Significance
This research identified a microstructural neural network biomarker for objective and accurate diagnosis of isolated dystonia based on the disorder pathophysiology using an advanced deep learning algorithm, DystoniaNet, and raw structural brain images of large cohorts of patients with isolated focal dystonia and healthy controls. DystoniaNet significantly outperformed shallow machine-learning pipelines and substantially exceeded the current agreement rates between clinicians, reaching an overall accuracy of 98.8% in diagnosing different forms of isolated focal dystonia. These results suggest that DystoniaNet could serve as an objective, robust, and generalizable algorithmic platform of dystonia diagnosis for enhanced clinical decision-making. Implementation of the identified biomarker for objective and accurate diagnosis of dystonia may be transformative for clinical management of this disorder.
Keywords: dystonia, biomarker, machine learning, brain MRI
Abstract
Isolated dystonia is a neurological disorder of heterogeneous pathophysiology, which causes involuntary muscle contractions leading to abnormal movements and postures. Its diagnosis is remarkably challenging due to the absence of a biomarker or gold standard diagnostic test. This leads to a low agreement between clinicians, with up to 50% of cases being misdiagnosed and diagnostic delays extending up to 10.1 y. We developed a deep learning algorithmic platform, DystoniaNet, to automatically identify and validate a microstructural neural network biomarker for dystonia diagnosis from raw structural brain MRIs of 612 subjects, including 392 patients with three different forms of isolated focal dystonia and 220 healthy controls. DystoniaNet identified clusters in corpus callosum, anterior and posterior thalamic radiations, inferior fronto-occipital fasciculus, and inferior temporal and superior orbital gyri as the biomarker components. These regions are known to contribute to abnormal interhemispheric information transfer, heteromodal sensorimotor processing, and executive control of motor commands in dystonia pathophysiology. The DystoniaNet-based biomarker showed an overall accuracy of 98.8% in diagnosing dystonia, with a referral of 3.5% of cases due to diagnostic uncertainty. The diagnostic decision by DystoniaNet was computed in 0.36 s per subject. DystoniaNet significantly outperformed shallow machine-learning algorithms in benchmark comparisons, showing nearly a 20% increase in its diagnostic performance. Importantly, the microstructural neural network biomarker and its DystoniaNet platform showed substantial improvement over the current 34% agreement on dystonia diagnosis between clinicians. The translational potential of this biomarker is in its highly accurate, interpretable, and generalizable performance for enhanced clinical decision-making.
Isolated dystonia is a rare neurological disorder of unknown pathophysiology, which causes involuntary muscle contractions leading to abnormal, typically patterned, twisting movements and postures. Symptoms of dystonia have a pervasive negative impact on patient's daily activities and quality of life, often leading to continuous stress, psychiatric comorbidities, social embarrassment, and occupational disability (1).
As the third most common movement disorder after essential tremor and Parkinson’s disease, isolated dystonia is estimated to affect up to 35.1 per 100,000 general population (2). Its exact incidence, however, is unknown because up to 50% of dystonia cases go misdiagnosed or underdiagnosed at their first encounter (3) and the average diagnostic delay extends up to 10.1 y, depending on the form of dystonia (4–10).
This significant diagnostic challenge is primarily associated with the absence of a biomarker for isolated dystonia, a defined and objective characteristic that is measured as an indicator of the common pathophysiological process for its accurate diagnosis (11). In the absence of a biomarker, there is no gold standard diagnostic test for dystonia, and current diagnostic recommendations remain formulated purely on clinical syndrome characteristics (12–15). Diagnosis is typically impacted by a number of factors, including phenotypical variability of the disorder, the circumstances of evaluation, the experience and expertise of the clinician, the psychological status of the patient, and nonneurological conditions that mimic dystonic symptoms (15, 16). Moreover, the predicted impact of current diagnostic recommendations in terms of their reliability, sensitivity, and specificity has not been established, while the validity of clinical diagnosis of dystonia without a biomarker could not be assessed. As a stark illustration, several studies repeatedly failed on the diagnostic consensus of isolated focal dystonia between clinicians, including laryngeal dystonia, cervical dystonia, blepharospasm, oromandibular dystonia, and writer’s cramp, with nil to a weak agreement at Cohen’s κ = 0.05 to 0.52 between the participating providers (14, 16, 17). Thus, the current diagnostic approach is open to bias, and a diagnostic consensus between clinicians is hard to achieve. Besides the disorder’s negative impact on the patient’s quality of life, the current suboptimal clinical care of dystonia in terms of its accurate and efficient diagnosis amounts to the continuous growth of healthcare costs from repeated office visits, redundant assessments, and professional disability (4–10). Importantly, delayed diagnosis results in deferred treatment.
Much of the challenges associated with the development of a diagnostic biomarker for isolated dystonia have been historically related to its unclear pathophysiology. Recent advances in brain imaging methodologies and analytical techniques have allowed the identification of widespread alterations of brain structure and function in this disorder, involving not only the basal ganglia and cerebellum but also higher-order motor and associative cortical regions (18). Different forms of dystonia have been characterized by similarities in large-scale disorganization of gray and white matter architecture, including abnormal distribution of influential regions of information transfer (hubs) in prefrontal, parietal, occipital cortices and thalamus and reduced connectivity of the sensorimotor and frontoparietal regions (19–23). This knowledge critically shifted our understanding of dystonia pathophysiology from a basal ganglia disorder to a large-scale neural network disorder and thrust the field toward probing these alterations as potential candidate markers for its diagnosis and treatment (24–28).
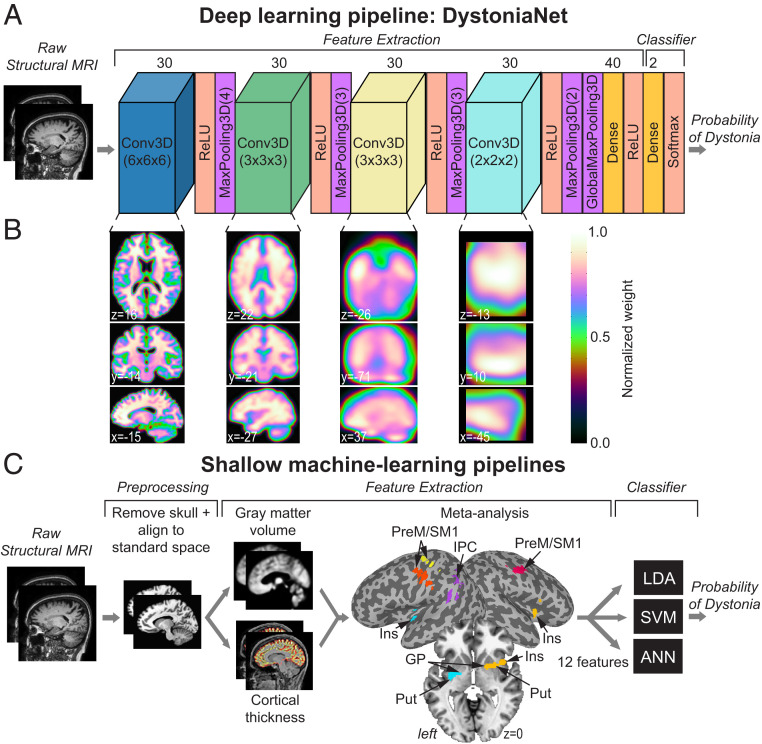
Building on the current knowledge of dystonia pathophysiology, we considered its whole-brain large-scale abnormalities for the development of a robust microstructural neural network biomarker of significant diagnostic potential. To reduce bias in the selection of neural network alterations as components of a candidate biomarker for dystonia diagnosis, we developed a deep learning platform based on the architecture of a 3D convolutional neural network, DystoniaNet (Fig. 1 A and B), which used a fully data-driven approach to automatically discover informative diagnostic features from raw structural brain images in a large dataset of 612 subjects. We assessed the performance and diagnostic potential of DystoniaNet in benchmark comparisons with three different shallow machine-learning pipelines, which used an alternative meta-analytical biomarker as an informative feature (Fig. 1C). We hypothesized that large-scale microstructural neural network alterations may be implemented, together with their algorithmic platform, DystoniaNet, as a biomarker for objective, accurate, fast, and cost-efficient diagnosis of isolated focal dystonia. We postulated that this diagnostic biomarker can be measured with sufficient precision and reliability across different forms of isolated dystonia to enhance its translation potential.
Fig. 1.
Deep learning and shallow machine-learning pipelines for diagnosis of isolated dystonia. (A) Raw structural brain MRIs were used with the deep learning pipeline (DystoniaNet). The architecture of DystoniaNet included four convolutional layers (Conv3D) for feature extraction from raw structural MR images, each followed by the rectified linear unit (ReLU) activation and maximum pooling (MaxPooling3D) layers. The kernel sizes of each Conv3D layer were 6 × 6 × 6, 3 × 3 × 3, 3 × 3 × 3, and 2 × 2 × 2 voxels, respectively. The kernel sizes of each MaxPooling3D layer were 4 × 4 × 4, 3 × 3 × 3, 3 × 3 × 3, and 2 × 2 × 2, respectively (given in the brackets). The global maximum pooling layer (GlobalMaxPooling3D) followed the fourth MaxPooling3D layer and was followed by a fully connected Dense layer of 40 filters. The classifier included dense layer of two filters with Softmax activation, with the probability of dystonia as the output. (B) Axial brain slices depict 2D visualization of the average 3D feature maps extracted from the corresponding Conv3D layers of DystoniaNet. The color bar shows the normalized weight of discriminative voxels learned by DystoniaNet based on the training set of 160 patients with laryngeal dystonia and 160 healthy controls. Coordinates are given in the AFNI standard Talairach–Tournoux space. (C) Shallow machine-learning pipelines show the steps for preprocessing of raw structural MRI (skull removal and alignment to the AFNI standard space), extraction of 12 features from gray matter volume and cortical thickness based on meta-analysis of neuroimaging literature in laryngeal dystonia, and their input into three machine-learning classifiers: linear discriminant analysis (LDA), support vector machine (SVM), and artificial neural network (ANN), with the probability of dystonia as the output. PreM, premotor cortex; SM1, primary sensorimotor cortex; IPC, inferior parietal cortex; Ins, insula; Put, putamen; GP, globus pallidus.
Results
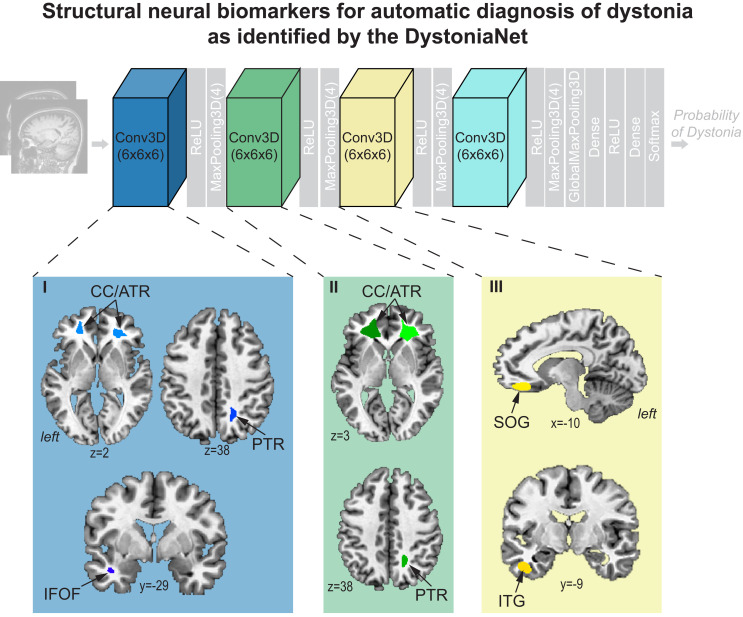
Based on a fully automated data-driven approach and without a priori knowledge, DystoniaNet identified a microstructural neural network biomarker for dystonia diagnosis using a training set of 160 patients with laryngeal dystonia and 160 healthy controls (Table 1). The multilayer components of this biomarker included the right posterior thalamic radiation of corona radiata and left inferior fronto-occipital fasciculus extending to uncinate fasciculus in the first layer of DystoniaNet, the bilateral corpus callosum extending to the anterior thalamic radiation of corona radiata in the first and second layers, and the left superior orbital and inferior temporal gyri in the third layer (Fig. 2 and Table 2). All of these identified brain regions have been previously reported to exhibit structural or functional alterations in different forms of dystonia (recent review in ref. 18). The pathophysiological relevance of this biomarker is reflected in its high generalizability for accurate diagnosis of dystonia across the phenotypical spectrum of this disorder, as described below.
Table 1.
Demographics of participants
| Participant groups | |||
| Training set | |||
| Number of participants | 160 healthy controls | 160 laryngeal dystonia | |
| Sex, F/M | 102/58 | 102/58 | |
| Mean age ± SD, y | 48.4 ± 11.6 | 49.5 ± 10.9 | |
| Mean dystonia duration ± SD, y | n/a | 13.3 ± 10.2 | |
| Scanner strength (3.0 T) | 160 | 160 | |
| First independent test set | |||
| Number of participants | 60 healthy controls | 60 laryngeal dystonia | |
| Sex, F/M | 42/18 | 57/3 | |
| Mean age ± SD, y | 62.3 ± 13.1 | 62.6 ± 4.8 | |
| Mean dystonia duration ± SD, y | n/a | 16.4 ± 12.6 | |
| Scanner strength (3.0 T) | 60 | 60 | |
| Second independent test set | |||
| Number of participants | 59 laryngeal dystonia | 54 blepharospasm | 59 cervical dystonia |
| Sex, F/M | 54/5 | 41/13 | 53/6 |
| Mean age ± SD, y | 67.4 ± 8.7 | 56.7 ± 15.6 | 52.1 ± 16.3 |
| Mean dystonia duration ± SD, y | 15.2 ± 8.6 | 7.1 ± 4.2 | 11.3 ± 7.8 |
| Scanner strength (3.0 T/1.5 T) | 59/0 | 19/35 | 27/32 |
n/a, not applicable.
Fig. 2.
Microstructural neural network biomarker for automatic diagnosis of isolated dystonia as identified by the DystoniaNet platform. Brain regions as components of the biomarker are identified by the first three convolutional layers of DystoniaNet for diagnostic classification. Brain regions in the fourth layer are not visualized due to low spatial resolution. Axial and sagittal brain slices depict 2D visualizations of the most discriminative features in the AFNI standard Talairach–Tournoux space. ReLU, rectified linear unit; CC/ATR, corpus callosum/anterior thalamic radiation of corona radiata; PTR, posterior thalamic radiation of corona radiata; IFOF, inferior fronto-occipital fasciculus; SOG, superior orbital gyrus; ITG, inferior temporal gyrus.
Table 2.
Informative features of deep and shallow machine learning pipelines
| Brain region | Center of cluster mass x, y, z | Cluster size voxels | ||
| Components of the microstructural neural network biomarker identified by DystoniaNet | ||||
| Layer 1 | ||||
| L corpus callosum extending to anterior thalamic radiation | −19, 37, 10 | 5091 | ||
| R corpus callosum extending to anterior thalamic radiation | 25, 34, 6 | 4214 | ||
| R posterior thalamic radiation | 17, −54, 41 | 545 | ||
| L inferior fronto-occipital fasciculus extending to uncinate fasciculus | −38, −5, −18 | 262 | ||
| Layer 2 | ||||
| L corpus callosum extending to anterior thalamic radiation | −18, 32, 10 | 7000 | ||
| R corpus callosum extending to anterior thalamic radiation | 24, 31, 5 | 5837 | ||
| R posterior thalamic radiation underlying superior parietal lobule | 18, −52, 36 | 689 | ||
| Layer 3 | ||||
| L superior orbital gyrus | −10, 35, −10 | 9669 | ||
| L inferior temporal gyrus | −41, −10, −26 | 1474 | ||
| Components of the meta-analytical biomarker identified from dystonia neuroimaging literature | ||||
| L insula/putamen/globus pallidus | −29, −8, −1 | 322 | ||
| R insula/putamen/globus pallidus | 34, 7, 1 | 245 | ||
| L inferior parietal cortex (area PF) | −50, −42, 26 | 301 | ||
| L premotor/primary sensorimotor cortex (areas 6, 4, 3, 1) | −48, −13, 34 / -37, -20, 51 | 266/221 | ||
| R premotor/primary sensorimotor cortex (areas 6, 4, 3, 1) | 48, −12, 34 | 273 | ||
L, left; R, right.
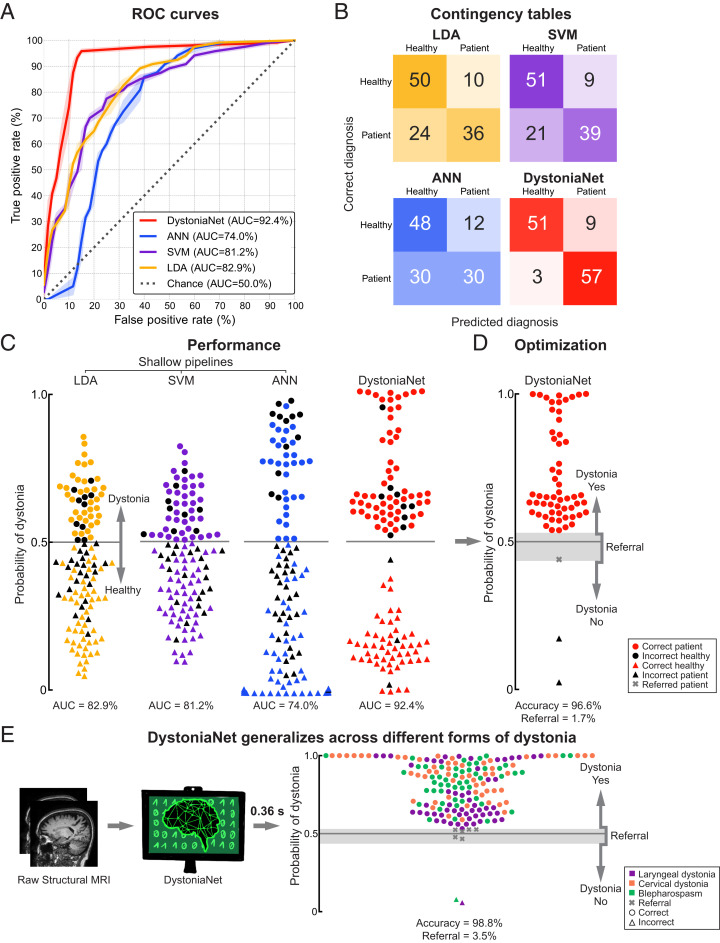
As shown in Fig. 3, using the first independent test set of 60 patients with laryngeal dystonia and 60 healthy controls, the DystoniaNet-based biomarker achieved an out-of-sample area under the curve (AUC) of 92.4% in discriminating patients from controls, with 95.0% sensitivity and 85.0% specificity (Fig. 3 A–C). The positive predictive value (PPV) was 86.4%, and the negative predictive value (NPV) was 94.4%. Following the optimization of DystoniaNet with the introduction of the dynamic range to manage its diagnostic uncertainty, its accuracy of dystonia diagnosis was 96.6%, with the referral of one patient (1.7%) for further examination (Fig. 3D). The average computational time of DystoniaNet for delivering the diagnostic decision was 0.36 s per subject.
Fig. 3.
Performance of deep learning and shallow machine-learning pipelines. (A) Receiver operating characteristic (ROC) curves for each pipeline using the first independent test set of 60 patients with laryngeal dystonia and 60 healthy controls. The area under the ROC curve (AUC) values for each pipeline are reported in the key. The dotted line represents the performance of a random classifier. (B) The corresponding contingency tables report the number of healthy controls and patients who are correctly and incorrectly classified by each pipeline. (C) The diagnostic performance of each pipeline in the first independent test set of 60 patients with laryngeal dystonia and 60 healthy controls. Each symbol represents a subject. Subjects classified as patients are represented by circles; subjects classified as healthy controls are represented by triangles. Colored symbols represent correct diagnosis; black symbols represent misclassifications. The y axis represents the probability of dystonia as assessed by each pipeline; the gray line represents the decision boundary. The corresponding AUC values are given for each pipeline. (D) Optimized DystoniaNet with a dynamic range to maximize diagnostic performance in 60 laryngeal dystonia patients of the first independent test set. The gray shading represents the area of uncertainty where DystoniaNet refers the subject (gray cross) for further examination. The y axis represents the probability of dystonia; the gray line represents the decision boundary. The corresponding accuracy and referral rate are reported. (E) Testing of generalizability of the DystoniaNet-derived biomarker in the second independent test set of 172 patients with different forms of dystonia, including 59 patients with laryngeal dystonia, 59 patients with cervical dystonia, and 54 patients with blepharospasm. The pipeline shows the steps from the use of raw structural MRI as input to the final optimized DystoniaNet, which processes data and outputs the final diagnostic decision as dystonia-yes, dystonia-no, or referral within 0.36 s for each subject. Each symbol represents a subject. Subjects classified as patients are represented by circles; misclassified subjects are represented by triangles; referrals are represented by crosses. The y axis represents the probability of dystonia; the gray line represents the decision boundary; the gray shading shows the area of diagnostic uncertainty (referral). The corresponding accuracy and referral rate are reported. Data are visualized using the Matplotlib library (63).
The performance of the microstructural neural network biomarker using the DystoniaNet pipeline was compared to the performance of the alternative meta-analytical biomarker using three different shallow machine-learning algorithms in the same training and test sets. The components of the meta-analytical biomarker included the bilateral insula extending to the putamen and globus pallidus, premotor and primary sensorimotor cortices, and left inferior parietal cortex (Fig. 1C and Table 2). The AUCs of the shallow pipelines were 82.9% for linear discriminant analysis (LDA), 81.2% for support vector machine (SVM), and 74.0% for one-layer artificial neural network (ANN; Fig. 3 A and C). McNemar’s tests showed that the diagnostic performance of the meta-analytical biomarker was not significantly different between the three shallow pipelines based on their AUCs (LDA vs. SVM P = 0.48; LDA vs. ANN P = 0.23; SVM vs. ANN P = 0.07). The sensitivity and specificity of the shallow pipelines were 60.0% and 83.3% for LDA, 65.0% and 85.0% for SVM, and 50.0% and 80.0% for ANN, respectively (Fig. 3B). The PPV and NPV were 78.3% and 67.6% for LDA, 81.2% and 70.8% for SVM, and 71.4% and 61.5% for ANN, respectively. While the meta-analytical biomarker achieved the above-the-chance AUCs in discriminating between patients and healthy controls, its specificity, sensitivity, PPVs, and NPVs were significantly lower compared to the DystoniaNet-based microstructural neural network biomarker (McNemar’s tests: DystoniaNet vs. LDA P = 6.8 × 10−4; DystoniaNet vs. SVM P = 2.1 × 10−3; DystoniaNet vs. ANN P = 2.8 × 10−6; Fig. 3 A–C).
The diagnostic generalizability and broader translational potential of the best-performing microstructural neural network biomarker and its DystoniaNet platform were further examined using the second independent test set of 172 patients with three different forms of isolated focal dystonia. DystoniaNet achieved overall accuracy of 98.8% with a stable diagnostic power across all cases (Fig. 3E). Specifically, it showed 98.2% accuracy in diagnosing laryngeal dystonia, 100% in diagnosing cervical dystonia, and 98.1% in diagnosing blepharospasm, while referring six patients (3.5%) for further examination.
The performance of the microstructural neural network biomarker and its DystoniaNet platform remained stable independent of the magnetic field strength (accuracy range: 98.0 to 100%), MRI scanner vendor (accuracy range: 96.9 to 100%), head coil (accuracy range: 95.2 to 100%), T1-weighted image acquisition sequence (accuracy range: 98.3 to 100%), or data collection site (accuracy range: 97.6 to 100%; SI Appendix, Fig. S1). Its high specificity was confirmed using a supplementary third independent dataset of 1,480 healthy controls (accuracy, 96.9%; referral rate, 2.6%; SI Appendix, Fig. S2E). Thus, our data show that the performance characteristics of the microstructural neural network biomarker and its advanced DystoniaNet platform are acceptable in terms of sensitivity, accuracy, specificity, and precision for diagnosis of isolated dystonia.
Discussion
At present, there is no biomarker or gold standard diagnostic test for dystonia, and there are no current technologies or approaches that address this problem (4–10). This presents a significant clinical challenge with a negative impact on patient's quality of life and healthcare costs. We demonstrate that our automatically defined microstructural neural network biomarker, together with its algorithmic platform, DystoniaNet, provides objective, accurate, fast, and cost-efficient diagnosis of isolated focal dystonia. Its translational value is in the streamlined, robust, explainable, and generalizable performance for delivering the diagnosis of dystonia compared to the current standard of care, which is based purely on the variable symptomatology of this disorder (14, 16). In particular, the overall 98.8% diagnostic accuracy of the microstructural neural network biomarker identified by DystoniaNet substantially exceeds the recently reported 34% diagnostic agreement rate between clinicians with different expertise based on extensive diagnostic workup and syndromic approach (14). The accuracy of the DystoniaNet-based biomarker is further enhanced by the built-in safeguards that are able to successfully identify uncertain cases and refer them for further evaluation, rather than provide a decision that may be prone to diagnostic errors. Furthermore, DystoniaNet shows an unprecedented speed in accurately diagnosing dystonia from individual raw structural brain MRI compared to the existing diagnostic delays of up to 10.1 y that require, on average, 3.95 office visits (4–10).
The performance and generalizability of DystoniaNet in delivering a highly accurate diagnosis across different forms of dystonia without any additional requirements for dystonia form-specific model retraining is based on its capability to identify the common features of disorder pathophysiology (18). Specifically, the biomarker components that were automatically learned by DystoniaNet as most informative for discriminating the disorder have been previously demonstrated to be aberrant in patients with dystonia. While gray matter changes appear to be more relevant to dystonia form-specific aberrations (29–32), white matter alterations across different forms of dystonia emerge as a more common feature of this disorder. Structural abnormalities in the corpus callosum were previously reported within the dystonia spectrum, including laryngeal dystonia, cervical dystonia, and blepharospasm, as well as focal hand dystonia, musician’s dystonia, X-linked dystonia-parkinsonism, and poststroke lingual dystonia (e.g., refs. 19, 20, 29, and 33–45). These alterations were related to the broader spread of cortical changes in dystonia and attributed to aberrant interhemispheric information transfer. Similarly, several studies reported abnormalities in the corona radiata, specifically involving its anterior and posterior thalamic radiations, pointing to altered structure and function of the thalamus within the aberrant basal ganglia-thalamo-cortical circuitry and its projecting targets in prefrontal and parietal cortical regions. The latter has also been shown to have abnormal functional activity and connectivity in patients with various forms of dystonia (19, 20, 39–42, 46). Other regions that were identified by DystoniaNet as informative features included inferior temporal and superior orbital gyri as well as the white matter pathways and the inferior fronto-occipital and uncinate fasciculi, connecting these cortical regions with the frontal, temporal, and occipital lobes (47). Alterations in these regions are thought to contribute to abnormal heteromodal sensorimotor processing and executive control of goal-oriented motor behaviors in patients with isolated dystonia (32, 33, 43–45). Taken together, pathophysiologically relevant alterations of white matter commissural, association, and projection fibers, as automatically detected by DystoniaNet, capture and reflect wider spread abnormalities in their target gray matter regions and likely represent a unifying neural biomarker of isolated dystonia.
Methodologically, the biomarker identification was possible due to the development, optimization, and validation of its advanced algorithmic platform, DystoniaNet. As the robustness of performance of deep learning algorithms depends on the availability of big data, it is not trivial to assemble such datasets for rare disorders like dystonia. The development of DystoniaNet was possible because of the continuous efforts to understand the neural pathophysiology of this disorder and the timely availability of the large (n = 392) research MRI dataset of carefully phenotyped patients with isolated focal dystonia for training and subsequent validations of this platform. In this regard, the DystoniaNet platform is unique because its highly accurate diagnostic decisions are built on the algorithm’s automatic selection of the pathophysiologically relevant biomarker from raw structural brain images, which are routinely acquired in the clinical setting and require no additional image processing prior to their input to this algorithm. This attribute is critical for the clinical applicability of DystoniaNet as a diagnostic test for informing the physician’s decision-making. Finally, the visualization of the identified microstructural neural network biomarker and the interpretability of DystoniaNet’s internal model for automated diagnostic decision-making offers a transparent and fair machine-learning platform and makes its translation to healthcare more compliant (48, 49).
As a comparison, shallow machine-learning algorithms require initial MR image preprocessing and a priori feature extraction and selection, all of which are laborious and computationally expensive procedures (24, 25). Importantly, the internal architecture of shallow ML models suffers from low complexity and sensitivity and limited classification accuracy, making these models unfeasible as clinically applicable automatic tools of dystonia diagnosis when compared to DystoniaNet (detailed in SI Appendix).
In conclusion, we present a microstructural neural network biomarker, which was discovered by a fully automated deep learning DystoniaNet algorithm from raw structural MR images and showed high diagnostic accuracy across different forms of isolated dystonia, independent of MRI hardware, acquisition protocol, or data collection site. It by far exceeded the currently existing clinical diagnostic workflow, both in terms of accuracy and time to diagnosis. Implementation of such an objective biomarker for dystonia diagnosis would be transformative for the clinical management of this disorder to reduce the rate of mis- and underdiagnoses and the overall cost associated with the wrong or delayed diagnosis, while, in parallel, accelerating the timely delivery of treatment. Future studies are warranted for the diagnostic assessment of DystoniaNet and its biomarker in an even broader phenotypical spectrum of dystonia, as well as across its genetic forms. Equally important, future research should focus on the extensions of DystoniaNet for comprehensive differential diagnosis of dystonia from other movement disorders and nonneurological conditions mimicking dystonia.
Materials and Methods
Study Participants.
A total of 612 subjects participated in the study, including 392 patients with isolated dystonia and 220 healthy controls (Table 1). Diagnosis of isolated focal dystonia (laryngeal dystonia, cervical dystonia, or blepharospasm) and the absence of other neurological disorders, including tics, dyskinesia, and other hyperkinetic movement disorders, or psychiatric and laryngeal problems was confirmed based on a detailed case history, physical examination, and neurological and laryngeal evaluations, as appropriate (demographics provided in Table 1 and SI Appendix). Tremor is known to co-occur in up to 70% of patients with dystonia (50–52), with dystonic tremor being considered as part of dystonia phenomenology (53). In line with this, 24.7% of patients in our cohort had concurrent dystonic tremor and 7.7% of patients had essential tremor. Because the overall co-occurrence of tremor was lower than generally reported (50–52), it was unlikely to significantly influence the identification of the dystonia-specific biomarker. Control subjects were healthy individuals without any past or present history of neurological, psychiatric, or laryngeal problems. The study was approved by the institutional review boards of the Icahn School of Medicine at Mount Sinai and Mass General Brigham Research Program. All subjects gave written informed consent before study participation.
Data Acquisition and Definition of Subject Cohorts.
Whole-brain T1-weighted MRI data were acquired on 3.0-T (n = 545) or 1.5-T (n = 67) scanners (detailed in SI Appendix, Tables S1 and S2). MRIs were used for the biomarker development and testing of the deep classification pipeline (DystoniaNet) and the three shallow machine-learning pipelines, including LDA, SVM, and ANN.
It is critical to train machine-learning algorithms on a well-characterized, balanced, homogeneous, large dataset in order to achieve their robust performance. This is especially important for rare diseases, such as dystonia, that lack an established biomarker. Our cohort of 160 patients with laryngeal dystonia and 160 age- and sex-matched healthy controls fit these characteristics best and was, therefore, chosen as a training set for the biomarker development and model training (Table 1). The subsequent biomarker and model testing and optimization were performed in the first independent test set of 60 patients with laryngeal dystonia (i.e., the same diagnosis as in the training set) and 60 healthy controls. Finally, validation of the biomarker performance was conducted in the second independent test set of 172 patients with three different forms of dystonia, including laryngeal dystonia, cervical dystonia, or blepharospasm, which allowed assessment of the generalizability of DystoniaNet across the dystonia phenotypic spectrum, various scanning hardware, MRI acquisition sequences, and data collection sites.
The age and sex of subjects in the training set were tightly matched between the patient and control groups (all P ≥ 0.37 based on D’Agostino–Pearson normality of data distribution and two-sample independent t tests), which prevented the machine-learning algorithms from learning the differences in these biological variables as false predictors of diagnosis. The remaining subjects were randomly assigned to the first and second independent test sets. As a result of this randomization, testing for the balance of age and sex in the first and second independent test sets was not performed (54). Nonetheless, because the internal parameters of all machine-learning algorithms were fixed during their development on the age- and sex-balanced training set, possible differences in sex or age distribution in the first and second independent test sets could not have an impact on the diagnostic performance of machine-learning algorithms. Moreover, randomization of subjects in the first and second independent test sets presented an opportunity to evaluate the performance and generalizability of the machine-learning algorithms in clinically realistic patient cohorts, as clinicians encountering dystonia patients have control over neither their demographics nor the clinical MRI acquisition parameters.
DystoniaNet Model Development.
DystoniaNet was developed to use raw structural brain MRI without any image preprocessing applied. As shown in Fig. 1A, the DystoniaNet deep classification pipeline included the Feature Extraction and Classifier components. For Feature Extraction, we first developed a shallow neural network architecture with one convolutional layer (Conv3D) and one dense layer (Dense) of 10 filters each and then increased the number of filters by 10 at each iteration, up to 100. Next, we added an additional convolutional layer and repeated the process. We trained our model after the addition of each new convolutional layer to assess its performance using 10% of training data that were set aside at each iteration. In this manner, the optimal architecture with four convolutional layers of 30 filters each and one dense layer of 40 filters was established. The kernel sizes of each convolutional layer were 6 × 6 × 6, 3 × 3 × 3, 3 × 3 × 3, and 2 × 2 × 2 voxels, respectively, to allow extraction of neural features at different resolutions in each of the four layers. The rectified linear unit (ReLU) activation and maximum pooling (MaxPooling3D) layers followed each convolutional layer to reduce the number of parameters in the model to the most relevant features. Specifically, the ReLU activation layer transformed the summed weighted input passed through the convolutional layer into the activation of a certain neuron of the network, while the maximum pooling operation calculated the maximum value in each patch of each feature map. The global maximum pooling layer (GlobalMaxPooling3D) followed the fourth maximum pooling layer and was introduced to allow the algorithm to classify input images of different sizes. The Classifier included the final dense layer of two filters with Softmax activation (i.e., normalized exponential function), which acted as a classifier to convert the output of this last layer into the probability of dystonia.
To visualize the biomarker components that were automatically identified by DystoniaNet, we computed four 3D average feature maps, representing the different levels of detail of each convolutional layer across 30 filters (Fig. 1B). The average 3D feature maps of the second, third, and fourth convolutional layers were zoomed using third-order spline interpolation to match the size of the standard Talairach-Tournoux space (SI Appendix). The significant clusters in each average feature map of the first, second, and third convolutional layers were localized by filtering out voxels with weights below 95% of the absolute maximum weight (Table 2). Due to the low spatial resolution of the fourth layer (Fig. 1B), significant clusters were not possible to visualize in this layer.
Benchmark Shallow Machine-Learning Pipelines.
The performance of DystoniaNet was compared to the benchmark performance of three different shallow machine-learning pipelines, including LDA, SVM, and one-layer ANN (Fig. 1C). LDA and SVM were used because both have previously shown a promising performance with up to 90.6% classification accuracy of laryngeal dystonia and cervical dystonia, respectively, based on resting-state functional MRI data (24, 25). One-layer ANN was used for its comparability with deep learning algorithms (details of shallow learning models provided in SI Appendix, SI Methods).
Due to their lower complexity and inability to handle both feature extraction/selection and classification compared to DystoniaNet, all three shallow machine-learning algorithms required predetermined input features. To determine a robust set of brain alterations as input features while controlling for single-study cohort- and methodology-related biases in feature extraction and selection, we conducted meta-analysis of the existing neuroimaging literature using the activation likelihood estimation (ALE). Because our training and first independent test sets included patients with laryngeal dystonia, meta-analysis was also performed on available imaging studies in laryngeal dystonia, as identified by a PubMed search in accordance with the Preferred Reporting Items for Systematic reviews and Meta-Analyses (PRISMA) guidelines (SI Appendix). This procedure identified a total of 221 clusters of structural (29, 32, 38, 55–58) and functional (5, 24, 46, 55–57, 59–62) abnormalities in a cumulative total of 1,084 patients and healthy controls across 18 studies (SI Appendix, Table S3). ALE meta-analysis found six significant clusters of common brain abnormalities across published studies (family-wise error [FWE] corrected P ≤ 0.05). The binary mask of these clusters was used to extract the average gray matter volume and cortical thickness from T1-weighted MR images in each subject, which were then used as predetermined input features for each LDA, SVM, and ANN algorithm (detailed in SI Appendix).
Deep Learning and Shallow Machine-Learning Training and Performance Evaluations.
Although the employed shallow and deep learning pipelines differed in their input features (i.e., meta-analytically defined for LDA, SVM, and ANN vs. fully data-driven for DystoniaNet) due to different requirements of their internal models, all pipelines were trained on the same training set and tested on the same first independent test set, which allowed direct comparisons of their performance. Specifically, the performance of each machine-learning pipeline was examined by computing the AUC, sensitivity, and specificity using the first independent test set of 60 patients with laryngeal dystonia and 60 healthy controls (Table 1). Statistical differences in performance between the four pipelines were examined using McNemar’s test at two-sided Bonferroni-corrected P ≤ 0.008 to adjust for multiple comparisons.
To optimize the performance and minimize the diagnostic errors of the best-performing DystoniaNet algorithm, a dynamic range was introduced using two optimal thresholds, ty and tn, which converted the output into three diagnostic decisions: dystonia-yes, dystonia-no, and referral. For a subject to be classified as having dystonia, the output of DystoniaNet had to exceed ty; for a subject to be classified as healthy, the output had to be lower than tn. If the output probability fell between tn and ty, the subject was classified as a referral for further evaluation. The referral rate was set to less than 10% of tested subjects (in this case, ty = 0.53 and tn = 0.44) to balance the compromise between the cost of misdiagnosis and the cost of additional evaluations while avoiding the deflation of true negatives. Optimized DystoniaNet with the dynamic range to manage its diagnostic uncertainty was retested in 60 patients with laryngeal dystonia of the first independent test set.
The final performance and diagnostic generalizability of the identified biomarker and its optimized DystoniaNet algorithmic platform were examined in the second independent test set of 172 patients with different forms of isolated focal dystonia, including 59 patients with laryngeal dystonia, 59 patients with cervical dystonia, and 54 patients with blepharospasm (Table 1). In addition, the second independent test set was used to evaluate the impact of the MRI scanner hardware, acquisition sequence, and data collection site on the diagnostic performance of optimized DystoniaNet (detailed in SI Appendix).
Supplementary Material
Acknowledgments
We thank Alexis Worthley, BA, Lena O’Flynn, BA, Sai Mupparaju, BA, and Azadeh Hamzehei Sichani, MA, for assistance with subject recruitment, data collection, and literature search; Demba Ba, PhD, for providing feedback on the first draft of the manuscript; Mert R. Sabuncu, PhD, for early suggestions on the development of the deep learning architecture; and Steven J. Frucht, MD, and Andrew Blitzer, MD, DDS, for referrals and clinical evaluation of participants. We acknowledge the use of the Medical Imaging Informatic Bench to Bedside (mi2b2) workbench for image retrieval. This research was supported by the NIH (R01DC011805, R01DC012545, R01NS088160), Amazon Web Services through the Machine Learning Research Award, and a charitable gift by Mr. and Mrs. Richardson. We acknowledge the support of NVIDIA Corporation with the donation of the Titan V GPU used for this research.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2009165117/-/DCSupplemental.
Data Availability.
All data relevant to clinical and research information of the datasets used in this study are included in the manuscript and supporting information. The datasets used to train and test the machine-learning algorithms are administered by Mass General Brigham. The dataset in its entirety is not currently publicly available, but a subset may be requested from the corresponding author, subject to the data user agreement and the approval by the Mass General Brigham Data and Tissue Sharing Committee.
Code Availability.
All machine-learning algorithms were developed based on publicly available Python libraries (https://numpy.org; https://scikit-learn.org/stable; https://keras.io; https://www.h5py.org). Materials and Methods, SI Appendix, and Fig. 1 contain all details on the architecture of used machine-learning algorithms needed to reproduce the results. The source code may be requested from the corresponding author, subject to the approval by the Mass General Brigham Data and Tissue Sharing Committee and the Mass General Brigham Innovation.
References
- 1.Sadnicka A., Kornysheva K., Rothwell J. C., Edwards M. J., A unifying motor control framework for task-specific dystonia. Nat. Rev. Neurol. 14, 116–124 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hellberg C., Alinder E., Jaraj D., Puschmann A., Nationwide prevalence of primary dystonia, progressive ataxia and hereditary spastic paraplegia. Parkinsonism Relat. Disord. 69, 79–84 (2019). [DOI] [PubMed] [Google Scholar]
- 3.Defazio G. et al., The Italian dystonia registry: Rationale, design and preliminary findings. Neurol. Sci. 38, 819–825 (2017). [DOI] [PubMed] [Google Scholar]
- 4.Creighton F. X. et al., Diagnostic delays in spasmodic dysphonia: A call for clinician education. J. Voice 29, 592–594 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Lima Xavier L., Simonyan K., The extrinsic risk and its association with neural alterations in spasmodic dysphonia. Parkinsonism Relat. Disord. 65, 117–123 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jinnah H. A. et al.; Dystonia Coalition Investigators , The focal dystonias: Current views and challenges for future research. Mov. Disord. 28, 926–943 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Macerollo A., Superbo M., Gigante A. F., Livrea P., Defazio G., Diagnostic delay in adult-onset dystonia: Data from an Italian movement disorder center. J. Clin. Neurosci. 22, 608–610 (2015). [DOI] [PubMed] [Google Scholar]
- 8.Powell A. T., Bidewell J. W., Walker A. C., Diagnosing idiopathic dystonia: Must it take so long? Aust. Health Rev. 18, 120–131 (1995). [PubMed] [Google Scholar]
- 9.Jog M. et al., Causes for treatment delays in dystonia and hemifacial spasm: A Canadian survey. Can. J. Neurol. Sci. 38, 704–711 (2011). [DOI] [PubMed] [Google Scholar]
- 10.Tiderington E. et al., How long does it take to diagnose cervical dystonia? J. Neurol. Sci. 335, 72–74 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Anonymous , BEST (Biomarkers, EndpointS, and other Tools), (Resource, Silver Spring, MD, 2016). [PubMed] [Google Scholar]
- 12.Defazio G. et al., Expert recommendations for diagnosing cervical, oromandibular, and limb dystonia. Neurol. Sci. 40, 89–95 (2019). [DOI] [PubMed] [Google Scholar]
- 13.Defazio G., Hallett M., Jinnah H. A., Berardelli A., Development and validation of a clinical guideline for diagnosing blepharospasm. Neurology 81, 236–240 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ludlow C. L. et al., Consensus-based attributes for identifying patients with spasmodic dysphonia and other voice disorders. JAMA Otolaryngol. Head Neck Surg. 144, 657–665 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Balint B. et al., Dystonia. Nat. Rev. Dis. Primers 4, 25 (2018). [DOI] [PubMed] [Google Scholar]
- 16.Logroscino G. et al., Agreement among neurologists on the clinical diagnosis of dystonia at different body sites. J. Neurol. Neurosurg. Psychiatry 74, 348–350 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McHugh M. L., Interrater reliability: The kappa statistic. Biochem. Med. (Zagreb) 22, 276–282 (2012). [PMC free article] [PubMed] [Google Scholar]
- 18.Simonyan K., Neuroimaging applications in dystonia. Int. Rev. Neurobiol. 143, 1–30 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fuertinger S., Simonyan K., Task-specificity in focal dystonia is shaped by aberrant diversity of a functional network kernel. Mov. Disord. 33, 1918–1927 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fuertinger S., Simonyan K., Connectome-Wide phenotypical and genotypical associations in focal dystonia. J. Neurosci. 37, 7438–7449 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Conte A. et al., Ten-year reflections on the neurophysiological abnormalities of focal dystonias in humans. Mov. Disord. 34, 1616–1628 (2019). [DOI] [PubMed] [Google Scholar]
- 22.Schirinzi T., Sciamanna G., Mercuri N. B., Pisani A., Dystonia as a network disorder: A concept in evolution. Curr. Opin. Neurol. 31, 498–503 (2018). [DOI] [PubMed] [Google Scholar]
- 23.Hanekamp S., Simonyan K., The large-scale structural connectome of task-specific focal dystonia. Hum. Brain Mapp. 41, 3253–3265 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Battistella G., Fuertinger S., Fleysher L., Ozelius L. J., Simonyan K., Cortical sensorimotor alterations classify clinical phenotype and putative genotype of spasmodic dysphonia. Eur. J. Neurol. 23, 1517–1527 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li Z. et al., Alterations of resting-state fMRI measurements in individuals with cervical dystonia. Hum. Brain Mapp. 38, 4098–4108 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rosset-Llobet J., Fàbregas-Molas S., Pascual-Leone Á., Effect of transcranial direct current stimulation on neurorehabilitation of task-specific dystonia: A double-blind, randomized clinical trial. Med. Probl. Perform. Art. 30, 178–184 (2015). [DOI] [PubMed] [Google Scholar]
- 27.Simonyan K., Frucht S. J., Blitzer A., Sichani A. H., Rumbach A. F., A novel therapeutic agent, sodium oxybate, improves dystonic symptoms via reduced network-wide activity. Sci. Rep. 8, 16111 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Merchant S. H. I. et al., The role of the inferior parietal lobule in writer’s cramp. Brain 143, 1766–1779 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ramdhani R. A. et al., What’s special about task in dystonia? A voxel-based morphometry and diffusion weighted imaging study. Mov. Disord. 29, 1141–1150 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Simonyan K., Cho H., Hamzehei Sichani A., Rubien-Thomas E., Hallett M., The direct basal ganglia pathway is hyperfunctional in focal dystonia. Brain 140, 3179–3190 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Black K. J. et al., Spatial reorganization of putaminal dopamine D2-like receptors in cranial and hand dystonia. PLoS One 9, e88121 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bianchi S., Fuertinger S., Huddleston H., Frucht S. J., Simonyan K., Functional and structural neural bases of task specificity in isolated focal dystonia. Mov. Disord. 34, 555–563 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bianchi S. et al., Phenotype- and genotype-specific structural alterations in spasmodic dysphonia. Mov. Disord. 32, 560–568 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pandey S., Tater P., Post-stroke lingual dystonia: Clinical description and neuroimaging findings. Tremor Other Hyperkinet. Mov. (N. Y.) 8, 610 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Colosimo C. et al., Diffusion tensor imaging in primary cervical dystonia. J. Neurol. Neurosurg. Psychiatry 76, 1591–1593 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brüggemann N. et al., Neuroanatomical changes extend beyond striatal atrophy in X-linked dystonia parkinsonism. Parkinsonism Relat. Disord. 31, 91–97 (2016). [DOI] [PubMed] [Google Scholar]
- 37.Fabbrini G. et al., Diffusion tensor imaging in patients with primary cervical dystonia and in patients with blepharospasm. Eur. J. Neurol. 15, 185–189 (2008). [DOI] [PubMed] [Google Scholar]
- 38.Kostic V. S. et al., Brain structural changes in spasmodic dysphonia: A multimodal magnetic resonance imaging study. Parkinsonism Relat. Disord. 25, 78–84 (2016). [DOI] [PubMed] [Google Scholar]
- 39.Gallea C., Horovitz S. G., Najee-Ullah M., Hallett M., Impairment of a parieto-premotor network specialized for handwriting in writer’s cramp. Hum. Brain Mapp. 37, 4363–4375 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dresel C. et al., Multiple changes of functional connectivity between sensorimotor areas in focal hand dystonia. J. Neurol. Neurosurg. Psychiatry 85, 1245–1252 (2014). [DOI] [PubMed] [Google Scholar]
- 41.Delnooz C. C., Pasman J. W., Beckmann C. F., van de Warrenburg B. P., Task-free functional MRI in cervical dystonia reveals multi-network changes that partially normalize with botulinum toxin. PLoS One 8, e62877 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Egger K. et al., Voxel based morphometry reveals specific gray matter changes in primary dystonia. Mov. Disord. 22, 1538–1542 (2007). [DOI] [PubMed] [Google Scholar]
- 43.Jahanshahi M., Torkamani M., The cognitive features of idiopathic and DYT1 dystonia. Mov. Disord. 32, 1348–1355 (2017). [DOI] [PubMed] [Google Scholar]
- 44.Mahajan A. et al., Cervical dystonia and executive function: A pilot magnetoencephalography study. Brain Sci. 8, 159 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Odergren T., Stone-Elander S., Ingvar M., Cerebral and cerebellar activation in correlation to the action-induced dystonia in writer’s cramp. Mov. Disord. 13, 497–508 (1998). [DOI] [PubMed] [Google Scholar]
- 46.Putzel G. G. et al., Polygenic risk of spasmodic dysphonia is associated with vulnerable sensorimotor connectivity. Cereb. Cortex 28, 158–166 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mori S. W., Wakana S., Poetscher L. M., van Zijl P. C. M., Atlas of the Human White Matter, (Elsevier, 2005). [Google Scholar]
- 48.Castelvecchi D., Can we open the black box of AI? Nature 538, 20–23 (2016). [DOI] [PubMed] [Google Scholar]
- 49.Topol E. J., High-performance medicine: The convergence of human and artificial intelligence. Nat. Med. 25, 44–56 (2019). [DOI] [PubMed] [Google Scholar]
- 50.Defazio G., Conte A., Gigante A. F., Fabbrini G., Berardelli A., Is tremor in dystonia a phenotypic feature of dystonia? Neurology 84, 1053–1059 (2015). [DOI] [PubMed] [Google Scholar]
- 51.Jankovic J., Leder S., Warner D., Schwartz K., Cervical dystonia: Clinical findings and associated movement disorders. Neurology 41, 1088–1091 (1991). [DOI] [PubMed] [Google Scholar]
- 52.Deuschl G., Bain P., Brin M., Consensus statement of the movement disorder society on tremor. Ad Hoc scientific committee. Mov. Disord. 13, 2–23 (1998). [DOI] [PubMed] [Google Scholar]
- 53.Bhatia K. P. et al.; Tremor Task Force of the International Parkinson and Movement Disorder Society , Consensus statement on the classification of tremors. From the task force on tremor of the international Parkinson and movement disorder society. Mov. Disord. 33, 75–87 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Senn S., Testing for baseline balance in clinical trials. Stat. Med. 13, 1715–1726 (1994). [DOI] [PubMed] [Google Scholar]
- 55.Kirke D. N. et al., Neural correlates of dystonic tremor: A multimodal study of voice tremor in spasmodic dysphonia. Brain Imaging Behav. 11, 166–175 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Simonyan K., Ludlow C. L., Abnormal structure-function relationship in spasmodic dysphonia. Cereb. Cortex 22, 417–425 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Termsarasab P. et al., Neural correlates of abnormal sensory discrimination in laryngeal dystonia. Neuroimage Clin. 10, 18–26 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Waugh J. L. et al., Thalamic volume is reduced in cervical and laryngeal dystonias. PLoS One 11, e0155302 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Battistella G., Simonyan K., Top-down alteration of functional connectivity within the sensorimotor network in focal dystonia. Neurology 92, e1843–e1851 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Haslinger B. et al., “Silent event-related” fMRI reveals reduced sensorimotor activation in laryngeal dystonia. Neurology 65, 1562–1569 (2005). [DOI] [PubMed] [Google Scholar]
- 61.Kiyuna A. et al., Brain activity in patients with adductor spasmodic dysphonia detected by functional magnetic resonance imaging. J. Voice 31, 379.e1-379.e11 (2017). [DOI] [PubMed] [Google Scholar]
- 62.Kiyuna A. et al., Brain activity related to phonation in young patients with adductor spasmodic dysphonia. Auris Nasus Larynx 41, 278–284 (2014). [DOI] [PubMed] [Google Scholar]
- 63.Hunter J. D., Matplotlib: A 2D graphics environment. Comput. Sci. Eng. 9, 90–95 (2007). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data relevant to clinical and research information of the datasets used in this study are included in the manuscript and supporting information. The datasets used to train and test the machine-learning algorithms are administered by Mass General Brigham. The dataset in its entirety is not currently publicly available, but a subset may be requested from the corresponding author, subject to the data user agreement and the approval by the Mass General Brigham Data and Tissue Sharing Committee.
All machine-learning algorithms were developed based on publicly available Python libraries (https://numpy.org; https://scikit-learn.org/stable; https://keras.io; https://www.h5py.org). Materials and Methods, SI Appendix, and Fig. 1 contain all details on the architecture of used machine-learning algorithms needed to reproduce the results. The source code may be requested from the corresponding author, subject to the approval by the Mass General Brigham Data and Tissue Sharing Committee and the Mass General Brigham Innovation.