Abstract
Human epidermal growth factor receptor 2 (HER-2)+ breast cancer has a high recurrence rate and a poor prognosis, with drug resistance contributing to disease progression. The present study aimed to establish a SKBR3 cell line with type II insulin-like growth factor receptor (IGR-IIR) gene site integration using the CRISPR/Cas9 system, and to provide a cell model for exploring the mechanism responsible for the effect of IGF-IIR on trastuzumab resistance in HER-2+ breast cancer cells. In the present study, six single guide (sg)RNA pairs according to the adeno-associated virus integration site 1 (AAVS1) gene sequence were designed and synthesized, and the Universal CRISPR Activity assay CRISPR/Cas9 rapid construction and activity detection kit was used to connect the annealed oligo with the pCS vector. The sgRNA with the highest efficiency was selected to construct a Cas9/sgRNA expression vector using AsiSI + Bstz17I restriction enzymes to cut IGF-IIR. The fragment was ligated into an human AAVS1-KI vector to construct the IGF-IIR targeting vector. The Cas9/sgRNA and IGF-IIR targeting vectors were electroporated into SKBR3 cells, screened using puromycin and identified via PCR, and the mixed cloned cells generated via IGF-IIR gene targeted integration were obtained. The semi-solid and limited dilution methods were used for monoclonal cell preparation, and the results revealed that a Cas9/sgRNA vector that targeted the AAVS1 was successfully constructed. sgRNA activity detection demonstrated that sgRNA2 had the highest efficiency, while enzyme digestion and sequencing confirmed that the IGF-IIR target vector was successfully constructed. The optimum conditions for electrotransfection were 1,200 V, 20 ms and 2 pulses, and the optimal screening concentration of puromycin was 0.5 µg/ml. Using these conditions, the IGF-IIR targeting vector and pCS-sgRNA2 plasmid were successfully transfected into SKBR3 cells, and PCR identification and sequencing verified the correct genotype of mixed clone fragments. The monoclonal cells proliferate slowly and gradually underwent apoptosis. Overall, the present study successfully obtained a mixed clone cell line with site-specific integration of the IGF-IIR gene at the AAVS1.
Keywords: CRISPR/Cas9, type II insulin-like growth factor receptor, breast cancer, knock-in, adeno-associated virus integration site 1
Introduction
According to 2018 global cancer statistics, there are 18.1 million new cancer cases, of which 9.5 million are in men and 8.6 million are in women. In 2018, ~2.1 million women were newly diagnosed with breast cancer, accounting for ~25% of all cancer cases in women, which far exceeded the proportion of other types of cancer (1). In patients with breast cancer, 15–30% of patients overexpress the human epidermal growth factor receptor-2 (HER-2) (2). HER-2+ breast cancer is considered the most dangerous subtype due to its invasiveness, poor differentiation, high risk of recurrence, insensitivity to conventional chemoradiotherapy and poor prognosis (3). Trastuzumab is currently the most effective targeted drug used to treat HER-2+ breast cancer, and it has greatly improved the survival rate of these patients (4). However, clinically, most patients who receive trastuzumab exhibit primary resistance during the initial stage of treatment, or, even if they initially respond to the drug, they will acquire resistance within 1 year of treatment, which decreases the treatment effectiveness and can result in disease progression (5). Therefore, the mechanism of trastuzumab resistance must be ascertained to increase the efficacy of targeted drug therapy and improve the prognosis and survival of patients with breast cancer.
Type II insulin-like growth factor receptor (IGF-IIR) is a member of the insulin-like growth factor (IGF) protein family that binds IGF-II. Since the IGF-IIR lacks tyrosine kinase activity, it can degrade IGF-II and reduce the binding of IGF-II and IGF-IR to promote anti-proliferative and pro-apoptotic activities; therefore, it is considered a tumor suppressor gene (6). On the other hand, IGF-IIR can bind proteins that contain a mannose 6-phosphate sugar group and participate in lysosomal transport (7). Overexpression of the IGF-IIR gene increases the secretion of lysosomal cathepsin D in MCF-7 breast cancer cells, promotes metastasis of breast cancer cells and decreases the disease-free survival time of patients (8). However, the effect of IGF-IIR on HER-2+ breast cancer prognosis remains unclear.
CRISPR/Cas9 technology is based on the short palindrome repeat sequences with regular intervals of clusters found in Archaea. Compared with traditional gene editing technologies, CRISPR/Cas9 can significantly improve the efficiency of gene integration and significantly shorten the homologous arm length of mediated homologous recombination, which is conducive to subsequent detection (9). For example, Ruan et al (10) used CRISPR/Cas9 technology to successfully knock in a 9.4-kb fragment of pig H11 site, with a homology arm of only 800 bp, which reduces the difficulty of amplification and the rate of mismatches. Moreover, it is simple to operate, has low cost and a short cycle time. On the other hand, adeno-associated virus integration site 1 (AAVS1), as a major hotspot for AAV integration, intron 1 of the protein phosphatase 1, is a regulatory subunit 12C (PPP1R12C) gene on human chromosome 19. This locus allows for stable, long-term transgene expression in numerous cell types, including embryonic stem cells. As disruption of PPP1R12C is not associated with any known diseases, the AAVS1 locus is often considered a safe harbor for transgene targeting (11). AAVS1 is an exemplary locus within the PPP1R12C gene that permits robust expression of CAG promoter-driven transgenes (11). In the present study, CRISPR/Cas9 technology was used to mediate IGF-IIR gene knock-in at the safe harbor of AAVS1 in order to construct a HER-2 positive breast cancer cell line SKBR3 that overexpresses IGF-IIR, providing a cell model to determine the influence of IGF-IIR overexpression and the potential to explore the resistance of HER-2+ breast cancer cells to trastuzumab.
Materials and methods
Materials
The SKBR3 cell line was purchased from BeNa Culture Collection; Beijing Beina Chunglian Biotechnology Research Institute. Cells were screened periodically for mycoplasma contamination using the Universal Mycoplasma Detection kit (American Type Culture Collection), which was used according to the manufacturer's protocol. The Universal CRISPR Activity (UCA)™ CRISPR/Cas9 rapid construction and activity detection kit contains three plasmids: Precut pCS, precut pUCA(Luc), and pCS-Positive, which can be used to construct Cas9/sgRNA, identify and screen sgRNA. As well as the human (h)AAVS1-KI vector, were purchased from Beijing Biocytogen Co., Ltd. The FlexiGene® DNA gene extraction kit was purchased from Qiagen GmbH, and the GeneRuler™ 1 kb plus DNA Ladder, T4 DNA ligase, EcoRI, ScaI, NcoI, BglII, Bstz17I+KpnI+ScaI and AsiSI+KpnI were purchased from Thermo Fisher Scientific, Inc. The TIANprep Mini Plasmid Kit and Top10 competent cells were purchased from Tiangen Biotech Co., Ltd. McCoy's 5A+10% FBS+1% L-G+1% NE A+1% SP+1% HEPES was purchased from Gibco; Thermo Fisher Scientific, Inc.
Cell culture
SKBR3 cells were removed from liquid nitrogen, rapidly thawed in a water bath at room temperature, and transferred to a 15-ml tube for centrifugation at 150 × g for 3 min at room temperature. After removing the supernatant, 1 ml complete medium (McCoy's 5A medium with 10% FBS, 1% L-G, 1% Non-Essential Amino Acids, 1% sodium pyruvate and 1% HEPES) was added to the pellet and mixed by pipetting, after which the cell suspension was inoculated in 10 ml complete medium. The cells were cultured at 37°C in a 5% CO2 incubator, and the medium was changed every other day. When the cells were in the logarithmic growth stage, the medium was discarded, and cells were washed with PBS once and digested using 0.25% trypsin at 37°C for 2 min. After gently pipetting, the medium was transferred to a new 15-ml tube for centrifugation at 150 × g for 3 min at room temperature, the supernatant was discarded, and 1 ml complete medium was added. After gentle pipetting to mix, the cells were counted and inoculated into 24-well plates at a concentration of 1×106 cells/well for further culture.
Construction of the Cas9/single guide (sg)RNA plasmid
Using NCBI Primer BLAST (http://www.ncbi.nim.nih.gov/BLAST/), primers were designed 400 bp upstream or downstream of the AAVS1 (upstream primer sequence, 5′-GCATCAAGCTTGGTACCGAT-3′ and downstream primer sequence, 5′-ACTTAATCGTGGAGGATGAT-3′). DNA from the SKBR3 HER-2+ breast cancer cells was extracted using the FlexiGene DNA gene extraction kit, and the target site sequence was amplified using this DNA as a template, aforementioned primers and PrimesSTAR HS DNA Polymerase, named MSD. PCR reaction conditions were pre-denaturation at 94°C for 5 min; 94°C denaturation for 30 sec, annealing at 67°C for 30 sec and extension at 68°C for 1 min, for 15 cycles; denaturation 94°C for 30 sec, annealing at 56°C for 30 sec and extension at 68°C for 1 min, for 25 cycles; extension at 68°C for another 10 min maintained at 4°C. Sequencing confirmed whether the target was consistent with the AAVS1 gene reference sequence in GenBank (https://www.ncbi.nlm.nih.gov/), and the sequencing was completed by Invitrogen; Thermo Fisher Scientific, Inc. The full-length genome sequence of human AAVS1 (Gene ID, 54776) was obtained from GenBank, and six pairs of complementary sgRNA sequences were designed at the AAVS1 target site using the sgRNA Guide Design Resources website (http://crispr.mit.edu/), where the following parameters were selected: CRISPOR tool, where the AAVS1 sequence was first copied; human species; and spCAS9 type. After submission, the sequence with the highest specificity scores were selected as sgRNAs and named sgRNA1-6, where the italic lowercase letters in Oligo are the sticky ends produced by annealing (Table I). According to the Universal CRISPR Activity (UCA)™ CRISPR/Cas9 rapid construction and activity detection kit manufacturer's protocol, sgRNA oligos were denatured-annealed to form a double chain and linked to the linearized pCS plasmid vector according to the manufacturer's protocol (Biocytogen). After confirming the sequence of the resulting construct, named pCS-sgRNA1-6, the activities of the sgRNAs were tested by the Universal CRISPR Activity (UCA)™ CRISPR/Cas9 rapid construction and activity detection kit to detect the level of luciferase activity. Luciferase activity is positively correlated with sgRNA activity. The most efficient sgRNA pair was selected for use in subsequent gene editing experiments.
Table I.
sgRNA design.
| sgRNA | Sequence (5′-3′) |
|---|---|
| sgRNA1 | F: cacc GGAAGGAGGAGGCCTAAGGA |
| R: aaac TCCTTAGGCCTCCTCCTTCC | |
| sgRNA2 | F: cacc GTCACCAATCCTGTCCCTAG |
| R: aaac CTAGGGACAGGATTGGTGAC | |
| sgRNA3 | F: cacc GGGGCCACTAGGGACAGGAT |
| R: aaac ATCCTGTCCCTAGTGGCCCC | |
| sgRNA4 | F: cacc GCACCCCACAGRGGGGCCACT |
| R: aaac AGTGGCCCCACTGTGGGGTGC | |
| sgRNA5 | F: cacc GTCCCCTCCACCCCACAGTG |
| R: aaac CACTGTGGGGTGGAGGGGAC | |
| sgRNA6 | F: cacc GGTTAATGTGGCTCTGGTTC |
| R: aaac GAACCAGAGCCACATTAACC |
F, forward; R, reverse; sgRNA, single guide RNA. The italic lowercase letters in oligo are the sticky ends produced by annealing.
Construction of the IGF-IIR target vector
The IGF-IIR coding sequence (CDS) (https://www.ncbi.nlm.nih.gov/nuccore/NM_000876.3) fragment was divided into two segments (A1 and A2), and A1 was digested using Bstz17I, KpnI and ScaI. After 30 min of enzyme digestion at 37°C, 1% agarose gel electrophoresis was performed, and 3,104-bp bands were recovered. A2 was digested using AsiSI and KpnI, with 4,386-bp bands recovered following electrophoresis. AsiSI and Bstz17I were used for enzyme digestion of the hAAVS1-KI vector (1 kb upstream homology arm, a strong promoter CAG, WPRE regulatory element, ploy A, puromycin resistance gene, 1 kb downstream homology arm), and 1% agarose gel electrophoresis was conducted to recover bands of ~11,232 bp. T4 DNA ligase was used to connect each recovered fragment. After connecting for 30 min at room temperature, the product was transformed into top 10 competent cells for 90 sec at 42°C and cultured overnight. Plasmids were extracted using the TIANprep Mini Plasmid kit, identified using EcoRI and ScaI, and NcoI and BglII restriction endonucleases, and subsequently sequenced by Invitrogen Shanghai Trading Co., Ltd. Sequencing primers for M13 were: Forward, is 5′-GTAAAACGACGGCCAGT-3′ and reverse, 5′-CAGGAAACAGCTATGAC-3′.
Cell transfection and screening
According to the Invitrogen Neon electroporation instrument and the matching electroporation kit (cat. no. MPK10096; Invitrogen; Thermo Fisher Scientific, Inc.) manufacturer's protocol, four sets of electrical parameters were used for cell transfection: i) 1,050 V, 30 ms and one pulse; ii) 1,000 V, 40 ms and one pulse; iii) 1,100 V, 30 ms and two pulses; and iv) 1,200 V, 20 ms and two pulses. Next, transfection efficiency was analyzed using flow cytometry (FACSCalibur; BD Bioscience) and FlowJo 7.6 analysis software, and detected the expression of GFP with a 488-nm laser. SKBR3 antibiotic screening concentrations were determined by treating cells with puromycin at concentrations of 0.2, 0.5, 1.0, 1.5 and 2.0 µg/ml at 37°C and observing cell death under an inverted phase contrast microscope at day 7. The IGF-IIR target vector and pCS-sgRNA plasmid were transferred to SKBR3 cells with a mass ratio of 1:2 (2.4 µg/100 µl:4.8 µg/100 µl) under optimal transfection conditions (1,200 V, 20 ms and 2 pulses), and then puromycin at the optimal concentration (0.5 µg/ml) was added after 2 days. The cells were cultured at 37°C in a 5% CO2 incubator, and the recovery medium used McCoy′5A+10%FBS+1%L-G+1% SP+1%NEAA+1%HEPES. After 7 days of drug exposure at 37°C, the cells were examined via PCR, and monoclonal cell screening was performed.
Identification of mixed clone genotypes
DNA was extracted from the mixed cells according to the FlexiGene® DNA gene extraction kit instructions and used for PCR. The PCR reaction conditions were as follows: Pre-denaturation at 94°C for 2 min; 98°C denaturation for 10 sec, annealing at 67°C for 30 sec, and extension at 68°C for 1 min for 15 cycles; denaturation at 98°C for 10 sec, annealing at 56°C for 30 sec and extension at 68°C for 1 min, for 25 cycles; extension at 68°C for another 10 min and maintained at 4°C. One primer (P1) targeted the lateral side of the left homologous arm, and another primer (P2) targeted the CAG promoter, resulting in an amplification product of 2,307 bp. Primer P3 targeted the IGF-IIR gene, and another primer (P4) targeted the poly-A tail. This primer pair was used to confirm integration of the exogenous gene IGF-IIR with an amplified product size of 8,555 bp. The P5 primer targeted the puromycin resistance gene, and primer P6 targeted the lateral side of the right homologous arm. This pair of primers was used to identify recombination in the right homologous arm, and produced an amplification product of 2,789 bp. Primer sequences are shown in Table II. After identification, single cell cloning was performed.
Table II.
Primer sequences used for PCR identification.
| Primer | Sequence (5′-3′) |
|---|---|
| P1 | CTATGCTGACACCCCGTCCCAGTC |
| P2 | TCGTTGGGCGGTCAGCCAGG |
| P3 | AGCTGAGAGTAGCACAATCTAGGCGTC |
| P4 | GAGCCTCTGCTAACCATGTTC |
| P5 | GCAACCTCCCCTTCTACGAGC |
| P6 | CTAAGAACTTGGGAACAGCCACAGC |
Single cell cloning
The mixed clones were centrifuged at 150 × g for 3 min at room temperature and added to the culture medium. A monoclonal preparation was obtained using the semi-solid and finite dilution methods (12,13). The semi-solid medium was thawed at 4°C overnight, left at room temperature for 10–15 min before the addition of 400 µl complete medium, subjected to vortex oscillation for 4–5 sec, and added to 6-well plates. The remaining semi-solid medium was added to the cell suspension, mixed and cultured in 6-well plates, which was placed in a 37°C incubator for 16 days. For selective cloning, 50 µl/well trypsin was added to 96-well plates, which were placed in an incubator at 37°C for digestion for 2 min. Subsequently, 100 µl complete medium was added to each well to terminate digestion, gently pipetting, and to initiate culture. The cell suspension was diluted to a concentration of 1 cell/100 µl with complete medium using the finite dilution method. After mixing, the cells were divided into 96-well plates with 100 µl complete medium added to each well for monoclonal culture.
Comparison between the proliferation of SKBR3 and mixed clone cells
A cell suspension of SKBR3 or mixed cloned cells was inoculated in a 96-well culture plate (6,000 cells/well) and cultured at 37°C in a 5% CO2 incubator, and observed the SKBR3 and mixed clone cells under an inverted phase contrast microscope. After culturing for 1–5 days, according to a Cell Counting Kit-8 (CCK-8) manufacturer's instructions (Beyotime Institute of Biotechnology), 10 µl/well CCK-8 reagent was added. After mixing, cells were incubated for 2 h at 37°C, and the absorbance was measured at 450 nm. The growth curves were plotted with the culture time as the abscissa and the absorbance as the ordinate.
Statistical analysis
The activity of the Cas9/sgRNA plasmid was determined using SPSS 22.0 software (IBM Corp.). The data obtained from three replicates were analysis of variance to determine the mean ± standard error of each group. The control group was used as the standard to calculate the relative sgRNA activity of each group.
Results
Plasmid construction and assessment of Cas9/sgRNA activity
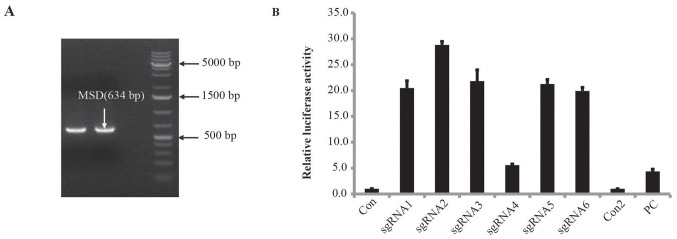
The SKBR3 genome was used as a template to amplify the target sequence AAVS1 with a band size of 634 bp, as shown in Fig. 1A. Sequencing results confirmed that the target sequence was consistent with the reference sequence in GenBank; therefore, sgRNAs were designed according to this sequence information (data not shown). Next, six 20-bp sgRNAs were designed to target the AAVS1 gene sequence and the PAM sequence was NGG (Table III). A Cas9/sgRNA plasmid acting on the AAVS1 gene was successfully constructed, and a UCA™ CRISPR/Cas9 activity detection kit was used to assess sgRNA activity. This assay revealed that sgRNA2 was the most efficient (Fig. 1B).
Figure 1.
Construction of the Cas9/sgRNA plasmid and activity assay. (A) The sgRNA target region was amplified by PCR. (B) Schematic diagram of sgRNA1-6 activity detection. sgRNA, single-guide RNA; Con, sgRNA blank control; PC, positive control; Con2, PC blank control; MSD, plasmid with reporter gene.
Table III.
sgRNA sequences.
| sgRNA sequence number | Sequence (5′-3′) | Length |
|---|---|---|
| sgRNA1 | ggaaggaggaggcctaagga tgg | 20 bp + PAM |
| sgRNA2 | gtcaccaatcctgtccctag tgg | 20 bp + PAM |
| sgRNA3 | ggggccactagggacaggat tgg | 20 bp + PAM |
| sgRNA4 | caccccacagtggggccact agg | 20 bp + PAM |
| sgRNA5 | gtcccctccaccccacagtg ggg | 20 bp + PAM |
| sgRNA6 | ggttaatgtggctctggttc tgg | 20 bp + PAM |
sgRNA, single guide RNA.
Enzyme digestion and identification of IGF-IIR target vectors
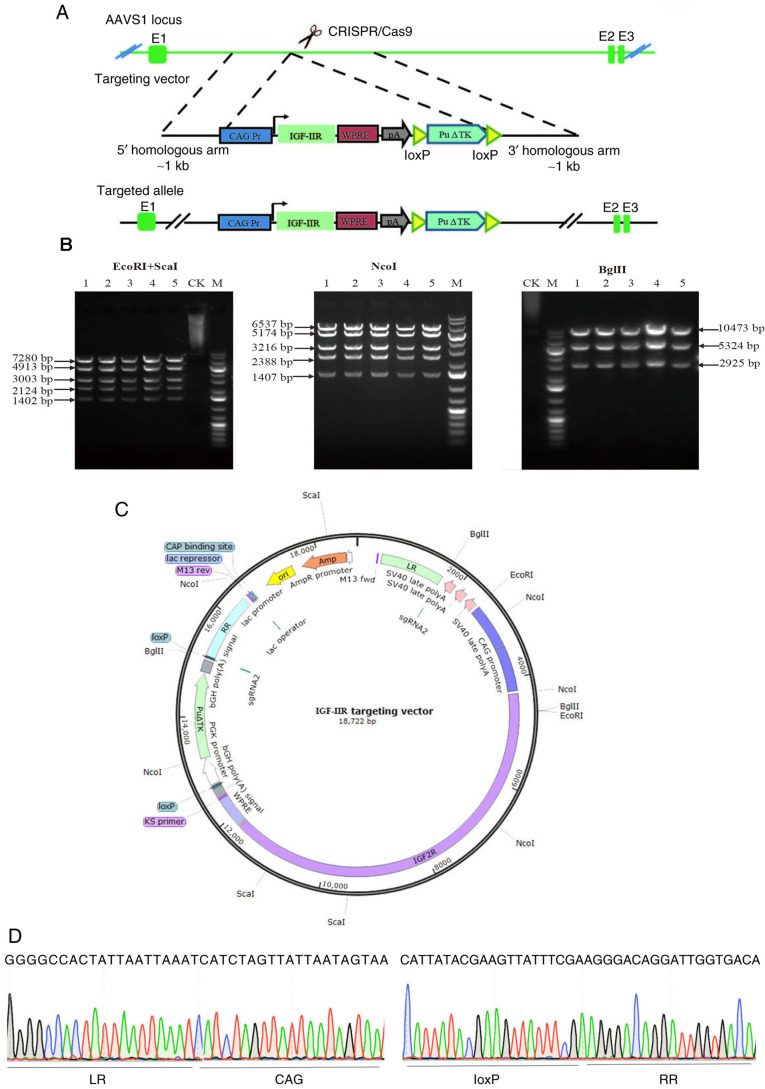
The target gene of IGF-IIR was digested using AsiSI and Bstz17I, and ligated into the hAAVS1-KI vector to construct the IGF-IIR target vector. The schematic diagram is displayed in Fig. 2A. IGF-IIR target vectors were identified by double enzyme digestion using EcoRI and Scal, and agarose gel electrophoresis revealed five bands with sizes of 7,280, 4,913, 3,003, 2,124 and 1,402 bp. After Ncol enzyme digestion, five bands with sizes of 6,537, 5,174, 3,216, 2,388 and 1,407 bp were observed. After BglII enzyme digestion, three bands of the expected sizes (10,473, 5,324 and 2,925 bp) were observed following electrophoresis (Fig. 2B). The restriction sites are shown in Fig. 2C. The IGF-IIR target vector was confirmed by sequencing (Fig. 2D).
Figure 2.
Schematic diagram of the IGF-IIR targeting vector, confirmation of the IGF-IIR targeting vector and target vector sequencing. (A) Schematic diagram of the IGF-IIR targeting vector, including the homologous arms, CAG promoter, IGF-IIR coding sequence, Woodchuck hepatitis virus post-transcriptional regulatory element, poly A and puromycin. (B) IGF-IIR targeting vector confirmed via enzyme digestion (plasmid numbers 1–5). (C) Schematic diagram of restriction sites of IGF-IIR targeting vector. (D) LR provides a partial sequencing diagram connecting the left homology arm to the CAG promoter; RR is a partial sequencing diagram connecting loxP to the right homology arm. CK, undigested control; M, 1-kb DNA ladder; AAVS1, adeno-associated virus integration site 1; IGF-IIR, type II insulin-like growth factor receptor; LR, upstream homology arm; RR, downstream homology arm.
Cell transfection and mixed clone genotyping
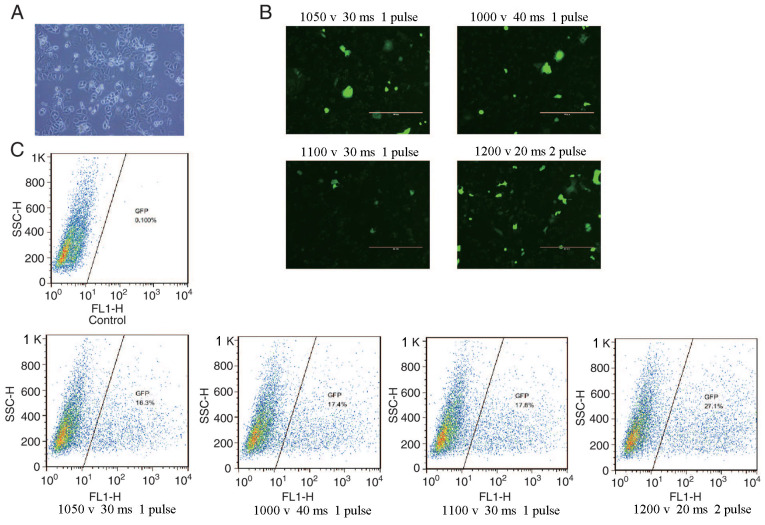
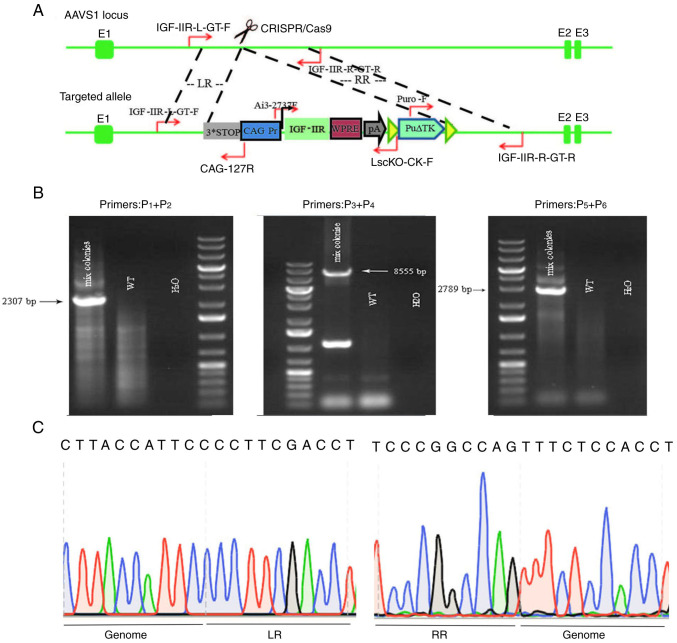
Flow cytometry was used to determine the transfection efficiency of green fluorescent protein particles under different electrotransfer parameters, as indicated in Fig. 3. The results revealed that the highest transfection efficiency was 27.1% obtained at 1,200 V, 20 ms and 2 pulses at 90% cell viability. The cells were treated with different concentrations of puromycin for 7 days, and the optimal concentration of puromycin was determined to be 0.5 µg/ml (data not shown). The pCS-sgRNA2 plasmid with the highest activity and IGF-IIR target vector were transferred to SKBR3 cells at 1,200 V, 20 ms and 2 pulses, and the cells were collected after screening with 0.5 µg/ml puromycin. The genome of the mixed cells was then extracted and used as a template for PCR identification. The identification method is shown in Fig. 4A. As expected, the bands of P1 and P2 corresponded to 2,307 bp, those of P3 and P4 to 8,555 bp, and those of P5 and P6 to 2,789 bp (Fig. 4B). The mixed clones were confirmed by sequencing (Fig. 4C). The results revealed homologous recombination of the IGF-IIR gene with the AAVS1 gene target, and single cell cloning was used for subsequent experiments.
Figure 3.
Cell culture and optimization of transfection conditions. (A) SKBR3 cell morphology under ×10 magnification by an inverted phase contrast microscope. (B) Green fluorescent protein plasmid transfection into SKBR3 cells using different transfection parameters. (C) Flow cytometry analysis of the transfection efficiency to determine the best transfection conditions. SSC, side scatter; H, height; FL1-H, FL1-height.
Figure 4.
Mixed clone identification procedure, determination of mixed clone genotypes and mixed clone fragment sequencing. (A) Mixed clone map showing primer positions for identification via PCR. IGF-IIR-L-GT-F represents primer P1 located upstream of LR. CAG-127R represents primer P2 located at the CAG promoter. Ai3-2737F represents primer P3 located at IGF-IIR. LscKO-CK-F represents primer P4 located at ploy A. Puro-F represents primer P5 located at the puromycin resistance gene. IGF-IIR-R-GT-R represents primer P6 located downstream of the RR. (B) Schematic diagram for PCR identification of three sets of primer mixed clones (WT cells were used as a negative control; H2O was used as a blank control). Mixed cloning primer P1 is located upstream of LR and P2 is located at the CAG promoter, amplifying a fragment of 2,307 bp; P3 is located at IGF-IIR and P4 is located at plot A, amplifying a fragment of 8,555 bp; P5 is located at the puromycin resistance gene and P6 is located downstream of the RR, amplifying a fragment of 2,789 bp. WT, wild-type; IGF-IIR, type II insulin-like growth factor receptor. (C) Schematic diagram of mixed clone sequencing; LR, upstream homology arm; RR, downstream homology arm.
Monoclonal preparation
After 19 days of culture in semi-solid medium, the cell mass was small, and cell proliferation was slow. After 22 days of culture, monoclonal cells were prepared using the limited dilution method. The number of cloned cells was low (~20), their proliferation was extremely slow, and gradually underwent apoptosis (data not shown). Therefore, the cells were not cultured further.
Proliferation of SKBR3 and mixed clone cells
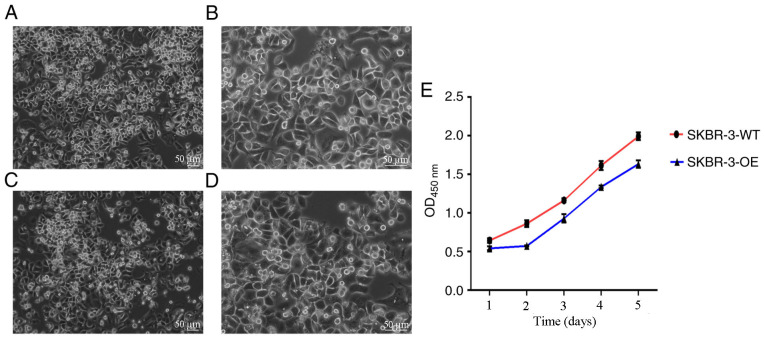
It was observed under an inverted phase contrast microscope that SKBR3 and mixed clone cells were full in morphology and uniform in proliferation. The CCK-8 method was used to detect cell proliferation of SKBR3 cells (SKBR-3-WT) and mixed clone cells (SKBR-3-OE). The results revealed that the proliferation of SKBR-3-OE was markedly lower compared with SKBR-3-WT (Fig. 5).
Figure 5.
Proliferation of SKBR3 and mixed clone cells. Morphology of SKBR3 cells under (A) ×10 and (B) ×20 magnification (scale bar, 50 µm). Morphology of mixed clone cells under (C) ×10 and (D) ×20 magnification (scale bar, 50 µm). (E) Growth curve of SKBR-3-OE and SKBR-3-WT cells. OD, optical density; WT, wild-type; SKBR-3-OE, mixed clone cells.
Discussion
IGF-IIR is a multifunctional receptor capable of binding to the mitotic peptide IGF-II and inhibiting tumor growth by degrading IGF-II to regulate its extracellular levels (14). According to previous studies, the loss of IGF-IIR heterozygosity at the 6q26-27 gene locus can result in the development of multiple types of tumor, including breast cancer, hepatocellular carcinoma and lung squamous cell carcinoma (8,15,16), confirming that IGF-IIR may be involved in inhibiting tumor growth. Furthermore, Lee et al (17) reported that in MDA-MB-231 breast cancer cells, increased IGF-IIR expression inhibited cell invasion and migration, and inhibited tumor growth in mouse models. Similarly, in vivo and study by Souza et al (18) have revealed that overexpression of IGF-IIR in breast cancer cells decreases the growth rate of cancer cells and in certain cases promotes cell death, while IGF-IIR silencing increases cell proliferation and survival. Devi et al (19) demonstrated that IGF-II can promote cell proliferation, inhibit apoptosis and enhance local infiltration and metastasis in breast tumors. Due to the lack of IGF-IIR tyrosine kinase activity, it cannot transmit intracellular mitotic signals. IGF-IR is mainly used as a buffer for the biological activity of IGF-II and its effects, and it can degrade IGF-II and reduce its interaction with IGF-IR combines to exert anti-proliferation and pro-apoptotic activity (20). Specifically, IGF-IR can activate the PI3K/Akt/MAPK/FAK signaling pathways to effectively regulate cell proliferation, anti-apoptosis mechanisms and drug resistance (21).
In this study, the molecular typing of SKBR3 cells is estrogen receptor (ER)−, progesterone receptor (PR)− and HER-23+, so they do not express ER or PR, but highly express the HER-2 gene (22). Using this cell type helps to exclude the impact of ER and PR on the experimental results. Additionally, the Cancer Cell Line Encyclopedia (https://portals.broadinstitute.org/ccle) and BioGPS websites (http://biogps.org/#goto=welcome) revealed that the low expression of IGF-IIR in SKBR3 cells is more conducive to our construction of over-expressing cells, and it is more conducive to compare the growth of wild-type SKBR3 and IGF-IIR over-expressed SKBR3 cells. Therefore, in the present study, CRISPR/Cas9 technology was used to mediate the site-specific knock-in of the IGF-IIR gene at the safe site of AAVS1 to construct a HER-2+ breast cancer SKBR3 cell line that overexpresses IGF-IIR, thereby providing the basis to investigate drug-resistance in breast cancer later, for example trastuzumab.
The CRISPR/Cas9 technology is based on clusters of regularly spaced short palindrome repeats found in Archaea. The CRISPR gene sequence was composed of Cas protein gene, leader sequence and CRISPR locus, of which the nuclease was Cas9 endonuclease, and DNA double-strand cutting was performed through identification of PAM. This technology recognizes DNA sequences with PAM sequences through sgRNA, and brings Cas9 nuclease to specific targets on the genome to complete the cleavage of DNA sequences at specific gene sites (23). This technology is simple and fast to operate, with low associated costs and higher efficiency compared with traditional gene editing technologies (9). van Diemen et al (24) used CRISPR/Cas9 technology to inhibit the three herpes viruses EBV, HSV and HCMV to prevent the replication of herpes viruses in latent infection and lytic infection models. In 2018, Ophinni et al (25) used CRISPR/Cas9 to target HIV-1 regulatory genes and successfully inhibited the replication of HIV-1 in human CD4+ T cell lines with persistent and latent infection. In terms of breast cancer, Feng et al (26) used the CRISPR/Cas9 system to construct an MDA-MB-231 cell line with PinX1 overexpression and knockout, which confirmed that PinX1 overexpression inhibits the proliferation and migration of human basal breast cancer. Low PinX1 expression is associated the malignant behavior of these cells. Mei et al (27) successfully designed an efficient multiplexed CRISPR/dCas9 system using CRISPR/Cas9 technology, and applied it to study the phylogenetic relationship between breast cancer subtypes driven by cancer stems, and clarified that Differentiating the treatment strategies of triple-negative breast cancer and HER-2 positive breast cancer. The CRISPR/Cas9 system has provided a plethora of ideal methods to improve cell and animal models for studying the functions of genes and biological progresses; therefore, it has greatly promoted the study of various diseases in vitro and in vivo (28).
In the present study, six sgRNAs at the AAVS1 target site with a length of ~20 bp were designed, and the PAM sequence was NGG. Using the CRISPOR tool to determine the specificity of the sgRNA sequences, the sequences with high specificity, low off-target effects and high activity were selected to ensure reliable research results. A strong CAG promoter was used to construct the IGF-IIR gene overexpression vector and the transfection conditions were optimized prior to electrotransfection. Four different sets of electrotransfection parameters were used to transfect the green fluorescent protein particles into SKBR3 cells. Flow cytometry analysis of the transfection efficiency under these different parameters revealed that the transfection efficiency was the highest using 1,200 V, 20 ms and 2 pulses at 90% cell viability. Therefore, the most successful transfection conditions were used for subsequent experiments to ensure the generation of a large number of transfected cells. After successful transfection, puromycin screening and PCR identification, a SKBR3 mixed clone cell line with IGF-IIR site-integrated integration at the AAVS1 site was successfully constructed.
By using the limited dilution method with monoclonal cells, there is essentially no damage done to the cells, the operation is simple, the technology is advanced, the cost is low, and no special antibody reagents or equipment are required (29). Furthermore, using a semi-solid medium for the cell growth medium allows for the formation of a single independent dispersed clone, which can prevent the clone from moving, thereby reducing the workload associated with the cell culture and experimental cycle procedure (30). Therefore, the present study used the aforementioned methods for monoclonal cell preparation. However, when selecting monoclonal cells, the proliferation was extremely slow and the cells were in poor condition. Moreover, since the monoclonal preparation method was performed manually, the damage to the cells was low, but the steps were cumbersome. Compared with the coordinated growth of mixed clone cells, the survival efficiency of a single cell is significantly reduced. It was demonstrated that the apoptosis of monoclonal cells is associated with the overexpression of IGF-IIR. IGF-IIR inhibits cell proliferation to a certain extent, leading to cell apoptosis. Future studies should aim to treat the successfully constructed IGF-IIR mixed-site cloned cell line with trastuzumab to observe the effects on cell proliferation, apoptosis, invasion and other characteristics, as well as to construct trastuzumab-resistant cells to investigate the effect of differential IGF-IIR expression on trastuzumab-resistant cells, and explore the role of IGF-IIR in the development of trastuzumab resistance.
In summary, the present study successfully generated a mixed clone cell line of the IGF-IIR gene at the AAVS1 using CRISPR/Cas9 gene editing technology which may be used to explore the effect of IGF-IIR on trastuzumab resistance in HER-2+ breast cancer cells.
Acknowledgements
Not applicable.
Funding
The present study was supported by the Science and technology project of Inner Mongolia autonomous region (grant no. 201802116).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
XM conceived and designed the study, guaranteed its integrity, performed the experiments and wrote the manuscript. RC and HX participated in the design of the experiments, performed the literature search, performed experimental studies and analyzed the data. ZC designed the study, provided financial support, analyzed and interpreted the data, and revised the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Liao N. HER2-positive breast cancer, how far away from the cure? On the current situation of anti-HER2 therapy in breast cancer treatment and survival of patients. Chin Clin Oncol. 2016;5:41. doi: 10.21037/cco.2016.05.10. [DOI] [PubMed] [Google Scholar]
- 3.Harbeck N. Advances in targeting HER2-positive breast cancer. Curr Opin Obstet Gynecol. 2018;30:55–59. doi: 10.1097/GCO.0000000000000431. [DOI] [PubMed] [Google Scholar]
- 4.Gianni L, Eiermann W, Semiglazov V, Lluch A, Tjulandin S, Zambetti M, Moliterni A, Vazquez F, Byakhov MJ, Lichinitser M, et al. Neoadjuvant and adju-vant trastuzumab in patients with HER2-positive locally advanced breast cancer (NOAH): Follow-up of a randomised controlled superiority trial with a parallel HER2-negative cohort. Lancet Oncol. 2014;15:640–647. doi: 10.1016/S1470-2045(14)70080-4. [DOI] [PubMed] [Google Scholar]
- 5.Zhou P, Jiang YZ, Hu X, Sun W, Liu YR, Liu F, Luo RC, Shao ZM. Clinicopathological characteristics of patients with HER2-positive breast cancer and the efficacy of trastuzumab in the People's Republic of China. Onco Targets Ther. 2016;9:2287–2295. doi: 10.2147/OTT.S97583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wong H, Leung R, Kwong A, Chiu J, Liang R, Swanton C, Yau T. Integrating molecular mechanisms and clinical evidence in the management of trastuzumab resistant or refractory Her-2(+) metastatic breast cancer. Oncologist. 2011;16:1535–1546. doi: 10.1634/theoncologist.2011-0165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pollak M. The insulin and insulin-like growth factor receptor family in neoplasia: An update. Nat Rev Cancer. 2012;12:159–169. doi: 10.1038/nrc3215. [DOI] [PubMed] [Google Scholar]
- 8.Hankins GR, De Souza AT, Bentley RC, Patel MR, Marks JR, Iglehart JD, Jirtle RL. M6P/IGF2 receptor: A candidate breast tumor suppressor gene. Oncogene. 1996;12:2003–2009. [PubMed] [Google Scholar]
- 9.Joung JK, Sander JD. TALENs: A widely applicable technology for targeted genome editing. Nat Rev Mol Cell Biol. 2013;14:49–55. doi: 10.1038/nrm3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ruan J, Li H, Xu K, Wu T, Wei J, Zhou R, Liu Z, Mu Y, Yang S, Ouyang H, et al. Highly efficient CRISPR/Cas9-mediated transgene knockin at the H11 locus in pigs. Sci Rep. 2015;5:14253. doi: 10.1038/srep14253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oceguera-Yanez F, Kim SI, Matsumoto T, Tan GW, Xiang L, Hatani T, Kondo T, Ikeya M, Yoshida Y, Inoue H, Woltjen K. Engineering the AAVS1 locus for consistent and scalable transgene expression in human iPSCs and their differentiated derivatives. Methods. 2016;101:43–55. doi: 10.1016/j.ymeth.2015.12.012. [DOI] [PubMed] [Google Scholar]
- 12.Davis JM, Pennington JE, Kubler AM, Conscience JF. A simple, single-step technique for selecting and cloning hybridomas for the production of monoclonal antibodies. Immunol Methods. 1982;50:161–171. doi: 10.1016/0022-1759(82)90222-8. [DOI] [PubMed] [Google Scholar]
- 13.Mao SJ, France DS. Enhancement of limiting dilution in cloning mouse myeloma-spleen hybridomas by human low density lipoproteins. J Immunol Methods. 1984;75:309–316. doi: 10.1016/0022-1759(84)90114-5. [DOI] [PubMed] [Google Scholar]
- 14.Iwamoto KS, Barber CL. Radiation-induced posttranscriptional control of M6P/IGF2r expression in breast cancer cell lines. Mol Carcinog. 2007;46:497–502. doi: 10.1002/mc.20303. [DOI] [PubMed] [Google Scholar]
- 15.O'Gorman DB, Weiss J, Hettiaratchi A, Firth SM, Scott CD. Insulin-like growth factor-II/mannose 6-phosphate receptor overexpression reduces growth of choriocarcinoma cells in vitro and in vivo. Endocrinology. 2002;143:4287–4294. doi: 10.1210/en.2002-220548. [DOI] [PubMed] [Google Scholar]
- 16.O'Gorman DB, Costello M, Weiss J, Firth SM, Scott CD. Decreased insulin-like growth factor-II/mannose 6-phosphate receptor expression enhances tumorigenicity in JEG-3 cells. Cancer Res. 1999;59:5692–5694. [PubMed] [Google Scholar]
- 17.Lee JS, Weiss J, Martin JL, Scott CD. Increased expression of the mannose 6-phosphate/insulin-like growth factor-II receptor in breast cancer cells alters tumorigenic properties in vitro and in vivo. Int J Cancer. 2003;107:564–570. doi: 10.1002/ijc.11453. [DOI] [PubMed] [Google Scholar]
- 18.Souza RF, Wang S, Thakar M, Smolinski KN, Yin J, Zou TT, Kong D, Abraham JM, Toretsky JA, Meltzer SJ. Expression of the wild-type insulin-like growth factor II receptor gene suppresses growth and causes death in colorectal carcinoma cells. Oncogene. 1999;18:4063–4068. doi: 10.1038/sj.onc.1202768. [DOI] [PubMed] [Google Scholar]
- 19.Devi GR, De Souza AT, Byrd JC, Jirtle RL, Macdonald RG. Altered ligand binding insulin-like growth facter2/mannose 6-phosphate receptor bearing missense mutations in human cancers. Cancer Res. 1999;59:4314–4319. [PubMed] [Google Scholar]
- 20.Haisa M. The type 1 insulin-like growth factor receptor signalling system and targeted tyrosine kinase inhibition in cancer. J Int Med Res. 2013;41:253–264. doi: 10.1177/0300060513476585. [DOI] [PubMed] [Google Scholar]
- 21.Pian L, Wen X, Kang L, Li Z, Nie Y, Du Z, Yu D, Zhou L, Jia L, Chen N, et al. Targeting the IGF 1R pathway in breast cancer using antisense inc RNA-mediated promoter cis competition. Mol Ther Nucleic Acids. 2018;12:105–117. doi: 10.1016/j.omtn.2018.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holliday DL, Speirs V. Choosing the right cell line for breast cancer research. Breast Cancer Res. 2011;13:215. doi: 10.1186/bcr2889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hussain W, Mahmood T, Hussain J, Ali N, Shah T, Qayyum S, Khan I. CRISPR/Cas system: A game changing genome editing technology, to treat human genetic diseases. Gene. 2019;685:70–75. doi: 10.1016/j.gene.2018.10.072. [DOI] [PubMed] [Google Scholar]
- 24.van Diemen FR, Kruse EM, Hooykaas MJ, Bruggeling CE, Schürch AC, van Ham PM, Imhof SM, Nijhuis M, Wiertz EJ, Lebbink RJ. CRISPR/Cas9-mediated genome editing of herpesviruses limits productive and latent infections. PLoS Pathog. 2016;12:e1005701. doi: 10.1371/journal.ppat.1005701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ophinni Y, Inoue M, Kotaki T, Kameoka M. CRISPR/Cas9 system targeting regulatory genes of HIV-1 inhibits viral replication in infected T-cell cultures. Sci Rep. 2018;8:7784. doi: 10.1038/s41598-018-26190-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Feng YZ, Zhang QY, Fu MT, Zhang ZF, Wei M, Zhou JY, Shi R. Low expression of PinX1 is associated with malignant behavior in basal-like breast cancer. Oncol Rep. 2017;38:109–119. doi: 10.3892/or.2017.5696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mei Y, Cai D, Dai X. Modulating cancer stemness provides luminal a breast cancer cells with HER2 positive-like features. J Cancer. 2020;11:1162–1169. doi: 10.7150/jca.37117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yoon DS, Kim YH, Jung HS, Paik S, Lee JW. Importance of Sox2 in maintenance of cell proliferation and multipotency of mesenchymal stem cells in low-density culture. Cell Prolif. 2011;44:428–440. doi: 10.1111/j.1365-2184.2011.00770.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Teh CZ, Wong E, Lee CY. Generation of monoclonal antibodies to human chorionic gonadotropin by a facile cloning procedure. J Appl Biochem. 1984;6:48–55. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.