Abstract
External apical root resorption is one of the most deleterious complications after orthodontic treatment. Studies to explain the causal relationship between orthodontic tooth movement and external apical root resorption have been inconclusive till date. Individual variations in external apical root resorption sometimes overshadow the treatment related factors which indicate genetic predisposition and/or multifactorial etiology. Mechanism of root resorption is not completely understood. Inflammatory root resorption induced by orthodontic treatment is a part of process of elimination of hyaline zone. An imbalance between bone resorption and deposition may contribute to root resorption by the cementoclasts/osteoclasts. This narrative review article explains the molecular pathway involved in external apical root resorption and also role of various genes involved at different level. It also reviews the literature published during the past 20 years concerning the association studies linking EARR to genetic polymorphisms. This literature review provides an insight into genetic predisposition of external apical root resorption that can be used in orthodontic practice to enable 'high-risk' subjects to be identified on the basis of their genetic information before orthodontic treatment is initiated.
Keywords: Etiology, external apical root resorption, genetic polymorphism
Background
EARR (External apical root resorption) is the reduction of root structure involving the apical region to the extent that it can be seen on standard radiographs.[1] Orthodontic force applied to teeth over a short period can produce resorption lacunae on the cementum root surface histologically which is a preliminary step towards EARR. When root resorption exceeds the reparative capacity of cementum, EARR is seen.[2]
Studies to explain the causal relationship between orthodontic tooth movement and external apical root resorption have been inconclusive till date. Maxillary incisors are the most commonly affected teeth by EARR during orthodontic treatment.[3,4] Occlusal forces have been presumably attributed as the causative factor for EARR.[5,6,7] In addition, Harris et al. studied the hypothesis of genetic influence on the EARR and found a great possibility of inheritance.[8]
The prevalence of EARR associated with orthodontic treatment greatly varies in the literature, depending on the methods used to determine it in the studies. More than one-third of patients undergoing orthodontic treatment have root resorption greater than 3 mm and severe root resorption (>5mm) has been found to occur in 2%-5% of the population.[4,9,10]
Risk factors for EARR
EARR is a complex phenomenon caused by a combination of poorly understood environmental and host factors. Biomechanical or orthodontic treatment-related risk factors may include treatment duration, type of orthodontic appliance, tooth extraction, intrusive movement, root torque, extensive tooth movement and force magnitude.[11] Orthodontic tooth movement or “biomechanics” has been found to account for approximately from one-tenth to one-third of total variation in EARR.[12]
There was considerable individual variation in the EARR associated with orthodontic treatment which overshadowed the force magnitude and force type, indicating an individual predisposition and multifactorial etiology.[4,8,13,14,15,16,17]
The role of genetic factors was first suggested by Newman in 1975.[15] Then in 1997, Harris et al. gave findings of his research and reported the involvement of genetic variation in EARR concurrent with orthodontic treatment through a heritability study.[8] Later, Hartsfield et al. confirmed that genetic variation was associated with as much as 50%-66% of the variations observed with EARR during orthodontic treatment.[4] Since then, various authors have studied the role of various genes in the occurrence of EARR.
The genetic aspect of EARR is of utmost importance to the general care physicians also. Various genetic markers involved in EARR are also associated with various systemic and bone metabolism disorders. They need to be aware of dental implications of such polymorphisms. Sometimes dental findings can prove to be very helpful in early diagnosis of systemic and bone metabolic disorders.
Materials and Methods
This article reviews the literature published during the past 20 years concerning the association studies linking EARR to genetic polymorphisms. The literature search was performed on PubMed and Science Direct using the following keywords: “external apical root resorption and gene and polymorphism”. All suitable articles were selected and their references were rechecked for any relevant articles overlooked during the electronic searches. Other relevant articles were handpicked from the institutional library. The research findings to date are discussed with emphasis on several candidate genes without constraints on population, sample size and treatment duration.
Main Text
Molecular and Genetic pathway of root resorption with orthodontic force
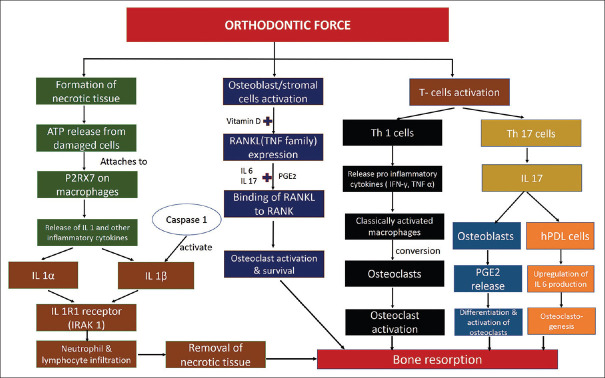
Orthodontic force application leads to a cascade of events in compression and tension region leading tooth movement. Figure 1 describes the molecular pathway during orthodontic tooth movement. This is mediated through two pathways: 1) activation of control of osteoclasts through the ATP/P2RX7/IL-1B inflammation modulation pathway; and 2) RANK/RANKL/OPG osteoclast activation control pathway.[18] Following section describes various mediators of tooth movement and their associated genetic polymorphisms responsible for external apical root resorption [Tables 1 and 2].
Figure 1.
Molecular pathway during orthodontic tooth movement
Table 1.
Genes involved in external apical root resorption
| Gene | Gene code | SNP | SNP location | Effect of variation | Exon/intron | Chromosome | Nucleotide | Amino acid | Type of variation |
|---|---|---|---|---|---|---|---|---|---|
| Interleukin-1 alpha (OMIM#147760) | IL-1A | rs1800587 | −889 | TT genotype (EARR causing) | - | 2q14.1 | C to T | - | Promoter variation |
| Interleukin-1 beta (OMIM#147720) | IL-1B | rs1143634 | +3953/+3954 | TT genotype (Increased IL-1B production) | Exon 5 | 2q14.1 | TTC to TTT | Phe105Phe | Synonymous variant |
| Interleukin-1 receptor antagonist (OMIM#147679) | IL-1RN | rs315952 rs419598 86 bp VNTR |
+390 +2018 |
CC genotype (EARR causing) TT genotype (EARR causing) Short allele (allele 2, 2 repeats) Long alleles (allele 4, 3 repeats) (allele 1, 4 repeats) (allele 3, 5 repeats) (allele 5, 6 repeats) |
Exon 4 Exon 2 Intron 2 |
2q14.1-2 | AGT to AGC GCT to GCC |
Ser133Ser Ala60Ala |
Synonymous variant |
| Interleukin-1 receptor-associated kinase (OMIM#300283) | IRAK1 | rs1059703 | +6434 | CC genotype (protective for EARR) | Exon 12 | Xq28 | TCG to TTG | Ser532Leu | Non-synonymous variant |
| Receptor activator of nuclear factor-κB (RANK) (OMIM#603499) | TNFRSF11A | rs1805034 D18S64 |
575 Microsatellite |
- | Exon 6 | 18q21.33 | GCG to GTG | Val192Ala | Non-synonymous variant |
| Osteoprotegerin (OMIM#602643) | TNFRSF11B (OPG) | rs2073618 rs3102735 |
+1181 −163 |
G allele was associated with EARR |
Exon 1 | 8q24.12 | AAC to AAG A to G |
Asn3Lsn | Non-synonymous variant Promoter variation |
| Osteopontin/ Bone sialoprotein 1 (BSP1/BNSP)/Secreted phosphoprotein 1 (SPP1) | OPN | rs11730582 rs9138 |
−443 +1239 |
A allele is EARR protective T allele is EARR protective |
5’ near gene 3’ UTR |
4q22.1 | T to C A to C |
- | Non-synonymous variant |
| Interleukin-6 (OMIM#147620) | IL-6 | rs1800796 | −634 | G allele (EARR causing) | - | 7p15.3 | C to G | - | Promoter variation |
| Interleukin-17A (OMIM#603149) | IL-17A | rs2275913 | −197 | - | - | 6p12.2 | G to A | - | Promoter variation |
| Purinergic-receptor-P2X, ligand-gated ion channel 7 (OMIM#602566) | P2RX7 | rs208294 rs1718119 rs2230912 |
+489 +1068 +1405 |
C allele (EARR causing) G allele (EARR causing) |
Exon 5 Exon 11 Exon 13 |
12q24.31 | CAT to TAT GCT to ACT CAG to CGG |
His155Tyr Ala348Thr Gln460Arg |
Non-synonymous variant |
| Interleukin-1beta converting enzyme/Caspase 1 (OMIM#147678) | CASP1 | rs530537 rs580253 rs554344 |
- | T allele (lower IL-1beta production) C allele (lower IL-1beta production) |
Intron 7 Exon 6 3’ UTR |
11q22.3 | T to C CTA 10370 TTA G to C |
Leu to Leu | Synonymous variant |
| Tumour necrosis factor alpha (OMIM#191160) | TNFα | rs1800629 | −308 | A allele (more TNFα production) | - | 6p21.33 | G to A | - | Promoter variation |
| Vitamin D receptor (OMIM#601769) | VDR | rs731236 | - | C allele (EARR protective) | Exon 9 | 12q13.11 | ATT to ATC | Ile352Ile | Synonymous variant |
Table 2.
Summary of various studies assessing association of different genepolymorphisms with external apical root resorption
| Author, Year, Journal | Country (Ethnicity) | Sample size | Gene and polymorphisms studied | Mean age | Treatment duration | Radiograph Technique | Result |
|---|---|---|---|---|---|---|---|
| Iglesias-Linares et al., 2017, AO | Spain (Caucasian) | 372 subjects (174 cases and 198 controls) treated with either fixed or removable appliances | IL-1β rs1143634 IL-1RN rs419598 Osteopontin (SPP1) rs11730582, rs9138 |
27.69+13.6 years (28.48+13.60 years 26.29+13.66 years) | Not mentioned | OPG | Predisposition to EARR is similar for invisalign and fixed appliances. Homozygous subjects (TT genotype of IL1RN, rs419598) were found to be three times more predisposed to experience EARR compared with the other genotypes. |
| Linhartova et al., 2017, OD | Czech Republic (Caucasian) | 30 cases (11 boys and 19 girls) with EARR in maxillary incisors/69 controls (26 boys and 43 girls) |
IL-17 rs2275913 Osteopontin (SPP1) rs11730582, rs9138 P2RX7 rs208294, rs1718119 TNFRSF11B rs3102735, rs2073618 |
14.6+3.2 years 15.2+5.3 years |
Not mentioned | OPG, Lateral Cephalogram |
CG haplotype (rs208294+rs1718119) was associated with EARR while no association was seen for individual SNP. |
| Pereira et al., 2016, OD | Portugal (Caucasian) | 195 (72 males and 123 females); six maxillary anterior teeth were assessed | IL-1β rs1143634 IL-1RN rs315952 IRAK1 rs1059703 |
17.24+6.8 years | 36+10 months | OPG | C allele of SNP rs1059703 (IRAK1) is protective for EARR. |
| Guo et al., 2016, AJODO | China (Han Chinese) | 174 patients (68 males and 106 females; left maxillary central incisors were monitored) | IL-1RN rs419598 IL-6 rs1800796 |
12-34 years | 20.55+6.54 months (14-28 months) |
CBCT | GC allele of SNP rs1800796 (IL-6) is associated increased risk for EARR. |
| Sharab et al., 2015, OCR | USA (Caucasian) | 67 cases (38 females, 29 males) with EARR of maxillary incisors/67 age- and sex-matched controls | P2RX7 (rs208294, rs1718119, rs2230912) CASP1 (rs530537, rs580253, rs554344) IL-1α rs1800587 (-889 C/T) IL-1β rs1143634 (+3953), IL-1RN rs419598 |
15.78+1.13 years 15.79+1.14 years |
2.49+0.10 years 1.97+1.14 years |
OPG, occlusal | CC or CT genotype of rs208294 (P2RX7) polymorphism is associated with EARR. |
| Lim et al., 2014, AJODO | Mice | Not applicable | Wnt signalling | Not applicable | Not applicable | Not applicable | Reduced wnt signaling causes spontaneous root resorption |
| Iglesias-Linares et al., 2014, OD | Spain (Caucasian) | 37 cases with (EARR>2mm) / 50 controls | Osteopontin rs9138, rs11730582 | 24.7+5.95 y 23.8+5.33 y |
27.5+8.3 months | Lateral cephalogram, OPG |
Allele 2/Allele 2 (CC genotype) in rs9138 rs11730582 (osteopontin gene) predispose to EARR secondary to orthodontic treatment. Allele 1(T) in rs9138 is protective for EARR. Allele 1(A) in rs11730582 is protective for EARR. |
| Pereira et al., 2014, OD | Portugal (Caucasian) | 195 cases (72 males and 123 females); six maxillary anterior teeth were assessed. | IL-1β rs1143634 (+3953) TNFRSF11A rs1805034 P2RX7 rs1718119 TNFRSF11B rs3102735 |
17.24+6.8 years | 36+10 months | OPG | GG genotype of rs1718119 (P2RX7 gene) is associated with EARR. |
| Linhartova et al., 2013, OD | Czech Republic (Caucasian) | 32 cases with EARR in maxillary incisors/74 controls without EARR |
IL-1α rs1800587 (−889) IL-1β rs1143634 (+3953) IL-1RN (86bp VNTR) |
15.0+4.1 years (cases) 15.2+5.3 years (controls) (15.2+5 years average) |
34.5+15.6 months | OPG, Lateral Cephalogram |
IL1RN*12, *22 genotypes and short allele *2 is associated with EARR in girls. |
| Iglesias-Linares et al., 2013, HH | Spain (Caucasian) | 93 patients (39 cases with EARR > 2mm in root-filled teeth/54 controls without EARR in root-filled teeth) | IL-1RN rs419598 (+2018) | 24 years 1 month+5 years 5 months |
27.21 months+4.9 months |
Lateral cephalogram, OPG | Genotype TT of IL-1RN is associated with EARR in root filled teeth |
| Iglesias Linares et al., 2012, OD | Spain (Caucasian) | 54 patients (25 cases with EARR>2mm/29 controls) |
IL-1α rs1800587 (−889) IL-1β rs1143634 (+3953) IL-1RN rs419598 (+2018) |
23.08+5.08 years | 31.1+6.4 months | Lateral cephalogram, OPG | Genotypes CC of rs1143634 (IL-1β) and TT of rs419598 (IL-1RN) are associated with EARR. |
| Iglesias Linares et al., 2012, IEJ | Spain (Caucasian) | 73 root canal filled teeth (35 with EARR>2 mm and 38 non EARR teeth)/73 vital control teeth (30 with EARR>2 mm and 43 non-EARR teeth) | IL-1α rs1800587 (-889) IL-1β rs1143634 (+3953) | 23+5 years per 8+9 months | 27+4.3 months | Lateral cephalogram, OPG | Homozygous (TT, rs1143634) subjects has two times risk of EARR in root-filled teeth as compared to vital teeth. |
| Iglesias Linares et al., 2012, JOE | Spain (Caucasian) | 93 RC treated teeth; 39 with EARR>2 mm/54 with EARR≤2mm |
IL-1α rs1800587 (-889) IL-1β rs1143634 (+3953) |
24 years 1 month + 5 years 5 months |
27.21 months+4.9 months |
Lateral cephalogram, OPG | Genotype TT of IL-1β is associated with EARR in endodontically treated teeth. |
| Fontana et al., 2012, AJODO | Brazil (Mixed whites) | 377 Class II div 2 individuals; group 1 (EARR≤1.43 mm; treated)=157; group 2 (EARR≤1.43 mm; treated)=175; group 3 (untreated)=35 | Vitamin D receptor rs731236 | 14.9 years | EARR measured at 6 months of treatment | IOPA | Allele C is weakly associated with protection against EARR |
| Tomoyasu et al., 2009, OW | Japanese | 54 Japanese (18 males and 36 females); 24 Han Chinese; 24 African Americans; 24 European Americans; and 24 Hispanics (maxillary and mandibular central incisors, mesial and distal roots of mandibular first molars were analysed) | IL-1β rs1143634 (+3953) | Males = 19 years, Females = 21 years | 3 years 1 month | Lateral cephalogram, OPG | rs1143634 SNP is not associated with EARR. |
| Gulden et al., 2009, JOO | Germany (Whites) | 49 cases (45 sporadic patients, 2 pairs of siblings)/40 controls | IL-1α rs1800587 (−889) IL-1β +3954 | >12 years | Not mentioned | OPG | Genotype TT (rs1800587) of IL-1α is associated with EARR. |
| Hartsfield, 2009, OCR | USA (Caucasian) | 135 cases (EARR>2mm in atleast one maxillary central incisor) | TNFRSF11B rs2073618 | 14.6+6.9 years | 1.6+0.5 years | G allele was associated with EARR. | |
| Lages et al., 2007, AJODO | Brazil (Brazilian whites) | 23 affected/38 controls (Maxillary incisors were evaluated) | IL-1β +3954 | 18.9+5.2 years | Not mentioned | IOPA | Allele C of IL-1β is associated with EARR |
| Al-Qawasmi et al., 2004, JMNI | Mice | Not applicable | IL-1β | Not applicable | Not applicable | Not applicable | IL-1β is significantly associated with orthodontic root resorption |
| Al-Qawasmi et al., 2003, AJODO | USA (Caucasian) | 35 families (118 persons: 73 siblings and 45 parents) | IL-1α rs1800587 (-889) IL-1β +3954 | 12.1+1.89 years | 2.82+1.09 years | Lateral cephalogram, OPG | Genotype CC of IL-1β is associated with EARR. |
| Al-Qawasmi et al., 2003, JDR | USA (Caucasian) | 38 families (124 persons: 79 siblings and 45 parents); EARR evaluated in Maxillary and mandibular central incisors and mandibular first molars. | TNSALP (AL215L) TNFα (-308) TNFRSF11A (D18S64) | 12.3+1.82 years | 2.77+1.13 years | Lateral cephalogram, OPG | Linkage of D18S64 with EARR in maxillary central incisors. |
*EARR: External apical root resorption, AO: The Angle Orthodontist, OD: Oral Diseases, AJODO: American Journal of Orthodontics and Dentofacial Orthopaedics, OCR: Orthodontics and Craniofacial Research, HH: Histology and Histopathology, IEJ: International Endodontic Journal, JOE: Journal of Endodontics, OW: Orthodontic Waves, JOO: Journal of Orofacial Orthopedics, JMNI: Journal of Musculoskeletal and Neuronal Interactions, JDR: Journal of Dental Research,IL-1α : Interleukin 1 alpha, IL-1β : Interleukin 1 beta, IL-1RN: Interleukin Receptor Antagonist, IL-17: Interleukin 17, P2RX7: Purinergicreceptor, TNFRSF: Tumor necrosis factor receptor superfamily, IRAK1- Interleukin 1 receptor associated kinase-1, IL-6:”Interleukin 6, CASP-1: Caspase 1, TNSALP: Tissue non-specific alkaline phosphatase, TNF α: Tumor necrosis factor alpha
Interleukin 1 (IL 1) family
IL1α and IL1β are encoded by distinct genes but bind to the same receptor IL 1R1.[19] IL1α precursor is constitutively present, fully active and is released from necrotic cells during early phases of inflammation. However, IL 1 β is inactive and is cleaved by caspase 1 to release the active cytokine in extracellular space.[20] The interleukin receptor antagonist, IL1ra is a unique, naturally occurring cytokine that inhibits IL1 activity by binding to IL1R1 receptors with high affinity thereby preventing signal transduction.[21]
IL-1β is a potent bone-resorptive cytokine and plays a key role in the complex pathways leading to root resorption.[4] IL-1B (+3954) polymorphism in orthodontically treated subjects plays a role in EARR and IL-1B allele is a risk factor of EARR.[22] A study by Bastos Lages et al.[23] also found similar results in 61 Brazilian orthodontic patients.IL-1B gene polymorphism, rs1143634 (C3954T) and EARR has frequently been found to be associated in several geographical patient populations[22,23,24] and in knock-out animal models.[25] Furthermore, this polymorphism was implicated in approximately 15% of the variation in EARR of maxillary central incisors of orthodontic patients.[22] However, some patient-based studies obscured this association[11,12,26,27,28]
Gulden et al.[26] found an association of IL-1A polymorphism (-889TT genotype) with root resorption in a German sample of 45 EARR patients and 40 anonymous controls. However, Sharab et al.,[12] Al Qawasmi et al.,[22] Iglesias-Linares et al.[24,29] and Linhartova et al.[30] in their studies found no significant association of IL-1A polymorphism with EARR.
IL-1ra might affect bone resorption during orthodontic treatment by blocking IL-1 from stimulating the osteoclasts and thus lead to root resorption. IL-1RN gene polymorphisms have been correlated with a high expression of IL-1ra and hence, with EARR.[31,32] Iglesias-Linares et al.[24] showed that the TT genotype of IL-1RN Single Nucleotide Polymorphism (SNP) rs419598 is highly and positively correlated with EARR.
IL1Receptor-associated kinase 1 (IRAK1)
IL1β roles are mainly dependent on IL-1 receptor 1 (IL1R1) binding. Activation of IL1R1 leads to the recruitment of adaptor molecules like IRAK1, with subsequent activation of signalling transduction pathways.[33] Hyper-phosphorylation of IRAK1, switch on the signal transduction pathway activation, and later, its ubiquitination and proteasome degradation switches off the signalling.[34] IRAK1 gene is located on chromosome X at position q28. IRAK1 polymorphism (rs1509703) has been associated with EARR by Pereira et al.
Interleukin-6 (IL-6)
IL-6 acts as a multifunctional cytokine with both inflammatory and anti-inflammatory effects. IL-6 and its receptors mainly activate two signal pathways: the JAK kinase, signal transduction and transcriptional activation (JAK/STAT) pathway and mitogen-activating protein kinases (MAPK) pathway.[35,36,37] IL-6 was observed to increase in the gingival crevicular fluid (GCF) and PDL in orthodontic patients. It was discerned from the studies that IL-6 is instrumental in local regulation of bone remodelling and acute inflammation at the commencement of orthodontic treatment.[38] Thus, Guo et al.[39] conducted a study where IL-6 SNP (rs1800796) with GC genotype was found to have greater root resorption than CC genotype.
Interleukin 17
Proinflammatory interleukin-17A (IL-17A) stimulated odontoclastogenesis and influenced the mRNA expression of RANKL from human dental pulp cells in vitro.[40] Thus, Hayashi et al.[41] suggested that T-helper 17 cells may aggravate the process of orthodontically induced inflammatory root resorption. Linhartova et al.[30] investigated IL-17A gene variability in postorthodontic EARR but found a non-significant association.
Purinergic receptor P2X, ligand gated ion channel 7 (P2RX7)
P2RX7 is expressed in osteoblasts and osteoclasts and seem to have a pro-osteogenic effect, activating osteoblast function and inducing osteoclast apoptosis. It also stimulates the release of inflammatory cytokines such as IL-1B by immune cells by acting through ATP/P2RX7/IL-1B inflammation modulation pathway. In the study by Sharab et al.[12] SNP, rs208294, located in the P2RX7 gene was found to be associated with EARR. Linhartova et al. in the recent study[42] revealed that variability in the P2RX7 gene and the length of orthodontic treatment may be important factors contributing to the etiopathogenesis of postorthodontic EARR.
Vitamin D receptor gene
Vitamin D is responsible for the regulation of certain genes at the transcription level, via interaction with the vitamin D receptor,[43] and influences host immune responses and aspects of bone development, growth, and homeostasis.[44] Vitamin D stimulates osteoclastogenesis acting through its nuclear receptor, Vitamin D receptor in immature osteoblast/stromal cells via RANKL/OPG regulatory pathway[45,46,47][Figure 1]. Also, Vitamin D enhanced IL-1β expression via a direct transcriptional mechanism during the inflammatory process.[48]
Polymorphisms in Vitamin D receptor gene have been associated with bone mineral density, bone turn over and diseases in which mineral loss is a cardinal sign. A study by Fontana et al. in the Brazilian population in 2012 reported that Vitamin D receptor TaqI (rs 731236) polymorphism is associated with EARR.[49]
Osteopontin (OPN) gene
Osteopontin is an acidic phosphorylated glycoprotein containing an Arg-Gly- Asp (RGD) motif, which can interact with various receptors including αv-β3 and other integrins and so cause the odontoclast to adhere to root surface at the onset of physiological or pathologic resorption.[50,51] OPN plays role in odontoclast activation during the root resorption process. Iglesias-Linares et al. showed that OPN gene (rs9138; 3'UTR and rs 11730582; 5' near region) are determinants of a genetic predisposition to suffer EARR secondary to orthodontic treatment.[52]
RANK/RANKL/OPG
Osteoblasts and stromal stem cells express receptor activator of NF-kappa B ligand (RANKL), which binds to its receptor activator of nuclear factor-kappa B (RANK, coded by TNFRSF11A gene), on the surface of osteoclasts and their precursors. Osteoprotegerin (OPG, coded for by the TNFRSF11B gene) is secreted by osteoblasts and osteogenic stromal stem cells and protects from excessive bone resorption by binding to RANKL and preventing it from binding to RANK.[53] Study by Hartsfield et al.[18] evaluated the association between single nucleotide polymorphism (SNP) rs2073618 of TNFRSF11B (OPG) gene and EARR in orthodontically treated patients and showed that OPG polymorphism accounted for approximately 8% of total EARR variation.
Wnt
Wnt signalling pathways are a group of signal transduction pathways made of proteins that pass signals into a cell through cell surface receptors.OPG/RANKL regulates bone formation, and studies in the literature suggest that disruptions in OPG/RANKL contribute to low bone mass phenotypes in humans[54] and root resorption as well.[55,56] The OPG/RANKL pathway, in turn, is mediated by Wnt signalling; a positive Wnt stimulus simultaneously downregulated RANKL and upregulates OPG, thus enhancing bone formation and inhibiting bone resorption.[57,58] Lim et al. in their study showed that genetically reducing Wnt signalling in the environment of the periodontal complex is sufficient to cause spontaneous root resorption.[59]
Discussion
The present review attempted to review genetic risk factors for orthodontic treatment-induced external apical root resorption. We located 21 studies on different genes, which might influence EARR [Table 2]. 19 were human studies and 2 were on mice.
Fifteen studies have been reported for IL1 gene involving association with EARR.[11,12,22,23,24,25,26,27,28,29,30,39,60,61,62] IL-1α has been investigated in seven studies[12,22,24,26,30,60,61] involving four ethnic groups (German, Czech, Hispanics and US Caucasians) where only German Caucasians with TT genotype of IL-1α was observed to have a significant association with orthodontically induced EARR. The occurrence of the rare genotypes of the SNP rs1800587 was associated with an almost fourfold increase in IL-1 protein levels in the gingival crevicular fluid.[26]
There were 13 studies[11,12,22,23,24,25,26,27,28,29,30,60,61] which investigated the association of IL-1B with EARR. 7 studies (3 Hispanics, 1 US, 1 Portugal, 1 Brazil and 1 mice knock out model) found an increased risk of IL-1B with EARR. 4 studies revealed CC genotype as associated with EARR[12,22,23,24] while 2 studies[60,61] on Root canal treated teeth in Hispanics showed association with TT genotype. These contrasting results may be pulp related as it is the major IL-1B producing. Other studies in the US, Portugal, Czech, Germany, Spain and Japan didn't find a significant association. Different results in different studies may be due to different allele frequencies in different ethnic groups and sample size variation. The minor allele frequency ofIL1B (+3954) is reported at 21%–29% in Caucasians, 14.7% in Hispanics and 5.6% in Japanese.[63] Failure to detect a statistically significant difference in frequency of IL1B polymorphism between EARR cases and control in Japanese is due to low minor allele frequency. C3954T polymorphism is present on exon and is synonymous (Phe105Phe) and does not result in a change in protein conformation but was observed to increase IL-1B production which caused relatively more catabolic bone modeling in the cortical bone interface of the periodontal ligament owing to the increased number of osteoclasts associated with higher levels of this cytokine. The variation might influence mRNA splicing, nuclear RNA stability, or, conceivably, levels of mRNA expression. Alternatively, the polymorphic site might be in strong linkage disequilibrium with another polymorphic site, within either the coding or the regulatory regions of these genes.[64] Al-Qawasmi et al.[22] further reported a greater increase in resistance to EARR with IL1 composite genotype involving the combined presence of T alleles of IL1A and IL1B. This expounded the stronger linkage of variation in IL-1B site as compared to the variation at the IL-1A marker and it further reflects the complex nature of these loci in determining susceptibility to orthodontically induced EARR.
Seven studies investigated the association of IL-1RN on EARR with 4 studies[24,29,30,62] (Czech, Spain) revealing a positive association and studies in US,[12] Portugal[28] and China[39] didn't find any association. A polymorphism in the second intron of IL1RN gene gives rise to VNTR (variable number tandem repeat) of 86 bp sequence and thus to different alleles. Atleast one copy of allele 2 is associated with decreased secretion of IL1ra and increased IL1B synthesis. IL1RN VNTR short allele with 2 repeats was associated with EARR in Czech females which might be due to more prevalence of this allele in women. This allele was controversial regarding the reporting of both increased and decreased IL1ra levels. Moreover, this VNTR polymorphism is strongly correlated with other IL1RN polymorphisms.[30] The + 2018T > C polymorphism in exon 2 of the gene is in complete linkage disequilibrium with a penta-allelic 86 bp VNTR polymorphisms in intron 2 of the gene. Genotype TT of this polymorphism is associated with EARR in both vital and root-filled teeth, which is consistent with an increase in IL1ra levels.[24,29,62] This genotype is not associated with EARR in Chinese[39] and the US.[12] C allele of rs315952 is associated with increased IL1ra levels but was not found to be significantly associated with EARR.[28]
IL-6 polymorphism is associated with EARR only in the Chinese population.[39] According to this study, root resorption of patients carrying the GC genotype was greater than that of patients carrying the CC genotype. Moreover, Gu et al. observed lesser blood IL-6 levels in patients with G allele as compared to C allele. This indicates that IL-6 has predominantly inflammatory action in orthodontic tooth movement with bone-resorbing effects thereby preventing EARR. However, Liu et al. detected IL-6 on both compression and tension sides of the teeth in mice after orthodontic force application. Hence, this association needs further validation by studies in other populations.
C-allele of IRAK1 (rs1509703) has been observed to have a protective effect against EARR whether in homozygous/heterozygous form in Portuguese Caucasians population.[28] T allele is the high-risk genotype for EARR and encodes leucine. As IRAK1 locus is influenced by X-inactivation, homozygotes CC have a similar expression of serine isoform as male hemizygotes for allele C. Due to skewed X chromosome inactivation; we may anticipate that some heterozygotes will also be protected. Similar studies in other ethnic populations should be performed to further validate the proven association.
A allele of IL-17A -197A/G (rs2275913) was observed to produce more IL-17 in vitro than those without the allele[65] and IL-17 plays a role in odontoclastogenesis.[40] But IL-17A (-197A/G; rs2275913; Promotor) was not observed to have a significant association with EARR in the Czech population.[42]
Different studies exploring the role of P2RX7 in EARR have yielded conflicting results. CC, CT genotype of rs208294 have been observed to be associated with EARR in Caucasians.[12] The gain of function variant of rs1718119 (GCT > ACT; Ala348Thr) has a protective role against EARR. Hence, the GG genotype is associated with EARR in the Portuguese population.[11] However, CG haplotype involving both SNPs was found to have an association with EARR in the Czech population. However, variant rs1718119 and another SNP rs2230912 were not found to be associated with EARR in Caucasians.[42] There may be more SNPs that may have an inductive/protective effect on EARR. Thus, whole gene sequencing is desired for exploring the role of P2RX7.
RANKL is a membrane-bound cytokine expressed in osteoblasts that induces osteoclast differentiation and activation, mediated by osteoclast expressed receptor RANK. RANK (TNFRSF11A; 18q21.2-21.3) SNP D18S64 was found to have association with EARR in US Caucasians[66] but rs1805034 did not associate with EARR in Portuguese.[11]
OPG is a decoy receptor for RANKL, inhibiting osteoclastogenesis and bone remodelling. Osteoprotegerin was not found to have an association with EARR in both conducted studies on Portuguese[11] and Czech Caucasians.[42] SNP rs3102735 (-163T/C; Promotor) in gene promoter region may affect transcription and has been associated with lower bone mineral density or higher frequency of fractures related to minor allele G. Its minor allele frequency has been observed to be low in Portuguese which might have resulted in non significant results. SNP rs2073618 (+1181 C/G; Lys3Asn; Exon1) and rs3102735 were not associated in Czech population.
Direct osteoclast activation by OPN has been found in Caucasians in the study by Iglesias-Linares et al.[29] This study suggests that inheritance of a specific allele of the OPN gene (rs9138 and rs11730582) could be a determinant of genetic susceptibility to EARR in this ethnic group. However, Linhartova et al. did not find any association of OPN with EARR in the Czech population.[42]
CASP1 did not show any significant association with EARR in a study conducted on US Caucasians.
Wnt showed a role in EARR in mice knockout study. It regulates cementum homeostasis and reduction in Wnt signaling has been associated with EARR.[59]
Tissue non-specific alkaline phosphatase, TNSALP (1p36.1-34) plays an important role in mineralization and cementum formation. Mice lacking functional TNSALP gene have defective acellular cementum formation and delayed tooth eruption. Tumor Necrosis Factor alpha, TNFα(6p21.3) levels elevate in gingival sulcus during orthodontic tooth movement. TNFα (-308, transcription start site) and TNSALP (AL215L) didn't show any significant association with EARR in US Caucasians. Al-Qawasmi et al. postulated that osteoclast maturation via RANK-mediated pathway is associated with EARR while that via TNF receptor is not associated with EARR.[66]
Vitamin D showed association with EARR in Brazilian population. Fontanna et al. suggested the association of Vitamin D receptor polymorphism (rs731236) with EARR. Genotype-containing C allele was found protective against root resorption in the Brazilian population.[49]
Population stratification and variation in frequencies of SNPs in different ethnicities must be taken into consideration. Genetic variants may interact with other variants in the same (or another) gene and environmental factors that influence the observed phenotype. Hence, a minor gene effect, ethnic diversity or linkage disequilibrium contributed to variations in different studies. Also, studies must be performed with sufficient statistical power to detect the association of a minor allele effect.
Presently studies apropos of orthodontic tooth movement have constraints (a) histologic analysis is restricted as teeth moved and/or surrounding bone cannot be analysed; (b) interindividual dissimilitude in mechanobiological responses; (c) not all factors influencing tooth movement have been unmasked. It is believed that once the molecular database is completed, the biomolecular therapeutic intervention will ensue which will pave our way to enhance the orthodontic tooth movement with minimal or no root resorption.
Recent literature suggests that EARR is multifactorial and clinical risk factors like maxillary premolar extraction, increased overjet and presence of dilacerated roots also pose high risk for development of EARR. Therefore, these factors must also be considered while doing orthodontic treatment.[67,68]
Conclusions
The outcome of this review shows that different gene polymorphisms may indicate the occurrence of EARR and also proposes certain recommendations for prospective researchers for further studies.
Timely and repeated estimation of cytokine levels at the site of localized force application must be performed concomitant with the determination of genetic polymorphisms to authenticate the proposed association between a potential mediator and EARR.
Genetics based studies along with other basic science research in the field might help to understand the exact nature of EARR, the influence of genetic inheritance and possibly lead to the prevention or even eradication of this phenomenon during orthodontic treatment.
Financial support and sponsorship
Nil.
Conflicts of interest
There are no conflicts of interest.
References
- 1.Brezniak N, Wasserstein A. Root resorption after orthodontic treatment Part 2 Literature review. Am J Orthod Dentofacial Orthop. 1993;103:138–46. doi: 10.1016/S0889-5406(05)81763-9. [DOI] [PubMed] [Google Scholar]
- 2.Kvam E. Scanning electron microscopy of tissue changes on the pressure surface of human premolars following tooth movement. Scand J Dent Res. 1972;80:357–68. doi: 10.1111/j.1600-0722.1972.tb00303.x. [DOI] [PubMed] [Google Scholar]
- 3.Brezniak N, Wasserstein A. Orthodontically induced inflammatory root resorption. Part II: Clinical aspects. Angle Orthod. 2002;72:180–4. doi: 10.1043/0003-3219(2002)072<0180:OIIRRP>2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 4.Hartsfield JK, Jr, Everett ET, Al-Qawasmi RA. Genetic factors in external apical root resorption and orthodontic treatment. Crit Rev Oral Bio Med. 2004;15:115–22. doi: 10.1177/154411130401500205. [DOI] [PubMed] [Google Scholar]
- 5.Rudolph CE. A comparative study in root resorption in permanent teeth. J Am Dent Assoc. 1936;23:822–6. [Google Scholar]
- 6.Harris EF, Butler MF. Patterns of incisor root resorption before and after orthodontic correction in cases with anterior open bites. Am J Orthod Dentofacial Orthop. 1992;101:112–9. doi: 10.1016/0889-5406(92)70002-R. [DOI] [PubMed] [Google Scholar]
- 7.Harris EF, Robinson QC, Woods MA. An analysis of causes of apical root resorption in patients not treated orthodontically. Quintessence Int. 1993;24:417–28. [PubMed] [Google Scholar]
- 8.Harris EF, Kineret SE, Tolley EA. A heritable component for external apical root resorption in patients treated orthodontically. Am J Orthod Dentofacial Orthop. 1997;111:301–9. doi: 10.1016/s0889-5406(97)70189-6. [DOI] [PubMed] [Google Scholar]
- 9.Taithongchai R, Sookkorn K, Killiany DM. Facial and dento-alveolar structure and the prediction of apical root shortening. Am J Orthod Dentofacial Orthop. 1996;110:296–302. doi: 10.1016/s0889-5406(96)80014-x. [DOI] [PubMed] [Google Scholar]
- 10.Killiany DM. Root resorption caused by orthodontic treatment: An evidence-based review of literature. Semin Orthod. 1999;5:128–33. doi: 10.1016/s1073-8746(99)80032-2. [DOI] [PubMed] [Google Scholar]
- 11.Pereira S, Lavado N, Nogueira L, Lopez M, Abreu J, Silva H. Polymorphisms of genes encoding P2X7R, IL-1B, OPG and RANK in orthodontic-induced apical root resorption. Oral Des. 2014;20:659–67. doi: 10.1111/odi.12185. [DOI] [PubMed] [Google Scholar]
- 12.Sharab LY, Morford LA, Dempsey J, Alencar-Falcao G, Mason A, Jacobson E, et al. Genetic and treatment related risk factors associated with external apical root resorption concurrent with orthodontia. Orthod Craniofac Res. 2015;18(Suppl 1):71–82. doi: 10.1111/ocr.12078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Massler M, Malone AJ. Root resorption in human permanent teeth: A roentgenographic study. Am J Orthod. 1954;40:619–33. [Google Scholar]
- 14.Massler M, Perreault J. Root resorption in the permanent teeth of young adults. J Dent Child. 1954;21:158–64. [Google Scholar]
- 15.Newman WG. Possible etiologic factors in external root resorption. Am J Orthod. 1975;67:522–39. doi: 10.1016/0002-9416(75)90298-5. [DOI] [PubMed] [Google Scholar]
- 16.Reitan K. Some factors determining the evaluation of forces in orthodontics. Am J Orthod. 1957;43:32–45. [Google Scholar]
- 17.Sameshima GT, Sinclair PM. Predicting and preventing root resorption: Part I. Diagnostic factors. Am J Orthod Dentofacial Orthop. 2001;119:505–10. doi: 10.1067/mod.2001.113409. [DOI] [PubMed] [Google Scholar]
- 18.Hartsfield JK., Jr Pathways in external apicalroot resorptionassociated with orthodontia. Orthod Craniofac Res. 2009;12:236–42. doi: 10.1111/j.1601-6343.2009.01458.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nicklin MJH, Weith A, Duff GW. A physical map of the region encompassing the human interleukin-1A, interleukin-1B, and interleukin-1 receptor antagonist genes. Genomics. 1994;19:382–4. doi: 10.1006/geno.1994.1076. [DOI] [PubMed] [Google Scholar]
- 20.Sutterwala FS, Ogura Y, Szczepanik M, Lara-Tejero M, Lichtenberger GS, Grant EP, et al. CriticalroleforNALP3/CIAS1/Cryopyrinininnateandadaptiveimmunitythrough itsregulationofcaspase-1. Immunity. 2006;24:317–27. doi: 10.1016/j.immuni.2006.02.004. [DOI] [PubMed] [Google Scholar]
- 21.Arend WP. Interleukin-1 receptor antagonist. A new member of interleukin 1 family. J Clin Invest. 1999;88:1445–51. doi: 10.1172/JCI115453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Al-Qawasmi RA, Hartsfield JK, Jr, Everett ET, Flury L, Liu L, Foroud TM, et al. Geneticpredisposition toexternalapicalroot resorption. Am J Orthod Dentofacial Orthop. 2003;123:242–52. doi: 10.1067/mod.2003.42. [DOI] [PubMed] [Google Scholar]
- 23.BastosLages EM, Drummond AF, Pretti H, Costa FO, Lages EJ, Gontijo AI, et al. Association of functional gene polymorphism IL-1beta in patients withexternalapicalroot resorption. Am J Orthod Dentofacial Orthop. 2009;136:542–6. doi: 10.1016/j.ajodo.2007.10.051. [DOI] [PubMed] [Google Scholar]
- 24.Iglesias-Linares A, Yanez-Vico RM, Ballesta-Mudarra S, Ortiz-Ariza E, Ortega-Rivera H, Mendoza-Mendoza A, et al. Postorthodontic external root resorption is associated with IL1 receptor antagonist gene variations. Oral Dis. 2012;18:198–205. doi: 10.1111/j.1601-0825.2011.01865.x. [DOI] [PubMed] [Google Scholar]
- 25.Al-Qawasmi RA, Hartsfield JK, Hartsfield JK, Jr, Everett ET, Weaver MR, Foroud TM, et al. Root resorption associated with orthodontic force in IL-1Beta knockout mouse. J Musculoskelet Neuronal Interact. 2004;4:383–5. [PubMed] [Google Scholar]
- 26.Gulden N, Eggermann T, Zerres K, Beer M, Meinelt A, Diedrich P. Imterleukin-1 polymorphism in relation to external apical root resorption. J Orofac Orthop. 2009;70:20–38. doi: 10.1007/s00056-009-8808-6. [DOI] [PubMed] [Google Scholar]
- 27.Tomoyasu Y, Yamaguchi T, Tajima A, Inoue I, Maki K. External apical root resorption and interleukin-1B gene polymorphism in japenese population. Orthod Waves. 2009;68:152–7. [Google Scholar]
- 28.Pereira S, Nogueira L, Canova F, Lopez M, Silva H. IRAK1 variant is protective for orthodontic-induced apical root resorption. Oral Dis. 2016;22:658–64. doi: 10.1111/odi.12514. [DOI] [PubMed] [Google Scholar]
- 29.Iglesias-Linares A, Sonnenberg B, Solano B, Yanez-Vico RM, Solano E, Lindauer SJ, et al. Orthodontically induced external apical root resorption in patients treated with fixed appliances vs removable aligners. Angle Orthod. 2017;87:3–10. doi: 10.2319/02016-101.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Linhartova P, Cernochova P, Izakovicova Holla L. IL1 gene polymorphisms in relation to external apical root resorption concurrent with orthodontia. Oral Dis. 2013;19:262–70. doi: 10.1111/j.1601-0825.2012.01973.x. [DOI] [PubMed] [Google Scholar]
- 31.Danis VA, Millington M, Hyland VJ, Grennan D. Cytokine production by normal human monocytes: Inter-subject variation and relationship to an IL-1 receptor antagonist (IL-1Ra) gene polymorphism. Clin Exp Immunol. 1995;99:303–10. doi: 10.1111/j.1365-2249.1995.tb05549.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hurme M, Santtila S. IL-1 receptor antagonist (IL-1Ra) plasma levels are co-ordinately regulated by both IL-1Ra and IL-1beta genes. Eur J Immunol. 1998;28:2598–602. doi: 10.1002/(SICI)1521-4141(199808)28:08<2598::AID-IMMU2598>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 33.Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2016;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 34.Gottipati S, Rao NL, Fung-Leung WP. IRAK1: A critical signaling mediator of innate immunity. Cell Signal. 2008;20:269–76. doi: 10.1016/j.cellsig.2007.08.009. [DOI] [PubMed] [Google Scholar]
- 35.Blanchard F, Duplomb L, Baud'huin M, Brounais F. The dual role of IL-6-type cytokines on bone remodeling and bone tumors. Cytokine Growth Factor Rev. 2009;20:19–28. doi: 10.1016/j.cytogfr.2008.11.004. [DOI] [PubMed] [Google Scholar]
- 36.Iwasaki K, Komaki M, Mimori K, Leon E, Izumi Y, Ishikawa I. IL-6 induces osteoblastic differentiation of periodontal ligament cells. J Dent Res. 2008;87:937–42. doi: 10.1177/154405910808701002. [DOI] [PubMed] [Google Scholar]
- 37.Kishimoto T. IL-6: From its discovery to clinical applications. Int Immunol. 2010;22:347–52. doi: 10.1093/intimm/dxq030. [DOI] [PubMed] [Google Scholar]
- 38.Madureira DF, de Albuquerque Taddei S, Abreu MHNG, Pretti H, Lages EMB, da Silva TA. Kinetics of Interleukin-6 and chemokine ligand 2 and 3 expression of periodontal tissues during orthodontic tooth movement. Am J Orthod Dentofacial Orthop. 2012;142:494–500. doi: 10.1016/j.ajodo.2012.05.012. [DOI] [PubMed] [Google Scholar]
- 39.Guo Y, He S, Gu T, Liu Y, Chen S. Genetic and clinical risk factors of root resorption associated with orthodontic treatment. Am J Orthod Dentofacial Orthop. 2016;150:283–9. doi: 10.1016/j.ajodo.2015.12.028. [DOI] [PubMed] [Google Scholar]
- 40.Nakano Y, Yamaguchi M, Shimizu M, Kikuta J, Yoshino T, Tanimoto Y, et al. Interleukin- 17 is involved in orthodontically induced inflammatory root resorption in dental pulp cells. Am J Orthod Dentofacial Orthop. 2015;148:302–9. doi: 10.1016/j.ajodo.2015.03.023. [DOI] [PubMed] [Google Scholar]
- 41.Hayashi N, Yamaguchi M, Nakajima R, Utsunomiya T, Yamamoto H, Kasai K. T-helper 17 cells mediate the osteo/odontoclastogenesis induced by excessive orthodontic forces. Oral Dis. 2012;18:375–88. doi: 10.1111/j.1601-0825.2011.01886.x. [DOI] [PubMed] [Google Scholar]
- 42.Linhartova PB, Cernochova P, Kastovsky J, Vrankova Z, Sirotkova M, Izakovicova Hola L. Genetic determinants and postorthodontic external apical root resorption in Czech children. Oral Diseases. 2017;23:29–35. doi: 10.1111/odi.12564. [DOI] [PubMed] [Google Scholar]
- 43.Sutton AL, MacDonald PN. Vitamin D: More than a “bone-a-fide” hormone. Mol Endocrinol. 2003;17:777–91. doi: 10.1210/me.2002-0363. [DOI] [PubMed] [Google Scholar]
- 44.Davideau JL, Lezot F, Kato S, Bailleul-Forestier I, Berdal A. Dental alveolar bone defects related to vitamin D and calcium status. J Steroid Biochem Mol Biol. 2004;89-90:615–8. doi: 10.1016/j.jsbmb.2004.03.117. [DOI] [PubMed] [Google Scholar]
- 45.Baldock PA, Thomas GP, Hodge JM, Baker SU, Dressel U, O'Loughlin PD, et al. Vitamin Daction and regulation of bone remodeling: Suppression of osteoclastogenesis by the mature osteoblast. J Bone Miner Res. 2006;21:1618–26. doi: 10.1359/jbmr.060714. [DOI] [PubMed] [Google Scholar]
- 46.Harada S, Mizoguchi T, Kobayashi Y, Nakamichi Y, Takeda S, Sakai S, et al. Dailyadministrationofeldecalcitol (ED-71), an activevitamin Danalog, increases bone mineral density by suppressing RANKL expression in mouse trabecular bone. J Bone Miner Res. 2012;27:461–73. doi: 10.1002/jbmr.555. [DOI] [PubMed] [Google Scholar]
- 47.Takahashi N, Udagawa N, Suda T. Vitamin D endocrine system and osteoclasts? BoneKEy Rep. 2014;3:495. doi: 10.1038/bonekey.2013.229. doi: 10.1038/bonekey. 2013.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Verway M, Bouttier M, Wang TT, Carrier M, Calderon M, An BS, et al. Vitamin D induces interleukin-1β expression: Paracrine macrophage epithelial signaling controls M. tuberculosis infection. PLoS Pathog. 2013;9:e1003407. doi: 10.1371/journal.ppat.1003407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fontana ML, de Souza CM, Bernardino JF, Hoette F, Hoette ML, Thum L, et al. Association analysis of clinical aspects and vitamin D receptor gene polymorphism with external apical root resorption in orthodontic patients. Am J Orthod Dentofacial Orthop. 2012;142:339–47. doi: 10.1016/j.ajodo.2012.04.013. [DOI] [PubMed] [Google Scholar]
- 50.Denhardt DT, Noda M, O'Regan AW, Pavlin D, Berman JS. Osteopontin as a means to cope with environmental insults: Regulation of inflammation, tissue remodeling, and cell survival. J Clin Invest. 2001;107:1055–61. doi: 10.1172/JCI12980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Weber GF. The metastasis gene osteopontin: A candidate target for cancer therapy. Biochim Biophys Acta. 2001;1552:61–85. doi: 10.1016/s0304-419x(01)00037-3. [DOI] [PubMed] [Google Scholar]
- 52.Iglesias-Linares A, Yanez-Vico RM, Moreno-Fernandez AM, Mendoza-Mendoza A, Orce-Romero A, Solano-Reina E. Osteopontin gene SNPs (rs9138, rs11730582) mediate susceptibility to external root resorption in orthodontic patients. Oral Dis. 2014;20:307–12. doi: 10.1111/odi.12114. [DOI] [PubMed] [Google Scholar]
- 53.Boyce BF, Xing L. The RANKL/RANK/OPG pathway. Curr Osteoporos Rep. 2007;5:98–104. doi: 10.1007/s11914-007-0024-y. [DOI] [PubMed] [Google Scholar]
- 54.Saki F, Karamizadeh Z, Nasirabadi S, Mumm S, McAlister WH, Whyte MP. Juvenile Paget's disease in an Iranian kindred with vitamin D deficiency and novel homozygous TNFRSF11B mutation. J Bone Miner Res. 2013;28:1501–8. doi: 10.1002/jbmr.1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tyrovola JB, Spyropoulos MN, Makou M, Perrea D. Root resorption and the OPG/RANKL/RANK system: A mini review. J Oral Sci. 2008;50:367–76. doi: 10.2334/josnusd.50.367. [DOI] [PubMed] [Google Scholar]
- 56.Abass SK, Hartsfield JK, Jr, Al-Qawasmi RA, Everett ET, Foroud TM, Roberts WE. Inheritance of susceptibility to root resorption associated with orthodontic force in mice. Am J Orthod Dentofacial Orthop. 2008;134:742–50. doi: 10.1016/j.ajodo.2007.04.035. [DOI] [PubMed] [Google Scholar]
- 57.Goldring SR, Goldring MB. Eating bone or adding it: The Wnt pathway decides. Nat Med. 2007;13:133–4. doi: 10.1038/nm0207-133. [DOI] [PubMed] [Google Scholar]
- 58.Glass DA, 2nd, Bialek P, Ahn JD, Starbuck M, Patel MS, Clevers H, et al. Canonical Wnt signaling in differentiated osteoblasts controls osteoclast differentiation. Dev Cell. 2005;8:751–64. doi: 10.1016/j.devcel.2005.02.017. [DOI] [PubMed] [Google Scholar]
- 59.Lim WH, Liu B, Hunter DJ, Cheng D, Mah S, Helms JA. Downregulation of Wnt causes root resorption. Am J Orthod DentofacialOrthop. 2014;146:337–45. doi: 10.1016/j.ajodo.2014.05.027. [DOI] [PubMed] [Google Scholar]
- 60.Iglesias-Linares A, Yanez-Vico RM, Ballesta S, Ortiz-Ariza E, Mendoza-Mendoza A, Perea E, et al. Interleukin-1 gene cluster SNPs (rs1800587, rs1143634) influences post-orthodontic root resorption in endodontic and their contralateral vital control teeth differently. Int Endod J. 2012;45:1018–26. doi: 10.1111/j.1365-2591.2012.02065.x. [DOI] [PubMed] [Google Scholar]
- 61.Iglesias-Linares A, Yanez-Vico RM, Ortiz-Ariza E, Ballesta S, Mendoza-Mendoza A, Perea E, Solano-Reina E. Postorthodontic external root resorption in root-filled teeth is influenced by Interleukin-1β polymorphism. J Endod. 2012;38:283–7. doi: 10.1016/j.joen.2011.12.022. [DOI] [PubMed] [Google Scholar]
- 62.Iglesias-Linares A, Yanez-Vico RM, Ballesta-Mudarra S, Ortiz-Ariza E, Mendoza-Mendoza A, Perea-Perez E, et al. Interleukin-1 receptor antagonist (IL1RN) genetic variations condition post-orthodontic external root resorption in endodontically treated teeth. Histol Histopathol. 2013;28:767–73. doi: 10.14670/HH-28.767. [DOI] [PubMed] [Google Scholar]
- 63.Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM, et al. dbSNP: The NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–11. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Diehl SR, Wang Y, Brooks CN, Burmeister JA, Califano JV, Wong S, et al. Linkage disequilibrium of interleukin-1 genetic polymorphisms with early onset periodontitis. J Periodontol. 1999;70:418–30. doi: 10.1902/jop.1999.70.4.418. [DOI] [PubMed] [Google Scholar]
- 65.Borilova Linhartova P, Kastovsky J, Lucanova S, Bartova J, Poskerova H, Vokurka J, et al. IL-17A gene variability in patients with type 1 diabetes mellitus and chronic periodontitis; its correlation with IL-17 levels and the occurrence of periodontopathic bacteria. Mediators Inflamm. 2016 doi: 10.1155/2016/2979846. doi: 101155/2016/2979846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Al-Qawasmi RA, Hartsfield JK, Jr, Everett ET, Flury LL, Liu L, Foroud TM, et al. Genetic predisposition to external apical root resorption in orthodontic patients: Linkage of chromosome-18 marker. J Dent Res. 2003;82:356–60. doi: 10.1177/154405910308200506. [DOI] [PubMed] [Google Scholar]
- 67.Fernandes LQP, Figueiredo NC, Montalvany Antonucci CC, Lages EMB, Andrade I, Jr, Capelli Junior J. Predisposing factors for external apical root resorption associated with orthodontic treatment. Korean J Orthod. 2019;49:310–8. doi: 10.4041/kjod.2019.49.5.310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sondeijker CFW, Lamberts AA, Beckmann SH, Kuitert RB, van Westing K, Persoon S, et al. Development of a clinical practice guideline for orthodontically induced external apical root resorption. Eur J Orthod. 2020;42:115–24. doi: 10.1093/ejo/cjz034. [DOI] [PMC free article] [PubMed] [Google Scholar]