaction in 12 paired PTC and adjacent nontumor tissues. *P < 0.05 vs. nontumor tissues (n = 12); (B) miR-188-5p expression in TPC-1, K1, and Nithy. Results are mean ± standard error (n = 3). *P < 0.05 vs. Nithy-ori 3-1. miR: microRNA; PTC: papillary thyroid carcinoma.
miR-188-5p Overexpression Suppresses PTC Cell Proliferation
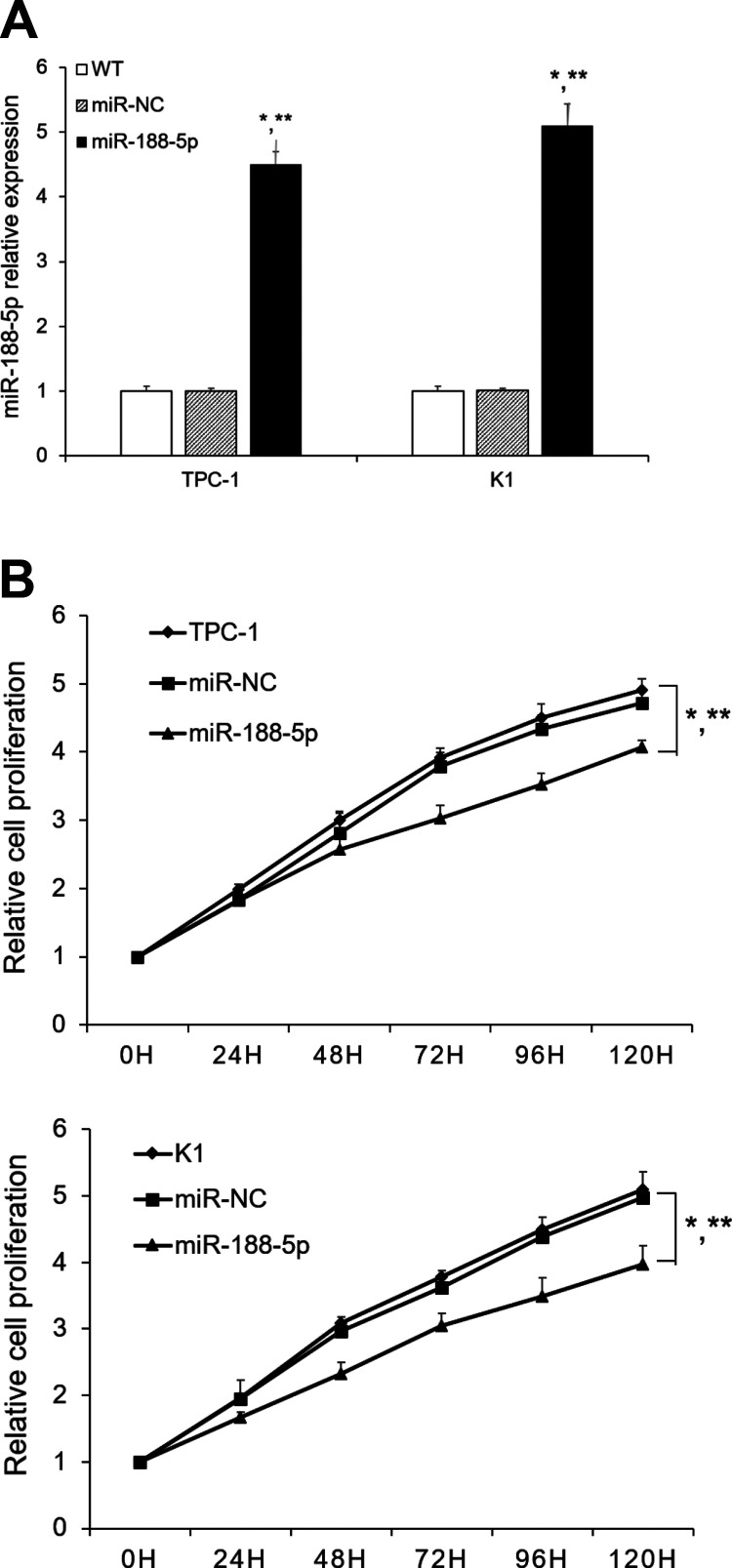
To explore the biological functions of miR-188-5p, we stably overexpressed the miR-188-5p in TPC-1 and K1. The overexpression of miR-188-5p in transfected cell lines was verified by qRT-PCR analysis. Cells transfected with miR-188-5p showed significantly increased expression of miR-188-5p than the wild type (WT) and cells transfected with an empty vector (Fig. 2A). As shown in Fig. 2B, overexpression of miR-188-5p significantly suppressed PTC cell proliferation compared with the WT and Ctrl group. The results revealed that miR-188-5p overexpression significantly suppressed the growth rates of PTC cells.
Fig. 2.
Effect of miR-188-5p on PTC cell proliferation. (A) PTC cells were infected with miR-188-5p lentivirus or miR-NC. miR-188-5p was determined by qualitative real-time polymerase chain reaction; (B) TPC-1/K1 cells or TPC-1/K1 infected with miR-188-5p or miR-NC cells were seeded into 96-well plates. Cyquant assay was performed to determine the cell proliferation. Results are mean ± standard error (n = 3). *P < 0.05 vs. WT; **P < 0.05 vs. miR-NC.
miR: microRNA; PTC: papillary thyroid carcinoma; WT: wild type
miR-188-5p Overexpression Inhibits PTC Cell Clonogenicity
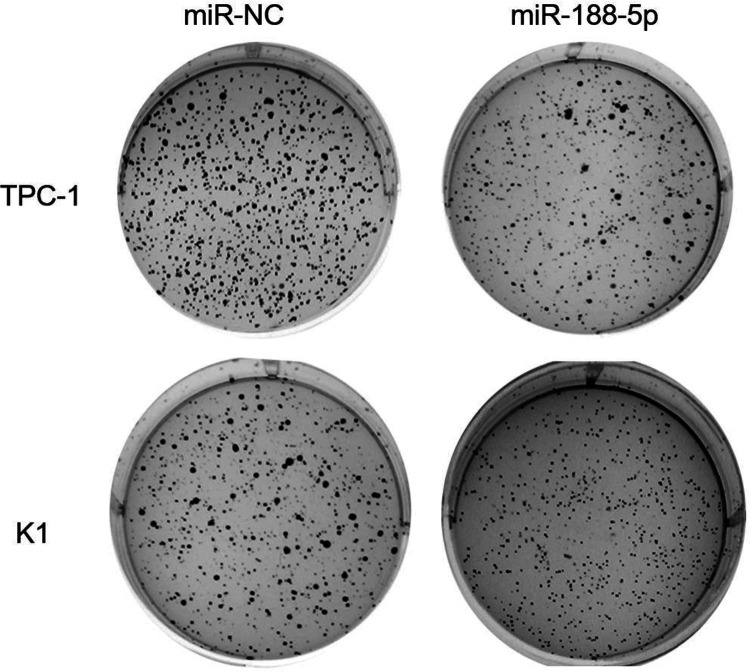
We next sought to establish whether miR-188-5p overexpression could enhance PTC cell clonogenicity. The colony formation efficiencies of transfected cell line were determined by soft agar culture. miR-188-5p overexpression significantly inhibited the anchorage-dependent growth of PTC cells (Fig. 3). Taken together, these results indicate that miR-188-5p has a growth-suppressive function in PTC cells.
Fig. 3.
Soft agar assay was performed to determine the anchorage-independent growth of TPC-1 and K1 cells after miR-188-5p overexpression.
FGF-5 is Upregulated in PTC Tumor Tissues
To unravel the relevant mechanism underlying miR-188-5p that contributed to the cell proliferation, we searched the potential functional target of miR-188-5p. FGF5 has already been identified as the direct target of miR-188-5p14. To further study the relationship between miR-188-5p and FGF5 in PTC, we detected the FGF mRNA and protein expression in 12 PTC tissues and the paired adjacent nontumor tissues. FGF5 mRNA levels were significantly upregulated in PTC tissues compared with the nontumor tissues (P < 0.05, Fig. 4A). After that, we determined the protein levels of FGF in PTC tumor tissues and nontumor tissues. Consistently, FGF5 protein levels were also higher in PTC tissues (Fig. 4B).
Fig. 4.
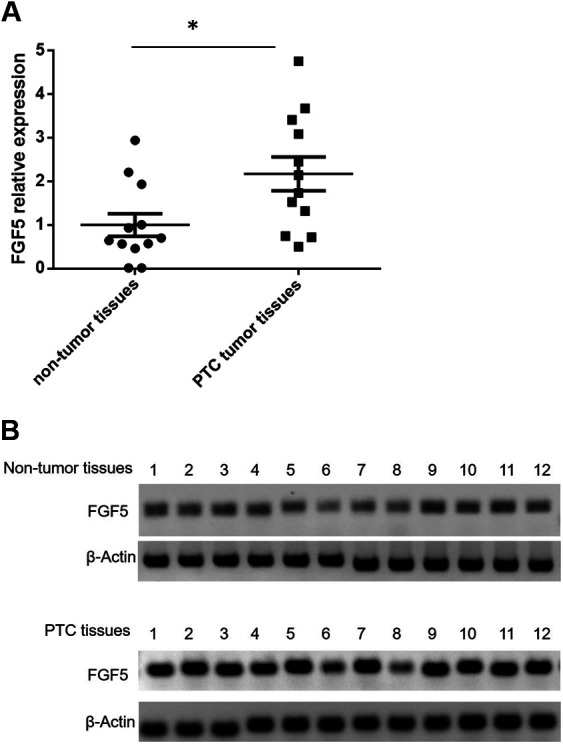
FGF5 expression was upregulated in PTC tissues. (A) FGF5 mRNA level was determined by qualitative real-time polymerase chain reaction in 12 paired PTC and adjacent nontumor tissues. *P < 0.05 vs. nontumor tissues (n = 12); (B) FGF5 protein expression level was determined by western blotting in 12 paired PTC and adjacent nontumor tissues. FGF5: fibroblast growth factor 5; PTC: papillary thyroid carcinoma.
FGF-5 is Repressed by miR-188-5p
Subsequently, the 3′-UTR luciferase assays confirmed that miR-188-5p directly binds FGF5 mRNA in PTC cells. MiR-188-5p significantly decreased the luciferase activities of FGF5 (P < 0.05, Fig. 5A). But we did not observe repressed luciferase activities of FGF5 by miR-Ctrl (Fig. 5A). In addition, the expression levels of FGF5 mRNA and protein were also significantly lower after miR-188-5p overexpression (P < 0.05, Fig. 5B, C). Collectively, these results imply that FGF5 is directly repressed by miR-188-5p in PTC cells.
Fig. 5.
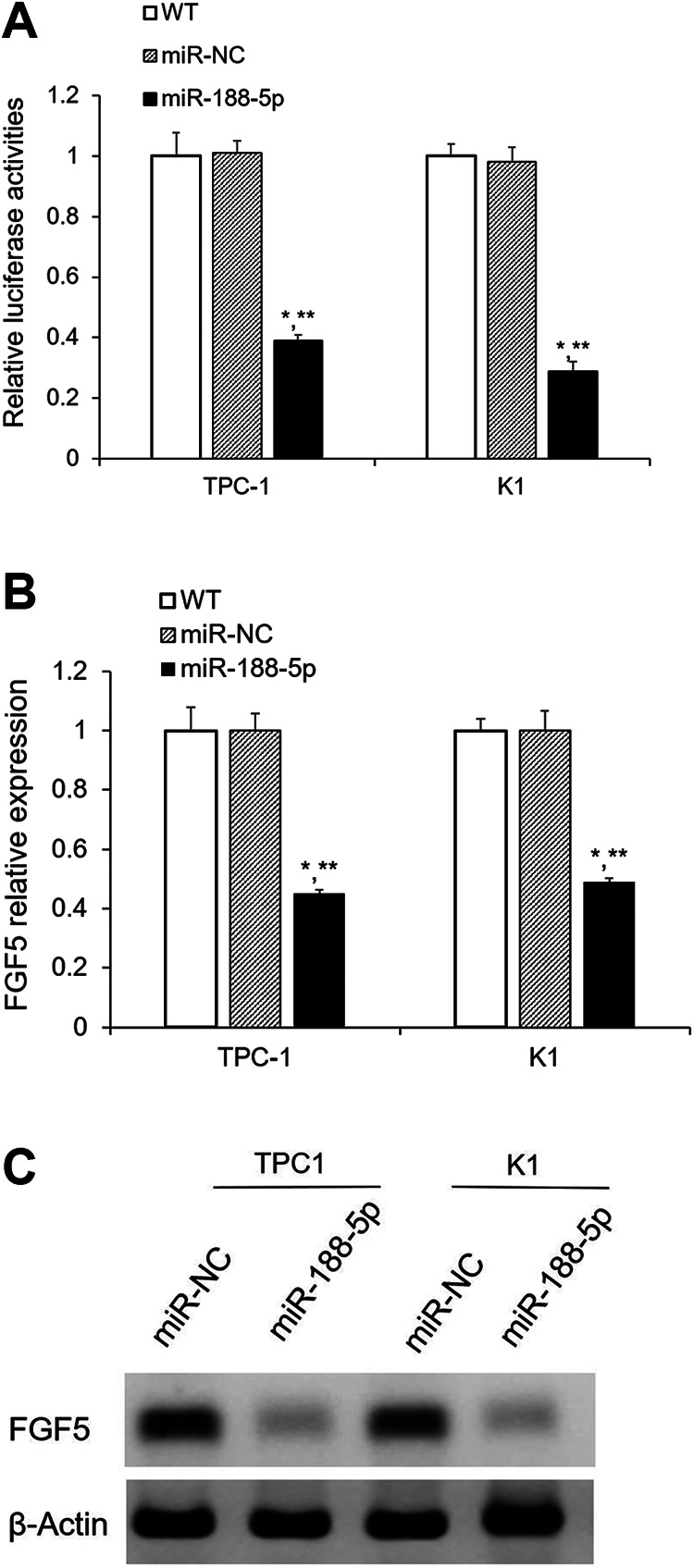
FGF5 is the direct target gene of miR-188-5p. (A) Confirm FGF5 is the direct target gene of miR-188-5p by dual-luciferase assay; (B) FGF5 mRNA level was significantly decreased after miR-188-5p overexpression, which was detected by qualitative real-time polymerase chain reaction; (C) FGF5 protein expression was remarkably decreased after miR-188-5p overexpression, as determined by western blotting. Results are mean ± standard error (n = 3). *P < 0.05 vs. WT; **P < 0.05 vs. miR-NC. FGF5: fibroblast growth factor 5; miR: microRNA; WT: wild type.
FGF5 Silencing Mimics the Effect of miR-188-5p Overexpression on PTC cells
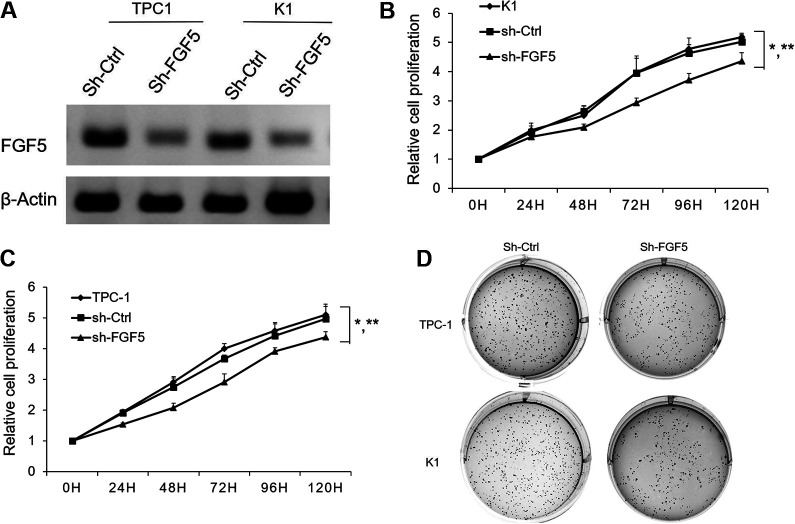
Silencing of FGF5 had the similar effects on cell proliferation as overexpression of miR-188-5p in PTC cells. FGF5 shRNA significantly reduced the expression of FGF5 protein in TPC-1 and K1 cells (Fig. 6A). FGF5 repression significantly decreased (P < 0.05) the rate of cell proliferation by Cyquant assay (Fig. 6B). Inhibition of FGF5 also leads to a reduction in anchorage-dependent growth of TPC and K1 cells (Fig. 6C). This result showed that miR-188-5p exerts tumor-suppressive function in PTC cells through directly regulating FGF5.
Fig. 6.
Effect of FGF5 silencing on PTC cell proliferation. (A) PTC cells were infected with sh-FGF5 lentivirus or sh-Ctrl. FGF5 protein expression was determined by western blotting; (B) TPC-1/K1 cells or TPC-1/K1 infected with sh-Ctrl or sh-FGF5 cells were seeded into 96-well plates. Cyquant assay was performed to determine the cell proliferation. Results are mean ± standard error (n = 3). *P < 0.05 vs. WT; **P < 0.05 vs. sh-FGF5. Soft agar assay was performed to determine the anchorage-independent growth of TPC-1 and K1 cells after FGF5 silencing. FGF5: fibroblast growth factor 5; PTC: papillary thyroid carcinoma; WT: wild type.
Discussion
MiRNAs are small noncoding RNAs that regulate protein expression. As a recognized cancer gene, the roles of miRNAs in cancer have been widely investigated, as well as the important roles in both the promotion and suppression of cell proliferation in many tumors15. Of them, the role of miR-188-5p in tumor progression associated with the cell proliferation has recently received much attention. Several studies have reported that miR-188-5p functions as a suppressor for cancer cell proliferation in various types of human tumors16. miR-188-5p has been reported to suppress gastric cancer cell proliferation and invasion via targeting ZFR9117. miR-188-5p has also been identified as a tumor suppressor for cell progression by targeting SOX4 in pediatric osteosarcoma18. However, little is known about the biological function and molecular mechanism of miR-188-5p in PTC. In our study, miR-188-5p was found to be downregulated in the PTC tissues compared with the nontumorous tissues. The PTC cell lines have lower expression level of miR-188-5p than the Nthy-ori 3-1 cells. Moreover, miR-188-5p overexpression was also found to suppress PTC cell proliferation. Subsequently, we also demonstrated that miR-188-5p suppressed FGF5 expression by directly binding to its 3′-UTR. These results indicated that miR-188-5p contributes to the tumor progression of PTC.
FGF5 has been identified to enhance proliferation of endothelial cells and colorectal carcinoma tumor cells8,19. Metzner demonstrated that the inhibition of FGF5 signaling is associated with reduced melanoma cell proliferation, and anchorage-independent growth20. Recently, downregulation of FGF5 has been reported to inhibit cell growth and invasion of human non-small-cell lung cancer cells9. In osteosarcoma, miR-567 has been found to inhibit cell proliferation, migration, and invasion by targeting FGF521. It has also been shown that the expression of FGF5 is required for the proliferation of hepatocellular carcinoma associated with miR-188-5p14. However, the biological function of FGF5 and the network with miRNAs still remain unclear. The upregulated expression of FGF5 in PTC tumor tissues found in our study is in line with these data. In addition, FGF5 was also shown as the functional target of miR-188-5p by the 3′ UTR luciferase reporter assay. Moreover, the mRNA and protein level of FGF5 were significantly decreased after miR-188-5p overexpression. The silencing of FGF5 was also demonstrated to have the similar effect on the cell proliferation as the overexpression of miR-188-5p. FGF5 silencing was observed to significantly inhibit the cell proliferation as well. The following in vivo experiments are needed to verify the in vitro cell model in the future. It is also worth to investigate the impeded effect change after cotransfection with miR-188-5p and FGF5, the cell metastasis, the gain function of FGF5, and the potential pathway of which activity was attenuated by miR-188-5p in the future experiments.
In summary, we identified the function of miR-188-5p, which is a novel overexpressed miRNA and contributes to the tumor progression by targeting FGF5. Our results highlight the therapeutic potential of miR-188-5p in PTC.
Acknowledgments
We thank the clinical laboratory of the Second Hospital of Wuhan Iron and Steel Group for the pathologic diagnosis and Acesci Biomed (Wuhan, China) for English editing.
Authors’ Contributions: WL conceived the study. WL, PZ, NG, and JA designed the experiments and wrote the manuscript. PZ completed the experiments. JL and HW collected the samples and discussed the results. MC, AI, and MK analyzed the data and revised the manuscript.
Ethical Approval: Ethical approval was obtained for all experimental procedures by the Ethics Committee of the Second Hospital of Wuhan Iron and Steel Group, Wuhan, China.
Statement of Human and Animal Rights: All procedures with patients in this study were conducted in accordance with the Human Ethics Committee of the Second Hospital of Wuhan Iron and Steel Group, Wuhan, China. This article does not contain any studies with animals.
Statement of Informed Consent: Verbal informed consent was obtained from the patients for their anonymized information to be published in this article.
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) received no financial support for the research, authorship, and/or publication of this article.
ORCID iD: Weibang Liu  https://orcid.org/0000-0001-6215-5583
https://orcid.org/0000-0001-6215-5583
References
- 1. Brito JP, Kim HJ, Han SJ, Lee YS, Ahn HS. Geographic distribution and evolution of thyroid cancer epidemic in South Korea. Thyroid. 2016;26(6):864–865. [DOI] [PubMed] [Google Scholar]
- 2. Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115–132. [DOI] [PubMed] [Google Scholar]
- 3. Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466(7308):835–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Kohlhapp FJ, Mitra AK, Lengyel E, Peter ME. MicroRNAs as mediators and communicators between cancer cells and the tumor microenvironment. Oncogene. 2015;34(48):5857–5868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75(5):843–854. [DOI] [PubMed] [Google Scholar]
- 6. Tan W, Liu B, Qu S, Liang G, Luo W, Gong C. MicroRNAs and cancer: key paradigms in molecular therapy. Oncol Lett. 2018;15(3):2735–2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kehler JS, David VA, Schaffer AA, Bajema K, Eizirik E, Ryugo DK, Hannah SS, O’Brien SJ, Menotti-Raymond M. Four independent mutations in the feline fibroblast growth factor 5 gene determine the long-haired phenotype in domestic cats. J Hered. 2007;98(6):555–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Allerstorfer S, Sonvilla G, Fischer H, Spiegl-Kreinecker S, Gauglhofer C, Setinek U, Czech T, Marosi C, Buchroithner J, Pichler J, Silye R, et al. FGF5 as an oncogenic factor in human glioblastoma multiforme: autocrine and paracrine activities. Oncogene. 2008;27(30):4180–4190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhou Y, Yu Q, Chu Y, Zhu X, Deng J, Liu Q, Wang Q. Downregulation of fibroblast growth factor 5 inhibits cell growth and invasion of human nonsmall-cell lung cancer cells. J Cell Biochem. 2019;120(5):8238–8246. [DOI] [PubMed] [Google Scholar]
- 10. Huang Y, Wang H, Yang Y. Expression of Fibroblast Growth Factor 5 (FGF5) and Its Influence on Survival of Breast Cancer Patients. Med Sci Monit. 2018;24:3524–3530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fu H, Tan J, Yin Q. Effects of recombinant adeno-associated virus-mediated CD151 gene transfer on the expression of rat vascular endothelial growth factor in ischemic myocardium. Exp Ther Med. 2015;9(1):187–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhang H, Qi S, Zhang T, Wang A, Liu R, Guo J, Wang Y, Xu Y. miR-188-5p inhibits tumour growth and metastasis in prostate cancer by repressing LAPTM4B expression. Oncotarget. 2015;6(8):6092–6104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Franken NA, Rodermond HM, Stap J, Haveman J, van Bree C. Clonogenic assay of cells in vitro. Nat Protoc. 2006;1(5):2315–2319. [DOI] [PubMed] [Google Scholar]
- 14. Fang F, Chang RM, Yu L, Lei X, Xiao S, Yang H, Yang LY. MicroRNA-188-5p suppresses tumor cell proliferation and metastasis by directly targeting FGF5 in hepatocellular carcinoma. J Hepatol. 2015;63(4):874–885. [DOI] [PubMed] [Google Scholar]
- 15. Chen D, Guo W, Qiu Z, Wang Q, Li Y, Liang L, Liu L, Huang S, Zhao Y, He X. MicroRNA-30d-5p inhibits tumour cell proliferation and motility by directly targeting CCNE2 in non-small cell lung cancer. Cancer Lett. 2015;362(2):208–217. [DOI] [PubMed] [Google Scholar]
- 16. Chai C, Wu H, Wang B, Eisenstat DD, Leng RP. MicroRNA-498 promotes proliferation and migration by targeting the tumor suppressor PTEN in breast cancer cells. Carcinogenesis. 2018;39(9):1185–1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Peng Y, Shen X, Jiang H, Chen Z, Wu J, Zhu Y, Zhou Y, Li J. miR-188-5p Suppresses Gastric Cancer Cell Proliferation and Invasion via Targeting ZFP91. Oncol Res. 2018;27(1):65–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Pan L, Meng L, Liang F, Cao L. miR188 suppresses tumor progression by targeting SOX4 in pediatric osteosarcoma. Mol Med Rep. 2018;18(1):441–446. [DOI] [PubMed] [Google Scholar]
- 19. Sonvilla G, Allerstorfer S, Stattner S, Karner J, Klimpfinger M, Fischer H, Grasl-Kraupp B, Holzmann K, Berger W, Wrba F, Marian B, et al. FGF18 in colorectal tumour cells: autocrine and paracrine effects. Carcinogenesis. 2008;29(1):15–24. [DOI] [PubMed] [Google Scholar]
- 20. Metzner T, Bedeir A, Held G, Peter-Vorosmarty B, Ghassemi S, Heinzle C, Spiegl-Kreinecker S, Marian B, Holzmann K, Grasl-Kraupp B, Pirker C, et al. Fibroblast growth factor receptors as therapeutic targets in human melanoma: synergism with BRAF inhibition. J Invest Dermatol. 2011;131(10):2087–2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Liu D, Zhang C, Li X, Zhang H, Pang Q, Wan A. MicroRNA-567 inhibits cell proliferation, migration and invasion by targeting FGF5 in osteosarcoma. EXCLI J. 2018;17:102–112. [DOI] [PMC free article] [PubMed] [Google Scholar]