Abstract
OBJECTIVE:
To measure the frequency of diseases related to latent tuberculosis infection (LTBI) and tuberculosis (TB), we assessed the agreement between diagnosis codes for TB or LTBI in electronic health records (EHRs) and insurance claims for the same person.
METHODS:
In a US population-based, retrospective cohort study, we matched TB-related Systematized Nomenclature of Medicine–Clinical Terms (SNOMED CT) EHR codes and International Statistical Classification of Diseases, 10th Revision, Clinical Modification (ICD-10-CM) claims codes. Furthermore, LTBI was identified using a published ICD-based algorithm and all LTBI- and TB-related SNOMED CT codes.
RESULTS:
Of people with the 10 most frequent TB-related claim codes, 50% did not have an exact-matched EHR code. Positive tuberculin skin test was the most frequent unmatched EHR code and people with the 10 most frequent TB EHR codes, 40% did not have an exact-matched claim code. The most frequent unmatched claim code was TB screening encounter. EHR codes for LTBI matched to claims codes for TB testing; pulmonary TB; and nonspecific, positive or adverse tuberculin reaction.
CONCLUSION:
TB-related EHR codes and claims diagnostic codes often disagree, and people with claims codes for LTBI have unexpected EHR codes, indicating the need to reconcile these coding systems.
Keywords: TB, latent tuberculosis, insurance, health, claims
Abstract
CONTEXTE:
On n’a pas évalué l’agrément entre codes diagnostiques de la tuberculose (TB) ou de l’infection latent à Mycobacterium tuberculosis (ITL) dans les dossiers de santé électroniques (EHR) et demandes de règlement pour le même patient.
MÉTHODE:
Etude rétrospective de cohorte en population aux Etats-Unis des EHR avec les données commerciales liées aux demandes de règlement. Athena® Software a été utilisé pour apparier les codes de la nomenclature systématisée de médecine (Systematized Nomenclature of Medicine–Clinical Terms, SNOMED CT) et d’Classification statistique internationale des maladies et des problèmes de santé connexes, 10e révision (ICD-10-CM) lies à la TB. De plus, l’ITL a été identifiée grâce à un algorithme publiée basée sur ICD et à tous les codes SNOMED CT liés à l’ITL et à la TB.
RÉSULTATS:
Parmi les personnes ayant les 10 codes ICD-10-CM les plus fréquents liés à la TB, 50% n’avaient pas de code SNOMED exactement apparié. Le test cutané à la tuberculine a été le code SNOMED le plus fréquemment non apparié. Parmi les personnes ayant les 10 codes ICD-10-CM les plus fréquents liés à la TB, 40% n’avaient pas de code ICD-10-CM exactement apparié. Le code ICD-10-CM non apparié le plus fréquent a été l’examen de dépistage pour la TB. Les codes SNOMED CT de l’ITL étaient appariés aux codes ICD-10-CM pour le test TB; la TB pulmonaire; et les effets secondaires non spécifiques, positifs ou indésirables, de la réaction tuberculinique.
CONCLUSION:
Les codes de diagnostic SNOMED CT et ICD-10-CM liés à la TB sont souvent en désaccord et les personnes ayant des codes ICD-10-CM pour l’ITL ont des codes SNOMED CT inattendus, témoignant d’un besoin de réconcilier ces systèmes de codage.
Abstract
MARCO DE REFERENCIA:
No se ha evaluado la concordancia de los códigos para la tuberculosis (TB) y la infección latente por Mycobacterium tuberculosis (ILT) en los registros sanitarios electrónicos (EHR) y los reclamos de gastos al seguro de enfermedad para la misma persona.
MÉTODO:
En un estudio de cohortes retrospectivo poblacional en los Estados Unidos, de los EHR que contaban con datos vinculados sobre los reclamos de gastos a las aseguradoras comerciales, se utilizó el programa Athena® Software para cotejar los códigos de los términos relacionados con la tuberculosis de la SNOMED CT (por Términos clínicos de la Nomenclatura sistematizada de la medicina) y de la Modificación clínica de la décima edición de la Clasificación Estadística Internacional de Enfermedades (ICD-10-CM). Además, se reconoció la ILT mediante el uso de un algoritmo publicado basado en la ICM y todos los códigos de la SNOMED CT relacionados con la ILT y la TB.
RESULTADOS:
De las personas con los diez códigos más frecuentes relacionados con la TB de la ICD-10-CM, el 50% no tenía un código SNOMED exacto correspondiente. La reacción tuberculínica positiva fue el código SNOMED CT que carecía con mayor frecuencia de un código emparejado. De las personas con los diez códigos más frecuentes relacionados con la TB de la SNOMED CT, el 40% no tenía un código ICD-10-CM exacto correspondiente. El código ICD-10-CM que con mayor frecuencia carecía de un código emparejado fue “examen de pesquisa especial para TB respiratoria”. Los códigos SNOMED CT para ILT correspondían a los códigos ICD-10-CM de pruebas para TB, TB respiratoria y reacción inespecífica, positiva o adversa a la prueba con tuberculina.
CONCLUSIÓN:
Con frecuencia, los códigos diagnósticos relacionados con la TB de la SNOMED CTy la ICD-10-CM difieren y las personas con códigos ICD-10-CM para ILT presentan códigos SNOMED CT imprevistos, lo cual destaca la necesidad de armonizar los sistemas de codificación.
TUBERCULOSIS (TB) IS AN AIRBORNE infectious disease caused by the bacterium Mycobacterium tuberculosis. Signs and symptoms of active pulmonary TB infection can include chronic cough, chest pain, fatigue, fever, weight loss, and others.1 Latent tuberculosis infection (LTBI) is a state of persistent immune response to stimulation by M. tuberculosis antigens without active TB. An estimated 5% of people in the United States have LTBI,2 whereas active TB has been estimated at 2.8 cases per 100 000 persons.3 Untreated, 5–10% of those with newly detected M. tuberculosis infection will experience active TB (caused by the bacterium’s reactivation) during the person’s lifetime.4 Although LTBI treatment is effective in preventing TB reactivation,5 the majority of US residents with LTBI are untreated6 for multiple reasons.7
Insurance claims data have been used to estimate the frequency of LTBI- and TB-related diagnosis.8 However, the clinical accuracy of these estimates might be decreased because of coding practices favoring conditions with the highest reimbursement.9 Because ICD-10-CM codes for LTBI were not used before release of the 2020 International Statistical Classification of Diseases, 10th Revision, Clinical Modification (ICD-10-CM),10 Stockbridge et al.11 published a claims-based algorithm for identifying LTBI cases. Alternative indicators of LTBI diagnoses that include information from electronic health records (EHRs) might be more accurate than diagnoses based entirely on claims data. To assess coding precision, we matched LTBI- and TB-related codes from EHRs and claims among a commercially insured US population.
METHODS
We conducted a US population-based, retrospective cohort study to analyse the IBM® MarketScan® Explorys® Claims-EHR Data Set (CED) (IBM Corporation, Armonk, NY, USA). This database contains EHRs of outpatient visits and inpatient stays during January 1, 2002–December 31, 2017 (from IBM Explorys® Universe12; IBM Corporation) linked with claims paid during January 1, 1999–September 30, 2018 (from IBM® MarketScan® Research Databases13 [IBM Corporation, Armonk, New York]) for approximately 5 million people. CED contains health insurance data for people with employer-sponsored private health insurance, their spouses, and their dependents who lived throughout the United States. Medical diagnoses, outpatient drug prescriptions, and the corresponding claims are linked by unique enrollee identification numbers, which facilitates longitudinal analyses. The data broker describes the potential benefits of the data source as 1) including a better understanding of LTBI and TB history, epidemiology, and progression; 2) determining the economic impact of LTBI or TB diagnoses and treatment for select populations; and 3) identifying the patients most likely to benefit from LTBI or TB treatment.
TB-related conditions in paid claims
We used the Tennessee TB Elimination Program’s TB ICD-10 Codes Cheat Sheet14 to identify TB-related ICD-10-CM codes. We excluded codes for symptoms and clinical encounters not specific for TB (Supplemental Table S1; all supplemental data located at: https://figshare.com/articles/Supplemental_files_for_Electronic_health_records_and_claims_diagnostic_code_agreement_for_tuberculosis/11344025).
LTBI in paid claims
We modified a previously defined LTBI claims algorithm11 to allow 12-months of claims adjudication instead of 6-months. Our algorithm identifies persons initiating isoniazid (INH) for LTBI treatment by using the National Drug Codes (NDC), Current Procedural Terminology (CPT) codes,15 Healthcare Common Procedure Coding System (HCPC)16 codes, ICD-10-CM,17 and the International Classification of Diseases Clinical Modifications, 9th Revision, diagnosis codes.17 Our algorithm required continuous enrollment for ≥1 year before the INH index date to ≥1 year after the INH index date to account for possible lags in EHR diagnosis reporting and outpatient prescription filling. We also updated relevant NDCs (Supplemental Table S2).
TB-related conditions in EHRs and in paid claims for the same person
To find TB-related conditions in EHR (Systematized Nomenclature of Medicine – Clinical Terms [SNOMED CT] codes, version 2019030118) in agreement with those in claims data (ICD-10-CM codes), we used Athena Software® (Athena Software, Waterloo, ON, Canada) for standardized vocabularies.19 SNOMED CT contains codes that indicate the connections among clinical events, symptoms, signs, tests, diagnoses, and other standard concepts. The codes are used to record the need for, the delivery of, and the results of clinical care, in standard terms that have the same meaning for all users, globally, of different electronic health record systems. ICD-10-CM contains codes for classifying diseases, conditions, and clinical procedures into standard categories that are used globally to support reimbursement for health insurance claims and to report vital events to public health authorities.20 We matched ICD-10-CM codes with the corresponding SNOMED CT codes. We used all matched SNOMED CT codes when >1 matched an ICD-10-CM code. Next, we searched the claims data for ICD-10-CM codes of interest. We restricted the search to persons with claims after October 1, 2015 (the date of official US ICD-10-CM adoption21) and to persons with continuous enrollment ≥1 year before the ICD-10-CM code of interest and who had SNOMED CT diagnosis data during the period of continuous enrollment (because not all persons with enrollment data have EHR data). We collected the top 250 SNOMED CT codes that occurred ≤1 year before the ICD-10-CM code of interest. We recorded the number of persons with matched SNOMED CT codes and the number of persons with the most frequent (on the basis of the number of persons with a particular SNOMED CT code) unmatched TB-related SNOMED CT codes.
To substantiate the ICD-10-CM to SNOMED CT code findings, we used Athena Software to map EHR-derived SNOMED CT codes to claims-derived ICD-10-CM codes of interest. For SNOMED CT codes with multiple matched ICD-10-CM codes, we selected the ICD-10-CM code on the TB ICD-10 Codes Cheat Sheet.14 We restricted our analysis to persons with continuous enrollment ≥1 year after the SNOMED CT code of interest from October 1, 2015, forward. We collected the top 250 ICD-10-CM codes that occurred ≤1 year after the SNOMED CT code of interest. We recorded the frequency of persons with matched ICD-10-CM diagnosis codes. We also recorded the most frequent TB-related code by person count for unmatched codes.
LTBI diagnoses in EHRs and in paid claims for the same person
We identified persons with LTBI in the EHRs by searching for LTBI-relevant SNOMED CT codes and validated the results by reverse mapping from SNOMED CT codes to codes for persons who met the LTBI algorithm. For persons who met the LTBI algorithm, we collected the top 250 SNOMED CT codes that occurred ≤1 year before the INH index date. We recorded all TB-related SNOMED CT codes by person count because ICD-10-CM codes for LTBI did not exist at the time of analysis.
For the LTBI and INH-LTBI algorithm SNOMED CT codes, we searched the EHRs from October 1, 2015, forward and restricted the search to continuous enrollment for ≥1 year after the SNOMED CT code of interest. We collected the top 250 ICD-10-CM diagnosis codes that occurred within 1 year after the SNOMED CT code of interest. By using Athena Software, we recorded counts of persons with the matched SNOMED CT code to the ICD-10-CM codes and the top unmatched ICD-10-CM TB-related diagnosis codes.
See Supplemental Figure S1 for a diagram of the look back or look forward times for the cohorts. Approval by an institutional review board was not required because data were collected and analyzed for this project as part of routine TB surveillance; therefore, the project is not considered research involving human subjects.
RESULTS
Of the 5 348 618 people with data in CED, 1 506 480 (28.2%) had EHR diagnosis and claims data that met the inclusion criteria. The characteristics of the final cohort (Supplement Table S3) included a mean age of 44 years and a female majority (57.0%; n = 859 173).
TB-related conditions in EHRs and in claims data for the same person
Half of the 10 most frequently used TB-related claims codes were for people who did not have a code in their EHR that matched exactly, but had other TB-related codes (Table 1). Of the unmatched SNOMED CT codes from the 10 most common claims codes, 40.0% (n = 4) of the codes were for a positive Mantoux TB test (SNOMED CT: 268376005). Similar results were noted for the full matching analysis of TB ICD-10-CM to SNOMED CT codes (Supplemental Table S4).
Table 1.
Number of people with the 10 most common TB-related ICD-10-CM claims codes and number and percentage of those with Athena-matched and most common unmatched SNOMED CT codes,* 2014–2017
| Description of ICD-10-CM (code) | Personsn† | Outcome of ICD-10-CM to SNOMED CT matching‡ | Description of matched and unmatched SNOMED CT (code) | Personsn | ICD-10 match %§ |
|---|---|---|---|---|---|
| Encounter for screening for respiratory TB (Z11.1) | 9196 | Matched | TB screening (171126009) | 0 | 0 |
| Most common unmatched | TB screening status (finding) (429599001) | 3201 | 34.8 | ||
| Nonspecific reaction to TST without active TB (R76.11) | 1741 | Matched | Nonspecific TST reaction (441846005) | 54 | 3.1 |
| Most common unmatched | Mantoux: positive (finding) (268376005) | 671 | 38.5 | ||
| Contact with and (suspected) exposure to TB (Z20.1) | 377 | Matched | Exposure to Mycobacterium tuberculosis (event) (444507004) | 0 | 0 |
| Most common unmatched | TB screening status (finding) (429599001) | 14 | 3.7 | ||
| Nonspecific reaction to cell-mediated immunity measurement of gamma interferon antigen response without active TB (R76.12) | 346 | Matched | Nonspecific TST reaction (441846005) | 124 | 35.8 |
| Most common unmatched | TB screening status (finding) (429599001) | 28 | 8.1 | ||
| Personal history of TB (Z86.11) | 268 | Matched | Past history of clinical finding (417662000) | 0 | 0 |
| Most common unmatched | Mantoux: positive (finding) (268376005) | 13 | 4.9 | ||
| TB of lung (A15.0) | 167 | Matched | Pulmonary TB (154283005) | 43 | 25.7 |
| Most common unmatched | Mantoux: positive (finding) (268376005) | 20 | 12.0 | ||
| TB of spine (Z18.01) | 95 | Matched | TB of vertebral column (35984006) | 15 | 15.8 |
| Most common unmatched | Pulmonary TB (154283005) | 3 | 3.2 | ||
| TB of skin and subcutaneous tissue (A18.4) | 48 | Matched | TB of skin and subcutaneous tissue (271423008) | 12 | 25.0 |
| Most common unmatched | TB of skin (disorder) (66986005) | 4 | 8.3 | ||
| Most common unmatched | TB of subcutaneous cellular tissue (disorder) (8250007) | 4 | 8.3 | ||
| Adverse effect of antimycobacterial drugs (T37.1×5¶) | 15 | Matched | Late effect of poisoning due to drug (14546008) | 0 | 0 |
| Matched | Antimycobacterial agent adverse reaction (293069003) | 0 | 0 | ||
| Most common unmatched | Infiltrative lung TB (disorder) (186175002) | 1 | 6.7 | ||
| Most common unmatched | Pulmonary TB (154283005) | 1 | 6.7 | ||
| Other respiratory TB (A15.8) | 15 | Matched | Respiratory TB (700272008) | 0 | 0 |
| Most common unmatched | Mantoux: positive (finding) (268376005) | 5 | 33.3 |
IBM® MarketScan® Explorys® Claims-EHR Data Set (CED) (IBM Corporation, Armonk, New YorkNY, USA) Database from October 1, 2015. TB codes from the Tennessee TB Elimination Program’s TB ICD-10 Codes Cheat Sheet.
Persons with indicated ICD-10-CM code with continuous enrollment and electronic health record diagnosis up to 1-year before ICD-10-CM code.
Matched refers to the SNOMED CT code identified by Athena® Software (Athena Software, Waterloo, ON, Canada) as equal to the ICD-10-CM code indicated. Most common unmatched refers to the most frequent or tied TB-related SNOMED CT code for persons with the listed ICD-10-CM code not matched by Athena Software.
Codes from the Tennessee TB Elimination Program’s TB ICD-10 Codes Cheat Sheet.
The appropriate seventh character is to be added to each code: A, initial encounter; D, subsequent encounter; and S, sequelae.
TB = tuberculosis; ICD-10-CM = International Statistical Classification of Diseases, 10th Revision, Clinical Modification; SNOMED CT = Systematized Nomenclature of Medicine–Clinical Terms; TST = tuberculin skin test; EHR = electronic health record.
LTBI diagnoses from INH-LTBI algorithm in EHRs for the same person
Among people who met the INH-LTBI algorithm (n = 1293), 423 (32.7%) had EHR data ≥1 year before the INH index date. The most frequent TB-related SNOMED CT code was adverse tuberculin reaction (25.3%; n = 107) (Figure). Two of the codes (pulmonary TB [SNOMED CT: 154283005] and nonspecific tuberculin test reaction [SNOMED CT: 441846005]) had matched claims codes. The other codes, adverse tuberculin reaction (SNOMED CT: 292093003), positive Mantoux result (SNOMED CT: 268376005), and TB screening status (SNOMED CT: 429599001) did not have matching EHR and claims codes.
Figure.
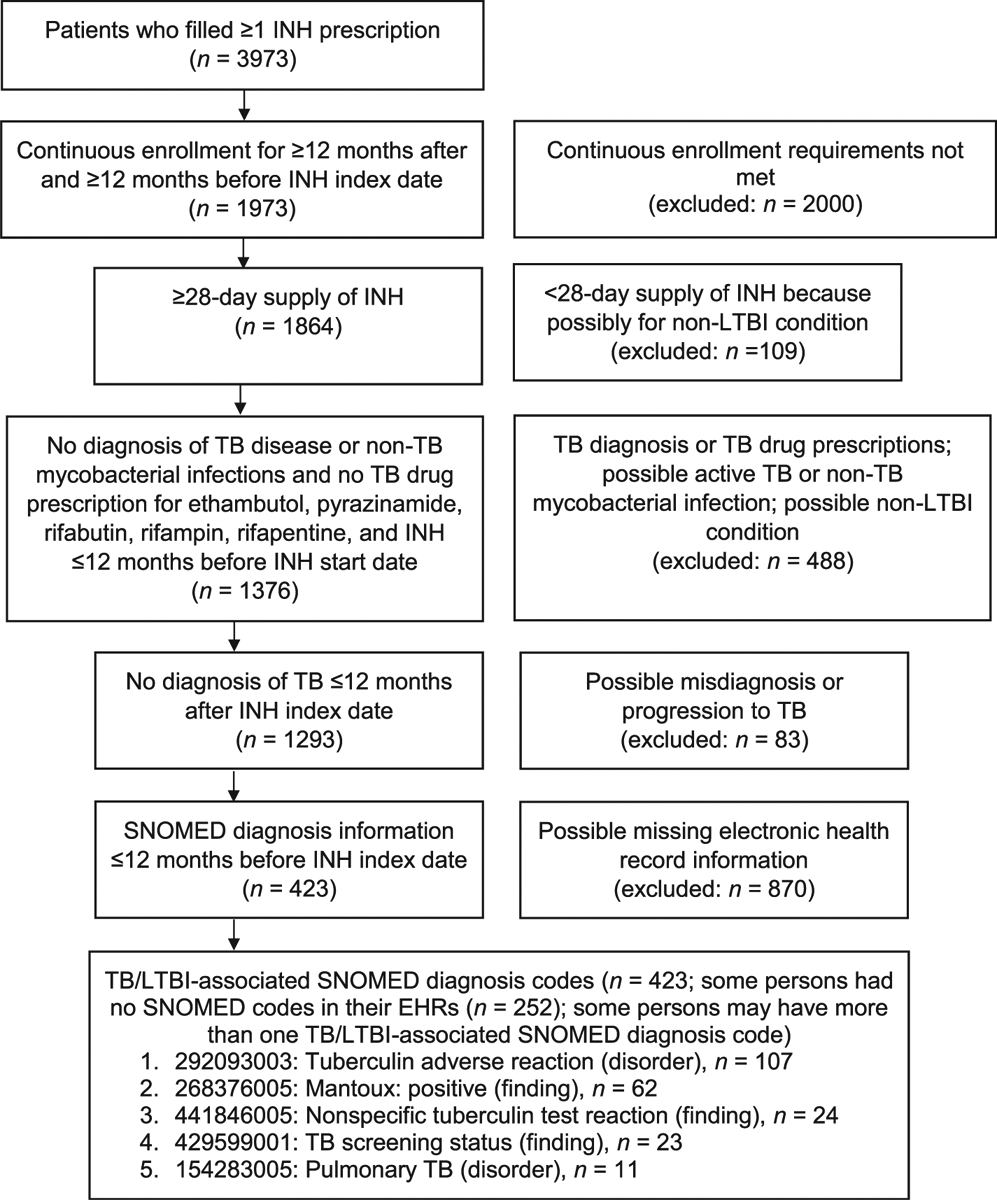
Patient inclusion and exclusion criteria for the LTBI treatment cohort and TB/LTBI-associated SNOMED CT diagnosis codes, 2002–2017 (n = 423). INH = isoniazid; LTBI = latent tuberculosis infection; TB = tuberculosis; SNOMED CT = Systematized Nomenclature of Medicine–Clinical Terms; EHR = electronic health record.
TB-related conditions defined by EHR codes identified by matches to claims codes
Of the top 10 most frequent SNOMED CT codes, 40.0% (n = 4) of the conditions did not have people with matching ICD-10-CM codes (Table 2). Of the unmatched TB claims codes among the top 10 SNOMED CT codes measured, the ICD-10-CM code encounter for TB screening (Z11.1) was the most frequent at 40.0% (n = 4).
Table 2.
Number of people with any of 10 most common TB-related conditions with SNOMED CT codes in EHRs and number and percentage of those with matched and most common unmatched ICD-10-CM codes in claims data,* 2015–2017
| Description (SNOMED CT code) | Personsn† | SNOMED CT to ICD-10-CM matched‡ or top unmatched TB code | Description (ICD-10-CM code§) | Personsn | ICD-10 match, %** |
|---|---|---|---|---|---|
| Late effect of poisoning due to drug (14546008) | 152 | Matched | Adverse effect of rifampicins (T36.6×5¶) | 0 | 0 |
| Matched | Adverse effect of antimycobacterial drugs (T37.1×5¶) | 0 | 0 | ||
| Pulmonary TB (154283005) | Most common unmatched | — | — | — | |
| 122 | Matched | TB of lung (A15.0) | 39 | 32.0 | |
| Most common unmatched | Nonspecific reaction to TST without active TB (R76.11) | 44 | 36.1 | ||
| Nonspecific TST reaction (441846005) | 116 | Matched | Nonspecific reaction to TST without active TB (R76.11) | 41 | 35.3 |
| Matched | Nonspecific reaction to cell-mediated immunity measurement of gamma interferon antigen response without active TB (R76.12) | 87 | 75.0 | ||
| Most common unmatched | Encounter for screening for respiratory TB (Z11.1) | 10 | 8.6 | ||
| Drug resistance (disorder) (31438003) | 45 | Matched | Resistance to single antimycobacterial drug (Z16.341) | 0 | 0 |
| Matched | Resistance to multiple antimycobacterial drugs (Z16.342) | 0 | 0 | ||
| Most common unmatched | — | — | — | ||
| TB of vertebral column (35984006) | 24 | Matched | TB of spine (A18.01) | 16 | 66.7 |
| Most common unmatched | TB of lung (A15.0) | 2 | 8.3 | ||
| Most common unmatched | Tuberculoma of brain and spinal cord (A17.81) | 2 | 8.3 | ||
| TB (56717001) | 16 | Matched | Other musculoskeletal TB (A18.09) | 0 | 0 |
| Matched | TB of other endocrine glands (A18.82) | 0 | 0 | ||
| Matched | TB of other sites (A18.89) | 0 | 0 | ||
| Most common unmatched | Nonspecific reaction to TST without active TB (R76.11) | 1 | 6.3 | ||
| Most common unmatched | Encounter for screening for respiratory TB (Z11.1) | 1 | 6.3 | ||
| TB of skin and subcutaneous tissue (271423008) | 8 | Matched | TB of skin and subcutaneous tissue (A18.4) | 3 | 37.5 |
| Most common unmatched | Encounter for screening for respiratory TB (Z11.1) | 4 | 50.0 | ||
| Antimycobacterial agent adverse reaction (293069003) | 2 | Matched | Adverse effect of antimycobacterial drugs (T37.1×5#) | 0 | 0 |
| Most common unmatched | Encounter for screening for respiratory TB (Z11.1) | 1 | 50.0 | ||
| Unmatched | TB of spine (A18.01) | 1 | 50.0 | ||
| Most common unmatched | Tuberculoma of brain and spinal cord (A17.81) | 1 | 50.0 | ||
| TB of heart (302131003) | 1 | Matched | TB of heart (A18.84) | 1 | 100 |
| Most common unmatched | — | — | — | ||
| Rifampicin adverse reaction (293075007) | 1 | Matched | Adverse effect of rifampicins (T36.6×5#) | 1 | 100 |
| Most common unmatch | — | — | — |
IBM® MarketScan® Explorys® Claims-EHR Data Set (CED) (IBM Corporation, Armonk, NY, USA) Database from October 1, 2015. TB codes from the Tennessee TB Elimination Program’s TB ICD-10 Codes Cheat Sheet.
With continuous enrollment and EHR diagnosis ≥12 months after SNOMED CTcode. Ten SNOMED CTcodes had an n = 1. To restrict to 10 codes for the table, we chose 2 of the 10 with the greatest n for persons with the SNOMED CT diagnosis (Supplementary Table S5).
Mapping conducted using Athena Software® (Waterloo, ON, Canada) (https://www.ohdsi.org/analytic-tools/athena-standardized-vocabularies/).
For SNOMED CTcodes with >1 standard to nonstandard matching ICD-10-CM codes, the ICD-10-CM code that matched in the TB ICD-10 codes cheat sheet was recorded as the mapping code.
Codes from the Tennessee TB Elimination Program’s TB ICD-10 Codes Cheat Sheet.
The appropriate seventh character is to be added to each code: A, initial encounter; D, subsequent encounter; and S, sequelae.
TB = tuberculosis; SNOMED CT = Systematized Nomenclature of Medicine–Clinical Terms; EHR = electronic health record; ICD-10-CM = International Statistical Classification of Diseases, 10th Revision, Clinical Modification; TST = tuberculin skin test.
Comparing LTBI definitions in EHRs with claims data
When we reversed mapped SNOMED CT to ICD-10-CM codes for the TB-related SNOMED CT codes for people who met the claims INH-LTBI algorithm, Athena Software matched 3 of the 9 codes (Table 3). The 3 matched ICD-10-CM codes were for TB of the lung (A15.0) or nonspecific reaction to TB tests (R76.11 and R76.12). The top unmatched ICD-10-CM codes were for an encounter for TB screening (Z11.1) and nonspecific reaction to tuberculin skin test without TB (R76.11).
Table 3.
Number of people with any of 10 most common LTBI-related conditions with SNOMED CT codes in EHRs and number and percentage of those with matched and most common unmatched ICD-10-CM codes in claims,* 2015–2017
| Description (SNOMED CT Code) | Personsn† | SNOMED CT to ICD-10-CM map, matched‡ or top unmatched TB code | Description (ICD-10-CM Code) | Personsn | ICD-10 match %§ |
|---|---|---|---|---|---|
| TB screening status (finding) (429599001) | 2817 | Match | —¶ | — | — |
| Most common unmatched | Encounter for screening for respiratory TB (Z11.1) | 2180 | 77.4 | ||
| Mantoux: positive (finding) (268376005) | 868 | Match | — | — | — |
| Most common unmatched | Nonspecific reaction to TST without active TB (R76.11) | 534 | 61.5 | ||
| TST adverse reaction (disorder) (292093003) | 320 | Match | — | — | — |
| Most common unmatched | Nonspecific reaction to TST without active TB (R76.11) | 214 | 66.9 | ||
| Pulmonary TB (154283005) | 122 | Matched | TB of lung (A15.0) | 39 | 32 |
| Most common unmatched | Nonspecific reaction to TST without active TB (R76.11) | 44 | 36.1 | ||
| Nonspecific TST reaction (441846005) | 116 | Matched | Nonspecific reaction to TST without active TB (R76.11) | 41 | 35.3 |
| Matched | Nonspecific reaction to cell-mediated immunity measurement of gamma interferon antigen response without active TB (R76.12) | 87 | 75 | ||
| Most common unmatched | Encounter for screening for respiratory TB (Z11.1) | 10 | 8.6 | ||
| Inactive TB of lung (428697002) | 0 | Match | — | — | — |
| Most common unmatched | — | — | — | ||
| Inactive TB (11999007) | 0 | Match | — | — | — |
| Most common unmatched | — | — | — | ||
| On TB chemoprophylaxis (414940009) | 0 | Match | — | — | — |
| Most common unmatched | — | — | — | ||
| Infection due to M. tuberculosis (373576009) | 0 | Match | — | — | — |
| Most common unmatched | — | — | — |
Latent TB SNOMED CT codes, including those generated from the isoniazid-latent TB infection claims algorithm.
With continuous enrollment and electronic health record diagnosis ≥12 months after SNOMED CT code.
Mapping conducted with Athena Software® (Waterloo, ON, Canada) (https://www.ohdsi.org/analytic-tools/athena-standardized-vocabularies/).
Codes from the Tennessee TB Elimination Program’s TB ICD-10 Codes Cheat Sheet.
— indicates the SNOMED CT term cannot be matched to ICD-10-CM code.
LTBI = latent TB infection; SNOMED CT = Systematized Nomenclature of Medicine–Clinical Terms; EHR = electronic health record; ICD-10-CM = International Statistical Classification of Diseases, 10th Revision, Clinical Modification; TST = tuberculin skin test; TB = tuberculosis.
When we reversed mapped SNOMED CT to ICD-10-CM codes for LTBI-related SNOMED CT codes, none of the LTBI description SNOMED CT codes had matching ICD-10-CM codes. We also did not find anyone within the EHR data set who had an LTBI-related SNOMED CT code.
DISCUSSION
We observed low levels of agreement between TB-related SNOMED CT codes from EHRs and ICD-10-CM codes for claims data. Specifically, we observed incongruent coding among people identified by an LTBI algorithm applied to claims data. The most frequent TB-related SNOMED CT code for persons identified by the LTBI algorithm was adverse tuberculin reaction, a relatively rare event.22 Some people with LTBI by claims data had a SNOMED CT code for pulmonary TB in their EHR, contradicting the LTBI claims data algorithm, which was intended to remove people with evidence of TB. Also, none of the SNOMED CT LTBI codes were used in EHRs, although claims data indicated treatment for LTBI.
We also observed incongruent coding among persons with TB-related claims (Table 1). Approximately half of people with TB-related ICD-10-CM codes in claims data did not have a SNOMED CT code from their EHR that Athena Software matched exactly. However, some of the people with unmatched claim codes had SNOMED CT codes that appeared similar. For example, the ICD-10-CM code for TB screening did not have anyone that matched exactly, instead the most commonly used TB-related SNOMED CT code was for TB screening status. Other people with unmatched claim codes had dissimilar or no TB-related SNOMED CT code. For example, people with the ICD-10-CM code other respiratory tuberculosis had Mantoux: positive as the most common TB-related SNOMED CT code.
Mismatched claims codes and EHR codes might result from: inadequate software translation between coding systems; SNOMED CT codes allowing for the use of compositional grammar when no 1:1 mapping exists; inadequate translation of EHRs to SNOMED CT codes; missing EHR data; missing claims data; or billing bias. Software for mapping between ICD-10-CM and SNOMED CT codes is improving through an iterative process with input from users.19,23 We used the Athena Software because it allowed translation from SNOMED CT to ICD-10-CM and ICD-10-CM to SNOMED CT, whereas the National Library of Medicine (US) software only translated from SNOMED CT to ICD-10-CM codes. We hope our study will encourage further improvements in TB-related coding.
Inadequate translations of EHR data to SNOMED CT codes might be from the natural language processing (NLP) system used. NLP uses computational methods to code unstructured text as an alternative to manual abstraction.24 Word sense disambiguation failures affect the feasibility of detecting LTBI or TB with NLP.25 For example, ambiguous terms (eg, suggests) applied to suspected or ruled-out cases have been reported to be translated into a disease diagnosis.26
Although continuous enrollment and EHR data were a requirement, people in our study might have received TB-related SNOMED CT codes from health care providers who did not contribute EHR data but submitted claims. Alternatively, ICD-10-CM codes might be missing when billing for conditions described in EHRs are not submitted or paid. Also, coding practices favoring conditions with the highest reimbursement might lead to ICD-10-CM coding inaccuracy.12
Strengths and limitations of this approach
Use of a large national data source of commercially insured patients with linked EHRs strengthens our study because it provides results regarding private-sector diagnoses for persons with TB-related conditions in the United States. Despite the large data source, however, TB-related codes were uncommon, and claims data directly linked to specific patient visits in EHRs were unavailable. Our requirement of continuous enrollment for 1 year partially mitigated the resulting information gap.
Information gaps existed for people who had sought coverage outside of EHR providers, covered services but without an insured amount, financial data that were withheld, or uncovered services. The data are subject to coding misclassification by health care providers, or administrators or by insurance administrators. Proximate event diagnoses were limited to the top 250 events. Because conditions of interest were uncommon, we were unable to create a validation data set for our reverse mapping. The data lack information regarding the intention of the listed diagnostic code, either as a rule-out or as a diagnosis code, resulting in the possibility of misclassification. Furthermore, incongruent coding among people identified by an LTBI algorithm applied to claims data could indicate the algorithm is not identifying true cases.
CONCLUSION
Our results indicate that further analyses of EHR data and claims coding for TB-related conditions is justified. However, because of the incongruent LTBI or TB codes used for SNOMED CT and ICD-10-CM, it is vital that continued efforts are made to improve the translation between physician notes, SNOMED CT and insurance claims data.
Supplementary Material
Acknowledgments
The authors specifically acknowledge the contributions of the Division of Health Informatics and Surveillance’s Partnership and Evaluation Branch at the Centers for Disease Control and Prevention (CDC; Atlanta, GA, USA) for providing financial support and technical assistance with CED data and C K Smith for editorial assistance.
Footnotes
Publisher's Disclaimer: Disclaimer: The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention.
Conflicts of interest: none declared.
References
- 1.Campbell IA, Bah-Sow O. Pulmonary tuberculosis: diagnosis and treatment. BMJ 2006; 332: 1194–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Miramontes R, Hill AN, Yelk Woodruff RS, et al. Tuberculosis infection in the United States: prevalence estimates from the National Health and Nutrition Examination Survey, 2011–2012. PLoS One 2015; 10: e0140881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Talwar A, Tsang CA, Price SF, et al. Tuberculosis—United States, 2018. Am J Transplant 2019; 19: 1582–1588. [Google Scholar]
- 4.World Health Organization. Global tuberculosis report, 2018. Geneva, Switzerland: WHO, 2018. [Google Scholar]
- 5.Efficacy of various durations of isoniazid preventive therapy for tuberculosis: five years of follow-up in the IUAT trial. International Union Against Tuberculosis Committee on Prophylaxis. Bull World Health Organ 1982; 60: 555–564. [PMC free article] [PubMed] [Google Scholar]
- 6.Bennett DE, Courval JM, Onorato I, et al. Prevalence of tuberculosis infection in the United States population: the national health and nutrition examination survey, 1999–2000. Am J Respir Crit Care Med 2008; 177 : 348–355. [DOI] [PubMed] [Google Scholar]
- 7.Hill L, Blumberg E, Sipan C, et al. Multi-level barriers to LTBI treatment: a research note. J Immigr Minor Health 2010; 12: 544–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Iqbal SA, Isenhour CJ, Mazurek G, et al. Factors associated with latent tuberculosis infection treatment failure among patients with commercial health insurance—United States, 2005–2016. J Public Health Manag Pract 2019. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Serden L, Lindqvist R, Rosen M. Have DRG-based prospective payment systems influenced the number of secondary diagnoses in health care administrative data? Health Policy 2003; 65: 101–107. [DOI] [PubMed] [Google Scholar]
- 10.Centers for Disease Control and Prevention/National Center for Health Statistics. 2020 release of ICD-10-CM. Hyattsville, MD, USA: US Department of Health and Human Services, CDC/NCHS, 2019. https://www.cdc.gov/nchs/icd/icd10cm.htm. Accessed November 2019. [Google Scholar]
- 11.Stockbridge EL, Miller TL, Carlson EK, et al. Tuberculosis prevention in the private sector: using claims-based methods to identify and evaluate latent tuberculosis infection treatment with isoniazid among the commercially insured. J Public Health Manag Pract 2018; 24: e25–e33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaelber DC, Foster W, Gilder J, et al. Patient characteristics associated with venous thromboembolic events: a cohort study using pooled electronic health record data. J Am Med Inform Assoc. 2012; 19: 965–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Adamson D, Chang S, Hansen LG. Health research data for the real world: the MarketScan databases. Thomson Medstat, 2008. http://patientprivacyrights.org/wp-content/uploads/2011/06/Thomson-Medstat-white-paper.pdf. Accessed November 2019. [Google Scholar]
- 14.Tennessee TB Elimination Program. TB ICD-10 codes cheat sheet; Nashville, TN, USA: Tennessee Department of Public Health, 2015. https://sntc.medicine.ufl.edu/Files/Resources/TB%20ICD-10%20Codes%20Cheat%20Sheet%20(TTBEP%2011-5-15).pdf. Accessed November 13, 2019. [Google Scholar]
- 15.American Medical Association. CPT 2017, professional edition. Chicago, IL, USA: American Medical Association Press, 2016. [Google Scholar]
- 16.American Medical Association. HCPCS level II 2018, professional edition. Chicago, IL: American Medical Association Press, 2017. [Google Scholar]
- 17.World Health Organization. International statistical classification of diseases and related health problems, 10th revision. Geneva, Switzerland: WHO, 2016. [Google Scholar]
- 18.SNOMED International. International Health Terminology Standards Organisation. London, UK: https://www.snomed.org/. Accessed November 2019. [Google Scholar]
- 19.Observational Health Data Sciences and Informatics (OHDSI). ATHENA standardized vocabularies. https://www.ohdsi.org/analytic-tools/athena-standardized-vocabularies/. Accessed November 2019.
- 20.Rodrigues JM, Schulz S, Mizen B, et al. Is the application of SNOMED CT concept model sufficiently quality assured? AMIA Annu Symp Proc 2017; 2017: 1488–1497. [PMC free article] [PubMed] [Google Scholar]
- 21.Centers for Disease Control and Prevention/National Center for Health Statistics. International classification of diseases, 10th revision, clinical modification (ICD-10-CM). Hyattsville, MD, USA: US Department of Health and Human Services, CDC/NCHS; https://www.cdc.gov/nchs/icd/icd10cm.htm. Accessed November 2019. [Google Scholar]
- 22.Nayak S, Acharjya B. Mantoux test and its interpretation. Indian Dermatol Online J 2012; 3): 2–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.US National Library of Medicine (NLM). Interactive map-assisted generation of ICD codes (I-MAGIC). Bethesda, MD, USA: NLM, https://imagic.nlm.nih.gov/imagic/code/map. Accessed November 2019. [Google Scholar]
- 24.Friedman C, Hripcsak G. Natural language processing and its future in medicine. Acad Med. 1999; 74(8): 890–895. [DOI] [PubMed] [Google Scholar]
- 25.Xu H, Markatou M, Dimova R, et al. Machine learning and word sense disambiguation in the biomedical domain: design and evaluation issues. BMC Bioinformatics 2006; 7: 334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carrell DS, Halgrim S, Tran DT, et al. Using natural language processing to improve efficiency of manual chart abstraction in research: the case of breast cancer recurrence. Am J Epidemiol 2014; 179(6): 749–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


