Abstract
DNA polymerase β (Pol β) is an essential mammalian enzyme involved in the repair of DNA damage during the base excision repair (BER) pathway. In hopes of faithfully restoring the coding potential to damaged DNA during BER, Pol β first uses a lyase activity to remove the 5′-deoxyribose phosphate moiety from a nicked BER intermediate, followed by a DNA synthesis activity to insert a nucleotide triphosphate into the resultant 1-nucleotide gapped DNA substrate. This DNA synthesis activity of Pol β has served as a model to characterize the molecular steps of the nucleotidyl transferase mechanism used by mammalian DNA polymerases during DNA synthesis. This is in part because Pol β has been extremely amenable to X-ray crystallography, with the first crystal structure of apoenzyme rat Pol β published in 1994 by Dr. Samuel Wilson and colleagues. Since this first structure, the Wilson lab and colleagues have published an astounding 267 structures of Pol β that represent different liganded states, conformations, variants, and reaction intermediates. While many labs have made significant contributions to our understanding of Pol β, the focus of this article is on the long history of the contributions from the Wilson lab. We have chosen to highlight select seminal Pol β structures with emphasis on the overarching contributions each structure has made to the field.
Introduction
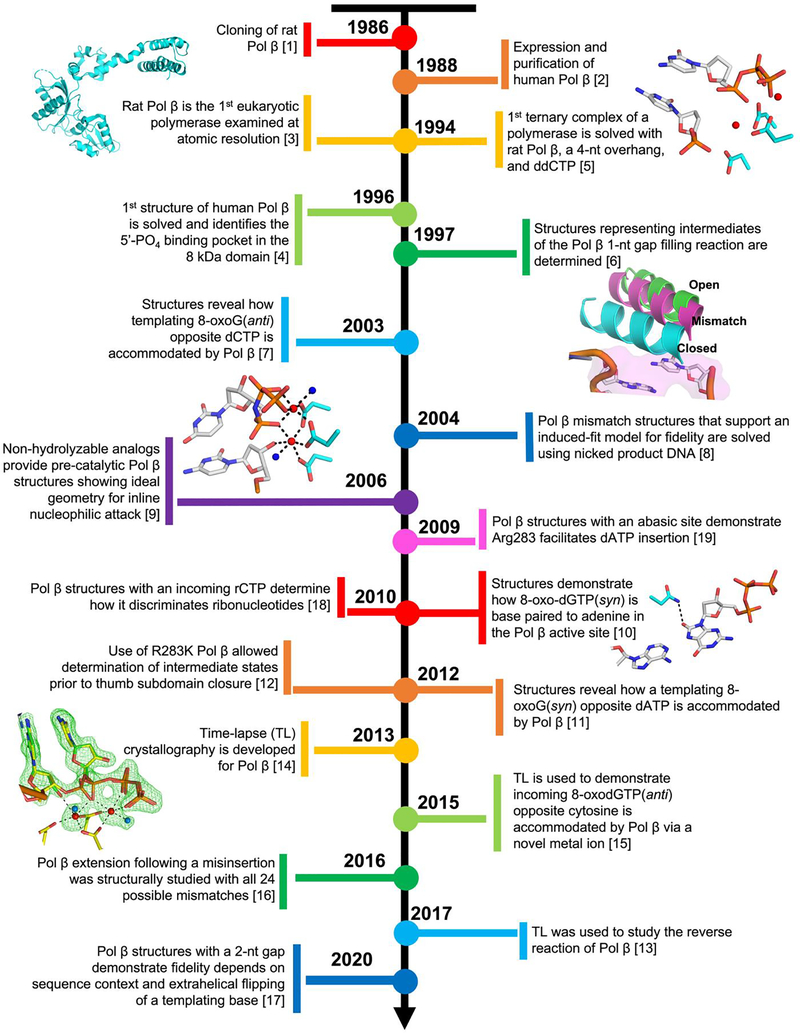
For over 40 years Dr. Samuel Wilson and colleagues have elegantly characterized DNA polymerase β (Pol β) at a biological and molecular level [20]. Marking the beginning of the structural characterization of Pol β, the Wilson lab cloned, over-expressed, and purified rat and human Pol β [1, 2]. Since then, Wilson and collaborators have gone on to publish over 200 articles on Pol β and will be forever associated with this powerful model mammalian DNA polymerase. Pol β is an essential mammalian enzyme that synthesizes DNA in short gaps during DNA repair. As the smallest eukaryotic DNA polymerase, equipped with the structural simplicity of only two domains and a low molecular weight, Pol β is an ideal candidate for X-ray crystallography. Consequently, Wilson and colleagues’ original ternary substrate Pol β complex structure was the first of any polymerase. This made Pol β the first mammalian DNA polymerase to be examined at atomic resolution [3, 5], and Pol β has continued to serve as a model DNA polymerase for structure function studies of the near-universal nucleotidyl transferase mechanism used by DNA polymerases during DNA synthesis. The adage that structure dictates function rings true, as the Wilson lab has significantly contributed to defining the molecular mechanism used by Pol β via numerous X-ray crystallization studies spanning several decades. The aim of this mini-review is to present a timeline (Figure 1) of select seminal Pol β structures contributed by Wilson and colleagues.
Figure 1.
A timeline of the seminal Pol β structures published by Wilson and colleagues [1–19].
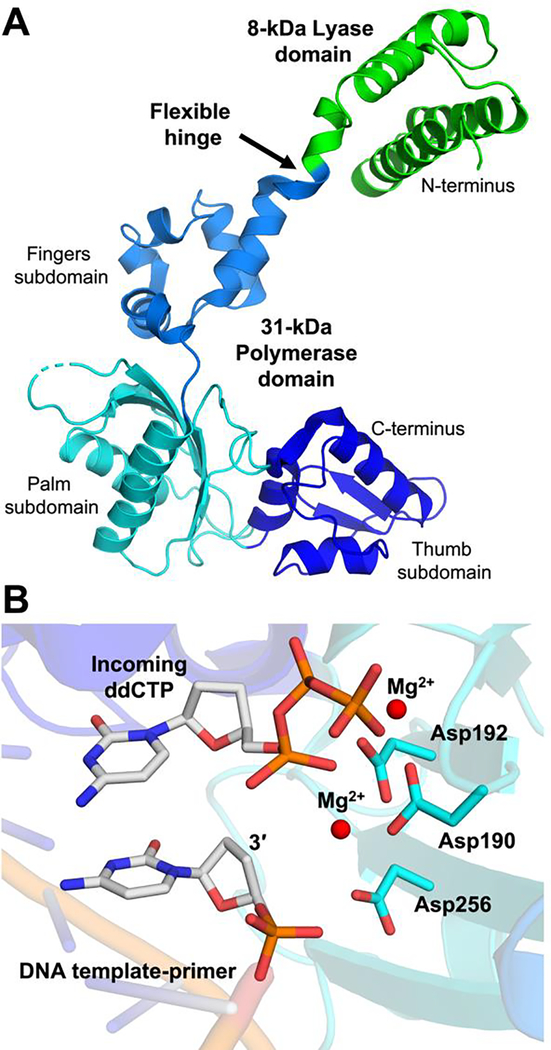
Pol β is composed of two specialized domains. The smaller 8 kDa N-terminal domain contains the 5′-deoxyribose phosphate (dRP) lyase activity, and the 31 kDa C-terminal domain contains the polymerase activity responsible for DNA synthesis (Figure 2A). These enzymatic functions represent two essential steps during the base excision repair (BER) pathway, which is responsible for excising and replacing incorrect (e.g. uracil) as well as damaged (e.g. abasic sites, 8-oxoguanine, 7-methlyguanine) bases generated from alkylation, oxidation, deamination, and depurination (for reviews see [21–24]). In 1994, the Wilson and Kraut labs collaborated to publish several structures of rat Pol β in two co-published Science articles [3, 5]. First, it was revealed that the 31 kDa polymerase domain is composed of fingers, palm, and thumb subdomains arranged to form a DNA binding channel reminiscent of the previously published polymerase domains of the Klenow fragment of E. coli DNA polymerase 1, HIV-1 reverse transcriptase, and bacteriophage T7 RNA polymerase [3]. The ternary substrate complex of Pol β exhibited characteristics similar to the model proposed for the nucleotidyl transferase reaction suggested by Beese and Steitz [25]. Additionally, an apoenzyme structure (Pol β alone) of the entire 39 kDa enzyme revealed the 8 kDa lyase domain to be disjointed from the 31 kDa C-terminal domain by a flexible hinge at the tip of the fingers subdomain (Figure 2A) [3].
Figure 2.
Structural snapshots of rat Pol β. A The extended structure of apoenzyme rat Pol β (PDB 1BPD) is shown with the 8 kDa lyase domain in green and the 31 kDa polymerase domain in blue. Domains, subdomains, and the flexible hinge are indicated. B The active site of the rat Pol β ternary complex (Pol β:DNA:ddCTP, PDB 2BPF) is shown. Key active site features are indicated.
The accompanying 1994 paper, also published in Science, provided the first ternary substrate complex: rat Pol β, a DNA template with a 4-nt overhang/dideoxy-terminated primer, and an incoming dideoxycytidine triphosphate (ddCTP) [5]. Of note, ddCTP is a nucleoside analog that targets the reverse transcriptase of human immunodeficiency virus (HIV) and is used to treat AIDS [26]. Once incorporated into a primer strand, ddC terminates DNA synthesis, thus preventing further elongation. This chain termination results in both its anti-HIV properties and its utility as a tool to obtain crystal structures of pre-catalytic polymerase ternary complexes (Pol β:DNA:dNTP). When these Pol β ternary complex structures are compared with the structure of the apo enzyme, the most apparent differences consist of large repositioning of the 8 kDa lyase domain, which goes from an extended conformation in the apoenzyme structure to a more compact donut-like conformation in the ternary complex structures. Closer examination of the Pol β ternary complex revealed that the active site clusters the attacking 3’-end of the elongating primer, ddCTP phosphates, and two Mg2+ ions around the catalytic triad, Asp190, Asp192, and Asp256 (Figure 2B). Importantly, these novel structural insights pointed toward a two-metal ion mechanism of nucleotidyl transfer that is utilized by all polymerases, in agreement with that postulated by other labs around the same time [27–33].
Just a few years later, in 1996, Kraut and Wilson again collaborated to obtain and report the first crystal structures of human Pol β and a 5-nt gapped substrate [4]. The structures exhibited a different space group with the polymerase domain bound to the blunt-end of the DNA while the lyase domain was complexed to the 5’-phosphate in the [5]. This binary DNA complex structure also revealed the thumb subdomain to be in an open position, compared to the rat ternary complex. Importantly, this led to a hypothesis that thumb subdomain closure upon nucleotide binding may represent the enigmatic rate-limiting conformational change that had been observed in pre-steady-state kinetic studies performed on several polymerases [34–40]. Moreover, both the lyase and polymerase domains were shown to contain Helix-hairpin-Helix (HhH) DNA-binding elements, providing insight into how these newly discovered motifs facilitate DNA binding for not only Pol β, but also a broad range of other DNA binding proteins [41]. Around the same time, Wilson and colleagues employed the human Pol β system to determine a ternary complex structure of the alanine mutant R283A (Arg283 is in hydrogen bonding distance to the templating nucleotide base) [42]. This structure, combined with fidelity assays and other minor groove DNA binding mutants, indicated that interactions between Pol β and the templating base are required for both high catalytic efficiency and nucleotide discrimination [42–44].
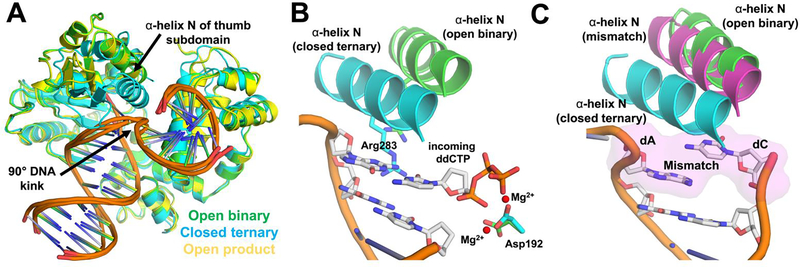
About a year later, new crystal structures captured several distinct steps in the 1-nt gap-filling reaction of human Pol β during BER [6]. These include a binary complex of Pol β bound to gapped DNA substrate, a ternary complex of Pol β bound to gapped DNA and an incoming ddCTP, and a Pol β binary product complex bound to nicked DNA (Figure 3A). These structures determined that upon binding ddCTP to the binary Pol β:DNA complex, the thumb subdomain rotates into a closed conformation to bring α-helix N in contact with the nascent base pair (Figure 3B). This movement of the thumb subdomain triggers additional conformational changes which assemble the active site for catalysis. In contrast to the previous rat ternary complex structure containing a DNA overhang and ddCTP [5], the 1-nt gapped DNA complex demonstrated that the 8 kDa domain binds to the 5’-phosphate in the DNA gap in a manner similar to that observed with the 5-nt gap and interacts with the thumb subdomain of the Pol β polymerase domain. Moreover, the nicked DNA product complex shows that the thumb subdomain returns to the open conformation, thus facilitating dissociation of Pol β from the 3’ nicked DNA (i.e. the Pol β reaction product, Figure 3A). These findings suggested that Pol β may enhance fidelity via an induced fit mechanism in which correct base pairing between the template and the incoming dNTP induces proper alignment of catalytic groups for catalysis, but incorrect base pairing misaligns reactive atoms. The structures also revealed that Pol β binds both gapped and nicked DNA with a 90° kink occurring precisely at the 5’- phosphodiester linkage of the templating residue (Figure 3A). If the DNA was not kinked in this way, contact between the thumb subdomain and dNTP-template base pair, presumably important for fidelity, would be impossible.
Figure 3.
Structures of human Pol β during 1-nt gap-filling support an induced-fit model for fidelity. A An overlay of the open binary complex of Pol β bound to gapped DNA (green, PDB 1BPX), a closed ternary complex of Pol β bound to gapped DNA and ddCTP (cyan, PDB 1BPY), and an open Pol β binary product complex bound to nicked DNA (yellow, PDB 1BPZ). α-helix N, and the 90° kink of the DNA are indicated. B A focused-view of the closed ternary structure (cyan protein, gray/orange DNA) showing rotation of α-helix N into a closed conformation to contact the nascent base pair and assemble the active site upon nucleotide binding. The position of α-helix N and Asp192 in the open binary conformation are indicated (green). C The structure of an A:C mismatched base pair is shown with a magenta surface representation for the mismatched base pairs (magenta protein and gray/orange DNA, PDB 1TV9). The position of α-helix N in the open binary (green) and closed ternary (cyan) conformations are indicated for comparison.
In 2003, the Wilson lab reported structures of Pol β with DNA mismatches in the active site to provide additional insights into DNA polymerase fidelity [8]. This was achieved utilizing nicked product DNA that allowed for the trapping of mispairs within the nascent base pair binding pocket. The position of the mismatch primer terminus in the nucleotide binding pocket deviated significantly from previous structures of the correct nucleotide bound to Pol β, mentioned above [6]. Specifically, these structures determined that mismatched base pairs lack the hydrogen bonding seen with matched base pairs, and alternatively the bases partially overlap and are stabilized by protein side-chain interactions. This staggered arrangement of the mispaired base pairs hinders the thumb subdomain repositioning, which is believed to be required for catalysis (Figure 3C). Instead, they observed a partially closed thumb subdomain conformation with a mismatched base pair, which is an intermediate position between the active closed (ternary with correct dNTP) and inactive open (binary) conformations. While previous observations merely suggested an induced-fit model for fidelity, these structures, specifically the altered protein/DNA interactions and conformations associated with the mismatched base pairs relative to those with a matched base pair in the active site, strongly supported the induced-fit model for fidelity.
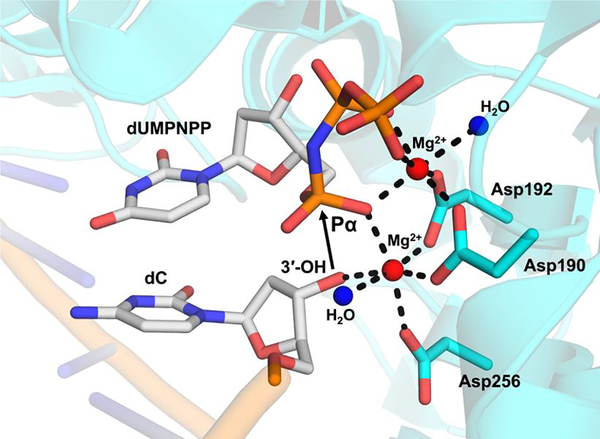
The Pol β structures representing reaction intermediates obtained prior to 2006 utilized DNA substrates lacking a primer terminal 3’-OH to prevent catalysis. This resulted in many of the structures also missing the catalytic Mg2+ ion and consequently they display distorted active site geometry. Therefore, the missing 3’-OH and catalytic metal ion placed limitations on the mechanistic insight each structural snapshot could provide. To overcome this limitation, in 2006 Wilson and colleagues were the first to utilize a non-hydrolyzable deoxynucleotide analog with a nitrogen substitution at the bridging oxygen between Pα and Pβ, dUMPNPP [9]. The resulting structure of a pre-catalytic intermediate provided direct evidence for the coordination of the 3’-OH of the primer terminus with the catalytic Mg2+ to promote an octahedral geometry facilitating inline nucleophilic attack on Pα (dNTP), Figure 4. Not only did this study provide new insights into the DNA polymerase mechanism, but it also provided a detailed analysis of polymerase active site metal coordination. For example, they showed that Na+ is able to bind in the catalytic binding site, distorting the active site geometry and influencing the sugar pucker of the primer terminus. This results in an increase in the distance between reactive atoms 3.4 Å (Mg2+) to 4.7 Å (Na+). Cumulatively, this elegant use of a non-hydrolyzable analog provided direct evidence for the role of the catalytic metal in establishing the position of the 3’-OH for nucleophilic attack.
Figure 4.
Non-hydrolyzable analog dUMPNPP ternary substrate complex of a pre-catalytic complex with ideal metal octahedral geometry (PDB 2FMS). The protein is represented in cyan, and DNA in gray/orange. Mg2+ octahedral geometry is indicated by dashed lines. The water molecules (blue spheres), Mg2+ ions (red spheres), and catalytic triad are indicated. An arrow is shown between the attacking 3’-OH of the primer terminus and Pα of dUMPNPP.
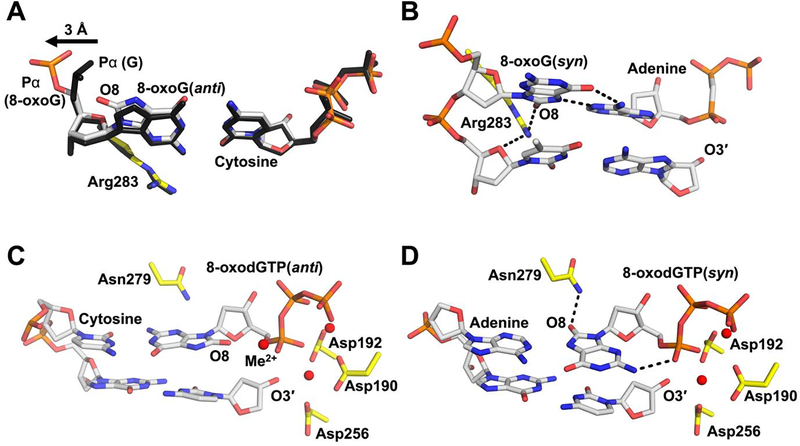
Wilson and colleagues have also made significant contributions to our understanding of how DNA polymerases accommodate and replicate past DNA damage. One of the most common forms of DNA damage encountered by a DNA polymerase is 8-oxo-7,8-dihydro-2’-deoxyguanosine (8-oxoG). 8-oxoG is generated by reactive oxygen species attack of the C8 position to generate an adducted oxygen at C8 and protonation of N7 [45]. This results in the ability of 8-oxoG to stably base pair with either cytosine in the anti-conformation or adenine in the syn-conformation [46]. Importantly, the mutagenic base pairing between 8-oxoG(syn) and adenine results in a G:C to T:A transversion mutation [47] and this mutation signature has been shown to be associated with multiple human diseases [21]. In the first seminal 8-oxoG study from the Wilson lab in 2003, they determined that when a templating 8-oxoG(anti) base pairs with an incoming dCTP the adducted oxygen at the C8 position (O8) is accommodated by the 5’-phosphate of the modified templating base flipping 180° to alleviate a clash between O8 of 8-oxoG(anti) and the non-bridging oxygens of the phosphate backbone (Figure 5A) [7].
Figure 5.
Pre-catalytic complexes showing the various strategies used by Pol β to accommodate the oxidized DNA lesion, 8-oxoG. A 8-oxoG(anti) in the templating position opposite dCTP (PDB 3RJI) is shown overlaid with the non-damaged guanine(anti) opposite dCTP (black, PDB 2FMP). B 8-oxoG(syn) in the templating position opposite dATP is shown (PDB 3RJF). C An incoming 8-oxodGTP(anti) opposite cytosine is shown with the additional metal near Pα indicated by Me2+ (PDB 4UBC). D An incoming 8-oxodGTP(syn) opposite adenine is shown (PDB code 4UAW). Metals are shown in red, Pol β in yellow, and DNA in gray/orange, except where noted.
In a follow-up 2012 study by the Wilson group, this dynamic flipping of the templating 8-oxoG phosphate backbone was observed in the absence of an incoming nucleotide (i.e. binary DNA complex). The resulting density for the templating nucleotide indicated that 8oxoG adopted both the anti- and syn-conformations [11]. This indicated that the nature of the incoming nucleotide (i.e. dATP or dCTP) can stabilize 8-oxoG in either the syn- or anti- conformation, respectively. Also, in this 2012 study, a ternary complex of Pol β:8-oxoG(syn):dATP was determined. In this structure Arg283 was observed within hydrogen bonding distance of O8 and was therefore proposed to stabilize the syn-conformation of 8-oxoG (Figure 5B). Cumulatively, these studies and others provided molecular insight into how 8-oxoG is accommodated in the active site of Pol β to alleviate steric clashes with the adducted oxygen at the C8 position during mutagenic and non-mutagenic bypass
Free deoxynucleotides are even more susceptible to nucleobase oxidation than duplex DNA [48, 49], and incorporation of oxidized nucleotide triphosphates into the genome by DNA polymerases during replication and repair leads to decreased genomic stability [15, 49–52]. One commonly generated damaged nucleotide is 8-oxo-dGTP. To understand how 8-oxo-dGTP is inserted into the genome, the Wilson lab determined the first structures of an incoming 8-oxo-dGTP base pairing to templating adenine [10] and cytosine [15] through two seminal publications in 2010 and 2015, respectively. The 2010 publication determined that when base pairing to adenine, the incoming 8-oxo-dGTP(syn) is stabilized through a hydrogen bonding interaction between Asn279 and O8, Figure 5D [10]. Additionally, there is an intramolecular hydrogen bond between N2 and the non-bridging oxygen on Pα. In a follow-up 2015 study, Wilson and colleagues observed that when 8-oxo-dGTP(anti) is opposite cytosine the potential clash between the adducted O8 and Pα is alleviated by an additional metal ion near Pα, Figure 5C [15]. This is in contrast to the displacement of the backbone phosphate observed with a templating 8-oxoG and likely results from the lack in flexibility around the triphosphate binding site within the Pol β active site (i.e. Pα of the incoming nucleotide must be precisely positioned). Overall, these structural snapshots provided insight into how modified nucleotides are accommodated in the polymerase active site to hasten catalysis.
To further probe the polymerase mechanism, in 2012 the Wilson lab published structures of intermediate nucleotide-binding states that occur prior to thumb subdomain closure [12]. These ternary complex structures of Pol β with correct and incorrect incoming nucleotides bound to the open conformation were determined using the R283K Pol β variant to destabilize the closed complex. Specifically, lysine substitution at position 283 disrupts interactions unique to the closed conformation, thus altering the equilibrium between the open and closed conformations of Pol β. These structures revealed several new things about the Pol β DNA synthesis reaction, including: 1) Watson-Crick hydrogen bonding between the incoming dNTP and templating base is assessed upon initial complex formation and; 2) the nucleotide bound states represent intermediate metal coordination states. In addition, the structures indicated that the triphosphate of the incoming dNTP requires rearrangement prior to thumb subdomain closure. These insights pointed to how Pol β deters misinsertion and increases fidelity prior to closure.
Adapting a new structural technique utilized by the labs of Wei Yang for DNA polymerase η and Katsuhiko Murakami for RNA polymerases [53, 54], the Wilson lab developed time-lapse crystallography with Pol β to further probe DNA polymerase fidelity [14]. This approach uses natural substrates to overcome the limitations of artificially trapping intermediates with nucleotide analogs such as a dideoxy-terminated primer or nonhydrolyzable incoming nucleotide. The result is high-resolution structural determinations representing snapshots before, during, and after dNTP insertion. Time-lapse crystallography was first used by the Wilson lab to capture and compare key steps throughout the reaction pathway for insertion of correct and incorrect nucleotides, published in 2013 in Cell [14, 55]. This set of fifteen unique structures allowed for the observation of previously unappreciated molecular adjustments at the active site that accelerate correct insertion and deter incorrect insertion. Importantly, these new insights into the mechanism of polymerase fidelity are consistent with an induced fit model. Specifically, binding of the correct incoming nucleotide promotes a closed conformation that is poised for catalysis. In contrast, binding of the incorrect nucleotide misaligns the active site and requires additional steps in the active site to promote catalysis. In addition, long incubation times indicated that pyrophosphate dissociation is coupled with thumb subdomain re-opening, and that this re-opening of the thumb subdomain occurs more rapidly after incorrect insertion, compared to correct insertion. This observation led to the hypothesis that rapid re-opening of Pol β following incorrect insertion may impact downstream repair events in large, multiprotein repair complexes such as those proposed to facilitate substrate channeling during BER.
For a misinserted nucleotide to become a base substitution error, it must be extended (i.e. further DNA synthesis) or ligated (i.e., a 3’-mispair in nicked DNA). To examine mispairs at the primer terminus affect correct nucleotide insertion, Wilson and colleagues analyzed an impressive twenty-three unique X-ray crystal structures of Pol β with terminal template-primer mismatches in both binary DNA and pre-catalytic ternary, utilizing non-hydrolyzable dUMPNPP, complexes [16]. Binary complex structures revealed unique, mispair-dependent distortions in the mismatched template strand. The binding of a correct incoming nucleotide to form a ternary pre-catalytic complex universally relieved the strain in the template while introducing new distortions in the primer terminus. Importantly, this distortion of the primer terminus discourages further extension, and provides an opportunity for an extrinsic proofreader to remove the mismatched base, enhancing overall fidelity. Accordingly, an induced fit mechanism also enhances the fidelity of mismatch extension in addition to nucleotide selection.
DNA polymerases also catalyze a reverse reaction, termed pyrophosphorolysis, that removes the DNA primer terminus to generate a dNTP. The Wilson lab determined that pyrophosphorolysis by Pol β to be inefficient and identified a pyrophosphate analog, where the bridging oxygen had been replaced with an imido group, that alters the reaction equilibrium to favor the reverse reaction [13]. Moreover, this analog, imidodiphosphate (PNP), results in a change in the rate-limiting step, so that the PNP-dependent reverse reaction is limited by chemistry and thus could be studied by time-lapse crystallography. Importantly, this 2017 study highlights that the equilibrium for the overall reaction depends on the nature of the DNA synthesis leaving group, indicating that terminal phosphate chemistry of an incoming nucleotide influences both chemical and conformational equilibria. Additionally, it provided a novel method to generate a pre-catalytic ternary substrate complex (Pol β:DNA:dNMPPNP) complex with two metals.
While this article has focused on the contributions from Dr. Samuel Wilson’s lab as part of a special issue dedicated to him, it is important that we acknowledge there have been many labs that have contributed structural insight to Pol β. It is increasingly rare that a lab can continually make such significant contributions within a single field, as the Wilson lab has been able to do through the structural biology of DNA polymerases. Moreover, as evidenced by the timeline presented as Figure 1, it is abundantly clear that their work has greatly expanded our understanding of the structure and function of DNA polymerases [60 this issue] and their roles during DNA repair [22]. Being at the forefront of polymerase mechanism biochemistry for so many decades, the Wilson group continually developed and refined new methods and tools necessary to answer pressing questions about the DNA polymerase mechanism and biology. These advances have now become commonplace in many X-ray crystallography studies outside of Pol β. Furthermore, the insight gained studying this model mammalian polymerase has provided a foundation for studying replicative, specialized, and novel DNA polymerases [56–59]. As a founding scientist of BER, Dr. Samuel Wilson has left his mark on the field of DNA repair and his scientific legacy will continue on through his ongoing publications, collaborations, and trainees.
Acknowledgements
We acknowledge the many groups that have made important contributions to Pol β enzymology, but were unable to highlight their work in this review, due to space limitations. We would like to acknowledge the many collaborators and colleagues that have contributed to the scientific successes of the Wilson laboratory (DNA Repair and Nucleic Acid Enzymology). We are also indebted to Mallory Smith and Matthew Schaich for their critical reading of the manuscript. This article was supported by National Institutes of Health R01-ES029203 and R35-GM128562 to B.D.F, and American Cancer Society PF-1815401-DMC to A.M.W.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Zmudzka BZ, SenGupta D, Matsukage A, Cobianchi F, Kumar P, Wilson SH, Structure of rat DNA polymerase beta revealed by partial amino acid sequencing and cDNA cloning, Proc Natl Acad Sci U S A, 83 (1986) 5106–5110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Abbotts J, SenGupta DN, Zmudzka B, Widen SG, Notario V, Wilson SH, Expression of human DNA polymerase beta in Escherichia coli and characterization of the recombinant enzyme, Biochemistry, 27 (1988) 901–909. [DOI] [PubMed] [Google Scholar]
- [3].Sawaya MR, Pelletier H, Kumar A, Wilson SH, Kraut J, Crystal structure of rat DNA polymerase beta: evidence for a common polymerase mechanism, Science, 264 (1994) 1930–1935. [DOI] [PubMed] [Google Scholar]
- [4].Pelletier H, Sawaya MR, Wolfle W, Wilson SH, Kraut J, Crystal structures of human DNA polymerase beta complexed with DNA: implications for catalytic mechanism, processivity, and fidelity, Biochemistry, 35 (1996) 12742–12761. [DOI] [PubMed] [Google Scholar]
- [5].Pelletier H, Sawaya MR, Kumar A, Wilson SH, Kraut J, Structures of ternary complexes of rat DNA polymerase beta, a DNA template-primer, and ddCTP, Science, 264 (1994) 1891–1903. [PubMed] [Google Scholar]
- [6].Sawaya MR, Prasad R, Wilson SH, Kraut J, Pelletier H, Crystal structures of human DNA polymerase beta complexed with gapped and nicked DNA: evidence for an induced fit mechanism, Biochemistry, 36 (1997) 11205–11215. [DOI] [PubMed] [Google Scholar]
- [7].Krahn JM, Beard WA, Miller H, Grollman AP, Wilson SH, Structure of DNA polymerase beta with the mutagenic DNA lesion 8-oxodeoxyguanine reveals structural insights into its coding potential, Structure, 11 (2003) 121–127. [DOI] [PubMed] [Google Scholar]
- [8].Krahn JM, Beard WA, Wilson SH, Structural insights into DNA polymerase beta deterrents for misincorporation support an induced-fit mechanism for fidelity, Structure, 12 (2004) 1823–1832. [DOI] [PubMed] [Google Scholar]
- [9].Batra VK, Beard WA, Shock DD, Krahn JM, Pedersen LC, Wilson SH, Magnesium-induced assembly of a complete DNA polymerase catalytic complex, Structure, 14 (2006) 757–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Batra VK, Beard WA, Hou EW, Pedersen LC, Prasad R, Wilson SH, Mutagenic conformation of 8-oxo-7,8-dihydro-2’-dGTP in the confines of a DNA polymerase active site, Nat Struct Mol Biol, 17 (2010) 889–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Batra VK, Shock DD, Beard WA, McKenna CE, Wilson SH, Binary complex crystal structure of DNA polymerase beta reveals multiple conformations of the templating 8-oxoguanine lesion, Proc Natl Acad Sci U S A, 109 (2012) 113–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Freudenthal BD, Beard WA, Wilson SH, Structures of dNTP intermediate states during DNA polymerase active site assembly, Structure, 20 (2012) 1829–1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Shock DD, Freudenthal BD, Beard WA, Wilson SH, Modulating the DNA polymerase beta reaction equilibrium to dissect the reverse reaction, Nat Chem Biol, 13 (2017) 1074–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Freudenthal BD, Beard WA, Shock DD, Wilson SH, Observing a DNA polymerase choose right from wrong, Cell, 154 (2013) 157–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Freudenthal BD, Beard WA, Perera L, Shock DD, Kim T, Schlick T, Wilson SH, Uncovering the polymerase-induced cytotoxicity of an oxidized nucleotide, Nature, 517 (2015) 635–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Batra VK, Beard WA, Pedersen LC, Wilson SH, Structures of DNA Polymerase Mispaired DNA Termini Transitioning to Pre-catalytic Complexes Support an Induced-Fit Fidelity Mechanism, Structure, 24 (2016) 1863–1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Howard MJ, Cavanaugh NA, Batra VK, Shock DD, Beard WA, Wilson SH, DNA polymerase beta nucleotide-stabilized template misalignment fidelity depends on local sequence context, J Biol Chem, 295 (2020) 529–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Cavanaugh NA, Beard WA, Wilson SH, DNA polymerase beta ribonucleotide discrimination: insertion, misinsertion, extension, and coding, J Biol Chem, 285 (2010) 24457–24465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Beard WA, Shock DD, Batra VK, Pedersen LC, Wilson SH, DNA polymerase beta substrate specificity: side chain modulation of the “A-rule”, J Biol Chem, 284 (2009) 31680–31689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Matsukage A, Bohn EW, Wilson SH, Multiple forms of DNA polymerase in mouse myeloma, Proc Natl Acad Sci U S A, 71 (1974) 578–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Whitaker AM, Schaich MA, Smith MR, Flynn TS, Freudenthal BD, Base excision repair of oxidative DNA damage: from mechanism to disease, Front Biosci (Landmark Ed), 22 (2017) 1493–1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Beard WA, Horton JK, Prasad R, Wilson SH, Eukaryotic Base Excision Repair: New Approaches Shine Light on Mechanism, Annu Rev Biochem, 88 (2019) 137–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Kim YJ, Wilson DM 3rd, Overview of base excision repair biochemistry, Curr Mol Pharmacol, 5 (2012) 3–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Wallace SS, Base excision repair: a critical player in many games, DNA Repair (Amst), 19 (2014) 14–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Beese LS, Steitz TA, Structural basis for the 3’−5’ exonuclease activity of Escherichia coli DNA polymerase I: a two metal ion mechanism, EMBO J, 10 (1991) 25–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Ueno T, Mitsuya H, Comparative enzymatic study of HIV-1 reverse transcriptase resistant to 2’,3’-dideoxynucleotide analogs using the single-nucleotide incorporation assay, Biochemistry, 36 (1997) 1092–1099. [DOI] [PubMed] [Google Scholar]
- [27].Burgers PM, Eckstein F, A study of the mechanism of DNA polymerase I from Escherichia coli with diastereomeric phosphorothioate analogs of deoxyadenosine triphosphate, J Biol Chem, 254 (1979) 6889–6893. [PubMed] [Google Scholar]
- [28].Steitz TA, A mechanism for all polymerases, Nature, 391 (1998) 231–232. [DOI] [PubMed] [Google Scholar]
- [29].Doublie S, Tabor S, Long AM, Richardson CC, Ellenberger T, Crystal structure of a bacteriophage T7 DNA replication complex at 2.2 A resolution, Nature, 391 (1998) 251–258. [DOI] [PubMed] [Google Scholar]
- [30].Kiefer JR, Mao C, Braman JC, Beese LS, Visualizing DNA replication in a catalytically active Bacillus DNA polymerase crystal, Nature, 391 (1998) 304–307. [DOI] [PubMed] [Google Scholar]
- [31].Brody RS, Frey PA, Unambiguous determination of the stereochemistry of nucleotidyl transfer catalyzed by DNA polymerase I from Escherichia coli, Biochemistry, 20 (1981) 1245–1252. [DOI] [PubMed] [Google Scholar]
- [32].Freemont PS, Friedman JM, Beese LS, Sanderson MR, Steitz TA, Cocrystal structure of an editing complex of Klenow fragment with DNA, Proc Natl Acad Sci U S A, 85 (1988) 8924–8928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Beese LS, Derbyshire V, Steitz TA, Structure of DNA polymerase I Klenow fragment bound to duplex DNA, Science, 260 (1993) 352–355. [DOI] [PubMed] [Google Scholar]
- [34].Patel SS, Wong I, Johnson KA, Pre-steady-state kinetic analysis of processive DNA replication including complete characterization of an exonuclease-deficient mutant, Biochemistry, 30 (1991) 511–525. [DOI] [PubMed] [Google Scholar]
- [35].Kuchta RD, Mizrahi V, Benkovic PA, Johnson KA, Benkovic SJ, Kinetic mechanism of DNA polymerase I (Klenow), Biochemistry, 26 (1987) 8410–8417. [DOI] [PubMed] [Google Scholar]
- [36].Wong I, Patel SS, Johnson KA, An induced-fit kinetic mechanism for DNA replication fidelity: direct measurement by single-turnover kinetics, Biochemistry, 30 (1991) 526–537. [DOI] [PubMed] [Google Scholar]
- [37].Erie DA, Hajiseyedjavadi O, Young MC, von Hippel PH, Multiple RNA polymerase conformations and GreA: control of the fidelity of transcription, Science, 262 (1993) 867–873. [DOI] [PubMed] [Google Scholar]
- [38].Spence RA, Kati WM, Anderson KS, Johnson KA, Mechanism of inhibition of HIV-1 reverse transcriptase by nonnucleoside inhibitors, Science, 267 (1995) 988–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Frey MW, Sowers LC, Millar DP, Benkovic SJ, The nucleotide analog 2-aminopurine as a spectroscopic probe of nucleotide incorporation by the Klenow fragment of Escherichia coli polymerase I and bacteriophage T4 DNA polymerase, Biochemistry, 34 (1995) 9185–9192. [DOI] [PubMed] [Google Scholar]
- [40].Werneburg BG, Ahn J, Zhong X, Hondal RJ, Kraynov VS, Tsai MD, DNA polymerase beta: pre-steady-state kinetic analysis and roles of arginine-283 in catalysis and fidelity, Biochemistry, 35 (1996) 7041–7050. [DOI] [PubMed] [Google Scholar]
- [41].Doherty AJ, Serpell LC, Ponting CP, The helix-hairpin-helix DNA-binding motif: a structural basis for non-sequence-specific recognition of DNA, Nucleic Acids Res, 24 (1996) 2488–2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Beard WA, Osheroff WP, Prasad R, Sawaya MR, Jaju M, Wood TG, Kraut J, Kunkel TA, Wilson SH, Enzyme-DNA interactions required for efficient nucleotide incorporation and discrimination in human DNA polymerase beta, J Biol Chem, 271 (1996) 12141–12144. [DOI] [PubMed] [Google Scholar]
- [43].Osheroff WP, Beard WA, Yin S, Wilson SH, Kunkel TA, Minor groove interactions at the DNA polymerase beta active site modulate single-base deletion error rates, J Biol Chem, 275 (2000) 28033–28038. [DOI] [PubMed] [Google Scholar]
- [44].Osheroff WP, Beard WA, Wilson SH, Kunkel TA, Base substitution specificity of DNA polymerase beta depends on interactions in the DNA minor groove, J Biol Chem, 274 (1999) 20749–20752. [DOI] [PubMed] [Google Scholar]
- [45].Culp SJ, Cho BP, Kadlubar FF, Evans FE, Structural and conformational analyses of 8-hydroxy-2’-deoxyguanosine, Chem Res Toxicol, 2 (1989) 416–422. [DOI] [PubMed] [Google Scholar]
- [46].Oda Y, Uesugi S, Ikehara M, Nishimura S, Kawase Y, Ishikawa H, Inoue H, Ohtsuka E, NMR studies of a DNA containing 8-hydroxydeoxyguanosine, Nucleic Acids Res, 19 (1991) 1407–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Grollman AP, Moriya M, Mutagenesis by 8-oxoguanine: an enemy within, Trends Genet, 9 (1993) 246–249. [DOI] [PubMed] [Google Scholar]
- [48].Topal MD, Baker MS, DNA precursor pool: a significant target for N-methyl-N-nitrosourea in C3H/10T1/2 clone 8 cells, Proc Natl Acad Sci U S A, 79 (1982) 2211–2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Kamiya H, Kasai H, Formation of 2-hydroxydeoxyadenosine triphosphate, an oxidatively damaged nucleotide, and its incorporation by DNA polymerases. Steady-state kinetics of the incorporation, J Biol Chem, 270 (1995) 19446–19450. [DOI] [PubMed] [Google Scholar]
- [50].Tsuzuki T, Egashira A, Igarashi H, Iwakuma T, Nakatsuru Y, Tominaga Y, Kawate H, Nakao K, Nakamura K, Ide F, Kura S, Nakabeppu Y, Katsuki M, Ishikawa T, Sekiguchi M, Spontaneous tumorigenesis in mice defective in the MTH1 gene encoding 8-oxo-dGTPase, Proc Natl Acad Sci U S A, 98 (2001) 11456–11461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Caglayan M, Horton JK, Dai DP, Stefanick DF, Wilson SH, Oxidized nucleotide insertion by pol beta confounds ligation during base excision repair, Nat Commun, 8 (2017) 14045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Hsu GW, Ober M, Carell T, Beese LS, Error-prone replication of oxidatively damaged DNA by a high-fidelity DNA polymerase, Nature, 431 (2004) 217–221. [DOI] [PubMed] [Google Scholar]
- [53].Nakamura T, Zhao Y, Yamagata Y, Hua YJ, Yang W, Watching DNA polymerase eta make a phosphodiester bond, Nature, 487 (2012) 196–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Chen Y, Basu R, Gleghorn ML, Murakami KS, Carey PR, Time-resolved events on the reaction pathway of transcript initiation by a single-subunit RNA polymerase: Raman crystallographic evidence, J Am Chem Soc, 133 (2011) 12544–12555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Freudenthal BD, Beard WA, Wilson SH, Watching a DNA polymerase in action, Cell Cycle, 13 (2014) 691–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Hoitsma NM, Whitaker AM, Schaich MA, Smith MR, Fairlamb MS, Freudenthal BD, Structure and function relationships in mammalian DNA polymerases, Cell Mol Life Sci, 77 (2020) 35–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Jamsen JA, Beard WA, Pedersen LC, Shock DD, Moon AF, Krahn JM, Bebenek K, Kunkel TA, Wilson SH, Time-lapse crystallography snapshots of a double-strand break repair polymerase in action, Nat Commun, 8 (2017) 253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Jain R, Aggarwal AK, Rechkoblit O, Eukaryotic DNA polymerases, Curr Opin Struct Biol, 53 (2018) 77–87. [DOI] [PubMed] [Google Scholar]
- [59].Sweasy JB, Lauper JM, Eckert KA, DNA polymerases and human diseases, Radiat Res, 166 (2006) 693–714. [DOI] [PubMed] [Google Scholar]
- [60].Beard WA, DNA Polymerase β: Closing the Gap between Structure and Function, DNA Repair (Amst.), (2020) in press. [DOI] [PMC free article] [PubMed] [Google Scholar]