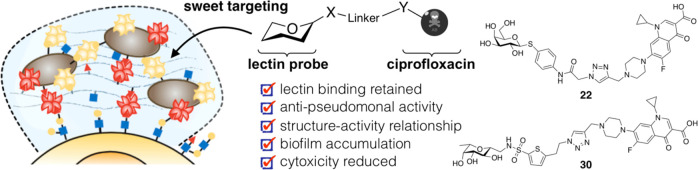
Abstract
Chronic infections by Pseudomonas aeruginosa are characterized by biofilm formation, which effectively enhances resistance toward antibiotics. Biofilm-specific antibiotic delivery could locally increase drug concentration to break antimicrobial resistance and reduce the drug’s peripheral side effects. Two extracellular P. aeruginosa lectins, LecA and LecB, are essential structural components for biofilm formation and thus render a possible anchor for biofilm-targeted drug delivery. The standard-of-care drug ciprofloxacin suffers from severe systemic side effects and was therefore chosen for this approach. We synthesized several ciprofloxacin-carbohydrate conjugates and established a structure–activity relationship. Conjugation of ciprofloxacin to lectin probes enabled biofilm accumulation in vitro, reduced the antibiotic’s cytotoxicity, but also reduced its antibiotic activity against planktonic cells due to a reduced cell permeability and on target activity. This work defines the starting point for new biofilm/lectin-targeted drugs to modulate antibiotic properties and ultimately break antimicrobial resistance.
Introduction
The Gram-negative, opportunistic pathogen Pseudomonas aeruginosa has become a serious threat1−3 for immunocompromised patients (e.g., geriatrics, untreated HIV patients,4,5 and cancer patients6) and people suffering from cystic fibrosis (CF). Severe infections with P. aeruginosa can lead to recurrent pneumonia, lung damage, and sepsis.7 Its intrinsic antimicrobial resistance and its ability to acquire further resistances, which often lead to multidrug-/extensively drug-resistant (MDR/XDR) strains, are major obstacles for therapeutic treatment.8 As a consequence, the WHO stated P. aeruginosa in 2017 to be a critical priority 1 pathogen, which increases research and therapeutic focus on this particular Gram-negative pathogen.9 The ability to colonize almost any part of the human body can lead to various infected tissues, e.g., chronic wound infections, catheter-associated urinary tract infections or pneumonia, and further challenges clinicians to find an appropriate antibiotic therapy. Additionally, pharmacokinetic properties such as tissue distribution, oral bioavailability, and others vary from antibiotic to antibiotic. Thus, not every drug can reach the specific site of infection. Further, high drug levels at sensitive tissues can lead to hazardous side effects, e.g., ototoxicity of many aminoglycosides or tendon rupture and neuropathy after extensive use of fluoroquinolones.
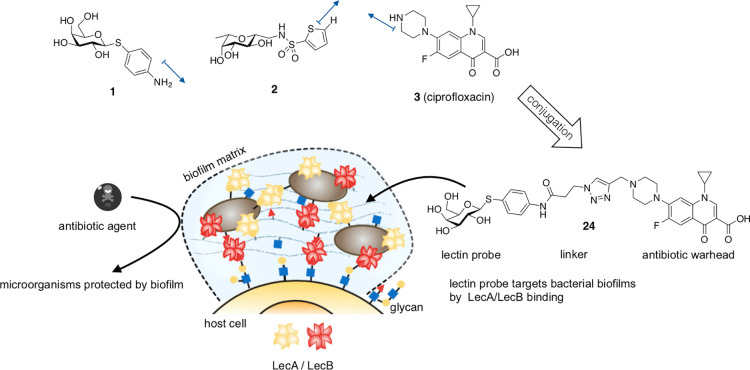
The ability to form biofilms is a hallmark of chronic P. aeruginosa infections. During this stage of living, the cells cluster together in a biofilm matrix and produce a highly impenetrable barrier against host immune defense or antibiotics.10,11 These biofilm cells can show an up to 1000-fold increase in resistance against antibiotic drugs.12 Despite the highly complex composition of the P. aeruginosa biofilm, the two quorum-sensing13 regulated extracellular virulence factors LecA14 and LecB15 (formerly called PA-IL and PA-IIL16−18) stand out. It is assumed that these Ca2+-dependent tetravalent proteins crosslink bacteria with the biofilm matrix as well as host tissue via glycan binding (Figure 1). It was shown that these carbohydrate-binding proteins (i.e., lectins), amongst other biological roles, are crucial for biofilm formation and its structural integrity by P. aeruginosa.(14,15) In the case of the d-mannose(d-Man)- and l-fucose(l-Fuc)-binding LecB, da Silva et al. recently showed that it organizes the localization of the exopolysaccharide Psl in the biofilm matrix.19 Further, both lectins also play roles in the direct infection process: LecB conveys virulence through carbohydrate-dependent inhibition of human ciliary beating,20 interference with repair of wounded tissues,21,22 and activation of B-cells.23 Next to its biofilm-related roles, it was shown that the d-galactose-binding LecA triggers host cell signaling pathways24 and mediates membrane invaginations after binding to its cellular receptor, the glycosphingolipid Gb3.25In vivo, both proteins are involved in the P. aeruginosa infection process and host colonization in a murine infection model.26,27 Interestingly, a study of P. aeruginosa infected CF patients and a case report on a pulmonary infected infant reported that the bacterial load in infected airways can be reduced by intrapulmonary application of fucose and galactose.28−30 Although P. aeruginosa is genetically highly diverse and adaptable,31,32 the protein sequence of LecA is highly conserved amongst clinical isolates. On the other hand, LecB does vary and can be clustered in either PAO1-like or PA14-like structures.33 However, both LecB variants bind to same glycosides, making the design of LecB-inhibitors against a wide range of clinical P. aeruginosa strain isolates possible.33,34
Figure 1.
The lectin inhibitors 1 and 2 are conjugated to the antibiotic ciprofloxacin (3) resulting in pathogen-specific, lectin-targeted antibiotics. These compounds target the biofilm-associated lectins LecA and LecB and therefore increase local antibiotic concentration at the site of infection, resulting in fewer side effects caused by unspecific distribution and tissue accumulation. Blue arrows display growth vectors used in this work.
Lectin-carbohydrate interactions are usually characterized by weak binding affinity, which Nature circumvents by multivalent presentation of ligand or receptor.35 Due to the high therapeutic interest, many compounds have been designed to inhibit LecA or LecB,36−38 most of them showing high affinity on the target in a multivalent fashion.39,40 Interestingly, LecB-directed multivalent molecules with nanomolar on-target activity required millimolar concentrations to inhibit biofilm formation of P. aeruginosa.(26) One possible explanation is the creation of additional crosslinks due to the protein’s and ligand’s multivalent structure, resulting in an undesired stabilization of the biofilm at therapeutic concentrations of the multivalent ligand.
We have previously identified monovalent LecB inhibitors, sulfonamide-capped mannosides, and C-glycosides combining pharmacophores of its natural ligands, fucose and mannose.41−43 Recently, we reported the first drug-like, oral bioavailable LecB inhibitor 1 and established its SAR.44,45 Glycomimetic 1 showed excellent binding affinity against LecB and inhibited biofilm formation in vitro at micromolar concentrations. In mice, high plasma and urine concentrations were obtained after oral application.
Whilst LecB can be inhibited with high affinity ligands, LecA only shows moderate binding affinity against monovalent galactose-based compounds.36−38,46 Instead of a multivalent ligand presentation, we circumvented the rapid dissociation of the ligand–receptor complex by introduction of a electrophilic warhead in the first covalent lectin inhibitor. After conjugation of this galactose-based epoxide to a fluorescent dye, we used the resulting LecA-targeted dye to stain P. aeruginosa biofilms in vitro, proposing its potential use as biofilm-recognizing diagnostic tools.47
Fluoroquinolone antibiotics are frequently used to treat a plethora of bacterial infections. The most common representative of this class is the drug ciprofloxacin, which is amongst other indications being used in cystic fibrosis-associated bronchopulmonary P. aeruginosa infections. Although fluoroquinolones were originally described to be pharmacologically safe, clinical phase IV studies revealed partially irreversible side effects like tendon ruptures or neuropathy, resulting from high tissue penetration and off-target effects. As a consequence, the fluoroquinolones have been categorized by drug agencies as high risk drugs and the U.S. Food and Drug Administration (FDA) issued a “black box” warning label,48 and the German Federal Institute for Drugs and Medical devices (BfArM) informed medical professionals about prescription restrictions in 2019.
Paul Ehrlich coined the concept of a “magic bullet”, describing molecules that would specifically target only pathogenic bacteria or tumor cells.49 One hundred fifty years later, this approach is on the way to become common therapeutic practice: Antibody-drug conjugates like trastuzumab-emtansine50 led to a great success in cancer therapy and are also being studied in antimicrobial research.51 Further, many antibiotic conjugates have been described so far, mainly targeting bacterial uptake mechanisms or non-targeted dual acting antibiotics (reviewed in refs (52, 53)). Interestingly, carbohydrate conjugates of ciprofloxacin were described to increase bacterial cell uptake via sugar transporters.54,55 Inspired by the successful detection of P. aeruginosa biofilms with LecA-directed dyes, we aimed to conjugate glycomimetics to ciprofloxacin in order to target the extracellular P. aeruginosa-specific, biofilm-related virulence factors LecA and LecB. By exploiting lectin accumulation in the P. aeruginosa biofilm, the targeted conjugates shall deliver their antibiotic cargo specifically to the site of infection. Thus, an enhanced local drug concentration could overcome antimicrobial resistance and lower nonspecific drug distribution, potentially reducing systemic side effects (Figure 1). Here, we report the synthesis of the first lectin-targeted antibiotic conjugates and their microbiological and biochemical evaluation. We describe an antimicrobial structure–activity relationship of these lectin binding conjugates and show their biofilm accumulation in vitro.
Results and Discussion
Design
The design of the lectin-targeted conjugates followed the established structure–activity relationships (SAR) of their individual components, i.e., targeting moiety and ciprofloxacin cargo.
The targeted lectins LecA and LecB both show shallow carbohydrate binding sites on their protein surfaces. As a consequence, linking a cargo to specific sites at the published probes without losing lectin inhibition activity was plausible. The SAR of d-galactose-based LecA inhibitors revealed β-linked aromatic aglycons to be vital for potent LecA inhibition. Further substitutions at the aromatic aglycon only result in minor changes in binding affinity.56−58 In the complex with LecA, the ligand’s surface-exposed phenyl aglycon reveals a potential growth vector for the conjugation of cargo to the para-position.59 As this linking strategy was used to stain P. aeruginosa biofilms in vitro,47 we decided to similarly link an antibiotic cargo, using 1 as a LecA targeting probe. To increase the metabolic stability, the O-glycosidic structure was replaced with a thioglycoside. The potent LecB inhibitor 2 displays a C-glycosidic hybrid structure, merging target interactions of d-mannose and l-fucose. The attachment of an aromatic sulfonamide addressed an additional subpocket on LecB.41−44 Analysis of the co-crystal structure of LecB in complex with 2 and extensive SAR studies45 revealed a potential growth vector on position 5 of the thiophene ring for subsequent conjugation to the antibiotic cargo.
Fluoroquinolones represent a highly active class of antibiotics, deriving from their predecessor nalidixic acid. The SAR of the fluoroquinolones60−63 is well described and exploited in several antimicrobial conjugates. Its main pharmacophore, 6-fluoro-quinolone-3-carboxylic acid, is essential for inhibition of its intracellular target, bacterial gyrase. Substitutions at position 7 mainly modify and fine-tune pharmacokinetic properties and strain specificity. In the case of ciprofloxacin, the presence of a piperazine increases anti-pseudomodal activity.64 We chose to derivatize the synthetically accessible secondary amine of the piperazine ring to a tertiary amine as this would result only in a smaller change of its physicochemical properties that influence porin-mediated bacterial cell uptake, as compared to, e.g., amide formation. Furthermore, analysis of the co-crystal structure65 of ciprofloxacin with the GyrA/GyrB heterodimer showed a possible growth vector at this position (Figure 1).
Copper-catalyzed Huisgen-type [3+2] cycloaddition of terminal alkynes and terminal azides was chosen as a convenient and modular way of linking both moieties. Further, we decided to analyze the impact of the linker length and flexibility on antibiotic activity by stepwise introduction of methylene spacers.
Synthesis
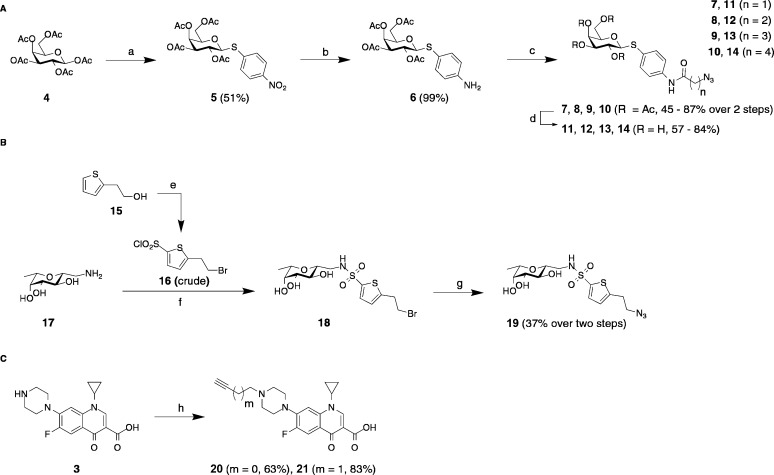
The LecA-targeting precursor 6 (Scheme 1) was synthesized in analogy to Casoni et al.66 Glycosylation of the acceptor para-nitrothiophenol with galactose pentaacetate (4) using BF3·Et2O as a Lewis acid resulted in thioglycoside 5 in 51% yield. Palladium-catalyzed hydrogenation gave the corresponding aniline 6 quantitatively. Compound 6 was then treated with various ω-bromo acylhalides followed by a nucleophilic substitution with sodium azide to the corresponding azides 7–10 in one pot. The usage of triethylamine during the amide coupling led to β-elimination in the case of the propionic acid derivative 7 or γ-lactam formation in the case of bromide 13, which could be circumvented by using potassium carbonate as a base. Deprotection of acetates 7–10 under Zemplén conditions resulted in the LecA-probes 11–14.
Scheme 1. Chemical Synthesis of the (A) LecA-Targeting (11–14) and (B) LecB-Targeting (19) Probes and (C) Alkyne Ciprofloxacin Derivatives 20 and 21.
Reagents and conditions: (a) p-nitrothiophenol, BF3·Et2O, CH2Cl2, 0 °C to r.t., 16 h; (b) H2, Pd/C, CH2Cl2, r.t., 24 h; (c) (i) Br(CH2)nCOHal, Et3N, or K2CO3, DMF, 0 °C to r.t., 1–4 h, (ii) NaN3, DMF, r.t., 4 h; (d) cat. NaOMe, MeOH, r.t., 1 h; (e) (i) PBr3, CH2Cl2, 0 °C to r.t., 1 h, (ii) HSO3Cl, CH2Cl2, 0 °C to r.t., 3 h; (f) crude 16, K2CO3, DMF, r.t., 5 h; (g) NaN3, DMF, r.t., 5 h; (h) propargylbromide or 4-bromo-but-1-yne, Et3N, DMF, 70 °C, 1–4 d.
Based on the results from the antimicrobial susceptibility testing (vide infra), we synthesized only one LecB probe (Scheme 1). β-C-glycoside 17 was synthesized as reported.42 Thiophene building block 16 was synthesized from 15 in two steps: The primary alcohol 15 was transformed to the corresponding bromide with phosphorous tribromide followed by chlorosulfonation of the thiophene in position 5 with chlorosulfonic acid. Crude sulfonylchloride 16 was reacted with amine 17 to yield sulfonamide 18. This intermediate was stirred with sodium azide to give compound 19 in an overall yield of 37% over two steps based on the amine starting material 17.
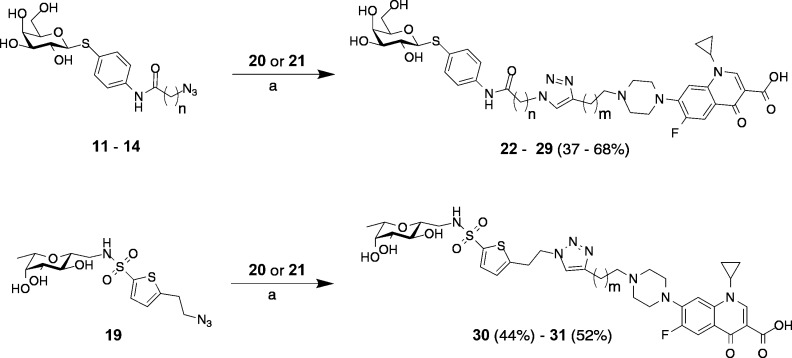
Alkylation of ciprofloxacin with propargyl bromide or 4-bromobut-1-yne in DMF at elevated temperatures yielded the corresponding terminal alkynes 20 and 21. Finally, copper-catalyzed 1,3-dipolar cycloaddition of alkynes 20 and 21 with azides 11–14 and 19 resulted in the lectin-targeted ciprofloxacin conjugates 22–31 (Scheme 2).
Scheme 2. Assembly of the Lectin-Targeted Ciprofloxacin Conjugates.
Reagents and conditions: (a) cat. CuSO4, cat. sodium ascorbate, DMF/H2O, r.t. 16 h, r.t. (for 11–14) or 40 °C (for 19).
Biophysical and Microbiological Evaluation
Competitive Lectin Binding Assay Based on Fluorescence Polarization
To analyze lectin binding of the targeted antibiotics, we quantified their binding affinity to LecA or LecB in the previously reported competitive binding assays.33,41,58
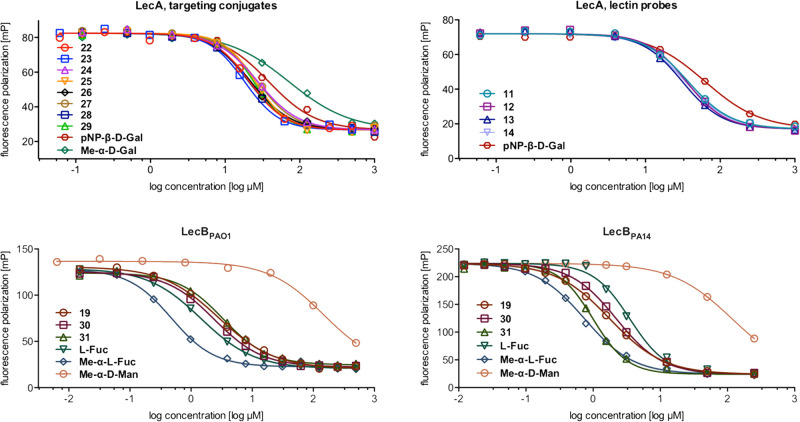
The binding affinity of the LecA-targeting conjugates 22–29 did not significantly differ from their corresponding lectin probes 11–14 (Figure 2 and Table 1), reaching IC50 values from 26 to 30 μM. Thus, they show an up to 2-fold increased inhibitory activity against LecA compared to p-nitrophenyl β-d-galactoside (pNP-β-d-Gal, IC50 = 52.7 ± 13 μM) and an up to 3-fold increase compared to methyl α-d-galactoside (Me-α-d-Gal, IC50 = 71.7 ± 16 μM), which served as reference compounds in this study.
Figure 2.
Competitive binding assay of lectin-targeted ciprofloxacin conjugates 22–31, lectin probes 11–14 and 19, and control compounds with LecA, LecBPAO1, and LecBPA14. One representative titration of triplicates on one plate is shown for each compound (IC50 in Table 1 and Ki in Table S1).
Table 1. Competitive Binding Assay of Lectin-Targeted Ciprofloxacin Conjugates and Control Compounds with LecA, LecBPAO1, and LecBPA14a.
| LecA | |||
|---|---|---|---|
| compound | n | m | IC50 ± s.d. [μM] |
| 11 | 1 | LecA-probes | 31.7 ± 11 |
| 12 | 2 | 30.9 ± 8.7 | |
| 13 | 3 | 31.1 ± 8.3 | |
| 14 | 4 | 29.9 ± 9.5 | |
| 22 | 1 | 0 | 30.4 ± 8.0 |
| 23 | 1 | 1 | 21.6 ± 5.5 |
| 24 | 2 | 0 | 32.2 ± 3.3 |
| 25 | 2 | 1 | 28.0 ± 1.8 |
| 26 | 3 | 0 | 27.3 ± 4.0 |
| 27 | 3 | 1 | 29.3 ± 3.7 |
| 28 | 4 | 0 | 28.3 ± 8.1 |
| 29 | 4 | 1 | 26.2 ± 2.4 |
| Me-α-d-Gal | controls | 71.7 ± 16 | |
| pNP-β-d-Gal | 52.7 ± 13 | ||
| LecBPAO1 | LecBPA14 | ||
|---|---|---|---|
| compound | m | IC50 ± s.d. [μM] | IC50 ± s.d. [μM] |
| 19 | LecB-probe | 3.91 ± 1.6 | 1.87 ± 0.21 |
| 30 | 0 | 2.37 ± 1.2 | 2.24 ± 0.23 |
| 31 | 1 | 2.53 ± 0.87 | 1.00 ± 0.06 |
| Me-α-d-Man | controls | 166 ± 22 | 101 ± 10 |
| l-Fuc | 2.63 ± 1.7 | 2.46 ± 0.33 | |
| Me-α-l-Fuc | 0.534 ± 0.07 | 0.79 ± 0.11 | |
Means and standard deviations were determined from a minimum of three independent experiments. Ki calculated from IC50 is shown in Table S1.
Competitive binding assays against LecBPAO1 (Figure 2 and Table 1) revealed IC50 values in the one digit micromolar range for LecB probe 19 (IC50 = 3.91 ± 1.6 μM) and its corresponding conjugates 30 and 31 (IC50 = 2.37 ± 1.2 and 2.53 ± 0.87 μM, respectively), which is in the range of l-fucose (IC50 = 2.63 ± 1.7 μM). The two glycosides, methyl α-d-mannoside (Me-α-d-Man) and methyl α-l-fucoside (Me-α-l-Fuc), which resemble terminal glycan structures recognized by LecB showed IC50 values of 166 ± 22 and 0.534 ± 0.07 μM, respectively. The inhibition assay on LecBPA14 showed similar trends (Table 1). As observed previously,33 LecBPA14 binds its ligands with higher affinity (e.g., IC50 of 1.00 μM vs 2.53 μM for compound 31). Since P. aeruginosa PA14 and PAO1 are representative for many clinical isolates, a broad range of P. aeruginosa strains can be targeted by these conjugates.
Comparing the conjugates with the unlinked lectin probes showed in all cases a comparable binding affinity. Further, all compounds showed better binding than Me-α-d-Gal (LecA) or Me-α-d-Man (LecB). Due to the highly optimized structure of the fucose-mannose pharmacophore, the LecB targeting compounds were comparably active on LecB as l-fucose. In conclusion, the topology of the carbohydrate binding sites in both proteins allowed the conjugation with an antibiotic cargo without influencing lectin binding.
Antibiotic Susceptibility Assay
The antibiotic activity of lectin-targeted ciprofloxacin conjugates 22–31 was tested against a panel of Gram-positive and Gram-negative bacteria (Table 2). The model organisms E. coli MG1655 (a common lab strain), E. coli DSM 1116 (an antibiotic susceptibility reference strain recommended by the DSMZ), and the Gram-positive Staphylococcus carnosus DSM 20501 were tested first to assess Gram-negative specific antibiotic activity and strain specificity. Afterward, the antibiotic activity against the two P. aeruginosa strains PA14 and PAO1 was studied. These two important reference strains represent a broad range of clinical isolates and are well studied in the literature.33 To determine the effect of the lectins’ presence on antibiotic activity, we used the lectin-deficient knockout mutants of P. aeruginosa PA14, i.e., PA14 ΔlecA and PA14 ΔlecB. Ciprofloxacin (3) and the synthetic intermediate 20 were used as reference compounds to study the effect of piperazine N-alkylation on antibiotic activity.
Table 2. Antibacterial Activity of Lectin Targeted Conjugates 22–31, 20, and Ciprofloxacin (3) against a Panel of Bacterial Organisms. LecA-targeting galactosides were generally more active than the LecB-targeting conjugates. A shorter linker length on the side of the antibiotic led to increased antimicrobial activitya.
| target:
LecA |
target: LecB |
references |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| compound | 22 | 23 | 24 | 25 | 26 | 27 | 28 | 29 | 30 | 31 | 20 | 3 |
| molecular mass [g/mol] | 739.8 | 753.8 | 753.8 | 767.8 | 767.8 | 781.9 | 781.9 | 795.9 | 761.8 | 775.9 | 369.4 | 331.3 |
| linker length n/m | 1/0 | 1/1 | 2/0 | 2/1 | 3/0 | 3/1 | 4/0 | 4/1 | -/0 | -/1 | 0 | |
| test organism | MIC [μg/mL] | |||||||||||
| E. coli K12 MG1655 | 2 | 8–16 | 2 | 16 | 1–2 | 16 | 2–4 | 16 | 8–16 | 16 | n.d. | <0.125 |
| E. coli DSM 1116 | 2–4 | 16 | 2–4 | 32 | 2–32 | 4–32 | 4–32 | 4–32 | 16–32 | 32 | n.d. | <0.125 |
| S. carnosus DSM 20501 | 32 | 64 | 32 | >64 | 16 | 64 | 8 | ≥64 | >64 | >64 | n.d. | <0.125 |
| P. aeruginosa PA14 wt | 16 | ≥64 | 8–16 | >64 | 8–16 | >64 | 32 | >64 | 64 | >64 | 2–4 | 0.025–0.1 |
| P. aeruginosa PA14 wt + 1 μg/mL PMBN | 4–16 | 16–64 | 8–16 | 32–64 | 4 | 32–64 | 2–8 | 32–64 | 64 | 64 | 0.025–0.5 | 0.025 |
| P. aeruginosa PA14 ΔlecA | 16–32 | ≥64 | 8–16 | >64 | 8–16 | >64 | 32 | >64 | ≥64 | >64 | 4–8 | 0.05–0.08 |
| P. aeruginosa PA14 ΔlecB | 16–32 | ≥64 | 8–32 | >64 | 8–16 | >64 | 32–64 | >64 | 64 | >64 | 4 | 0.05–0.08 |
| P. aeruginosa PAO1 wt | 16–32 | >64 | 16 | >64 | 16–32 | >64 | 32–64 | >64 | ≥64 | >64 | 4–8 | 0.025–0.08 |
| P. aeruginosa PAO1 wt + 1 μg/mL PMBN | 4–8 | 32–64 | 4–8 | 32–64 | 4–8 | 32–64 | 8–16 | 32–64 | 32–64 | ≥64 | 1–2 | 0.025–0.05 |
Data is presented as minimal inhibitory concentration (MIC) range from at least three independent experiments. Molar MIC is given in Table S2. n.d. = not determined.
Ciprofloxacin is known to be particularly active against Gram-negative compared to Gram-positive organisms. Both E. coli strains showed higher susceptibility against the ciprofloxacin conjugates than the Gram-positive organism S. carnosus. Comparing both E. coli strains, the antibiotic susceptibility reference strain (DSM 1116) showed similar or slightly higher MIC values (Table 2).
Compared to E. coli, P. aeruginosa PA14 and PAO1 both showed lower susceptibility against all compounds tested, which was expected due to the well-known increased intrinsic antimicrobial resistance of P. aeruginosa. It was also observed that the clinical isolate PAO1 was similarly or slightly less susceptible than the clinical isolate PA14. Importantly, some of the lectin-targeted conjugates reached antibiotic activity down to 8 μg/mL against planktonic P. aeruginosa (Table 2).
Comparing the MIC values amongst the different conjugates and the reference compounds 20 and ciprofloxacin (3), we observed a structure–activity relationship: Conjugates containing galactosides as lectin-targeting probes showed higher antimicrobial activity than LecB-targeting compounds, which are based on a C-glycosidic hybrid structure. It has been previously postulated that galactosides are recognized by the bacterial sugar uptake machinery,54,55 which would result in an active transportation over the Gram-negative cell wall. A comparative study by O’Shea and Moser68 on commonly used antibiotics showed that especially P. aeruginosa active compounds have clogD values of <0. LogD calculation (data not shown) of all conjugates 22–31 and 20 revealed positive values, which could explain the reduction in antimicrobial activity with respect to ciprofloxacin (3) showing a clogD of <0.
Further, a decreased linker length between triazole and ciprofloxacin (entitled m in the structure drawings) amplified the antibiotic activity in all cases, independent of the carbohydrate probe or microorganism tested. This effect becomes most evident in case of E. coli K12 MG1655, where an up to 8-fold increase in MIC could be observed (e.g., 24 vs 25, Table 2). We assume that changing the distance between the tertiary amine and the electron-withdrawing triazole affects the amine’s basicity, which is believed to play a role in porin diffusion.67 The parent drug ciprofloxacin reached MIC values of 0.025–0.1 μg/mL against P. aeruginosa, while the propargylated derivative 20 showed MIC values of 2–4 μg/mL against P. aeruginosa PA14 and 4–8 μg/mL against P. aeruginosa PAO1, thereby reaching the concentration range of the most potent conjugates. As alkylation of ciprofloxacin alone already led to a significant decrease in activity, conjugation at the secondary amine in the piperazine ring is most likely responsible for the decreased antibiotic activity.60−62
Regarding total linker size, increasing length resulted in higher MIC values (e.g., 22 vs 29), which can be explained by a size exclusion effect of outer membrane porins. It is believed that these barrel-formed, hydrophilic channels play crucial roles for membrane permeation of hydrophilic compounds and are limited to a certain molecular weight or three-dimensional molecular structure.67,68 Further, the introduction of additional methylene groups results in an increased number of rotatable bonds and increased lipophilicity, which is also described to reduce bacterial cell uptake.67,68 We compared retention times from reversed-phase HPLC analyses as a surrogate parameter for lipophilicity (Table S4 and Figure S4). Two trends were observed that correlated with the antimicrobial activity assays: (i) In general, all galactose-based conjugates showed lower retention times than the C-glycosides indicative for higher polarity, and (ii) the stepwise introduction of methylene groups in both linkers led to a stepwise increase in retention times indicating higher lipophilicity, which correlated with the reduced antimicrobial activity. Only the shortest galactose-based conjugates 22 and 23 (n = 1, m = 0 or 1, respectively) showed retention times slightly higher than expected in their series, which may be a result of an intramolecular hydrogen bonding between the amide NH and the central nitrogen atom of the triazole for n = 1 altering their conformation and thus their physicochemical properties. We observed that the most anti-Pseudomonas active compound 24 showed the lowest retention time amongst the conjugates. Thus, we conclude that the conjugates’ lipophilicity is an important parameter for antimicrobial activity. Ciprofloxacin (3) was eluted much earlier than all conjugates, reflecting its higher hydrophilicity.
Polymyxin B nonapeptide (PMBN) is a membrane-active antimicrobial compound that is used at sub-MIC concentrations to increase outer membrane permeability. Without being lethal to the microbe, this can provide information on bacterial cellular uptake of antimicrobial drugs. In our studies, all conjugates, except 24 and 30, benefit from the presence of the permeabilizer at least twofold (e.g., 26, Table 2). Interestingly, the MIC of reference compound 20 was increased most and reached high antimicrobial activity approximating ciprofloxacin. Thus, the drop in antibiotic activity for the conjugates can partially be explained by decreased cell wall permeability, as a consequence of derivatization of the secondary amine. As expected, unmodified ciprofloxacin benefitted only marginally by the addition of PMBN.
Gyrase-Dependent DNA Supercoiling Inhibition Assay
The antimicrobial susceptibility assays revealed a decrease in antibiotic activity after conjugation (Table 2). We showed that this decrease is most likely caused by a reduced bacterial cellular uptake as shown by the co-incubation experiments with membrane permeabilizer. However, the addition of PMBN did not result in MIC values comparable to ciprofloxacin, suggesting that further features are affected by conjugation of ciprofloxacin to the lectin probes. Thus, we investigated the compounds’ ability to inhibit bacterial gyrase, the target of ciprofloxacin.
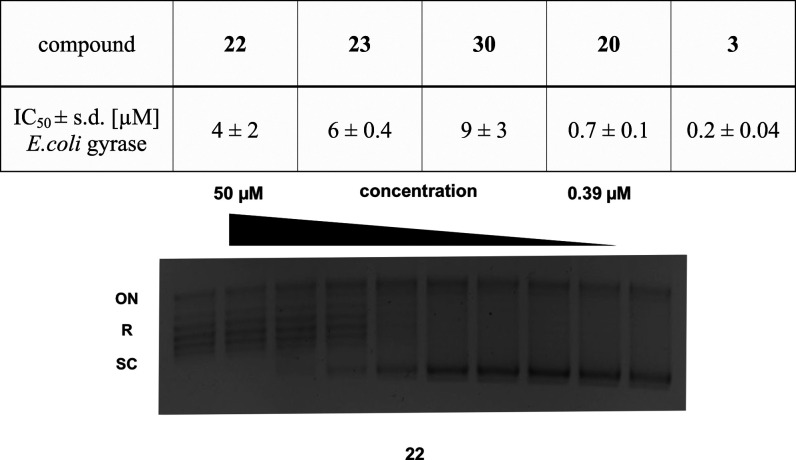
We compared the gyrase inhibition activity of three conjugates (22, 23, and 30), while the propargylated ciprofloxacin derivative 20 and unmodified ciprofloxacin (3) were used as controls (Figure 3). Gyrase-inhibition leads to a reduction of supercoiled DNA, which can be visualized by gel electrophoresis. Ciprofloxacin was the most active compound, reaching full inhibition of plasmid supercoiling in the nanomolar range. Compound 20 (IC50 = 0.7 ± 0.1 μM) was less active than ciprofloxacin; however, it still showed an IC50 in the nanomolar range, suggesting that modification in this region of the molecule as concluded from the crystal structure analysis is indeed possible. The lectin-targeting conjugates were also potent inhibitors of gyrase supercoiling activity in the single digit micromolar range, although they were not as potent as reference compounds 20 and 3. This decrease in activity explains why the compounds did not reach the antibiotic activity of N-propargyl ciprofloxacin (20) after membrane permeabilization with PMBN.
Figure 3.
Effect of 20, 22, 23, 30, and ciprofloxacin (3) on gyrase-catalyzed DNA supercoiling. Propargylation (20) decreased the inhibitory concentration only by a factor of 3.5 compared to 3. Gyrase inhibition as a putative mode of action was confirmed as all conjugates inhibit gyrase-catalyzed DNA supercoiling. Mean and standard deviations were determined from three independent experiments. A representative titration of E. coli gyrase with 22 in a supercoiling inhibition assay is shown. Controls: plasmid without gyrase and inhibitor (leftmost band) and plasmid with gyrase and without inhibitor (rightmost band). ON, open circular/nicked plasmid; R, relaxed topoisomers; SC, supercoiled topoisomers of E. coli DNA.
P. aeruginosa Biofilm Accumulation Assay
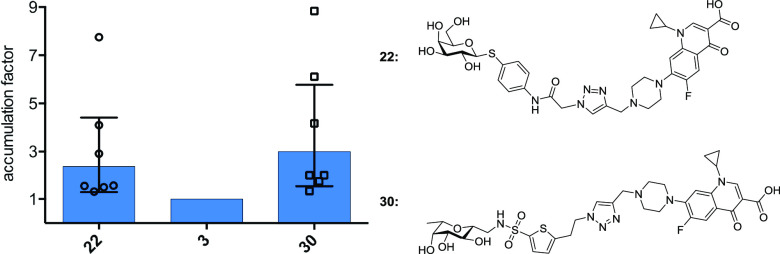
Since the carbohydrate-ciprofloxacin conjugates 22–31 bind their respective lectins in a competitive binding assay, we investigated the ability of two representative lectin-targeting conjugates to accumulate in P. aeruginosa biofilms in vitro (Figure 4).
Figure 4.
Accumulation of 22 (targeting LecA) and 30 (targeting LecB) in P. aeruginosa PAO1 biofilm relative to ciprofloxacin (3). Each data point reflects the relative accumulation compared to ciprofloxacin of a single independent assay with at least three technical replicates. Bars show geometric mean and 95% confidence interval (see the Supporting Information for more detailed information, Figure S2).
For this purpose, biofilms were grown on peg lids in a 96-well format that allows incubation and washing steps in a batch format. After 24 h of bacterial growth, P. aeruginosa PAO1 formed a visible biofilm on the pegs, which was used for compound accumulation assays. After one washing step to remove planktonic bacteria, the biofilm was immersed for 10 min into solutions containing two lectin-targeting conjugates (22 and 30) or ciprofloxacin (3) at 100 μM. After a subsequent washing step to remove an unspecifically bound compound, the biofilm was disrupted and the amount of bound compound was quantified by LC-MS/MS.
Although the assay showed variation in absolute compound binding between biological replicates (Figure S2), we observed an obvious trend: the lectin targeted conjugates reached higher concentrations in the bacterial biofilm than the unmodified ciprofloxacin, independent of their lectin targeting moiety (Figure 4). These results are fundamental for the future development of further biofilm targeting antibiotic conjugates.
In Vitro Early ADMET
Metabolic stability of two representative conjugates (22 and 30) and ciprofloxacin (3) as the parent molecule was studied in vitro against human plasma, human liver microsomes, and mouse liver microsomes (Table 3). The data reveals high metabolic stability in all matrices tested: half-life in human plasma was above 150 min for all compounds and microsomal clearance by mouse and human liver microsomes was very low on the lectin-targeting compounds. Both conjugates showed clearance of 10 μL/min/mg protein by human liver microsomes, reaching the assay’s lower limit. Against mouse liver microsomes, compound 22 also reached the assay limit of 10 μL/min/mg protein, whereas 30 was slightly less stable (CLMIC = 15 μL/min/mg protein) but still classified in the most stable category of this assay (≤15 μL/min/mg protein). Thus, the compounds are considered metabolically stable, fitting the molecular design approach as S-/C-glycosides. Both conjugates showed higher plasma protein binding than ciprofloxacin (69 ± 7% for 22, 75 ± 10% for 30 vs 33 ± 2% for 3).
Table 3. Early ADMET Data on Two Representative Lectin-Targeted Conjugates (22 and 30) and Ciprofloxacin (3): All Compounds Were Metabolically Stable in Human Plasma and Microsomal Fractions. Cytotoxicity was reduced compared to ciprofloxacina.
| metabolic
stability |
|
|||||
|---|---|---|---|---|---|---|
| t1/2 [min] | CLMIC [μL/min/mg protein] |
plasma protein binding [%] | cytotoxicity @ 100 μM [% inhibition] |
|||
| compound | human plasma | MLM | HLM | human plasma | HEK293 | A549 |
| 22 | >150 | 10 | 10 | 69 ± 7 | 8 ± 4 | 5 ± 22 |
| 30 | >150 | 10 | 15 | 75 ± 10 | 11 ± 12 | –9 ± 15 |
| 3 | >150 | n.d. | n.d. | 33 ± 2 | 48 ± 5 | 18 ± 11 |
Data is presented as mean and standard deviation from at least two independent experiments (exception: one experiment for CLMIC data). MLM, mouse liver microsomes; HLM, human liver microsomes; n.d., not determined.
Acute cytotoxicity was tested against a human embryonic kidney cell line (HEK 293) and adenocarcinoma human alveolar basal epithelial cells (A549). Compounds 22 and 30 showed no cytotoxicity at 100 μM after 48 h incubation, whereas ciprofloxacin showed detectable cytotoxicity (48 ± 5% inhibition) against HEK 293 cells (Table 3). Furthermore, penetration over cultured human airway epithelial cells (Calu-3 HTB-55) was assessed in vitro via a Transwell system to analyze the compounds ability to permeate over mammalian cell membranes. No detectable permeation (apical to basal) was observed for compounds 22 and 30 after 4 h, while 10% ciprofloxacin was permeated after 4 h (data not shown). The low acute toxicity against human alveolar basal epithelial cells and the low lung cell permeation suggest the possibility of pulmonal application routes for patients suffering from cystic fibrosis.
Conclusions and Outlook
Biofilms present a hallmark in chronic P. aeruginosa infections. The ability to protect against the host immune system and antibiotic treatment renders this chemo-mechanic barrier as a strong virulence factor. Notably, it is not advisable to solely focus MIC optimization on planktonic cells during the development of new antibiotics but rather to find new therapeutic strategies. As an example, Müsken et al. showed that biofilm susceptibility of clinical P. aeruginosa isolates cannot be deduced from commonly studied phenotypes like MIC or minimal bactericidal concentration values.69 Delivering antibiotics specifically to the site of infection could decrease potential side effects and enhance efficacy. In this work, we developed and characterized the first P. aeruginosa lectin-targeted antibiotic conjugates. Based on our previous work, we conjugated ciprofloxacin to LecA and LecB probes and varied the linker length.
The antibiotic conjugates showed effective lectin binding against LecA and both LecB variants from P. aeruginosa PAO1 and PA14, which represent a broad range of clinical isolates of P. aeruginosa. A structure–activity relationship regarding the antimicrobial activity of the synthesized conjugates could be established. In general, a shorter spacer between triazole and antibiotic as well as a d-galactose-based lectin probe was preferred. The observed reduction in antibiotic activity could be rationalized due to a higher molecular weight, decreasing the ability to penetrate the Gram-negative cell wall. Comparison with N-propargylated ciprofloxacin showed, that alkylation of the secondary amine of the piperazine ring already resulted in a decreased antibiotic activity. Further, we proved the inhibition of gyrase-catalyzed DNA supercoiling as the conjugates’ antimicrobial mode of action.
In the first P. aeruginosa biofilm accumulation assay, we observed an enrichment of lectin-targeting conjugates compared to ciprofloxacin, which could compensate for the decrease in antimicrobial activity. Since cytotoxicity of both conjugates was decreased compared to ciprofloxacin especially against kidney cells, and the biofilm accumulation was achieved, a reduction of the severe systemic side effects of ciprofloxacin is possible. Further, in vitro metabolism assays showed good metabolic stability supporting the conjugates’ design as S- or C-glycosides.
This work reports the first P. aeruginosa biofilm-targeted antibiotics and analyzes their properties on lectin binding, antimicrobial activity, target inhibition, and biofilm enrichment. In vitro studies revealed a reduced cytotoxicity of the conjugates compared to the parent drug ciprofloxacin. Future work will address the improvement of antimicrobial activity of the antibiotic conjugates. Our modular synthesis allows the conjugation of lectin probes to other antibiotics, leading to future generations of biofilm targeting antibiotics.
Experimental Section
Chemical Synthesis
Thin layer chromatography (TLC) was performed on Silica Gel 60 coated aluminum sheets containing a fluorescence indicator (Merck KGaA, Darmstadt, Germany) and developed under UV light (254 nm) and aqueous KMnO4 solution or a molybdate solution (a 0.02 M solution of ammonium cerium sulfate dihydrate and ammonium molybdate tetrahydrate in aqueous 10% H2SO4). Self-packed Silica Gel 60 columns (60 Å, 400 mesh particle size, Fluka, for normal-phase liquid chromatography) or Chromabond Flash RS15 C18 ec columns (Macherey-Nagel, Düren, Germany, for reversed-phase liquid chromatography), and a Teledyne Isco Combiflash Rf200 system were used for preparative medium pressure liquid chromatography (MPLC). Nuclear magnetic resonance (NMR) spectroscopy was performed on a Bruker Avance III 500 UltraShield spectrometer at 500 MHz (1H) or 126 MHz (13C). Chemical shifts are given in parts per million (ppm) and were calibrated on residual solvent peaks as an internal standard. Multiplicities were specified as s (singlet), d (doublet), t (triplet), q (quartet), or m (multiplet). The signals were assigned with the help of 1H,1H COSY, and DEPT-135-edited 1H,13C HSQC experiments. Assignment numbering of the C-glycoside atoms and groups corresponds to the numbering in fucose. Assignment numbering of the galactoside atoms and groups corresponds to the numbering in galactose. Assignment numbering of the ciprofloxacin atoms and groups corresponds to the numbering in ciprofloxacin (cipro).70 Commercial chemicals and solvents were used without further purification. Deuterated solvents were purchased from Eurisotop (Saarbrücken, Germany). Ciprofloxacin and polymyxin B nonapeptide·HCl (PMBN) was purchased from Sigma-Aldrich (purity ≥98%, HPLC, Merck KGaA, Darmstadt, Germany), and ciprofloxacin·HCl was purchased from Cayman Chemical (Ann Arbor, Michigan, USA). If not stated otherwise, the purity of the final compounds was further analyzed by HPLC-UV, and all UV active compounds had a purity of at least 95%. Chromatographic separation was performed on a Dionex Ultimate 3000 HPLC (Thermo Scientific, Germany) with UV detection at 254 nm using a RP-18 column (100/2 Nucleoshell RP18plus, 2.7 μm, from Macherey-Nagel, Germany) as a stationary phase. LCMS-grade distilled MeCN and double distilled H2O were used as mobile phases containing formic acid (0.1% v/v). In a gradient run, an initial concentration of 5% MeCN in H2O was increased to 95% during 7 min at a flow rate 600 μL/min. The injection volume was 4 μL of 1 mM compound in H2O/DMSO = 100:1. UPLC-HRMS for key compounds were obtained using a RP-18 column (EC 150/2 Nucleodur C18 Pyramid, 3 μm, from Macharey-Nagel, Germany) and a Q Exactive Focus Orbitrap spectrometer (Thermo Scientific, Germany). The data was analyzed using Xcalibur data acquisition and interpretation software (Thermo Scientific, Germany).
General procedure (i) for amide couplings of 6: Aniline 6 and K2CO3 (2 eq.) were dispersed in dry DCM (0.1 M) and cooled (0 °C). The corresponding (ω-bromo)acylhalide was added dropwise under vigorous stirring. After stirring for 15 min, the reaction was allowed to warm to r.t. and stirred for 1–4 h until full conversion as monitored by TLC (PE:EtOAc) or HPLC-MS. The reaction was quenched with ice-cold water. The organic phase was washed with brine, and combined organic layers were dried over anhydrous Na2SO4. After filtration, the solvent was evaporated in vacuo.
General procedure (ii) for SN2 reactions with NaN3 toward 7–10: The crude starting material was dissolved in dry DMF (0.1 M). A 5 eq. solution of NaN3 was added, and the reaction was stirred at r.t. until completion (monitored by HPLC-MS). Then, the reaction was diluted with an excess of water and extracted with EtOAc (3x). The combined organic layers were washed with half satd. brine and dried over anhydrous Na2SO4. After filtration, the solvent was evaporated in vacuo and the products were purified by MPLC (PE:EtOAc, 30–80%).
General procedure (iii) for the Zemplén deprotection of 7–10: The starting material was suspended in dry MeOH (0.1 M) and a freshly prepared solution of NaOMe in MeOH (1 M) was added dropwise to 10 mol %. The reaction was stirred for 1–2 h until the disappearance of the starting material, monitored by TLC (PE:EtOAc, 4:6). Then, the reaction was diluted with MeOH and neutralized with Amberlite IR-120 H+ exchange resin. The resin was filtered off, and the solvent was evaporated in vacuo. Purification was performed by reversed-phase MPLC (MeCN:H2O, 10–20%, 0.1% formic acid). The solvent was removed by lyophilization.
General procedure (iv) for the copper-catalyzed click reaction toward conjugates 22–31: Alkyne (1.1 eq.) and azide (1 eq.) were dissolved in 1 mL of dry DMF (purged with argon). CuSO4·7H2O (10 mol %) and sodium ascorbate (20 mol %) were added as aqueous solutions from freshly prepared stock solutions (100 mM). The mixture was stirred at r.t. or 40 °C for 16–24 h. Reaction progress was monitored by HPLC-MS. After full conversion, the solvents were evaporated in vacuo followed by purification via RP-MPLC (MeCN:H2O, 10–20%, 0.1% formic acid). The solvent was removed by lyophilization.
p-Nitrophenyl 2,3,4,6-Tetra-O-acetyl-β-d-thiogalactopyranoside (5)
Galactose pentaacetate (4, 2.0 g, 5.1 mmol, 1 eq.) and p-nitrothiophenol (2.4 g, 15.3 mmol, 3 eq.) were dissolved in 20 mL dry CH2Cl2 in a heat-dried flask under a N2 atmosphere. The mixture was cooled (0 °C), and BF3·Et2O (3.2 mL, 25.5 mmol, 5 eq.) was added dropwise under vigorous stirring. Afterward, the reaction was allowed to warm to r.t. and stirred overnight (17 h). Reaction progress was monitored by TLC (Tol:EtOAc, 9:1). After consumption of the starting material, the reaction was poured on ice water. The organic phase was isolated and washed with aq. satd. NaHCO3. The combined organic layers were washed with half satd. brine and dried over anhydrous Na2SO4. After filtration, the solvent was removed in vacuo. Purification by MPLC (SiO2, EtOAc in toluene, 5–20%) gave the product as a pale yellow amorphous solid (1.3 g, 51%). 1H NMR (500 MHz, CHCl3-d) δ 8.16 (d, J = 8.8 Hz, 2H, Ar–H), 7.61 (d, J = 8.8 Hz, 2H, Ar-H), 5.47 (d, J = 3.2 Hz, 1H, glyco-H-4), 5.29 (t, J = 10.0 Hz, 1H, glyco-H-2), 5.10 (dd, J = 9.9, 3.3 Hz, 1H, glyco-H-3), 4.86 (d, J = 10.0 Hz, 1H, glyco-H-1), 4.21 (dd, J = 11.5, 7.2 Hz, 1H, glyco-H-6), 4.14 (dd, J = 11.5, 5.8 Hz, 1H, glyco-H-6′), 4.04 (t, J = 6.5 Hz, 1H, glyco-H-5), 2.35 (s, 3H, Ac–CH3), 2.16 (s, 3H, Ac–CH3), 2.09 (s, 3H, Ac–CH3), 2.08 (s, 3H, Ac–CH3), 1.99 (s, 3H, Ac–CH3); 13C NMR (126 MHz, CHCl3-d) δ 170.44 (C=O), 170.15 (C=O), 170.08 (C=O), 169.50 (C=O), 146.96 (Ar–C), 142.52 (Ar–C), 130.52 (Ar–C), 123.97 (Ar–C), 84.97 (glyco-C-1), 74.97 (glyco-C-5), 71.85 (glyco-C-3), 67.20 (glyco-C-4), 66.84 (glyco-C-2), 61.81 (glyco-C-6), 20.88 (Ac–CH3), 20.84 (Ac–CH3), 20.79 (Ac–CH3), 20.68 (Ac–CH3). LR-MS: m/z = 503.16, [M + Na]+. Spectroscopic data is in accordance with the literature.71
p-Aminophenyl 2,3,4,6-Tetra-O-acetyl-β-d-thiogalactopyranoside (6)
Compound 6 was synthesized according to Casoni et al.:66p-nitrophenyl 2,3,4,6-tetra-O-acetyl-β-d-galactothiopyranoside (5, 1.0 g, 2.06 mmol, 1 eq.) was dissolved in 70 mL of dry DCM and Pd/C (50 mg, 5 wt %) was added. The reaction vessel was flushed several times with hydrogen and subsequently stirred under a hydrogen atmosphere (1 bar) for 48 h. The reaction was followed by TLC (PE:EtOAc, 1:1). After completion, the reaction was filtered over celite. The solvent was removed in vacuo, and the pure product was obtained as a pink amorphous solid (903 mg, 96%), which was used without further purification in the next step. 1H NMR (500 MHz, DMSO-d6) δ 7.18–7.08 (m, 2H, ArH), 6.57–6.48 (m, 2H, ArH), 5.39 (s, 2H, NH2), 5.25 (dd, J = 3.5, 1.0 Hz, 1H, glyco-H-4), 5.18 (dd, J = 9.7, 3.5 Hz, 1H, glyco-H-3), 4.93 (t, J = 9.9 Hz, 1H, glyco-H-2), 4.78 (d, J = 10.0 Hz, 1H, glyco-H-1), 4.21–4.13 (m, 1H, glyco-H-5), 4.11–3.93 (m, 2H, glyco-H-6), 2.09 (s, 3H, Ac-CH3), 2.06 (s, 3H, Ac-CH3), 2.00 (s, 3H, Ac–CH3), 1.90 (s, 3H, Ac–CH3). 13C NMR (126 MHz, DMSO-d6) δ 169.93 (C=O), 169.84 (C=O), 169.46 (C=O), 169.17 (C=O), 149.42 (ArC), 135.15 (ArC), 115.20 (ArC), 114.07 (ArC), 86.07 (glyco-C-1), 73.25 (glyco-C-5), 71.20 (glyco-C-3), 67.62 (glyco-C-4), 67.24 (glyco-C-2), 61.66 (glyco-C-6), 20.63 (Ac–CH3), 20.51 (Ac–CH3), 20.40 (Ac–CH3), 20.36 (Ac–CH3). LR-MS: m/z = 456.2, [M + H]+.
p-(α-Azidoacetamido)phenyl 2,3,4,6-Tetra-O-acetyl-β-d-thiogalactopyranoside (7)
7 was synthesized starting from 6 in two chemical steps in analogy to Casoni et al.:66 Aniline 6 (300 mg, 0.66 mmol, 1 eq.) and triethylamine (140 μL, 1.01 mmol, 1.6 eq.) were dissolved in 6 mL of dry DCM. The solution was cooled (0 °C), and bromoacetylbromide (86 μL, 0.99 mmol, 1.5 eq.) was added dropwise under vigorous stirring. The reaction was stirred for 1 h followed by TLC (PE:EtOAc, 7:3). After completion, the mixture was quenched with ice water. The organic phase was washed with aq. satd. NH4Cl (3x), water (2x), and brine (1x) and dried over anhydrous Na2SO4. After filtration, the solvent was removed in vacuo to yield the crude intermediate as an oil (370 mg), which was transformed according to general procedure ii. Product 7 was obtained as a white amorphous solid (283.2 mg, 80% over two steps). 1H NMR in accordance with the literature66 (500 MHz, CHCl3-d) δ 8.04 (s, 1H, Amide-NH), 7.52 (s, 4H, ArH), 5.41 (d, J = 3.0 Hz, 1H, glyco-H-4), 5.20 (t, J = 9.9 Hz, 1H, glyco-H-2), 5.04 (dd, J = 9.9, 3.3 Hz, 1H, glyco-H-3), 4.65 (d, J = 10.0 Hz, 1H, glyco-H-1), 4.18 (dd, J = 11.3 Hz, overlaps with 4.16, 1H, glyco-H-6), 4.16 (s, 2H, CH2N3), 4.11 (dd, J = 11.3, 6.3 Hz, 1H, glyco-H-6′), 3.92 (t, J = 6.6 Hz, 1H, glyco-H-5), 2.12 (s, 3H, Ac-CH3), 2.10 (s, 3H, Ac-CH3), 2.05 (s, 3H, Ac-CH3), 1.97 (s, 3H, Ac-CH3). 13C NMR (126 MHz, CHCl3-d) δ 170.54 (C=O), 170.33 (C=O), 170.20 (C=O), 169.55 (C=O), 164.64 (C=O), 137.21 (ArC), 134.29 (ArC), 128.05 (ArC), 120.34 (ArC), 86.79 (glyco-C-1), 74.60 (glyco-C-5), 72.12 (glyco-C-3), 67.32 (glyco-C-4), 61.72 (glyco-C-2), 53.11 (glyco-C-6), 53.07 (CH2N3, extracted from HSQC), 21.01 (Ac–CH3), 20.85 (Ac–CH3), 20.81 (Ac–CH3), 20.73 (Ac–CH3). LR-MS: m/z = 539.1, [M + H]+.
p-(β-Azidopropamido)phenyl 2,3,4,6-Tetra-O-acetyl-β-d-thiogalactopyranoside (8)
The title compound was synthesized starting from 6 (300 mg, 0.66 mmol, 1 eq.) according to general procedures i and ii and was obtained as a white amorphous solid over two chemical steps (316 mg, 87%). However, the elimination product could not be separated, resulting in a <10% contamination of the corresponding Michael-acceptor side product (quantified by 1H NMR). 1H NMR (500 MHz, CHCl3-d) δ 7.49 (s, 4H, ArH), 7.44 (s, 1H, NH), 6.44 (d, J = 16.9 Hz, 1H, −COCHCH2, from impurity), 6.24 (dd, J = 16.8, 10.3 Hz, 1H, −COCHCH–H, from impurity), 5.80 (d, J = 10.2 Hz, 1H, −COCHCH–H′, from impurity), 5.40 (d, J = 3.1 Hz, 1H, glyco-H-4), 5.19 (t, J = 9.9 Hz, 1H, glyco-H-2), 5.03 (dd, J = 9.9, 3.1 Hz, 1H, glyco-H-3), 4.63 (d, J = 9.9 Hz, 1H, glyco-H-1), 4.17 (dd, J = 11.3, 6.9 Hz, 1H, glyco-H-6), 4.10 (dd, J = 11.5, 6.3 Hz, 1H, glyco-H-6′), 3.90 (t, J = 6.5 Hz, 1H, glyco-H-5), 3.72 (t, J = 6.2 Hz, 2H, COCH2), 2.60 (t, J = 6.2 Hz, 2H, CH2N3), 2.11 (s, 3H, Ac–CH3), 2.10 (s, 3H, Ac–CH3), 2.05 (s, 3H, Ac–CH3), 1.97 (s, 3H, Ac–CH3). 13C NMR (126 MHz, CHCl3-d) δ 170.45 (C=O), 170.25 (C=O), 170.10 (C=O), 169.48 (C=O), 168.22 (C=O), 137.92 (ArC), 134.23 (ArC), 127.27 (ArC), 120.14 (ArC), 86.78 (glyco-C-1), 74.43 (glyco-C-5), 72.00 (glyco-C-3), 67.24 (glyco-C-4), 67.20 (glyco-C-2), 61.58 (glyco-C-6), 47.24 (COCH2CH2N3), 36.96 (COCH2CH2N3), 20.88 (Ac-CH3), 20.72 (Ac-CH3), 20.67 (Ac-CH3), 20.60 (Ac-CH3). LR-MS: m/z = 553.1, [M + H]+.
p-(γ-Azidobutyramido)phenyl 2,3,4,6-Tetra-O-acetyl-β-d-thiogalactopyranoside (9)
The title compound was synthesized starting from 6 (300 mg, 0.66 mmol, 1 eq.) according to general procedures i and ii and was obtained as a white amorphous solid over two chemical steps (296 mg, 79%). 1H NMR (500 MHz, CHCl3-d) δ 7.48 (s, 4H, Ar–H), 7.44 (s, 1H, NH), 5.40 (d, J = 3.4 Hz, 1H, glyco-H-4), 5.20 (t, J = 9.9 Hz, 1H, glyco-H-2), 5.02 (dd, J = 10.0, 3.3 Hz, 1H, glyco-H-3), 4.62 (d, J = 9.9 Hz, 1H, glyco-H-1), 4.17 (dd, J = 11.3, 6.9 Hz, 1H, glyco-H-6), 4.09 (dd, J = 11.3, 6.3 Hz, 1H, glyco-H-6′), 3.90 (t, J = 6.6 Hz, 1H, glyco-H-5), 3.41 (t, J = 6.4 Hz, 2H, COCH2), 2.47 (t, J = 7.1 Hz, 2H, CH2N3), 2.12 (s, 3H, Ac-CH3), 2.10 (s, 3H, Ac–CH3), 2.04 (s, 3H, Ac–CH3), 2.00 (t, J = 6.8 Hz, 2H, −CH2−), 1.97 (s, 3H, Ac-CH3). 13C NMR (126 MHz, CHCl3-d) δ 170.57 (C=O), 170.35 (C=O), 170.26 (C=O), 170.21 (C=O), 169.61 (C=O), 138.25 (ArC) , 134.31 (ArC), 127.14 (ArC), 120.11 (ArC), 86.95 (glyco-C-1), 74.52 (glyco-C-5), 72.10 (glyco-C-3), 67.36 (glyco-C-4), 67.32 (glyco-C-2), 61.68 (glyco-C-6), 50.78 (COCH2), 34.26 (CH2N3), 24.66 (CH2), 20.99 (Ac–CH3), 20.82 (Ac–CH3), 20.77 (Ac–CH3), 20.71 (Ac–CH3). LR-MS: m/z = 567.1, [M + H]+.
p-(δ-Azidovalerylamido)phenyl 2,3,4,6-Tetra-O-acetyl-β-d-thiogalactopyranoside (10)
The title compound was synthesized starting from 6 (300 mg, 0.66 mmol, 1 eq.) according to general procedures i and ii and was obtained as a white amorphous solid over two chemical steps (327 mg, 85%). 1H NMR (500 MHz, CHCl3-d) δ 7.48 (s, 4H, Ar–H), 7.24 (s, 1H, NH), 5.40 (d, J = 3.2 Hz, 1H, glyco-H-4), 5.19 (t, J = 9.9 Hz, 1H, glyco-H-2), 5.03 (dd, J = 9.9, 3.3 Hz, 1H, glyco-H-3), 4.63 (d, J = 10.0 Hz, 1H, glyco-H-1), 4.17 (dd, J = 11.3, 6.9 Hz, 1H, glyco-H-6), 4.10 (dd, J = 11.3, 6.3 Hz, 1H, glyco-H-6′), 3.90 (t, J = 6.6 Hz, 1H, glyco-H-5), 3.34 (t, J = 6.7 Hz, 2H, −COCH2−), 2.41 (t, J = 7.3 Hz, 2H, −CH2N3), 2.11 (s, 3H, Ac–CH3), 2.10 (s, 3H, Ac–CH3), 2.05 (s, 3H, Ac–CH3), 1.97 (s, 3H, Ac–CH3), 1.82 (p, J = 7.4 Hz, 2H, −CH2CH2N3), 1.74–1.64 (p, 2H, −COCH2CH2−). 13C NMR (126 MHz, CHCl3-d) δ 170.69 (C=O), 170.56 (C=O), 170.36 (C=O), 170.21 (C=O), 169.59 (C=O), 138.34 (ArC), 134.39 (ArC), 127.03 (ArC), 120.05 (ArC), 86.99 (glyco-C-1), 74.55 (glyco-C-5), 72.13 (glyco-C-3), 67.37 (glyco-C-4), 67.33 (glyco-C-2), 61.70 (glyco-C-6), 51.31 (CO–CH2−), 37.07 (−CH2–N3), 28.43 (−COCH2CH2−), 22.75 (−CH2CH2N3), 21.01 (Ac–CH3), 20.85 (Ac–CH3), 20.80 (Ac–CH3), 20.72 (Ac-CH3). LR-MS: m/z = 581.2, [M + H]+.
p-(α-Azidoacetamido)phenyl-β-d-thiogalactopyranoside (11)
The title compound was synthesized from 7 (275 mg, 0.51 mmol, 1 eq.) according to general procedure iii and was obtained as a white solid (142 mg, 75%). 1H NMR (500 MHz, MeOH-d4) δ 7.54 (s, 4H, ArH), 4.51 (d, J = 9.7 Hz, 1H, glyco-H-1), 4.01 (s, 2H, -CH2N3), 3.89 (d, J = 3.2 Hz, 1H, glyco-H-4), 3.76 (dd, J = 11.5, 6.8 Hz, 1H, glyco-H-6), 3.70 (dd, J = 11.5, 5.2 Hz, 1H, glyco-H-6′), 3.62–3.52 (m, 2H, glyco-H-2 + glyco-H-5), 3.49 (dd, J = 9.2, 3.3 Hz, 1H, glyco-H-3). 13C NMR (126 MHz, MeOH-d4) δ 168.47 (C=O), 138.61 (ArC), 133.58 (ArC), 130.91 (ArC), 121.59 (ArC), 90.50 (glyco-C-1), 80.61 (glyco-C-5), 76.30 (glyco-C-3), 70.93 (glyco-C-2), 70.40 (glyco-C-4), 62.60 (glyco-C-6), 53.26 (−CH2N3). HR-MS calcd [C14H17N4O6S]−: 369.0874, found 369.0877.
p-(β-Azidopropamido)phenyl-β-d-thiogalactopyranoside (12)
The title compound was synthesized from 8 (309 mg, 0.56 mmol, 1 eq.) according to general procedure iii and was obtained as a white solid (216 mg, 54%) .1H NMR (500 MHz, MeOH-d4) δ 7.53 (d, J = 1.1 Hz, 4H, ArH), 4.50 (d, J = 9.6 Hz, 1H, glyco-H-1), 3.88 (d, J = 2.5 Hz, 1H, glyco-H-4), 3.76 (dd, J = 11.5, 6.8 Hz, 1H, glyco-H-6), 3.70 (dd, J = 11.5, 5.3 Hz, 1H, glyco-H-6′), 3.64 (t, J = 6.4 Hz, 2H, −COCH2−), 3.60–3.52 (m, 2H, glyco-H-2 + glyco-H-5), 3.48 (dd, J = 9.2, 3.3 Hz, 1H, glyco-H-3), 2.63 (t, J = 6.4 Hz, 2H, −CH2N3). 13C NMR (126 MHz, MeOH-d4) δ 171.27 (C=O), 139.20 (ArC), 133.68 (ArC), 130.38 (ArC), 121.39 (ArC), 90.59 (glyco-C-1), 80.61 (glyco-C-5), 76.33 (glyco-C-3), 70.93 (glyco-C-2), 70.43 (glyco-C-4), 62.63 (glyco-C-6), 48.43 (−COCH2−), 37.08 (−CH2N3). HR-MS calcd [C15H19N4O6S]−: 383.1031, found 383.1036.
p-(γ-Azidobutyramido)phenyl-β-d-thiogalactopyranoside (13)
The title compound was synthesized from 9 (296 mg, 0.52 mmol, 1 eq.) according to general procedure iii and was obtained as a white solid in 81% yield. 1H NMR (500 MHz, MeOH-d4) δ 7.52 (s, 4H, Ar-H), 4.50 (d, J = 9.6 Hz, 1H, glyco-H-1), 3.88 (d, J = 3.2 Hz, 1H, glyco-H-4), 3.76 (dd, J = 11.5, 6.8 Hz, 1H, glyco-H-6), 3.70 (dd, J = 11.5, 5.3 Hz, 1H, glyco-H-6′), 3.61–3.52 (m, 2H, glyco-H-2 + glyco-H-5), 3.48 (dd, J = 9.2, 3.3 Hz, 1H, glyco-H-3), 3.39 (t, J = 6.7 Hz, 2H, −COCH2−), 2.47 (t, J = 7.3 Hz, 2H, −CH2N3), 1.94 (p, J = 7.0 Hz, 2H, −CH2−). 13C NMR (126 MHz, MeOH-d4) δ 173.37 (C=O), 139.37 (ArC), 133.70 (ArC), 130.21 (ArC), 121.38 (ArC), 90.62 (glyco-C-1), 80.62 (glyco-C-5), 76.34 (glyco-C-3), 70.94 (glyco-C-2), 70.43 (glyco-C-4), 62.63 (glyco-C-6), 51.92 (−COCH2−), 34.73 (-CH2N3), 25.94 (−CH2−). HR-MS calcd [C16H21N4O6S]−: 397.1187, found 397.1189.
p-(δ-Azidovalerylamido)phenyl-β-d-thiogalactopyranoside (14)
The title compound was synthesized from 10 (327 mg, 0.56 mmol) according to general procedure iii and was obtained as a white solid (235 mg, 84%). 1H NMR (500 MHz, MeOH-d4) δ 7.52 (s, 4H, ArH), 4.49 (d, J = 9.6 Hz, 1H, glyco-H-1), 3.88 (dd, J = 3.3, 0.8 Hz, 1H, glyco-H-4), 3.76 (dd, J = 11.5, 6.8 Hz, 1H, glyco-H-6), 3.70 (dd, J = 11.5, 5.3 Hz, 1H, glyco-H-6′), 3.62–3.50 (m, 2H, glyco-H-2 + glyco-H-5), 3.48 (dd, J = 9.2, 3.3 Hz, 1H, glyco-H-3), 3.34 (t, J = 6.7 Hz, 2H, −COCH2−), 2.40 (t, J = 7.4 Hz, 2H, −CH2N3), 1.82–1.72 (m, 2H, −CH2CH2N3), 1.70–1.60 (m, 2H, −COCH2CH2-). 13C NMR (126 MHz, MeOH-d4) δ 174.06 (C=O), 139.39 (ArC), 133.72 (ArC), 130.18 (ArC), 121.37 (ArC), 90.62 (glyco-C-1), 80.62 (glyco-C-5), 76.34 (glyco-C-3), 70.94 (glyco-C-2), 70.43 (glyco-C-4), 62.63 (glyco-C-6), 52.16 (−COCH2−), 37.27 (−CH2N3), 29.45 (−COCH2CH2-), 23.99 (−CH2CH2N3). HR-MS calcd [C17H23N4O6S]−: 411.1344, found 411.1350.
N-Propargyl-ciprofloxacin (20)
The title compound was synthesized in analogy to McPherson et al.:72 Ciprofloxacin (500 mg, 1.5 mmol, 1 eq.) was dispersed in 10 mL of dry DMF together with Et3N (310 μL, 2.25 mmol, 1.5 eq.) and propargyl bromide (250 μL, 2.25 mmol, 1.5 eq.). The mixture was stirred at 90 °C for 24 h, and further equivalents of Et3N (309 μL, 2 mmol, 2 eq.) and propargyl bromide (250 μL, 2 mmol, 2 eq.) were added stepwise until the disappearance of the starting material, monitored by TLC (DCM:MeOH, 9:1). The reaction was poured on ice water. After filtration, the precipitate was redissolved and purified by MPLC (DCM:MeOH, 1–10%) to yield the title product as a beige amorphous solid (353 mg 64%). 1H NMR (500 MHz, CHCl3-d) δ 14.99 (br s, 1H, COOH), 8.77 (s, 1H, ArH-2), 8.02 (d, J = 13.0 Hz, 1H, ArH-5), 7.37 (d, J = 6.7 Hz, 1H, ArH-8), 3.55 (br s, 1H, cPr-H), 3.43 (s, 2H, HCCCH2−), 3.41 (br s, 4H, 2x piperazine-CH2−), 2.84 (br s, 4H, 2x piperazine-CH2′ −), 2.33 (s, 1H, alkyne-H), 1.39 (d, J = 6.3 Hz, 2H, cPr-CH2), 1.20 (br s, 2H, cPr-CH2′). 13C NMR (126 MHz, CHCl3-d) δ 177.28 (C4=O), 167.17 (COOH), 153.82 (d, J = 251.4 Hz, cipro-C-6), 147.61 (cipro-C-2), 145.86 (d, J = 10.1 Hz, cipro-C-7), 139.21 (cipro-C-8a), 120.14 (d, J = 7.6 Hz, cipro-C-4a), 112.67 (d, J = 23.4 Hz cipro-C-5), 108.35 (cipro-C-3), 105.04 (d, J = 2.4 Hz, cipro-C-8), 74.23 (HCCCH2−), 51.52 (HCCCH2−), 49.67 (piperazine), 46.95 (piperazine), 35.42 (cPr-CH), 8.39 (cPr-CH2), −HCCCH2– (not observed). HR-MS calcd [C20H21FN3O3]+: 370.1561, found 370.1552.
N-Butynyl-ciprofloxacin (21)
Ciprofloxacin (500 mg, 1.5 mmol, 1 eq.) was dissolved in dry DMF and heated to 70 °C. Over 72 h, Et3N (1512 μL, 10.5 mmol, 7 eq.) and 4-bromo-1-butyne (982 μL, 10.5 mmol, 7 eq.) were added portionwise in 1 eq. steps until the disappearance of the starting material, monitored by TLC (DCM:MeOH, 9:1). The reaction was poured on ice-cold water. After precipitation, the precipitate was purified by MPLC (DCM:MeOH, 1–10%) to yield the product as a beige amorphous solid (245 mg, 43%). 1H NMR (500 MHz, DMSO-d6) δ 15.22 (br s, 1H, COOH), 8.66 (s, 1H, ArH-2), 7.89 (d, J = 13.3 Hz, 1H, ArH-5), 7.56 (d, J = 7.3 Hz, 1H, ArH-8), 3.85–3.77 (br s, 1H, cPr-H), 3.32 (br s, 4H, 2x piperazine-CH2−), 2.81 (s, 1H, HCCCH2CH2−), 2.64 (br s, 4H, piperazine-CH2−), 2.56 (t, J = 7.2 Hz, 2H, RR′NCH2CH2CCH −), 2.38 (t, J = 6.2 Hz, 2H, RR′NCH2CH2CCH −), 1.31 (q, J = 6.0 Hz, 2H, cPr-CH2−), 1.17 (br s, 2H, cPr-CH2′−). 13C NMR (126 MHz, DMSO-d6) δ 176.40 (C4=O), 166.01 (COOH), 153.04 (d, J = 249.4 Hz, cipro-C-6), 148.05 (cipro-C-2), 145.22 (cipro-C-7), 139.20 (cipro-C-8a), 118.63 (cipro-C4a), 110.94 (d, J = 23.0 Hz, cipro-C-5), 106.74 (cipro-C-3), 106.44 (cipro-C-8), 83.16 (HCCCH2CH2−), 71.87 (HCCCH2CH2−), 56.26 (HCCCH2CH2−), 51.98 (piperazine), 49.41 (piperazine), 49.38 (piperazine), 35.88 (cPr-CH), 16.19 (HCCCH2CH2−), 7.59 (cPr-CH2). HR-MS calcd [C21H23FN3O3]+: 384.1718, found 384.1711.
Gal-ciprofloxacin Conjugate 22 (n = 1, m = 0)
The title compound was synthesized from 11 (20 mg, 0.054 mmol, 1 eq.) and 20 (40 mg, 0.108 mmol, 2 eq.) according to general procedure iv and was obtained as a beige amorphous solid (22 mg, 55%). 1H NMR (500 MHz, DMSO-d6) δ 15.22 (br s, 1H, COOH), 10.51 (s, 1H, −CONH−), 8.65 (s, 1H, cipro-ArH-2), 8.06 (s, 1H, triazoleH), 7.88 (d, J = 13.3 Hz, 1H, cipro-ArH-5), 7.55 (d, J = 7.4 Hz, 1H, cipro-ArH-8), 7.52 (d, J = 8.6 Hz, 2H, Phenyl-H), 7.43 (d, J = 8.7 Hz, 2H, Phenyl-H), 5.32 (s, 2H, −HNCO-CH2-triazole), 5.11 (br s, 1H, OH), 4.85 (br s, 1H, OH), 4.62 (br s, 1H, OH), 4.48 (d, J = 9.4 Hz, 1H, glyco-H-1), 4.44 (br s, 1H, OH), 3.81 (s, 1H, cPr-H), 3.70 (s, 2H, −triazole-CH2-N-cipro), 3.69 (br s, 1H, glyco-H-4), 3.53–3.45 (m, 2H, glyco-H-6 + H-6′), 3.43 (glyco-H-2, extracted from HSQC), 3.38 (glyco-H-5, extracted from HSQC), 3.33 (glyco-H-3, extracted from HSQC), 3.33 (2x piperazine-CH2, extracted from HSQC), 2.65 (s, 4H, 2x piperazine-CH2), 1.31 (d, J = 6.5 Hz, 2H, cPr-CH2), 1.17 (br s, 2H, cPr-CH2′). 13C NMR (126 MHz, DMSO-d6) δ 176.41 (cipro-C4=O), 166.05 (COOH), 164.34 (C=O), 153.06 (d, J = 249.3 Hz, cipro-C-6), 148.03 (cipro-C-2), 145.23 (d, J = 10.1 Hz, cipro-C-7), 142.84 (triazole-C), 139.24 (cipro-C-8a), 137.09 (phenyl-C), 131.00 (phenyl-C), 129.46 (phenyl-C), 125.67 (triazole-CH), 119.60 (phenyl-C), 118.59 (d, J = 7.5 Hz, cipro-C-4a), 110.98 (d, J = 23.4 Hz, cipro-C-5), 106.75 (cipro-C-3), 106.43 (d, J = 3.9 Hz, cipro-C-8), 88.17 (glyco-C-1), 79.22 (glyco-C-5), 74.72 (glyco-C-3), 69.26 (glyco-C-2), 68.40 (glyco-C-4), 60.63 (glyco-C-6), 52.29 (−triazole-CH2-N-cipro), 52.17 (−HNCO-CH2-triazole), 51.83 (piperazine), 49.40 (piperazine), 35.92 (cPr-CH), 7.61 (cPr-CH2). HR-MS calcd [C34H39FN7O9S]+: 740.2509, found 740.2500.
Gal-ciprofloxacin Conjugate 23 (n = 1, m = 1)
The title compound was synthesized from 11 (20 mg, 0.054 mmol, 1 eq.) and 21 (41 mg, 0.108 mmol, 2 eq.) according to general procedure iv and was obtained as a beige amorphous solid (15 mg, 37%). 1H NMR (500 MHz, DMSO-d6) δ 15.23 (br s, 1H, COOH), 10.50 (s, 1, CONH), 8.66 (s, 1H, cipro-ArH-2), 7.94 (s, 1H, triazole-H), 7.90 (d, J = 13.3 Hz, 1H, cipro-ArH-5), 7.57 (d, J = 6.6 Hz, 1H, cipro-ArH-8), 7.52 (d, J = 8.4 Hz, 2H, Phenyl-H), 7.42 (d, J = 8.7 Hz, 2H, Phenyl-H′), 5.28 (s, 2H, −HNCOCH2−), 4.48 (d, J = 9.4 Hz, 1H, glyco-H-1), 3.82 (s, 1H, cPr-H), 3.69 (d, J = 2.8 Hz, 1H, glyco-H-4), 3.49 (glyco-H-6 + H-6′, extracted from HSQC), 3.43 (glyco-H-2, extracted from HSQC), 3.38 (glyco-H-5, extracted from HSQC), 3.35 (2x piperazine-CH2, extracted from HSQC), 3.33 (glyco-H-3, extracted from HSQC), 2.88 (t, J = 7.5 Hz, 2H, −triazole-CH2CH2NRR′), 2.68 (br s, 6H, 2x piperazine-CH2 + −triazole-CH2CH2NRR′), 1.31 (d, J = 6.0 Hz, 2H, cPr-CH2), 1.18 (br s, 2H, cPr-CH2′). 13C NMR (126 MHz, DMSO-d6) δ 176.42 (cipro-C4=O), 166.06 (COOH), 164.39 (C=O), 153.09 (d, J = 249.8 Hz, cipro-C-6), 148.05 (cipro-C-2), 145.27 (d, J = 9.9 Hz, cipro-C-7), 144.96 (triazole-C), 139.26 (cipro-C-8a), 137.11 (phenyl-C), 131.00 (phenyl-C), 129.44 (phenyl-C), 124.00 (triazole-CH), 119.59 (phenyl-C), 118.58 (d, J = 8.0 Hz, cipro-C-4a), 110.99 (d, J = 23.1 Hz, cipro-C-5), 106.76 (cipro-C-3), 106.38 (d, J = 3.1 Hz, cipro-C-8), 88.17 (glyco-C-1), 79.21 (glyco-C-5), 74.72 (glyco-C-3), 69.25 (glyco-C-2), 68.39 (glyco-C-4), 60.63 (glyco-C-6), 57.29 (linker-CH2), 52.27 (piperazine), 52.18 (linker-CH2), 49.43 (piperazine), 35.92 (cPr-CH), 22.97 (linker-CH2), 7.62 (cPr-CH2). HR-MS calcd [C35H41FN7O9S]+: 754.2665, found 754.2658.
Gal-ciprofloxacin Conjugate 24 (n = 2, m = 0)
The title compound was synthesized from 12 (20 mg, 0.052 mmol, 1 eq.) and 20 (20 mg, 0.054 mmol, 1 eq.) according to general procedure iv and was obtained as a beige amorphous solid (26 mg, 66%). 1H NMR (500 MHz, DMSO-d6) δ 15.23 (br s, 1H, COOH), 10.09 (s, 1H, CONH), 8.66 (s, 1H, cipro-ArH-2), 7.97 (s, 1H, triazole-H), 7.90 (d, J = 13.3 Hz, 1H, cipro-ArH-5), 7.53 (d, J = 7.4 Hz, 1H, cipro-ArH-8), 7.48 (d, J = 8.6 Hz, 2H, phenyl-H), 7.37 (d, J = 8.5 Hz, 2H, phenyl-H′), 5.07 (br s, 1H, OH), 4.84 (br s, 1H, OH), 4.65 (t, J = 6.6 Hz, 2H, −NHCOCH2CH2−), 4.60 (br s, 1H, OH), 4.43 (d, J = 9.2 Hz, 1H, glyco-H-1), 4.43 (br s, 1H, OH), 3.90–3.79 (br s, 1H, cPr-H), 3.67 (s, 1H, glyco-H-4), 3.64 (s, 2H, −triazole-CH2-NRR′), 3.47 (glyco-H-6 + H-6′, extracted from HSQC), 3.39 (glyco-H-2, extracted from HSQC), 3.35 (glyco-H-5, extracted from HSQC), 3.30 (glyco-H-3, extracted from HSQC), 3.29–3.25 (m, 4H, 2x piperazine-CH2), 2.96 (t, J = 6.6 Hz, 2H, −NHCOCH2CH2−), 2.61–2.57 (m, 4H, 2x piperazine-CH2′), 1.34–1.25 (m, 2H, cPr-CH2), 1.18–1.15 (m, 2H, cPr-CH2′). 13C NMR (126 MHz, DMSO-d6) δ 176.43 (cipro-C4=O), 168.22 (COOH), 166.07 (C=O), 153.07 (d, J = 250.0 Hz, cipro-C-6), 148.07 (cipro-C-2), 145.23 (d, J = 10.1 Hz, cipro-C-7), 142.88 (triazole-C), 139.25 (cipro-C-8a), 137.65 (phenyl-C), 131.13 (phenyl-C), 128.70 (phenyl-C), 124.23 (triazole-CH), 119.48 (phenyl-C), 118.61 (d, J = 7.5 Hz, cipro-C-4a), 110.99 (d, J = 22.9 Hz, cipro-C-5), 106.77 (cipro-C-3), 106.40 (d, J = 2.5 Hz, cipro-C-8), 88.28 (glyco-H-1), 79.19 (glyco-H-5), 74.73 (glyco-H-3), 69.24 (glyco-H-2), 68.36 (glyco-H-4), 60.60 (glyco-H-6), 52.30 (linker-CH2), 51.80 (piperazine), 49.39 (piperazine), 45.58 (linker-CH2), 36.56 (linker-CH2), 35.91 (cPr-CH), 7.61 (cPr-CH2). HR-MS calcd [C35H41FN7O9S]+: 754.2665, found 754.2657.
Gal-ciprofloxacin Conjugate 25 (n = 2, m = 1)
The title compound was synthesized from 12 (30 mg, 0.078 mmol, 1 eq.) and 21 (33 mg, 0.086 mmol, 1.1 eq.) according to general procedure iv as and was obtained a beige amorphous solid (35 mg, 58%). 1H NMR (500 MHz, DMSO-d6) δ 15.23 (br s, 1H, COOH), 10.07 (s, 1H, CONH), 8.66 (s, 1H, cipro-ArH-2), 7.91 (d, J = 13.3 Hz, 1H, cipro-ArH-5), 7.87 (s, 1H, triazole-H), 7.55 (d, J = 7.3 Hz, 1H, cipro-ArH-8), 7.49 (d, J = 8.7 Hz, 2H, phenyl-H), 7.38 (d, J = 8.8 Hz, 2H, phenyl-H), 5.06 (br s, 1H, OH), 4.83 (br s, 1H, OH), 4.61 (t, J = 6.7 Hz, 2H, −NHCOCH2CH2– + OH), 4.43 (d, J = 9.3 Hz, 1H, glyco-H-1), 4.42 (s, 1H, OH), 3.83 (s, 1H, OH), 3.68 (s, 1H, glyco-H-4), 3.48 (ddd, J = 10.8, 6.5, 5.5 Hz, 2H), 3.40 (t, J = 6.3 Hz, 2H, glyco-H-2), 2.95 (t, J = 6.6 Hz, 2H, −NHCOCH2CH2−), 2.82 (t, J = 7.5 Hz, 2H, −triazole-CH2CH2NRR′), 2.65 (br s, 6H, 2x piperazine-CH2 + −triazole-CH2CH2NRR′), 1.31 (q, J = 7.1 Hz, 2H, cPr-CH2), 1.20–1.16 (m, 2H, cPr-CH2). 13C NMR (126 MHz, DMSO-d6) δ 176.37 (cipro-C4=O), 168.15 (C=O), 165.98 (COOH), 153.02 (d, J = 248.9 Hz, cipro-C-6), 148.02 (cipro-C-2), 145.16 (d, J = 10.5 Hz, cipro-C-7), 144.88 (cipro-C-7), 139.21 (cipro-C-8a), 137.65 (phenyl-C), 131.05 (phenyl-C), 128.66 (phenyl-C), 122.58 (triazole-CH), 119.38 (phenyl-C), 118.55 (d, J = 7.4 Hz, cipro-C-4a), 110.96 (d, J = 23.1 Hz, cipro-C-5), 106.73 (cipro-C-3), 106.32 (d, J = 3.8 Hz, cipro-C-8), 88.26 (glyco-C-1), 79.17 (glyco-C-5), 74.69 (glyco-C-3), 69.19 (glyco-C-2), 68.33 (glyco-C-4), 60.57 (glyco-C-6), 57.18 (linker-CH2), 52.16 (piperazine), 49.35 (piperazine), 45.35 (linker-CH2), 36.48 (linker-CH2), 35.88 (cPr-CH), 22.87 (linker-CH2), 7.58 (cPr-CH2). HR-MS calcd [C36H43FN7O9S]+: 768.2822, found 768.2822.
Gal-ciprofloxacin Conjugate 26 (n = 3, m = 0)
The title compound was synthesized from 13 (30 mg, 0.075 mmol, 1 eq.) and 20 (31 mg, 0.083 mmol, 1.1 eq.) according to general procedure iv and was obtained as a beige amorphous solid (30 mg, 52%). 1H NMR (500 MHz, DMSO-d6) δ 15.21 (br s, 1H, COOH), 9.96 (s, 1H, CONH), 8.65 (s, 1H, cipro-ArH-2), 8.06 (s, 1H, triazole-H), 7.89 (d, J = 13.3 Hz, 1H, cipro-ArH-5), 7.54 (d, J = 7.3 Hz, 1H, cipro-ArH-8), 7.51 (d, J = 8.6 Hz, 2H, phenyl-H), 7.39 (d, J = 8.8 Hz, 2H, phenyl-H), 5.06 (br s, 1H, OH), 4.84 (br s, 1H, OH), 4.60 (br s, 1H, OH), 4.44 (d, J = 9.4 Hz, 1H, glyco-H-1), 4.42 (OH, extracted from COSY) 4.41 (t, J = 6.9 Hz, 1H, −NHCOCH2CH2CH2−), 3.81 (s, 1H, cPr-H), 3.68 (s, 1H, glyco-H-4), 3.66 (s, 2H, −triazol-CH2-NRR′), 3.56–3.44 (m, 2H, glyco-H-6 + H-6′), 3.41 (t, J = 6.3 Hz, 1H, glyco-H-5), 3.37 (glyco-H-2, extracted from HSQC), 3.32 (glyco-H-3, extracted from HSQC), 3.32 (2x piperazine-CH2, extracted from HSQC) 2.64 (br s, 4H, 2x piperazine-CH2), 2.33 (t, J = 7.2 Hz, 2H, −NHCOCH2CH2CH2−), 2.13 (tt, J = 7.1 Hz, 2H, −NHCOCH2CH2CH2−), 1.33–1.27 (m, 2H, cPr-CH2), 1.23–1.14 (br s, 2H, cPr-CH2’). 13C NMR (126 MHz, DMSO-d6) δ 176.36 (cipro-C4=O), 170.11 (C=O), 165.96 (COOH), 153.01 (d, J = 249.7 Hz, cipro-C-6), 148.00 (cipro-C-2), 145.17 (d, J = 10.1 Hz, cipro-C-7), 143.11 (triazole-C), 139.19 (cipro-C-8a), 137.97 (phenyl-C), 131.15 (phenyl-C), 128.26 (phenyl-C), 123.81 (triazole-CH), 119.34 (phenyl-C), 118.56 (d, J = 7.5 Hz, cipro-C-4a), 110.94 (d, J = 23.1 Hz, cipro-C-5), 106.72 (cipro-C-3), 106.36 (d, J = 3.7 Hz, cipro-C-8), 88.34 (glyco-C-1), 79.17 (glyco-C-5), 74.70 (glyco-C-3), 69.20 (glyco-C-2), 68.34 (glyco-C-4), 60.58 (glyco-C-6), 52.41 (linker-CH2), 51.87 (piperazine), 49.39 (piperazine), 48.84 (linker-CH2), 35.85 (cPr-CH), 32.91 (linker-CH2), 25.51 (linker-CH2), 7.57 (cPr-CH2). HR-MS calcd [C36H43FN7O9S]+: 768.2822, found 768.2815.
Gal-ciprofloxacin Conjugate 27 (n = 3, m = 1)
The title compound was synthesized from 13 (30 mg, 0.075 mmol, 1 eq.) and 21 (32 mg, 0.083 mmol, 1.1 eq.) according to general procedure iv and was obtained as a beige amorphous solid (31 mg, 53%). 1H NMR (500 MHz, DMSO-d6) δ 15.22 (br s, 1H, COOH), 9.94 (s, 1H, CONH), 8.66 (s, 1H, cipro-ArH-2), 7.93 (s, 1H, triazole-H), 7.90 (d, J = 13.3 Hz, 1H, cipro-ArH-5), 7.56 (d, J = 7.2 Hz, 1H, cipro-ArH-8), 7.51 (d, J = 8.6 Hz, 2H, phenyl-H), 7.38 (d, J = 8.8 Hz, 2H, phenyl-H), 5.06 (br s, 1H, OH), 4.83 (br s, 1H, OH), 4.59 (br s, 1H, OH), 4.44 (d, J = 9.4 Hz, 1H, glyco-H-1), 4.42 (br s, 1H, OH), 4.38 (t, J = 6.8 Hz, 2H, −NHCOCH2CH2CH2−), 3.82 (br s, 1H, cPr-H), 3.68 (s, 1H, glyco-H-4), 3.53–3.44 (m, 2H, glyco-H-6 + H-6′), 3.41 (t, J = 6.3 Hz, 1H, glyco-H-5), 3.37 (glyco-H-2 ,extracted from HSQC), 3.34 (2x piperazine-CH2, extracted from HSQC), 3.32 (glyco-H-3) 2.84 (t, J = 7.6 Hz, 2H, −triazole-CH2CH2NRR′), 2.67 (br s, 6H, 2x piperazine-CH2 + −triazole-CH2CH2NRR′), 2.31 (t, J = 7.3 Hz, 2H, −NHCOCH2CH2CH2−), 2.11 (tt, J = 8.1, 7.5 Hz, 2H, −NHCOCH2CH2CH2−), 1.36–1.28 (m, 2H, cPr-CH2), 1.21–1.13 (m, 2H, cPr-CH2). 13C NMR (126 MHz, DMSO-d6) δ 176.37 (cipro-C4=O), 170.11 (CO), 165.97 (COOH), 153.03 (d, J = 249.5 Hz, cipro-C-6), 148.01 (cipro-C-2), 145.19 (d, J = 9.9 Hz, cipro-C-7), 145.05 (triazole-C), 139.20 (cipro-C-8a), 137.95 (phenyl-C), 131.12 (phenyl-C), 128.27 (phenyl-C), 122.21 (triazole-CH), 119.33 (phenyl-C), 118.56 (d, J = 8.0 Hz, cipro-C-4a), 110.94 (d, J = 23.1 Hz, cipro-C-5), 106.73 (cipro-C-3), 106.35 (d, J = 3.2 Hz, cipro-C-8), 88.33 (glyco-C-1), 79.17 (glyco-C-5), 74.70 (glyco-C-3), 69.20 (glyco-C-2), 68.34 (glyco-C-4), 60.58 (glyco-C-6), 57.21 (linker-CH2), 52.19 (piperazine), 49.40 (piperazine), 48.74 (linker-CH2), 35.87 (cPr-CH), 32.90 (linker-CH2), 25.53 (linker-CH2), 22.98 (linker-CH2), 7.58 (cPr-CH2). HR-MS calcd [C37H45FN7O9S]+: 782.2987, found 782.2965.
Gal-ciprofloxacin Conjugate 28 (n = 4, m = 0)
The title compound was synthesized from 14 (30 mg, 0.073 mmol, 1 eq.) and 20 (30 mg, 0.08 mmol, 1.1 eq.) according to general procedure iv and was obtained as a beige amorphous solid (25 mg, 43%). 1H NMR (500 MHz, DMSO-d6) δ 15.21 (br s, 1H, COOH), 9.93 (s, 1H, CONH), 8.65 (s, 1H, cipro-ArH-2), 8.05 (s, 1H, triazole-H), 7.89 (d, J = 13.3 Hz, 1H, cipro-ArH-5), 7.54 (d, J = 8.1 Hz, 1H, cipro-ArH-5), 7.51 (d, J = 8.5 Hz, 2H, phenyl-H), 7.38 (d, J = 8.4 Hz, 2H, phenyl-H), 5.06 (br s, 1H, OH), 4.84 (br s, 1H, OH), 4.59 (br s, 1H, OH), 4.43 (d, J = 9.4 Hz, 2, glyco-H-1 + OH), 4.37 (t, J = 6.9 Hz, 2H, −NHCOCH2CH2CH2CH2−), 3.81 (s, 1H, cPr-H), 3.68 (br s, 1H, glyco-H-4), 3.65 (s, 2H, −triazole-CH2CH2NRR′), 3.56–3.44 (m, 2H, glyco-H-6 + H-6′), 3.41 (d, J = 6.0 Hz, 19H), 3.40 (glyco-H-5, extracted from HSQC), 3.37 (glyco-H-2, extracted from HSQC), 3.32 (2x piperazine-CH2, extracted from HSQC), 2.63 (br s, 4H, 2x piperazine-CH2′), 2.34 (t, J = 7.4 Hz, 2H, −NHCOCH2CH2CH2CH2−), 1.86 (tt, J = 7.1 Hz, 2H, −NHCOCH2CH2CH2CH2−), 1.55 (tt, J = 7.5 Hz, 2H, −NHCOCH2CH2CH2CH2−), 1.39–1.26 (m, 2H, cPr-CH2), 1.22–1.12 (m, 2H, cPr-CH2). 13C NMR (126 MHz, DMSO-d6) δ 176.36 (cipro-C4=O), 170.82 (C=O), 165.96 (COOH), 153.01 (d, J = 249.9 Hz, cipro-C-6), 148.01 (cipro-C-2), 145.17 (d, J = 9.9 Hz, cipro-C-7), 143.02 (triazole-C), 139.19 (cipro-C-8a), 138.03 (phenyl-C), 131.15 (phenyl-C), 128.20 (phenyl-C), 123.77 (triazole-CH), 119.31 (phenyl-C), 118.56 (d, J = 7.8 Hz, cipro-C-4a), 110.94 (d, J = 23.2 Hz, cipro-C-5), 106.72 (cipro-C-3), 106.37 (d, J = 3.0 Hz, cipro-C-8), 88.35 (glyco-C-1), 79.17 (glyco-C-5), 74.69 (glyco-C-3), 69.20 (glyco-C-2), 68.33 (glyco-C-4), 60.57 (glyco-C-6), 52.40 (linker-CH2), 51.86 (piperazine), 49.39 (piperazine), 49.03 (linker-CH2), 35.85 (cPr-CH), 35.59 (linker-CH2), 29.34 (linker-CH2), 22.01 (linker-CH2), 7.57 (cPr-CH2). HR-MS calcd [C37H45FN7O9S]+: 782.2987, found 782.2972.
Gal-ciprofloxacin Conjugate 29 (n = 4, m = 1)
The title compound was synthesized from 14 (30 mg, 0.073 mmol, 1 eq.) and 21 (56 mg, 0.146 mmol, 2 eq.) according to general procedure iv and was obtained as a beige amorphous solid (28 mg, 48%). 1H NMR (500 MHz, DMSO-d6) δ 15.23 (br s, 1H, COOH), 9.93 (s, 1H, CONH), 8.66 (s, 1H, cipro-ArH-2), 7.93–7.86 (m, 2H, triazole-H + cipro-ArH-5), 7.55 (d, J = 7.2 Hz, 1H, cipro-ArH-8), 7.50 (d, J = 8.6 Hz, 2H, phenyl-H), 7.37 (d, J = 8.6 Hz, 2H, phenyl-H′), 5.07 (br s, 1H, OH), 4.84 (br s, 1H, OH), 4.62 (br s, 1H, OH), 4.43 (d, J = 9.2 Hz, 2H, glyco-H-1 + OH), 4.34 (t, J = 6.9 Hz, 2H, −NHCOCH2CH2CH2CH2−), 3.81 (br s, 1H,), 3.68 (s, 1H, glyco-H-4), 3.49 (glyco-H-6 + H-6′, extracted from HSQC), 3.41 (glyco-H-5, extracted from HSQC), 3.37 (glyco-H-2, extracted from HSQC), 3.34 (2x piperazine-CH2, extracted from HSQC), 3.32 (glyco-H-3, extracted from HSQC), 2.84 (t, J = 7.5 Hz, 2H, −triazole-CH2CH2NRR′), 2.67 (br s, 6H, 2x piperazine-CH2 + −triazole-CH2CH2NRR′), 2.33 (t, J = 7.3 Hz, 2H, −NHCOCH2CH2CH2CH2−), 1.83 (tt, J = 6.9 Hz, 2H, −NHCOCH2CH2CH2CH2−), 1.54 (tt, J = 7.4 Hz, 2H, −NHCOCH2CH2CH2CH2−), 1.36–1.24 (m, 2H, cPr-CH2), 1.25–1.06 (br s, 2H, cPr-CH2′). 13C NMR (126 MHz, DMSO-d6) δ 176.41 (cipro-C4=O), 170.89 (C=O), 166.04 (COOH), 153.07 (d, J = 249.6 Hz, cipro-C-6), 148.04 (cipro-C-2), 145.23 (d, J = 9.9 Hz, cipro-C-7), 144.99 (triazole-C), 139.24 (cipro-C-8a), 138.05 (phenyl-C), 131.19 (phenyl-C), 128.23 (phenyl-C), 122.23 (triazole-CH), 119.35 (phenyl-C), 118.58 (d, J = 7.5 Hz, cipro-C-4a), 110.98 (d, J = 23.2 Hz, cipro-C-5), 106.75 (cipro-C-3), 106.36 (d, J = 3.0 Hz, cipro-C-8), 88.37 (glyco-C-1), 79.19 (glyco-C-5), 74.72 (glyco-C-3), 69.24 (glyco-C-2), 68.38 (glyco-C-4), 60.62 (glyco-C-6), 57.25 (linker-CH2), 52.21 (piperazine), 49.41 (piperazine), 49.00 (linker-CH2), 35.90 (cPr-CH), 35.66 (linker-CH2), 29.40 (linker-CH2), 22.99 (linker-CH2), 22.07 (linker-CH2), 7.61 (cPr-CH2). HR-MS calcd [C37H45FN7O9S]+: 796.3135, found 796.3128.
5-(2′-Bromoethyl)thiophene-2-sulfonyl Chloride (16)
16 was synthesized in two chemical steps: thiopheneethanol 15 (1.0 mL, 9.0 mmol, 1 eq.) was dissolved in 40 mL of dry CH2Cl2. The solution was cooled (0 °C), and a solution of PBr3 (846 μL, 9.0 mmol, 1 eq.) in dry CH2Cl2 was added dropwise under vigorous stirring; the reaction was stirred for 1 h until full transformation, monitored by TLC (PE:EtOAc, 95:5). The reaction was quenched with ice water. The organic phase was washed with water (2x), aq. half satd. Na2CO3 (2x), and brine and dried over anhydrous Na2SO4. The organic phase was reduced in vacuo and filtered over silica. After evaporation of the solvent in vacuo crude 2-(2′-bromoethyl)thiophene was obtained as a yellow oil (490 mg, 28%). 1H NMR (500 MHz, CHCl3-d) δ 7.20 (dd, J = 5.1, 1.2 Hz, 1H, ArH-5), 6.97 (dd, J = 5.1, 3.5 Hz, 1H, ArH-4), 6.90 (dd, J = 3.4, 1.0 Hz, 1H, ArH-3), 3.58 (t, J = 7.4 Hz, 2H, −CH2CH2Br), 3.38 (t, J = 7.5 Hz, 2H, −CH2CH2Br). 2-(2′-Bromoethyl)thiophene (255 mg, 1.33 mmol, 1 eq.) was dissolved in 10 mL of dry CH2Cl2, and the mixture was cooled (0 °C). HSO3Cl (266 μL, 4 mmol, 3 eq.) was dissolved in 5 mL of dry CH2Cl2 and added dropwise to the starting material under vigorous stirring. The reaction was stirred 1 h until full transformation, monitored by TLC (PE:EtOAc, 95:5). The reaction was quenched with ice water. The aqueous phase was extracted with CH2Cl2 (3x). The combined organic phases were washed with half satd. brine (x) and brine (1x) and dried over anhydrous Na2SO4. The solvent was evaporated in vacuo to obtain the crude product as a dark yellow oil (261 mg).
β-l-Fucopyranosyl-1-methylamine (17)
β-l-Fucopyranosyl-1-nitromethane was synthesized according to Phiasivongsa et al.73 with subsequent reduction to the amine as previously described in Sommer et al.42 NMR in agreement with literature data.42
N-β-l-Fucopyranosylmethyl-2-(5-(2′-azidoethyl)thiophene)sulfonamide (19)
β-l-Fucopyranosyl-1-methylamine (17, 128 mg, 0.60 mmol, 1 eq.) and K2CO3 (166 mg, 1.2 mmol, 2 eq.) were dispersed in 6 mL of dry DMF and cooled to 0 °C. Crude 2-chlorosulfonyl-5-(2′-bromoethyl)thiophene (261 mg, 0.90 mmol) was dissolved in 6 mL of dry DMF and added dropwise to the starting material under vigorous stirring. The reaction was stirred for 3 h until full conversion, as monitored by TLC (MeOH:EtOAc:aq. NH4OH 25%, 4:4:2). After quenching with water, the aqueous phase was extracted with EtOAc (4x). The combined organic layers were washed with half satd. brine (3x) and brine (1x) and dried over anhydrous Na2SO4. After filtration, the solvent was evaporated in vacuo and the crude material (191 mg) was dissolved in 10 mL of dry DMF. NaN3 (143 mg, 2.2 mmol) was added, and the mixture was stirred for 3 h. After full transformation (monitored by HPLC-MS), the reaction was diluted with water and extracted with EtOAc (3x). The combined organic layers were washed with half satd. brine (3x) and satd. brine (1x) and dried over anhydrous Na2SO4. After filtration, the solvent was evaporated in vacuo, and the product was purified by MPLC (DCM:MeOH, 1–11%) to yield the target compound as a white amorphous solid (141 mg, 60% after three chemical steps, 8% impurity of the corresponding alkyl chloride, determined by 1H NMR). 1H NMR (500 MHz, MeOH-d4) δ 7.46 (d, J = 3.8 Hz, 1H, Ar-H), 6.97 (d, J = 3.7 Hz, 1H, Ar-H), 3.65–3.57 (m, 3H, −CH2CH2N3 + H-4), 3.50 (q, J = 7.0 Hz, 1H, H-5), 3.45–3.34 (m, 3H, −CH2N– + H-2), 3.17 (td, J = 9.1, 8.6, 2.4 Hz, 1H, H-1), 3.12 (t, J = 6.6 Hz, 2H, −CH2CH2N3), 3.06 (dd, J = 12.9, 7.2 Hz, 1H, −CH2′N −), 1.20 (d, J = 6.5 Hz, 3H, H-6). 13C NMR (126 MHz, MeOH-d4) δ 149.48 (Ar-C), 141.03 (Ar-C), 132.98 (Ar-C), 127.32 (Ar-C), 79.55 (glyco-C-2), 76.37 (glyco-C-3), 75.57 (glyco-C-5), 73.61 (glyco-C-4), 69.74 (glyco-C-1), 53.08 (glyco-C-2), 45.75 (linker-CH2), 30.71 (linker-CH2), 17.07 (glyco-C-6). HR-MS calcd [C13H19N4O6S2]−: 391.0751, found 391.0759.
Hybrid-Ciprofloxacin Conjugate 30 (m = 0)
The title compound was synthesized from 19 (35 mg, 0.09 mmol, 1 eq.) and 20 (35 mg, 0.095 mmol, 1.1 eq.) according to general procedure iv and was obtained as a beige amorphous solid (30 mg, 44%). 1H NMR (500 MHz, DMSO-d6) δ 15.23 (br s, 1H, -COOH), 8.66 (s, 1H, cipro-H-2), 7.98 (s, 1H, triazole-H), 7.90 (d, J = 13.3 Hz, 1H, cipro-H-5), 7.66 (t, J = 5.9 Hz, 1H, −NHSO2−), 7.55 (d, J = 7.4 Hz, 1H, cipro-H-8), 7.37 (d, J = 3.7 Hz, 1H, thienyl-H), 6.89 (d, J = 3.8 Hz, 1H, thienyl-H), 4.80 (br s, 1H, OH), 4.65 (t, J = 6.7 Hz, 1H, thiophene-CH2CH2-triazole), 4.59 (br s, 1H, OH), 4.28 (d, J = 5.5 Hz, 1H, OH), 3.86–3.77 (m, 1H, cPr-H), 3.63 (s, 2H, triazole-CH2-NRR′), 3.47 (t, J = 6.7 Hz, 2H, thiophene-CH2CH2-triazole), 3.39 (s, 1H, glyco-H-4), 3.37, 3.25–3.18 (m, 2H, −CH2NHSO2– + glyco-H-3), 3.14 (t, J = 9.3 Hz, 1H, glyco-H-2), 3.01 (td, J = 8.8, 2.3 Hz, 1H, glyco-H-1), 2.73 (ddd, J = 13.4, 8.4, 5.6 Hz, 1H, −CH2NHSO2−), 2.59 (2.63–2.56 m, 4H, 2x piperazine-CH2), 1.39–1.30 (m, 2H, cPr-CH2), 1.20–1.14 (m, 4H, cPr-CH2), 1.07 (d, J = 6.4 Hz, 3H, glyco-H-6). 13C NMR (126 MHz, DMSO-d6) δ 176.38 (cipro-C4=O), 165.98 (COOH), 153.02 (d, J = 249.2 Hz, cipro-C-6), 147.98 (cipro-C-2), 146.07 (Ar-C), 145.23 (d, J = 10.1 Hz, cipro-C-7), 143.18 (triazole-C), 139.65 (Ar-C), 139.23 (cipro-C-8a), 131.14 (Ar-C), 126.59 (Ar-C), 124.22 (triazole-CH), 118.56 (d, J = 7.4 Hz, cipro-C-4a), 110.95 (d, J = 22.9 Hz, cipro-C-5), 106.72 (cipro-C-3), 106.34 (d, J = 2.6 Hz, cipro-C-8), 78.24 (glyco-C-2), 74.64 (glyco-C-3), 73.64 (glyco-C-5), 71.56 (glyco-C-4), 68.30 (glyco-C-1), 52.41 (linker-CH2), 51.88 (piperazine), 50.01 (linker-CH2), 49.40 (piperazine), 44.74 (glyco-CH2), 35.90 (cPr-CH), 29.97 (linker-CH2), 16.93 (glyco-C-6), 7.57 (cPr-CH2). HR-MS calcd [C33H41FN7O9S2]+: 762.2386, found 762.2382.
Hybrid-Ciprofloxacin Conjugate 31 (m = 1)
The title compound was synthesized from 19 (56 mg, 0.14 mmol, 1 eq.) and 21 (59 mg, 0.15 mmol, 1.1 eq.) according to general procedure iv and was obtained as a beige amorphous solid (57 mg, 52%). 1H NMR (500 MHz, DMSO-d6) δ 15.23 (s, 1H, COOH), 8.67 (s, 1H, cipro-ArH-2), 7.93–7.89 (m, 2H, triazole-H + cipro-ArH-5), 7.68 (t, J = 5.9 Hz, 1H, −NHSO2−), 7.58 (d, J = 7.4 Hz, 1H, cipro-ArH-8), 7.38 (d, J = 3.7 Hz, 1H, thienyl-H), 6.89 (d, J = 3.8 Hz, 1H, thienyl-H), 4.82 (br s, 1H, OH), 4.62 (t, J = 6.9 Hz, 2H, thiophene-CH2CH2-triazole + OH), 4.29 (s, 1H, OH), 3.84 (s, 1H, cPr-H), 3.44 (t, J = 6.9 Hz, 2H, thiophene-CH2CH2-triazole), 3.40 (s, 1H, glyco-H-4), 3.37 (1H, glyco-H-5, extracted from HSQC), 3.35 (4H, 2x piperazine-CH2), 3.28–3.20 (m, 2H, −CH2NSO2– + glyco-H-3), 3.15 (t, J = 9.3 Hz, 1H, glyco-H-2), 3.02 (td, J = 8.8, 2.3 Hz, 1H, glyco-H-1), 2.85 (t, J = 7.5 Hz, 2H, −triazole-CH2CH2NRR′), 2.74 (ddd, J = 13.6, 8.4, 5.7 Hz, 1H, −CH2′NSO2−), 2.68 (br s, 6H, 2x piperazine-CH2 + −triazole-CH2CH2NRR′), 1.91 (s, 0H), 1.35–1.29 (m, 2H, cPr-CH2), 1.22–1.16 (m, 2H, cPr-CH2), 1.08 (d, J = 6.4 Hz, 3H, glyco-H-6). 13C NMR (126 MHz, DMSO-d6) δ 176.37 (cipro-C4=O), 165.97 (COOH), 153.03 (d, J = 250.0 Hz, cipro-C-6), 148.02 (cipro-C-2), 146.17 (Ar-C), 145.17 (d, J = 10.4 Hz, cipro-C-7), triazole-C not found, 139.66 (Ar-C), 139.21 (cipro-C-8a), 131.18 (Ar-C), 126.45 (Ar-C), 122.47 (triazole-CH), 118.57 (d, J = 7.4 Hz, cipro-C-4a), 110.95 (d, J = 23.1 Hz, cipro-C-5), 106.73 (cipro-C-3), 106.37 (d, J = 2.5 Hz, cipro-C-8), 78.24 (glyco-C-2), 74.65 (glyco-C-3), 73.63 (glyco-C-5), 71.57 (glyco-C-4), 68.31 (glyco-C-1), 57.16 (linker-CH2), 52.15 (piperazine), 49.88 (linker-CH2), 49.32 (piperazine), 44.74 (glyco-CH2), 35.89 (cPr-CH), 29.99 (linker-CH2), 22.87 (linker-CH2), 16.93 (glyco-C-6), 7.59 (cPr-CH2). HR-MS calcd [C34H43FN7O9S2]+: 776.2542, found 776.2538.
Competitive Binding Assays
LecA (According to Joachim et al.58)
A serial dilution of the test compounds was prepared in TBS/Ca (8.0 g/L NaCl, 2.4 g/L Tris, 0.19 g/L KCl, 0.15 g/L CaCl2·2H2O), with 30% DMSO as a co-solvent. A concentrated solution of LecA was diluted in TBS/Ca together with the fluorescent reporter ligand (N-(fluorescein-5-yl)-N′-(β-d-(m-aminophenyl)galactopyranosyl)thiocarbamide) to yield concentrations of 40 μM and 20 nM, respectively. A 10 μL solution of this mix was added to 10 μL serial dilutions of the test compounds in a black 384-well microtiter plates (Greiner Bio-One, Germany, cat. no. 781900) in triplicate. After centrifugation (2680 rcf, 1 min, r.t.), the reactions were incubated for 30–60 min at r.t. in a humidity chamber. Fluorescence (excitation 485 nm, emission 535 nm) was measured in parallel and perpendicular to the excitation plane on a PheraStar FS plate reader (BMG Labtech GmbH, Germany). The measured intensities were reduced by the values of only LecA in TBS/Ca, and fluorescence polarization was calculated. The data were analyzed with the MARS Data Analysis Software (BMG Labtech GmbH, Germany) and fitted according to the four-parameter variable slope model. Bottom and top plateaus were fixed according to the control compounds in each assay (p-nitrophenyl)-β-d-galactoside), and the data was reanalyzed with these values fixed. A minimum of three independent measurements on three plates was performed for each inhibitor.
LecB (LecBPAO1 According to Hauck et al.41 and LecBPA14 According to Sommer et al.33)
A serial dilution of the test compounds was prepared in TBS/Ca, with 10% DMSO as a co-solvent. A concentrated solution of LecB PAO1 or PA14 was diluted in TBS/Ca together with the fluorescent reporter ligand (N-(fluorescein-5-yl)-N′-(α-l-fucopyranosyl ethylene)thiocarbamide) to yield concentrations of 300 nM and 20 nM, respectively. A 10 μL solution of this mix was added to 10 μL serial dilutions of the test compounds in a black 384-well microtiter plates (Greiner Bio-One, Germany, cat. no. 781900) in triplicate. After centrifugation (2680 rcf, 1 min, r.t.), the reactions were incubated for 4–8 h at r.t. in a humidity chamber. Fluorescence was measured and analyzed as for LecA. Bottom and top plateaus were fixed according to the control compound in each assay (l-fucose), and the data were reanalyzed with these values fixed. A minimum of three independent measurements on three plates was performed for each inhibitor.
Gyrase Supercoiling Inhibition
The assay was performed with the E. coli gyrase supercoiling kit (Inspiralis, Norwich, UK) according to the manufacturer’s instructions. All pipetting steps before the reaction was started were performed on ice. A serial dilution of the test compounds was prepared in 5% DMSO in water. A mix of relaxed pBR322 DNA (5.5 μg), 66 μL assay buffer (5x), and 192.5 μL water was prepared. 3 μL of the dilution series (or 3 μL 5% DMSO in water for control reactions) was added. 10 U gyrase (2 μL, 5 U/μL) was diluted in 28 μL dilution buffer. 3 μL of the gyrase (1 U) solution was added to the reaction mixtures. For the negative control, 3 μL of dilution buffer was added instead of the enzyme. The reaction was incubated for 30 min at 37 °C. The reactions were stopped by the addition of 30 μL of STE-buffer (40% (m/v) sucrose, 100 mM Tris·HCl, pH 8, 10 mM EDTA, pH 8, 0.5 mg/mL bromophenol blue) and 30 μL of CHCl3/isoamyl alcohol (24:1) and vortexing. After centrifugation (17,600 rcf, 1 min, 4 °C), 50 μL of the aqueous layer was loaded on an agarose gel (1%, Tris-EDTA-acetate buffer). The gel was run for 3 h at 85 V, and DNA was visualized afterward by staining with ethidium bromide. Agarose gels were digitalized using the E-box VX2 gel documentation instrument (Vilber, Eberhardzell, Germany). The fluorescence intensity of each supercoiled band was quantified using ImageJ (Version 1.52a, National Institute of Health, USA). The data were analyzed using GraphPad Prism (Version 6.0 h, GraphPad Software, USA) and fitted against inhibitor concentration according to the four-parameter variable slope model to determine IC50 values. Bottom plateaus were fixed to 0. A minimum of three different experiments was performed for each inhibitor.
Bacterial Strain List
All microorganisms were obtained from the German Collection of Microorganisms and Cell Cultures (DSMZ) and the American Type Culture Collection (ATCC) or were part of our internal strain collection. The following strains were used: Escherichia coli DSM 1116 (source: Rolf Müller, HIPS), Escherichia coli K12 MG1655 (source: Winfried Boos, Universität Konstanz), Staphylococcus carnosus DSM 20501 (source: Rolf Müller, HIPS), Pseudomonas aeruginosa PA14 wt (DSM 19882), Pseudomonas aeruginosa PAO1 wt (DSM 19880), Pseudomonas aeruginosa PA14 ΔlecA (Wagner et al., in preparation), and Pseudomonas aeruginosa PA14 ΔlecB (Wagner et al., in preparation).
Antibiotic Susceptibility (MIC Assay)
The antibiotic activity of the synthesized conjugates was determined by broth microdilution assay based on the EUCAST guidelines, according to Wiegand, Hilpert, and Hancock.74 Serial dilutions in sterile Müller-Hinton broth II (Fluka analytical, cat. no. 90922: 17.5 g/L casein acid hydrolysate, 3 g/L beef extract, 1.5 g/L starch, supplemented with 20–25 mg/L Ca2+ and 10–15 mg/L Mg2+, pH 7.3) of the conjugates 21–31 and 20 were prepared from 100 mM DMSO stocks (for ciprofloxacin (3), a 10 mM aq. stock of ciprofloxacin·HCl was used), in sterile 96-well plates, yielding a concentration range from 128 to 0.125 μg/mL (12.8–0.0125 for ciprofloxacin). Bacterial strains were streaked on LB-agar plates (1% agar) from glycerol stocks and incubated at 37 °C overnight. Colonies were picked from plate and dispersed in fresh Müller-Hinton broth II (MHB II) to yield an OD600 of 0.08–0.13. This dispersion was diluted 1:100 in fresh MHB II, which was then used for the assay to achieve a final inoculum of 5 × 105 CFU/mL. If indicated, PMBN was added to this inoculum at 2 μg/mL. A 50 μL inoculum was mixed with 50 μL of the serial dilution in the corresponding well of the 96-well plate. The plates were incubated at 37 °C for 18–20 h in a humid incubator. Growth inhibition was assessed by visual inspection, and the given MIC values are the lowest concentration of the antibiotic at which there was no visible growth.
Biofilm Accumulation Assay
Bacterial precultures of P. aeruginosa PAO1 were prepared in 10 mL of LB and grown at 37 °C and 180 rpm overnight. The bacterial precultures were diluted in fresh LB to 50 mL and centrifuged (5925 rcf, 10 min, r.t.). The supernatant was discarded, and the pellet was resuspended and washed in 50 mL of fresh LB and centrifuged again (5925 rcf, 10 min, r.t.). The supernatant was discarded and the pellet was again resuspended in fresh LB to yield an OD600 of 0.1. Then, 150 μL of this inoculum were transferred to each well of a 96-well MBEC assay plate (SKU: 19113, Category: Well Base, Innovotech Inc., Canada). The outer wells were filled with 150 μL of sterile LB as a control. Plates were incubated at 37 °C, 125 rpm, and 75% humidity for 24 h. Compound solutions (170 μL, 200 μM, 1% DMSO) in phosphate-buffered saline pH 7.4, supplemented with 100 μM CaCl2 (PBS/Ca) were dispensed in a 96-well plate (cat. no. 167008, Nunc MicroWell 96-Well Microplates, Thermo Scientific) in quintuplicate on plates. Each peg of the biofilm covered peg lid was washed in 200 μL of PBS/Ca in a 96-well plate (Nunc) for 1 min at r.t. and then incubated with the compound solution for 5 or 10 min at 37 °C, 80 rpm under humid conditions. After the incubation step, the pegs were again washed with 200 μL of PBS/Ca in a 96-well format for 30 s at r.t. and transferred to a last 96-well plate (Nunc) filled with 170 μL PBS/Ca per well. The plate was sealed with parafilm and sonicated for 15 min using an ultrasound bath. A 100 μL sample of each well was transferred to a vial and treated with 100 μL of MeCN (spiked with 1.5 μM diphenhydramine·HCl as an internal standard). After centrifugation (17,600 rcf, 10 min, 4 °C), the compound concentration in the supernatant was determined by LC-MS/MS. Fresh calibration curves for each compound were prepared in the same matrix for each experiment. In each assay, the accumulation factor relative to ciprofloxacin was determined. Statistical analysis (unpaired t-test) was performed using the GraphPad-Prism QuickCalcs online tool (https://www.graphpad.com/quickcalcs/contMenu/).
LC-MS/MS
LC-MS/MS analysis was performed on an Ultimate 3000 system (degasser, pump, autosampler, column compartment) equipped with a Nucleodur C18 Pyramid column (150 × 2 mm, 3 μm, Macherey-Nagel, Düren, Germany) coupled to a TSQ Quantum Access MAX (Thermo Fisher Scientific, Waltham MA) with the following gradient conditions: A, water (0.1% formic acid); B, acetonitrile (0.1% formic acid); flow 0.600 mL/min; 90% A for 1.0 min; 90–5% A in 0.7 min; 5% A for 1.8 min; equilibration at 90% A for 1.0 min. MS was operated in positive SRM mode with the following mass transitions:
Diphenhydramin (IS): 256.04–164.90; 256.04–166.90.
Ciprofloxacin (3): 332.063–230.908; 332.063–244.968; spray voltage: 4001 V, vaporizer temperature: 420 °C, sheath gas pressure: 50 psi, ion sweep pressure: 2.5 psi, aux gas pressure: 30 psi, capillary temperature: 260 °C, tube lens offset: 97 V, skimmer offset: 0 V, collision pressure: 1.5 mTorr, collision energy: 36 eV (230.908), 23 eV (244.968).
(21): 740.140–559.933; 740.140–577.966; spray voltage: 3000 V, vaporizer temperature: 470 °C, sheath gas pressure: 60 psi, ion sweep pressure: 0 psi, aux gas pressure: 55 psi, capillary temperature: 296 °C, tube lens offset: 99 V, skimmer offset: 0 V, collision pressure: 1.5 mTorr, collision energy: 36 eV (559.933), 27 eV (577.966).
(29): 762.124–726.026; 762.124–744.061; spray voltage: 4500 V, vaporizer temperature: 223 °C, sheath gas pressure: 60 psi, ion sweep pressure: 0 psi, aux gas pressure: 55 psi, capillary temperature: 284 °C, tube lens offset: 99 V, skimmer offset: 0 V, collision pressure: 1.5 mTorr, collision energy: 33 eV (726.026), 29 eV (744.061).
Cytotoxicity (MTT Assay, According to Haupenthal et al.75)
HEK293 or A549 cells (2 × 105 cells per well) were seeded in 24-well, flat-bottom plates. Culturing of cells, incubations, and OD measurements were performed as described with small modifications. Twenty-four hours after seeding the cells, the incubation was started by the addition of compounds in a final DMSO concentration of 1%. The living cell mass was determined after 48 h in a PHERAstar microplate reader (BMG Labtech, Ortenberg, Germany). Two independent measurements were performed for each compound.
Microsomal Stability
Microsomal stability was performed as previously described in Sommer et al.44
Plasma Protein Binding
Plasma protein binding was measured with a rapid equilibrium dialysis assay plate (Thermo Fisher Scientific, Waltham MA). On one side of the membrane, 150 μL of human plasma (seralab-BioIVT, West Sussex United Kingdom) and 150 μL of PBS pH 7.4 (Gibco Thermo Fisher Scientific, Waltham MA) were added to the well; on the other side, 550 μL of PBS was added to the well. The compound was added to a final concentration of 1 μM to the plasma-containing well. The plate was closed and incubated in an orbital shaker at 37 °C for 6 h at 750 rpm. Samples of 10 μL from each well were taken at 0, 5, and 6 h and mixed with 90 μL of ice-cold acetonitrile with internal standard diphenhydramine (1 μM). The concentration of compound in the supernatant was analyzed with LC-MS/MS. Plasma protein binding was calculated from the concentration difference between the wells. Five and 6 h samples were compared to ensure equilibrium. Warfarin was used as an assay control.
Human Plasma Stability
Compound stability in plasma was measured by incubation with plasma and LC-MS/MS quantification of the remaining compound. A 195 μL solution of human plasma (seralab-BioIVT, West Sussex, United Kingdom) was incubated with 5 μL of compound (40 μM stock) at 37 °C for 0, 5, 60, and 150 min. Then, 800 μL of ice-cold acetonitrile containing internal standard diphenhydramine (1 μM) was added. The concentration of remaining compound in the supernatant was determined via LC-MS/MS measurement. Procaine was used as an activity control of plasma metabolism.
Cell Permeability
Permeability of the compound was assessed in vitro with Calu-3 HTB-55 cell line (ATCC). Cells were cultivated in minimum essential medium supplemented with Earle’s salts, l-glutamine, 10% FCS, 1% non-essential amino acids (NEAA), and 1 mM sodium pyruvate. Passages between 35 and 55 were used, and the medium was changed every 2–3 days. For experiments, cells were harvested using Trypsin/EDTA and 1 × 105 cells seeded on Transwell inserts 3460. Cells were grown in an air–liquid interface beginning at day 3 and used for transport studies on days 11–13. TEER values exceeded 300 Ω·cm2 before beginning transport studies. For experiments, Krebs-Ringer solution with 1% BSA was used and cells were accommodated to the buffer for at least 1 h with no decrease in TEER. Samples (200 μL) were taken in regular intervals from the apical side (time intervals of 0, 20, 40, 60, 90, 120, 180, and 240 min) and replenished with fresh buffer. TEER was monitored during the experiment, and epithelial barriers were considered compromised if the TEER fell below 300 Ω·cm2 during 4 h of experiment duration. Fluorescein sodium salt and ciprofloxacin·HCl were used as a control. A 50 μL sample was mixed with 150 μL of ice-cold acetonitrile containing internal standard diphenhydramine (1 μM), and the concentration of compound was analyzed with LC-MS/MS.
Acknowledgments
The authors are thankful to Prof. Dr. Rolf Müller and Dr. Jennifer Hermann (HIPS) for scientific discussions and for providing the bacterial strains S. carnosus DSM 20501 and E. coli DSM 1116. We are grateful to Dr. Thomas Ryckmans (F. Hoffmann la Roche, Basel) for metabolic stability assays against human liver microsomes and mouse liver microsomes, Tabea Wittmann and Dennis Jener (HIPS) for plasma stability, plasma protein binding, and cytotoxicity assays, and Justus Horstmann (HIPS) for cell permeation measurements. A.T. acknowledges financial support from the Helmholtz Association (VH-NG-934), the European Research Council (ERC Starting Grant, Sweetbullets), and DZIF.
Glossary
List of Abbreviations
- XDR
extensively drug-resistant
- WHO
World Health Organization
- P. aeruginosa
Pseudomonas aeruginosa
- MLM
mouse liver microsomes
- HLM
human liver microsomes
- PE
petroleum ether
- CLMIC
microsomal clearance
- cPr
cyclopropyl
- rcf
relative centrifugal force
- LR-MS
low-resolution mass spectrometry
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.jmedchem.0c00856.
1H and 13C NMR spectra of new compounds; gyrase supercoiling inhibition assay gels; P. aeruginosa PAO1 biofilm accumulation raw data; lectin inhibition Ki values calculated from IC50; antibiotic susceptibility in molar concentration; key compounds and intermediates as SMILES; purity of key compounds 11–14, 19–31 by HPLC-UV; and retention times and a representative chromatogram of conjugates 22–31 and ciprofloxacin (3) from slow gradient HPLC runs for lipophilicity comparison (PDF)
Molecular formula strings for all new compounds and key compounds (5–14, 16, and 19–31) (CSV)
Author Contributions
J.M. synthesized conjugates and individual building blocks. D.H. synthesized compound 17. J.M. and E.Z. performed lectin inhibition assays. J.M. performed gyrase supercoiling inhibition, antibiotic susceptibility, and biofilm accumulation assays. T.R. analyzed data for metabolic stability in human plasma, plasma protein binding, and acute cytotoxicity. S.W. provided conceptual advice and analyzed the data. J.M. and A.T. conceived the study. J.M. and A.T. wrote the paper with input from all coauthors.
The authors declare no competing financial interest.
Supplementary Material
References
- Rice L. B. Federal funding for the study of antimicrobial resistance in nosocomial pathogens: no ESKAPE. J. Infect. Dis. 2008, 197, 1079–1081. 10.1086/533452. [DOI] [PubMed] [Google Scholar]
- Boucher H. W.; Talbot G. H.; Bradley J. S.; Edwards J. E.; Gilbert D.; Rice L. B.; Scheld M.; Spellberg B.; Bartlett J. Bad bugs, no drugs: no ESKAPE! An update from the Infectious Diseases Society of America. Clin. Infect. Dis. 2009, 48, 1–12. 10.1086/595011. [DOI] [PubMed] [Google Scholar]
- Rice L. B. Progress and challenges in implementing the research on ESKAPE pathogens. Infect. Control Hosp. Epidemiol. 2010, 31, S7–S10. 10.1086/655995. [DOI] [PubMed] [Google Scholar]
- Meynard J.-L.; Barbut F.; Guiguet M.; Batissel D.; Lalande V.; Lesage D.; Guiard-Schmid J.-B.; Petit J.-C.; Frottier J.; Meyohas M.-C. Pseudomonas aeruginosa infection in human immunodeficiency virus infected patients. J. Infect. 1999, 38, 176–181. 10.1016/S0163-4453(99)90247-5. [DOI] [PubMed] [Google Scholar]
- Rizzi E. B.; Schininà V.; Bordi E.; Buontempo G.; Narciso P.; Bibbolino C. HIV-related bronchopulmonary infection by pseudomonas aeruginosa in the HAART era: radiological findings. Acta Radiol. 2006, 47, 793–797. 10.1080/02841850600827569. [DOI] [PubMed] [Google Scholar]
- Bodey G. P. Pseudomonas aeruginosa infections in cancer patients: have they gone away?. Curr. Opin. Infect. Dis. 2001, 14, 403–407. 10.1097/00001432-200108000-00001. [DOI] [PubMed] [Google Scholar]
- Hauser A. R.; Rello J.. Severe infections caused by Pseudomonas aeruginosa; Springer Science & Business Media: Boston, MA, 2012. [Google Scholar]
- Poole K. Pseudomonas aeruginosa: resistance to the max. Front. Microbiol. 2011, 2, 65. 10.3389/fmicb.2011.00065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Available at: https://www.who.int/news-room/detail/27-02-2017-who-publishes-list-of-bacteria-for-which-new-antibiotics-are-urgently-needed
- Suci P. A.; Mittelman M. W.; Yu F. P.; Geesey G. G. Investigation of ciprofloxacin penetration into Pseudomonas aeruginosa biofilms. Antimicrob. Agents Chemother. 1994, 38, 2125–2133. 10.1128/AAC.38.9.2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flemming H.-C.; Wingender J. The biofilm matrix. Nat. Rev. Microbiol. 2010, 8, 623–633. 10.1038/nrmicro2415. [DOI] [PubMed] [Google Scholar]
- Davies D. Understanding biofilm resistance to antibacterial agents. Nat. Rev. Drug Discovery 2003, 2, 114–122. 10.1038/nrd1008. [DOI] [PubMed] [Google Scholar]
- Winzer K.; Falconer C.; Garber N. C.; Diggle S. P.; Camara M.; Williams P. The Pseudomonas aeruginosa lectins PA-IL and PA-IIL are controlled by quorum sensing and by RpoS. J. Bacteriol. 2000, 182, 6401–6411. 10.1128/JB.182.22.6401-6411.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diggle S. P.; Stacey R. E.; Dodd C.; Cámara M.; Williams P.; Winzer K. The galactophilic lectin, LecA, contributes to biofilm development in Pseudomonas aeruginosa. Environ. Microbiol. 2006, 8, 1095–1104. 10.1111/j.1462-2920.2006.001001.x. [DOI] [PubMed] [Google Scholar]
- Tielker D.; Hacker S.; Loris R.; Strathmann M.; Wingender J.; Wilhelm S.; Rosenau F.; Jaeger K.-E. Pseudomonas aeruginosa lectin LecB is located in the outer membrane and is involved in biofilm formation. Microbiology 2005, 151, 1313–1323. 10.1099/mic.0.27701-0. [DOI] [PubMed] [Google Scholar]
- Gilboa-Garber N. Pseudomonas aeruginosa lectins. Methods Enzymol. 1982, 83, 378–385. 10.1016/0076-6879(82)83034-6. [DOI] [PubMed] [Google Scholar]
- Gilboa-Garber N.; Mizrahi L.; Garber N. Mannose-binding hemagglutinins in extracts of Pseudomonas aeruginosa. Can. J. Biochem. 1977, 55, 975–981. 10.1139/o77-145. [DOI] [PubMed] [Google Scholar]
- Gilboa-Garber N. Purification and properties of hemagglutinin from Pseudomonas aeruginosa and its reaction with human blood cells. Biochim. Biophys. Acta, Gen. Subj. 1972, 273, 165–173. 10.1016/0304-4165(72)90204-8. [DOI] [PubMed] [Google Scholar]
- da Silva D. P.; Matwichuk M. L.; Townsend D. O.; Reichhardt C.; Lamba D.; Wozniak D. J.; Parsek M. R. The Pseudomonas aeruginosa lectin LecB binds to the exopolysaccharide Psl and stabilizes the biofilm matrix. Nat. Commun. 2019, 10, 2183. 10.1038/s41467-019-10201-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adam E. C.; Mitchell B. S.; Schumacher D. U.; Grant G.; Schumacher U. Pseudomonas aeruginosa II lectin stops human ciliary beating: therapeutic implications of fucose. Am. J. Respir. Crit. Care Med. 1997, 155, 2102–2104. 10.1164/ajrccm.155.6.9196121. [DOI] [PubMed] [Google Scholar]
- Landi A.; Mari M.; Kleiser S.; Wolf T.; Gretzmeier C.; Wilhelm I.; Kiritsi D.; Thünauer R.; Geiger R.; Nyström A.; Reggiori F.; Claudinon J.; Römer W. Pseudomonas aeruginosa lectin LecB impairs keratinocyte fitness by abrogating growth factor signalling. Life Sci. Alliance 2019, 2, e201900422 10.26508/lsa.201900422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cott C.; Thuenauer R.; Landi A.; Kühn K.; Juillot S.; Imberty A.; Madl J.; Eierhoff T.; Römer W. Pseudomonas aeruginosa lectin LecB inhibits tissue repair processes by triggering β-catenin degradation. Biochim. Biophys. Acta, Mol. Cell Res. 2016, 1863, 1106–1118. 10.1016/j.bbamcr.2016.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm I.; Levit-Zerdoun E.; Jakob J.; Villringer S.; Frensch M.; Übelhart R.; Landi A.; Müller P.; Imberty A.; Thuenauer R.; Claudinon J.; Jumaa H.; Reth M.; Eibel H.; Hobeika E.; Römer W. Carbohydrate-dependent B cell activation by fucose-binding bacterial lectins. Sci. Signaling 2019, 12, eaao7194 10.1126/scisignal.aao7194. [DOI] [PubMed] [Google Scholar]
- Zheng S.; Eierhoff T.; Aigal S.; Brandel A.; Thuenauer R.; de Bentzmann S.; Imberty A.; Römer W. The Pseudomonas aeruginosa lectin LecA triggers host cell signalling by glycosphingolipid-dependent phosphorylation of the adaptor protein CrkII. Biochim. Biophys. Acta, Mol. Cell Res. 2017, 1864, 1236–1245. 10.1016/j.bbamcr.2017.04.005. [DOI] [PubMed] [Google Scholar]
- Eierhoff T.; Bastian B.; Thuenauer R.; Madl J.; Audfray A.; Aigal S.; Juillot S.; Rydell G. E.; Muller S.; de Bentzmann S.; Imberty A.; Fleck C.; Romer W. A lipid zipper triggers bacterial invasion. Proc. Natl. Acad. Sci. U. S. A. 2014, 111, 12895–12900. 10.1073/pnas.1402637111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boukerb A. M.; Rousset A.; Galanos N.; Méar J.-B.; Thépaut M.; Grandjean T.; Gillon E.; Cecioni S.; Abderrahmen C.; Faure K.; Redelberger D.; Kipnis E.; Dessein R.; Havet S.; Darblade B.; Matthews S. E.; de Bentzmann S.; Guéry B.; Cournoyer B.; Imberty A.; Vidal S. Antiadhesive properties of glycoclusters against Pseudomonas aeruginosa lung infection. J. Med. Chem. 2014, 57, 10275–10289. 10.1021/jm500038p. [DOI] [PubMed] [Google Scholar]
- Chemani C.; Imberty A.; de Bentzmann S.; Pierre M.; Wimmerová M.; Guery B. P.; Faure K. Role of LecA and LecB lectins in Pseudomonas aeruginosa-induced lung injury and effect of carbohydrate ligands. Infect. Immun. 2009, 77, 2065–2075. 10.1128/IAI.01204-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Bismarck P.; Schneppenheim R.; Schumacher U. Successful treatment of Pseudomonas aeruginosa respiratory tract infection with a sugar solution - a case report on a lectin based therapeutic principle. Klin. Paediatr. 2001, 213, 285–287. 10.1055/s-2001-17220. [DOI] [PubMed] [Google Scholar]
- Hauber H.-P.; Schulz M.; Pforte A.; Mack D.; Zabel P.; Schumacher U. Inhalation with fucose and galactose for treatment of Pseudomonas aeruginosa in cystic fibrosis patients. Int. J. Med. Sci. 2008, 5, 371–376. 10.7150/ijms.5.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucior I.; Abbott J.; Song Y.; Matthay M. A.; Engel J. N. Sugar administration is an effective adjunctive therapy in the treatment of Pseudomonas aeruginosa pneumonia. Am. J. Physiol.: Lung Cell. Mol. Physiol. 2013, 305, L352–L363. 10.1152/ajplung.00387.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klockgether J.; Cramer N.; Wiehlmann L.; Davenport C. F.; Tümmler B. Pseudomonas aeruginosa genomic structure and diversity. Front. Microbiol. 2011, 2, 150. 10.3389/fmicb.2011.00150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dötsch A.; Schniederjans M.; Khaledi A.; Hornischer K.; Schulz S.; Bielecka A.; Eckweiler D.; Pohl S.; Häussler S. The Pseudomonas aeruginosa transcriptional landscape is shaped by environmental heterogeneity and genetic variation. MBio 2015, 6, e00749 10.1128/mBio.00749-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer R.; Wagner S.; Varrot A.; Nycholat C. M.; Khaledi A.; Häussler S.; Paulson J. C.; Imberty A.; Titz A. The virulence factor LecB varies in clinical isolates: consequences for ligand binding and drug discovery. Chem. Sci. 2016, 7, 4990–5001. 10.1039/C6SC00696E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boukerb A. M.; Decor A.; Ribun S.; Tabaroni R.; Rousset A.; Commin L.; Buff S.; Doléans-Jordheim A.; Vidal S.; Varrot A.; Imberty A.; Cournoyer B. Genomic rearrangements and functional diversification of lecA and lecB lectin-coding regions impacting the efficacy of glycomimetics directed against Pseudomonas aeruginosa. Front. Microbiol. 2016, 7, 811. 10.3389/fmicb.2016.00811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varki A.; Etzler M. E.; Cummings R. D.; Esko J. D.. Discovery and classification of glycan-binding proteins. In Essentials of Glycobiology; 2nd edition, Eds: Varki A.; Cummings R. D.; Esko J. D.; Freeze H. H.; Stanley P.; Bertozzi C. R.; Hart G. W.; Etzler M. E.. Cold Spring Harbor Laboratory Press: 2009, Chapter 26. Available at: http://www.ncbi.nlm.nih.gov/books/NBK1923/. [PubMed] [Google Scholar]
- Calvert M. B.; Jumde V. R.; Titz A. Pathoblockers or antivirulence drugs as a new option for the treatment of bacterial infections. Beilstein J. Org. Chem. 2018, 14, 2607–2617. 10.3762/bjoc.14.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meiers J.; Siebs E.; Zahorska E.; Titz A. Lectin antagonists in infection, immunity, and inflammation. Curr. Opin. Chem. Biol. 2019, 53, 51–67. 10.1016/j.cbpa.2019.07.005. [DOI] [PubMed] [Google Scholar]
- Wagner S.; Sommer R.; Hinsberger S.; Lu C.; Hartmann R. W.; Empting M.; Titz A. Novel strategies for the treatment of Pseudomonas aeruginosa infections. J. Med. Chem. 2016, 59, 5929–5969. 10.1021/acs.jmedchem.5b01698. [DOI] [PubMed] [Google Scholar]
- Cecioni S.; Imberty A.; Vidal S. Glycomimetics versus multivalent glycoconjugates for the design of high affinity lectin ligands. Chem. Rev. 2015, 115, 525–561. 10.1021/cr500303t. [DOI] [PubMed] [Google Scholar]
- Bernardi A.; Jiménez-Barbero J.; Casnati A.; De Castro C.; Darbre T.; Fieschi F.; Finne J.; Funken H.; Jaeger K.-E.; Lahmann M.; Lindhorst T. K.; Marradi M.; Messner P.; Molinaro A.; Murphy P. V.; Nativi C.; Oscarson S.; Penadés S.; Peri F.; Pieters R. J.; Renaudet O.; Reymond J.-L.; Richichi B.; Rojo J.; Sansone F.; Schäffer C.; Turnbull W. B.; Velasco-Torrijos T.; Vidal S.; Vincent S.; Wennekes T.; Zuilhof H.; Imberty A. Multivalent glycoconjugates as anti-pathogenic agents. Chem. Soc. Rev. 2013, 42, 4709–4727. 10.1039/C2CS35408J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauck D.; Joachim I.; Frommeyer B.; Varrot A.; Philipp B.; Möller H. M.; Imberty A.; Exner T. E.; Titz A. Discovery of two classes of potent glycomimetic inhibitors of Pseudomonas aeruginosa LecB with distinct binding modes. ACS Chem. Biol. 2013, 8, 1775–1784. 10.1021/cb400371r. [DOI] [PubMed] [Google Scholar]
- Sommer R.; Exner T. E.; Titz A. A biophysical study with carbohydrate derivatives explains the molecular basis of monosaccharide selectivity of the Pseudomonas aeruginosa lectin LecB. PLoS One 2014, 9, e112822 10.1371/journal.pone.0112822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer R.; Hauck D.; Varrot A.; Wagner S.; Audfray A.; Prestel A.; Möller H. M.; Imberty A.; Titz A. Cinnamide derivatives of d-mannose as inhibitors of the bacterial virulence factor LecB from Pseudomonas aeruginosa. ChemistryOpen 2015, 4, 756–767. 10.1002/open.201500162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommer R.; Wagner S.; Rox K.; Varrot A.; Hauck D.; Wamhoff E.-C.; Schreiber J.; Ryckmans T.; Brunner T.; Rademacher C.; Hartmann R. W.; Brönstrup M.; Imberty A.; Titz A. Glycomimetic, orally bioavailable LecB inhibitors block biofilm formation of Pseudomonas aeruginosa. J. Am. Chem. Soc. 2018, 140, 2537–2545. 10.1021/jacs.7b11133. [DOI] [PubMed] [Google Scholar]
- Sommer R.; Rox K.; Wagner S.; Hauck D.; Henrikus S. S.; Newsad S.; Arnold T.; Ryckmans T.; Brönstrup M.; Imberty A.; Varrot A.; Hartmann R. W.; Titz A. Anti-biofilm agents against Pseudomonas aeruginosa: a structure-activity relationship study of C-glycosidic LecB inhibitors. J. Med. Chem. 2019, 62, 9201–9216. 10.1021/acs.jmedchem.9b01120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cioci G.; Mitchell E. P.; Gautier C.; Wimmerová M.; Sudakevitz D.; Pérez S.; Gilboa-Garber N.; Imberty A. Structural basis of calcium and galactose recognition by the lectin PA-IL of Pseudomonas aeruginosa. FEBS Lett. 2003, 555, 297–301. 10.1016/S0014-5793(03)01249-3. [DOI] [PubMed] [Google Scholar]
- Wagner S.; Hauck D.; Hoffmann M.; Sommer R.; Joachim I.; Müller R.; Imberty A.; Varrot A.; Titz A. Covalent lectin inhibition and application in bacterial biofilm imaging. Angew. Chem., Int. Ed. 2017, 56, 16559–16564. 10.1002/anie.201709368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanne J. H. FDA adds ‘black box’ warning label to fluoroquinolone antibiotics. BMJ 2008, 337, a816. 10.1136/bmj.a816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz R. S. Paul Ehrlich’s magic bullets. N. Engl. J. Med. 2004, 350, 1079–1080. 10.1056/NEJMp048021. [DOI] [PubMed] [Google Scholar]
- Barok M.; Joensuu H.; Isola J. Trastuzumab emtansine: mechanisms of action and drug resistance. Breast Cancer Res. 2014, 16, 209. 10.1186/bcr3621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehar S. M.; Pillow T.; Xu M.; Staben L.; Kajihara K. K.; Vandlen R.; DePalatis L.; Raab H.; Hazenbos W. L.; Morisaki J. H.; Kim J.; Park S.; Darwish M.; Lee B.-C.; Hernandez H.; Loyet K. M.; Lupardus P.; Fong R.; Yan D.; Chalouni C.; Luis E.; Khalfin Y.; Plise E.; Cheong J.; Lyssikatos J. P.; Strandh M.; Koefoed K.; Andersen P. S.; Flygare J. A.; Tan M. W.; Brown E. J.; Mariathasan S. Novel antibody-antibiotic conjugate eliminates intracellular S. aureus. Nature 2015, 527, 323–328. 10.1038/nature16057. [DOI] [PubMed] [Google Scholar]
- Zhang G.-F.; Liu X.; Zhang S.; Pan B.; Liu M.-L. Ciprofloxacin derivatives and their antibacterial activities. Eur. J. Med. Chem. 2018, 146, 599–612. 10.1016/j.ejmech.2018.01.078. [DOI] [PubMed] [Google Scholar]
- Klahn P.; Brönstrup M. Bifunctional antimicrobial conjugates and hybrid antimicrobials. Nat. Prod. Rep. 2017, 34, 832–885. 10.1039/C7NP00006E. [DOI] [PubMed] [Google Scholar]
- Milner S. J.; Carrick C. T.; Kerr K. G.; Snelling A. M.; Thomas G. H.; Duhme-Klair A.-K.; Routledge A. Probing bacterial uptake of glycosylated ciprofloxacin conjugates. ChemBioChem 2014, 15, 466–471. 10.1002/cbic.201300512. [DOI] [PubMed] [Google Scholar]
- Howse G. L.; Bovill R. A.; Stephens P. J.; Osborn H. M. I. Synthesis and antibacterial profiles of targeted triclosan derivatives. Eur. J. Med. Chem. 2019, 162, 51–58. 10.1016/j.ejmech.2018.10.053. [DOI] [PubMed] [Google Scholar]
- Rodrigue J.; Ganne G.; Blanchard B.; Saucier C.; Giguère D.; Shiao T. C.; Varrot A.; Imberty A.; Roy R. Aromatic thioglycoside inhibitors against the virulence factor LecA from Pseudomonas aeruginosa. Org. Biomol. Chem. 2013, 11, 6906–6918. 10.1039/c3ob41422a. [DOI] [PubMed] [Google Scholar]
- Kadam R. U.; Garg D.; Schwartz J.; Visini R.; Sattler M.; Stocker A.; Darbre T.; Reymond J.-L. CH-π ‘T-shape’ interaction with histidine explains binding of aromatic galactosides to Pseudomonas aeruginosa lectin LecA. ACS Chem. Biol. 2013, 8, 1925–1930. 10.1021/cb400303w. [DOI] [PubMed] [Google Scholar]
- Joachim I.; Rikker S.; Hauck D.; Ponader D.; Boden S.; Sommer R.; Hartmann L.; Titz A. Development and optimization of a competitive binding assay for the galactophilic low affinity lectin LecA from Pseudomonas aeruginosa. Org. Biomol. Chem. 2016, 14, 7933–7948. 10.1039/C6OB01313A. [DOI] [PubMed] [Google Scholar]
- Kadam R. U.; Bergmann M.; Hurley M.; Garg D.; Cacciarini M.; Swiderska M. A.; Nativi C.; Sattler M.; Smyth A. R.; Williams P.; Cámara M.; Stocker A.; Darbre T.; Reymond J.-L. A Glycopeptide dendrimer inhibitor of the galactose-specific lectin LecA and of Pseudomonas aeruginosa biofilms. Angew. Chem., Int. Ed. 2011, 50, 10631–10635. 10.1002/anie.201104342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu D. T.; Fernandes P. B. Structure-activity relationships of the fluoroquinolones. Antimicrob. Agents Chemother. 1989, 33, 131–135. 10.1128/AAC.33.2.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gootz T. D.; Brighty K. E. Fluoroquinolone antibacterials: SAR, mechanism of action, resistance, and clinical aspects. Med. Res. Rev. 1996, 16, 433–486. . [DOI] [PubMed] [Google Scholar]
- Idowu T.; Schweizer F. Ubiquitous nature of fluoroquinolones: the oscillation between antibacterial and anticancer activities. Antibiotics 2017, 6, 26. 10.3390/antibiotics6040026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham T. D. M.; Ziora Z. M.; Blaskovich M. A. T. Quinolone antibiotics. Med. Chem. Commun. 2019, 10, 1719–1739. 10.1039/C9MD00120D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koga H.; Itoh A.; Murayama S.; Suzue S.; Irikura T. Structure-activity relationships of antibacterial 6,7- and 7,8-disubstituted 1-alkyl-1,4-dihydro-4-oxoquinoline-3-carboxylic acids. J. Med. Chem. 1980, 23, 1358–1363. 10.1021/jm00186a014. [DOI] [PubMed] [Google Scholar]
- Mustaev A.; Malik M.; Zhao X.; Kurepina N.; Luan G.; Oppegard L. M.; Hiasa H.; Marks K. R.; Kerns R. J.; Berger J. M.; Drlica K. Fluoroquinolone-gyrase-DNA complexes: two modes of drug binding. J. Biol. Chem. 2014, 289, 12300–12312. 10.1074/jbc.M113.529164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casoni F.; Dupin L.; Vergoten G.; Meyer A.; Ligeour C.; Géhin T.; Vidal O.; Souteyrand E.; Vasseur J.-J.; Chevolot Y.; Morvan F. The influence of the aromatic aglycon of galactoclusters on the binding of LecA: a case study with O-phenyl, S-phenyl, O-benzyl, S-benzyl, O-biphenyl and O-naphthyl aglycons. Org. Biomol. Chem. 2014, 12, 9166–9179. 10.1039/C4OB01599A. [DOI] [PubMed] [Google Scholar]
- Richter M. F.; Drown B. S.; Riley A. P.; Garcia A.; Shirai T.; Svec R. L.; Hergenrother P. J. Predictive compound accumulation rules yield a broad-spectrum antibiotic. Nature 2017, 545, 299–304. 10.1038/nature22308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Shea R.; Moser H. E. Physicochemical properties of antibacterial compounds: implications for drug discovery. J. Med. Chem. 2008, 51, 2871–2878. 10.1021/jm700967e. [DOI] [PubMed] [Google Scholar]
- Müsken M.; Klimmek K.; Sauer-Heilborn A.; Donnert M.; Sedlacek L.; Suerbaum S.; Häussler S. Towards individualized diagnostics of biofilm-associated infections: a case study. NPJ Biofilms Microbiomes 2017, 3, 22. 10.1038/s41522-017-0030-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziȩba A.; Maślankiewicz A.; Sitkowski J. 1H, 13C and 15N NMR spectra of ciprofloxacin. Magn. Reson. Chem. 2004, 42, 903–904. 10.1002/mrc.1468. [DOI] [PubMed] [Google Scholar]
- Driguez H.; Szeja W. Facile synthesis of 1,2-trans-nitrophenyl-1-thioglycopyranosides. Synthesis 1994, 1994, 1413–1414. 10.1055/s-1994-25704. [DOI] [Google Scholar]
- McPherson J. C. III; Runner R.; Buxton T. B.; Hartmann J. F.; Farcasiu D.; Bereczki I.; Rőth E.; Tollas S.; Ostorházi E.; Rozgonyi F.; Herczegh P. Synthesis of osteotropic hydroxybisphosphonate derivatives of fluoroquinolone antibacterials. Eur. J. Med. Chem. 2012, 47, 615–618. 10.1016/j.ejmech.2011.10.049. [DOI] [PubMed] [Google Scholar]
- Phiasivongsa P.; Samoshin V. V.; Gross P. H. Henry condensations with 4,6-O-benzylidenylated and non-protected d-glucose and L-fucose via DBU-catalysis. Tetrahedron Lett. 2003, 44, 5495–5498. 10.1016/S0040-4039(03)01328-5. [DOI] [Google Scholar]
- Wiegand I.; Hilpert K.; Hancock R. E. W. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat. Protoc. 2008, 3, 163–175. 10.1038/nprot.2007.521. [DOI] [PubMed] [Google Scholar]
- Haupenthal J.; Baehr C.; Zeuzem S.; Piiper A. RNAse A-like enzymes in serum inhibit the anti-neoplastic activity of siRNA targeting polo-like kinase 1. Int. J. Cancer 2007, 121, 206–210. 10.1002/ijc.22665. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.