A new putative copper chaperone localizes in the nucleus and improves plant defense response in Arabidopsis thaliana.
Keywords: Copper chaperone, plant immunity, PR1, Pseudomonas syringae pv tomato DC3000, salicylic acid, TGA2
Abstract
Copper is essential for many metabolic processes but must be sequestrated by copper chaperones. It is well known that plant copper chaperones regulate various physiological processes. However, the functions of copper chaperones in the plant nucleus remain largely unknown. Here, we identified a putative copper chaperone induced by pathogens (CCP) in Arabidopsis thaliana. CCP harbors a classical MXCXXC copper-binding site (CBS) at its N-terminus and a nuclear localization signal (NLS) at its C-terminus. CCP mainly formed nuclear speckles in the plant nucleus, which requires the NLS and CBS domains. Overexpression of CCP induced PR1 expression and enhanced resistance against Pseudomonas syringae pv. tomato DC3000 compared with Col-0 plants. Conversely, two CRISPR/Cas9-mediated ccp mutants were impaired in plant immunity. Further biochemical analyses revealed that CCP interacted with the transcription factor TGA2 in vivo and in vitro. Moreover, CCP recruits TGA2 to the PR1 promoter sequences in vivo, which induces defense gene expression and plant immunity. Collectively, our results have identified a putative nuclear copper chaperone required for plant immunity and provided evidence for a potential function of copper in the salicylic pathway.
Introduction
Copper ions are cofactors for many enzymes in biological processes including electron transport, hormone perception, and free radical elimination (Pilon et al., 2006; Robinson and Winge, 2010). Copper exists as Cu2+ or Cu1+, between which redox cycling triggers production of toxic hydroxyl radicals and severe damage to lipids, proteins, DNA, and other biomolecules (Robinson and Winge, 2010). Thus, copper chaperones, a set of soluble copper-binding proteins, sequestrate copper and limit the exposure of free copper to the cytoplasm. Moreover, copper chaperones are responsible for intracellular copper trafficking to their specific target copper proteins or compartments (Harrison et al., 1999). Copper chaperones usually harbor a classical MXCXXC metal-binding motif that is required for their physiological functions (Harrison et al., 1999).
The Arabidopsis genome encodes seven copper chaperones, namely the copper chaperone for superoxide dismutase (CCS), antioxidant protein1 (ATX1), ATX1-like copper chaperone (CCH), cytochrome c oxidase 11 (COX11), COX17, and two homologs of the yeast copper chaperone (HCC1 and HCC2) (Himelblau et al., 1998; Chu et al., 2005; Puig et al., 2007; Burkhead et al., 2009; Attallah et al., 2011). In Arabidopsis, CCS delivers copper to Cu/Zn superoxide dismutases (SODs) in the cytoplasm, chloroplast, and peroxisome, respectively (Kliebenstein et al., 1998; Burkhead et al., 2009). ATX1 and CCH share high sequence homology with the yeast ATX1 protein and both can complement the yeast atx1 mutant, but they have distinct properties and functions in copper homeostasis (Puig et al., 2007; Shin et al., 2012). ATX1 contributes to tolerance of both excess and deficient copper through its copper-binding MXCXXC motif (Shin and Yeh, 2012; Shin et al., 2012). In contrast, the biological role of CCH in the copper homeostasis process has not been well defined (Shin and Yeh, 2012; Shin et al., 2012). In addition, copper chaperones COX11, COX17, HCC1, and HCC2 play important roles in mitochondrial respiration (Balandin and Castresana, 2002; Burkhead et al., 2009; Attallah et al., 2011). Although copper chaperones have been extensively studied, much information is still lacking, particularly concerning whether copper chaperones and copper are imported into the nucleus and mediate plant defense responses.
Salicylic acid (SA), one of the major plant hormones, plays a central role in various plant immune response to pathogens, including pathogen-associated molecular pattern (PAMP)-triggered immunity (PTI), effector-triggered immunity (ETI), and systemic acquired resistance (SAR) (Seyfferth and Tsuda, 2014; Yan and Dong, 2014). SAR is an SA-induced broad-spectrum defense mechanism, in which the non-expressor of pathogenesis-related gene 1 (NPR1) is a key regulator (Cao et al., 1994; Fu and Dong, 2013). As the SA concentration increases upon pathogen infection or exogenous application of SA, oligomerized NPR1 is reduced to monomers in the cytosol and translocated into the nucleus. and mediates transcriptomic changes (Kinkema et al., 2000; Mou et al., 2003; Wang et al., 2006). Since it lacks a known DNA-binding domain, NPR1 regulates pathogenesis-related (PR) gene expression through interaction with nuclear transcription factors, such as TGAs and NIMINs (Zhang, 1999; Després et al., 2000; Weigel et al., 2001). The expression of PR genes is induced by SA, which is used as the best understood readout for SAR (Uknes et al., 1992). The induction of the PR1 gene by SA is dependent on the presence of the activation sequence-1 (as-1) element in the promoter regions of the PR1 gene (Qin et al., 1994). In healthy cells, the as-1 element specifically binds to the TGA transcription factors, leading to repressed transcription of PR1 under a normal SA concentration (Zhang et al., 2003; Rochon et al., 2006). However, upon pathogen-induced SA accumulation, NPR1 interacts with TGA2 and forms an enhancesome that activates PR1 transcription (Boyle et al., 2009). In the transcriptionally active NPR1–TGA2 complex, the N-terminal BTB domain of NPR1 interacts with and negates the function of the TGA2 repression domain (Boyle et al., 2009). In addition, the oxidation of Cys521 and Cys529 in the C-terminal transactivation domain of NPR1 is required for the NPR1–TGA2 enhancesome (Rochon et al., 2006).
Here, we report that a putative copper chaperone is an SA-responsive gene and induced by infections with plant pathogens, including plant viruses and bacteria. Unlike previously identified plant copper chaperones located in the cytoplasm, plasmids, and mitochondria, CCP harbors a nuclear localization signal (NLS) at its C-terminus that mediates its nuclear translocation. We further found that CCP could interact with TGA2 in vivo and in vitro, thereby inducing SA-mediated defense signaling.
Materials and methods
Construction of plasmids
The CCP ORF was inserted into the pMD19-T vector to generate pMD-CCP, which served as a template to generate pMD-CCPmCBS (amino acid changes C13G/C16G) and pMD-CCPmNLS (K61E, K62E, and F65E) using the QuikChange II kit (Stratagene, La Jolla, CA, USA). To generate pMDC32-CCP-Flag, pMDC32-CCPmCBS-Flag, pMDC32-CCPmNLS-Flag, and pMDC32-TGA2-Flag for transgenic or co-immunoprecipition assays, the CCP, CCPmCBS, CCPmNLS, and TGA2 cDNAs were fused to the N-terminus of three glycine residues and the Flag tag (3× Flag) of the binary vector pMDC32-3×Flag, a modified version of pMDC32 (Curtis and Grossniklaus, 2003).
To determine the expression pattern of the CCP gene, the genomic region from 2.5 kb upstream of the start codon to the stop codon of the CCP gene was engineered into the pBI101 vector (Clontech). For subcellular localization, the CCP ORF and derivatives were amplified from pMD19T-CCP and derivatives, and then inserted into pGDG (Goodin et al., 2002). To generate pGFP-GUS-CCP and mutants, the β-glucuronidase (GUS) ORF was amplified from the pBI101 plasmid and was introduced in-frame to the position between green fluorescent protein (GFP) and CCP.
To construct Arabidopsis thaliana ccp mutant lines, two 19 bp sequences (see Supplementary Table S1 at JXB online) specifically targeting the 5' coding region of CCP were cloned into the pHEC401 vector as described previously (Xing et al., 2014; Zhang et al., 2019). All primers used for plasmid constructions are listed in Supplementary Table S1, and all the plasmids were verified by sequencing.
Plant materials and growth conditions
For transient expression assays, H2B–red fluorescent protein (RFP) transgenic (Martin et al., 2009) or wild-type Nicotiana benthamiana plants were used for agroinfiltration as described previously (Fang et al., 2019). Arabidopsis thaliana seeds were incubated in 10% bleach for 15 min and washed with sterilized water five times, and then were plated on Murashige and Skoog (MS) medium. After vernalization at 4 °C for 3 d, plates were transferred into chambers with the condition of 23 °C and a 16 h light/8 h dark period for 10 d. Seedlings were then transferred into soil and grown in a room at 23 °C with a 10 h light/14 h dark cycle.
Bacterial and viral inoculation assay
For bacterial infection assay, two fully grown leaves of 4-week-old Arabidopsis plants were infiltrated with Pseudomonas syringae pv. tomato DC3000 (Pst DC3000) strains as described previously (Li et al., 2019). Pst DC3000 was cultured in King’s B liquid medium, washed in 10 mM MgCl2 twice, and diluted to OD600=0.0002 for infiltration. Infiltrated leaves were harvested for bacterial growth detection using colony-forming units per cm2. Bacterial growth was assessed in three independent experiments. For virus infection assay, three fully grown leaves of 4-week-old Arabidopsis plants were mechanically inoculated with Cucumber mosaic virus (CMV; 20 ng μl–1) as described previously (Zhang et al., 2019). Systemically infected leaves were collected for RNA analyses.
GUS staining
GUS staining assays were performed as described previously (Guo et al., 2018). Briefly, plant tissues were treated with X-Gluc solution (3 mM 5-bromo-4-chloro-3-indolyl-β-d-glucuronide, 10 mM potassium ferricyanide, 0.1% Triton X-100, 10 mM potassium ferrocyanide, 50 mM NaH2PO4 buffer, pH 7.0) at 37 °C overnight, washed in an ethanol series, and photographed using a digital camera.
Real-time quantitative reverse transcription–PCR (RT–PCR)
Total RNAs were extracted from A. thaliana tissues using TRIzol reagent following the manufacturer’s instructions (Invitrogen). Total RNA was treated with RNase-free DNase I (Takara) and served as template for reverse transcription using the M-MLV reverse transcriptase (Promega). Real-time quantitative PCRs were performed to examine accumulation of CCP and PR1 mRNA with SsoFast EvaGreen Supermix (Bio-Rad). The ACTIN2 gene served as an internal control. Specific primers are listed in Supplementary Table S1.
Identification of terminal sequences of the CCP cDNA
3' and 5' RACE were performed as described previously (Yan et al., 2015). Briefly, RNA samples were isolated from SA-treated A. thaliana plants, enriched by oligo(dT) beads, and used to identify two terminal sequences of the CCP mRNA with a SMARTer RACE 5′/3' Kit (Clontech, https://www.takarabio.com/). Amplified PCR products were cloned into pMD-19T for sequencing. Gene-specific primers for 5′/3' RACE are listed in Supplementary Table S1.
Protein subcellular localization
For subcellular localization, the CCP cDNA and derivatives were cloned into pGDG (Goodin et al., 2002), and the resulting plasmids were agroinfiltrated into H2B transgenic N. benthamiana leaves. At 2 days post-inoculaton (dpi), localization images were taken with a confocal laser scanning microscope (Zeiss LSM710). In addition, pGDG-CCP and derivatives were transformed into Arabidopsis protoplasts as described previously (Yoo et al., 2007). The transformed Arabidopsis protoplasts were incubated at 20 °С for 18 h and were visualized by a Zeiss LSM710.
Protein interaction assays
Bimolecular fluorescence complementation (BiFC) assays were performed to examine the interaction of CCP and TGA2 as described previously (Zhang et al., 2017). CCP, TGA2 cDNA fragments, their derivatives, and the Rubisco gene (Rub) were introduced into the BiFC vectors pSPYNE-35S or pSPYCE-35S (Walter et al., 2004). The recombinant BiFC plasmids and the Tomato bushy stunt virus P19 plasmid were transformed into A. tumefaciens EHA105 strains and co-infiltrated into N. benthamiana leaves. At 2 dpi, epidermal cells of infiltrated patches were monitored for yellow fluorescent protein (YFP) fluorescence by a Zeiss LSM710 confocal microscope (Carl Zeiss).
In vivo co-immunoprecipitation (Co-IP) of CCP and TGA2 was performed as described previously (Zhang et al., 2017). TGA2-Flag was co-expressed with GFP, CCP–GFP, or CCPmCBS–GFP in N. benthamiana leaves through agroinfitration-mediated transient expression assays. Co-infiltrated leaf tissues were homogenized in Co-IP buffer [0.1% Triton X-100, 10% glycerol, 25 mM Tris–HCl (pH 7.5), 2% PVP-40, 200 mM NaCl, 1 mM EDTA, 10 mM DTT, and protease inhibitor cocktail]. After centrifugation, the resulting supernatants were mixed with anti-Flag M2 affinity gel (Sigma, https://www.sigmaaldrich.com/) at 4 °C for 5 h. The immunoprecipitated products were washed five times and boiled in SDS buffer for protein detection with corresponding antibodies.
For pull-down assays, TGA2 and GFP cDNAs were cloned into pGEX-4T-1, and the resulting recombinant plasmids were introduced into Escherichia coli (BL21 strain) for expression of GST–TGA2 and GST–GFP proteins, respectively. CCP and CCPmCBS cDNAs were engineered into pET30a for expression of His-CCP and His-CCPmCBS, respectively. Glutathione S-transferase (GST)–TGA2, GST–GFP, His-CCP, and His-CCPmCBS were expressed and purified according to the manufacturer’s instruction. For in vitro binding assays, 2 μg of bait proteins (GST–TGA2 or GST–GFP) and 2 μg of prey proteins (6× His-CCP and mutant) were incubated with GST beads in 500 μl of binding buffer [50 mM Tris–HCl (pH 6.8), 150 mM NaCl, 0.2% glycerol, 5 mM DTT, protease inhibitor cocktail, and 0.6% Triton X-100] at 4 °С for 6 h. After washing six times with binding buffer, the pull-down proteins were analyzed by immunoblotting using anti-His and GST antibody, respectively.
EMSAs
For EMSAs, CCP and NPR1 were cloned into pGEX-4T-1 and the resulting recombinant plasmids were introduced into E. coli (BL21 strain) for expression of GST–CCP and GST–NPR1 proteins, respectively. EMSAs were performed using the LightShift Chemiluminescent EMSA Kit following the manufacturer’s instructions (Thermo Scientific, https://www.thermofisher.com/). Briefly, the LS7 probe (5'-TATTTTACTTACGTCATAGATGTGGCGGCA-3' annealed to 5'-TGCCGCCACATCTATGACGTAAGTAAAATA-3') was labeled with biotin. A total of 500 ng of recombinant GST-tagged proteins were mixed with 4 fmol of biotin-labeled LS7 probe in 20 μl reaction mixtures containing 1 μl of dI–dC, 1 μl of 50% glycerol, and 10× binding buffer for 20 min at 25 °С, and then separated on 6% native polyacrylamide gels. The labeled probes were detected according to the EMSA kit instructions.
Yeast complementation
The yeast complementation assays were performed as described previously (Abdel-Ghany et al., 2005). Briefly, the Saccharomyces cerevisiae lys7 mutant was generated by replacing the Lys7 gene with the LEU2 gene in the yeast BY4741 strain. The CCP cDNA and derivatives were cloned into pGPD-416-1 and the resulting plasmids were transformed into the lys7 mutant. The SC minimal medium lacking leucine and uracil was used to select positive transformed colonies. For complementation assays, serial dilutions of transformed yeasts were plated on SC minimal medium lacking leucine and uracil with or without menadione (Sigma) at 30 °C for 3 d.
ChIP assays
ChIP assays were performed as described previously (Wang et al., 2019). Briefly, Arabidopsis transgenic lines CCPOE and CCPmCBS/OE were grown in a room at 23 °C with a 10 h light/14 h dark cycle for ChIP. Fully expanded leaves of 4-week-old seedlings were inoculated with Pst DC3000 and harvested at 1 dpi for ChIP assays. The inoculated leaves (2 g) were treated with 1% formaldehyde to cross-link the protein–DNA complexes under vacuum. Then, the samples were isolated, sonicated, and centrifuged at 12 000 g at 4 °C for 10 min. The supernatant was transferred into new tubes and incubated with anti-Flag affinity beads. The precipitated DNA samples were quantified by quantitative PCR using the primers listed in Supplementary Table S1.
Results
Identification of a putative copper chaperone induced by plant pathogens
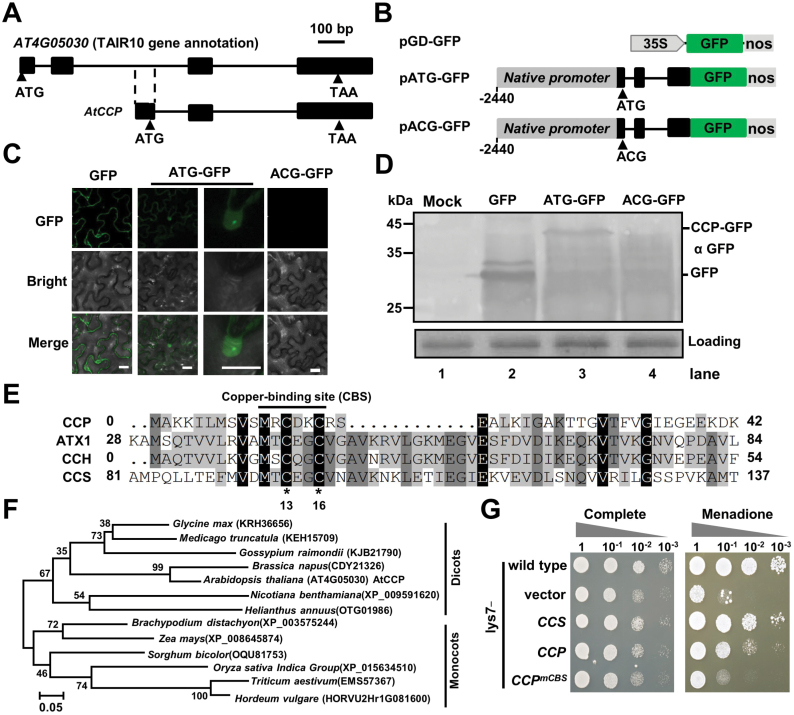
To investigate host transcriptome response to virus infection, total RNA from CMV-infected A. thaliana leaves or mock-treated leaves was used for transcriptome analysis. Among up-regulated genes, the AT4G05030 mRNA accumulated to a low level in mock-treated leaves, but was induced by CMV infection (Supplementary Fig. S1A). Moreover, 5′ and 3' RACE assays revealed that the AT4G05030 cDNA in vivo corresponds to part of the second intron, and third and fourth exons of the TAIR10 gene annotated as AT4G05030 cDNA (Fig. 1A; Supplementary Fig. S1B).
Fig. 1.
Identification of a novel copper chaperone in Arabidopsis. (A) Genome structure of AT4G05030 from TAIR10 gene annotation (upper) or the identified CCP cDNA (bottom). Exons and introns are indicated by black boxes and lines, respectively. The start codon (ATG) and stop codon (TAA) of the predicted small ORF are indicated by arrowheads. (B) Illustration of binary vectors for expressing GFP or GFP-fused proteins in N. benthamiana leaves. In pATG-GFP, the DNA genome fragment between 2440 bp upstream of the CCP start codon to the end of the CCP ORF was amplified from Col-0 and fused to the N-terminus of GFP. In pACG-GFP, the predicted start codon ATG of pATG-GFP was mutated to ACG. (C) Confocal micrographs examining GFP, ATG–GFP, or ACG–GFP expression in agroinfiltrated N. benthamiana leaves at 3 dpi. Scale bar=20 μm. (D) Western blotting analyses showing accumulation of GFP, ATG–GFP, or ACG–GFP in the samples of (C) with anti-GFP antibody. Buffer-infiltrated leaves served as a mock control. The Coomassie brilliant blue staining was used as the protein loading control. (E) Alignments of Arabidopsis CCP, ATX1, CCH, and CCS. The conserved copper-binding site (CBS) is indicated by a line. (F) Phylogenetic trees of plant CCP orthologs based on amino acid sequences. Accession numbers or locus are shown. Scale bar=0.05 amino acid substitutions per site. (G) Complementation of the growth phenotype of the S. cerevisiae lys7 mutant cells transformed with empty plasmid (vector), or plasmids expressing Arabidopsis CCS, CCP, or CCPmCBS. Wild-type S. cerevisiae and complemented lys7 mutant cells were assayed for growth on complete medium or medium with 25 μM menadione. Cells were spotted in 10-fold serial dilutions starting at OD600=0.1, and incubated at 30 °C for 3 d. (This figure is available in color at JXB online.)
Interestingly, a small ORF was predicted from the identified AT4G05030 cDNA (Fig. 1A; Supplementary Fig. S1C). To confirm the ORF translation, the AT4G05030 genomic region from 2440 bp upstream of the start codon to the end of the ORF was fused to the GFP N-terminus to generate pATG-GFP (Fig. 1B). Moreover, the pACG-GFP mutant was generated by replace the new start codon ATG with ACG (Fig. 1B). Agrobacterium harboring the pGD-GFP, pATG-GFP, or pACG-GFP plasmids was agroinfiltrated into N. benthamiana leaves. At 5 dpi, GFP fluorescent speckles were observed in the nuclei of leaf epidermal cells infiltrated with pATG-GFP, but not in those infiltrated with pACG-GFP (Fig. 1C). Western blotting analyses revealed that the ATG–GFP protein was larger than GFP protein (Fig. 1D), indicating that the AT4G05030 small ORF was translated in vivo.
The small ORF of AT4G05030 encodes a 77 amino acid polypeptide harboring a classical copper-binding motif, MXCXXC, that is conserved in well-studied copper chaperones, such as ATX1, CCH, and CCS (Fig. 1E). Thus, AT4G05030 probably encodes a putative CCP. Blast searches reveal that many monocot and dicot plant species harbor a single CCP ortholog (Fig. 1F), implying a conserved function of CCP orthologs in plants. However, all CCP orthologs have not been characterized previously.
To investigate whether CCP is a functional homolog of the yeast copper chaperone Ccs1/Lys7, Arabidopsis CCS and CCP were expressed in the yeast lys7 mutant (Supplementary Fig. S2). The functional complementation assay was performed by analyzing the growth phenotype on complete medium or medium containing the superoxide generator menadione. As expected, ectopic expression of Arabidopsis CCS completely rescued SOD activity of the lys7 mutant, whereas expression of CCP partially rescued SOD activity (Fig. 1G). In contrast, ectopic expression of the CCPmCBS mutant harboring substitution of the conserved Cys13 and Cys16 with glycine could not rescue the SOD activity in the lys7 mutant (Fig. 1G). These results suggest that CCP can partially protect the lys7 mutant from reactive oxygen toxicity in a copper binding-dependent manner.
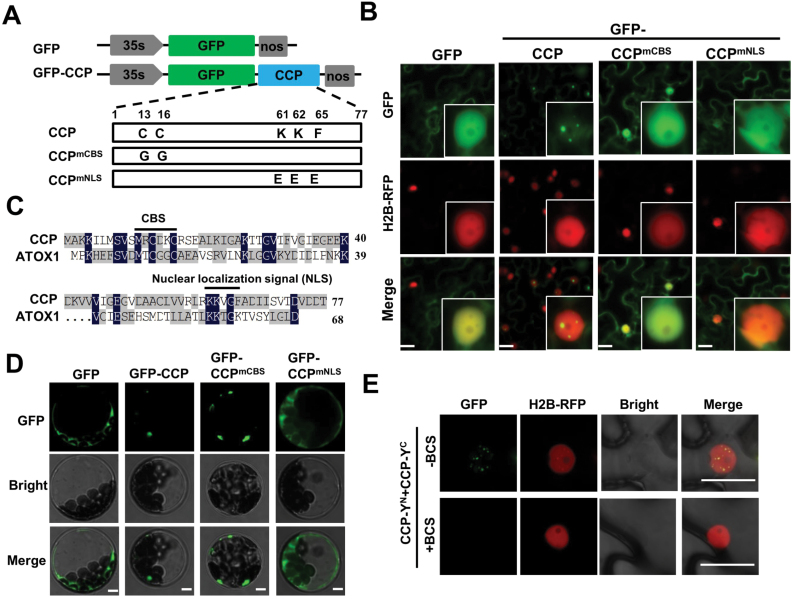
The nuclear translocation of CCP requires the CBS and NLS motifs
To determine CCP subcellular localization, Agrobacterium harboring plasmids for expression of GFP–CCP or GFP (Fig. 2A) were infiltrated into leaves of H2B–RFP transgenic N. benthamiana plants (Martin et al., 2009). At 3 dpi, confocal examinations revealed that free GFP localized evenly in the cytoplasm and nucleus (Fig. 2B; Supplementary Fig. S3A). In contrast, GFP–CCP predominantly formed fluorescent nuclear speckles in the nucleus (Fig. 2B; Supplementary Fig. S3A). The mammalian antioxidant-1 (ATOX1) protein has 68 amino acids and an NLS at the C-terminus (Fig. 2C) (Itoh et al., 2008; Muller ansd Klomp, 2009). Amino acid sequence alignment revealed that a KKVGF motif at the CCP C-terminus might be a potential NLS (Fig. 2C).
Fig. 2.
Subcellular localization of CCP and derivative mutants. (A) Diagram illustrating GFP–CCP, GFP–CCPmCBS, and GFP-CCPmNLS. Mutated amino acids are indicated in boxes. (B) Confocal micrographs showing the subcellular localization of GFP, GFP–CCP, GFP–CCPmCBS, and GFP–CCPmNLS in agroinfiltrated leaves of transgenic N. benthamiana plants with H2B–RFP, a nuclear marker. Photographs were taken at 3 dpi. Scale bar=20 μm. (C) Alignment of CCP and mammalian ATOX1. A copper-binding site (CBS) and a predicted nuclear localization signal (NLS) are indicated by lines. (D) Fluorescence micrographs showing subcellular localization of GFP (control), GFP–CCP, GFP–CCPmCBS, and GFP–CCPmNLS in Arabidopsis protoplasts. Scale bar=5 μm. (E) BiFC analyzing the CCP self-interaction in epidermal cells of N. benthamiana leaves treated with buffer or BCS, a copper chelator. Scale bar=5 μm. (This figure is available in color at JXB online.)
To determine the requirement for the CBS and NLS motifs for subcellular localization, GFP–CCPmCBS and GFP–CCPmNLS were further expressed in H2B–RFP transgenic N. benthamiana plants (Fig. 2A). Western blotting revealed expression of GFP, GPF–CCP, GFP–CCPmCBS, and GFP–CCPmNLS in infiltrated leaves (Supplementary Fig. S3). Confocal examination revealed that GFP–CCPmCBS and GFP–CCPmNLS localized evenly in the cytoplasm and nucleus, which is distinct from the nuclear speckles of GFP–CCP (Fig. 2B). Consistently, GFP–CCP fluorescence was observed mainly in the nuclei of transfected Arabidopsis protoplasts, whereas GFP, GFP–CCPmCBS, and GFP–CCPmNLS fluorescence was observed in the cytoplasm of transfected protoplasts (Fig. 2D). To rule out non-specific diffusion of the small GFP–CCP protein into the nucleus, CCP, CCPmCBS, and CCPmNLS were fused with GFP–GUS and expressed in H2B–RFP transgenic N. benthamiana leaves. As expected, GFP fluorescence from GFP–GUS–CCP was predominantly present in the nuclei (Supplementary Fig. S4), whereas GFP–GUS–CCPmCBS and GFP–GUS–CCPmNLS were limited in the cytoplasm (Supplementary Fig. S4). These results clearly indicate that both the CBS and NLS motifs are required for CCP nuclear localization.
To explore whether self-interactions of CCP were required for CCP nuclear localization, CCP, CCPmNLS, and CCPmCBS were fused to the N (YN) or C (YC) halves of sYFP, and then co-expressed in N. benthamiana leaves. BiFC assays revealed that self-interactions of CCP and CCPmNLS were detected in the nuclei and cytoplasm, respectively (Supplementary Fig. S5A). In contrast, self-interaction of CCPmCBS was not detected although all the proteins were expressed (Supplementary Fig. S5A, C). The YN- or YC-fused transcription factor ELONGATED HYPOCOTYL 5 (HY5) (Oyama et al., 1997) served as a negative control that did not exhibit BiFC fluorescence with other combinations (Supplementary Fig. S5C). We further treated the CCP-YN/CCP-YC co-infiltrated leaves with buffer or the copper chelator bathocuproine sulfonate (BCS) (Itoh et al., 2008). The CCP-YN/CCP-YC interaction in the nucleus was nearly abolished in BCS-treated leaves compared with the control treatment (Fig. 2E). In addition, the GFP–CCP-expressing leaves were further treated with mock buffer or BCS, showing that the cytoplasmic localization of GFP–CCP increased compared with the mock control (Supplementary Fig. S6). Collectively, these results indicate that CBS-dependent self-interaction is required for CCP nuclear localization.
Overall, these results indicate that both the CBS and NLS motifs are required for CCP nuclear localization. The N. benthamiana CCP ortholog (NbCCP) had similar nuclear translocation and an NLS (Supplementary Fig. S7A). However, Arabidopsis ATX1 and CCH did not localize in the plant nucleus (Supplementary Fig. S7B), which was in agreement with previous studies (Puig et al., 2007). To ensure non-specific diffusion of the small GFP–ATX1/CCH protein into the nucleus, GFP–GUS–ATX1 and GFP–GUS–CCH were expressed in H2B–RFP transgenic N. benthamiana leaves, showing that these fusion proteins were exclusively in the cytoplasm (Supplementary Fig. S7B). Western blotting revealed expression of GFP–ATX1, CCH, GUS–ATX1, and GUS–CCH in H2B–RFP transgenic N. benthamiana leaves (Supplementary Fig. S7C). Thus, the nuclear localization of CCP is a unique feature among the identified plant copper chaperones.
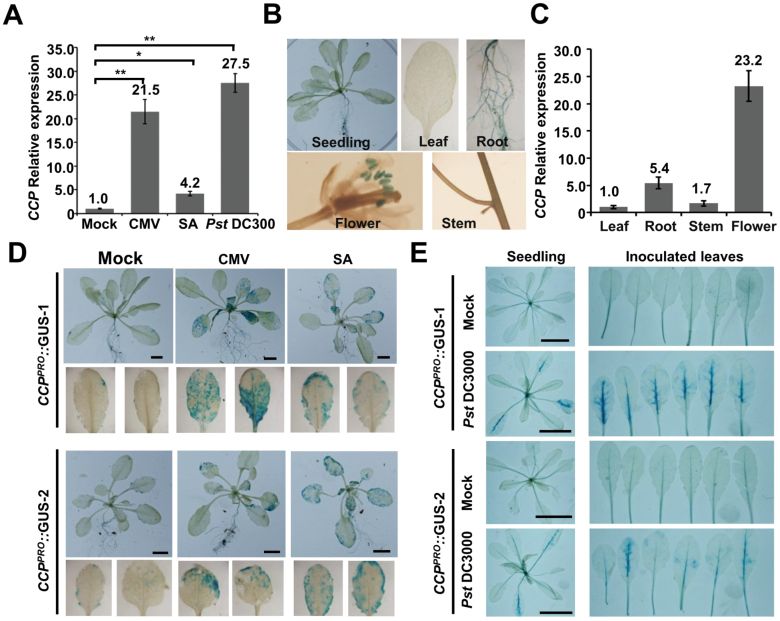
CCP is induced by plant pathogen infections and SA treatment
To determine CCP response to pathogen infections, Col-0 leaves were treated by mock buffer, CMV, Pst DC3000, or SA, and then collected for quantitative RT–PCR. Compared with mock treatment, CMV, SA, and Pst DC3000 treatment induced CCP accumulation by ~21.5-, 4.2-, and 27.5-fold (Fig. 3A). Furthermore, transgenic Col-0 plants expressing GUS under control of the CCP native promoter were generated to investigate the CCP tissue expression pattern. GUS staining results revealed that GUS activity was detected at higher levels in roots and flowers than in leaves and stems of healthy CCPPRO::GUS transgenic plants (Fig. 3B), which was further verified by quantitative RT–PCR assays (Fig. 3C). Two independent CCPPRO::GUS transgenic lines were infected with CMV or treated with SA, revealing that GUS expression was induced by CMV infection and SA treatment (Fig. 3D). Similarly, Pst DC3000 inoculation resulted in increased GUS activity in infiltrated leaves, especially in the veins (Fig. 3E). However, we found that the CCP gene was suppressed but not induced by excess copper (Supplementary Fig. S8), which is consistent with the CCH gene (Shin et al., 2012). Collectively, our findings demonstrate that CCP is transcriptionally induced by pathogen infections and SA treatment.
Fig. 3.
The tissue expression pattern of Arabidopsis CCP. (A) Quantitative RT–PCR analyzing CCP mRNA accumulation in systemic leaves of CMV-infected plants at 7 dpi, SA-treated leaves at 3 dpi, and Pst DC300-inoculated leaves at 1 dpi. Accumulation of CCP mRNA in mock-treated leaves was set as one unit. Numbers above columns are mean values of three independent experiments. Error bars represent the SEM. *P-value <0.05; **P-value <0.01. (B) Histochemical GUS activities in different tissues of CCPPRO::GUS transgenic plants. (C) Relative accumulation of the CCP mRNA in different tissues of Col-0 plants. Accumulation of CCP mRNA in leaves was set as one unit. Numbers above columns are mean values of three independent experiments. Error bars represent the SEM of three independent experiments. (D) GUS activity in two independent CCPPRO::GUS transgenic lines treated with buffer, CMV infection, or SA. Detached systemically infected leaves are shown in the bottom panels. Scale bar=5 cm. (E) GUS activity in CCPPRO::GUS-1 and CCPPRO::GUS-2 after mock treatment or Pst DC3000 infection at 1 dpi. Detached inoculated leaves are shown in the right-hand panels. Scale bar=3 cm. (This figure is available in color at JXB online.)
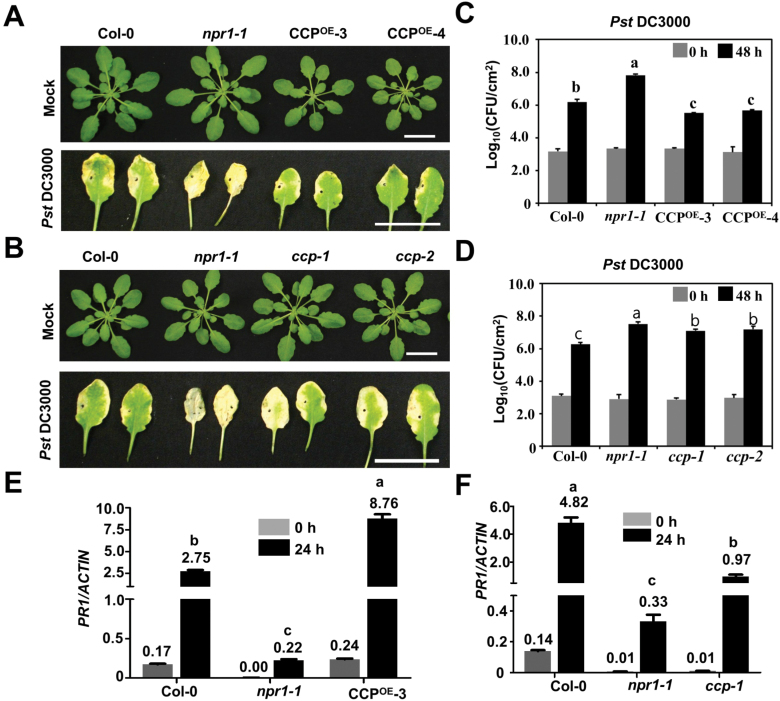
CCP enhances plant resistance to bacterial pathogens
We next examined whether CCP is involved in resistance against bacterial pathogens. To this end, we generated CCP-overexpressing transgenic lines (CCPOE) under control of the Cauliflower mosaic virus (CaMV) 35S promoter, in which increased accumulation of CCP was verified by RT–PCR (Supplementary Fig. S9A). Accumulation of the CCP protein in CCPOE-3 and CCPOE-4 was confirmed by western blotting analyses for functional experiments (Supplementary Fig. S9B). In addition, we generated two ccp mutants using the CRISPR/Cas9 [clustered regularly interspaced palindromic repeats (CRISPR)/CRISPR-associated protein 9] system with the guide RNA targeting the downstream region of the start codon (Supplementary Fig. S10). The resulting ccp-1 and ccp-2 contain a 445 bp deletion and a 1 bp insertion, respectively (Supplementary Fig. S10).
Rosette leaves of 4-week-old Col-0, CCPOE-3, CCPOE-4, ccp-1, ccp-2, and npr1-1 were infiltrated with Pst DC3000. At 0 and 48 h post-inoculation (hpi), infiltrated leaves were collected for assessing bacterial growth by counting colony-forming units. The npr1-1 mutant allowed much higher accumulation of Pst DC3000 at 48 hpi and exhibited enhanced chlorotic symptoms at 72 hpi compared with Col-0 leaves (Fig. 4A–C). The infiltrated leaves of CCPOE-3 and CCPOE-4 exhibited milder symptoms (Fig. 4A), whereas more severe chlorotic symptoms were observed in the ccp-1 and ccp-2 mutant leaves (Fig. 4B). Consistently, CCPOE-3 and CCPOE-4 allowed a lower level of Pst DC3000, while the ccp-1 and ccp-2 mutants supported an increased level of bacteria compared with Col-0 plants (Fig. 4C, D). Consistently, compared with the infected Col-0 leaves, PR1 mRNA accumulated to a substantially higher level in CCPOE-3 leaves (Fig. 4E) and a reduced level in ccp-1 leaves (Fig. 4F). Collectively, CCP enhances plant defense against Pst DC3000.
Fig. 4.
CCP improves plant resistance against bacterial pathogens. (A) Morphology and symptom development of wild-type Col-0, npr1-1, CCPOE-3-, and CCPOE-4 leaves inoculated with buffer or Pst DC3000 at 72 hpi. Scale bar=2 cm. (B) Morphology and symptom development of wild-type Col-0, npr1-1, ccp-1, and ccp-2 leaves inoculated with buffer or Pst DC3000 at 72 hpi. Scale bar=2 cm. (C) Growth of Pst DC3000 on infiltrated leaves of Col-0, npr1-1, CCPOE-3, and CCPOE-4 plants at 0 and 48 hpi. Four-week-old plants were infiltrated with Pst DC3000 (OD600=0.0002). CFU, colony-forming units. (D) Growth of Pst DC3000 on infiltrated leaves of Col-0, npr1-1, ccp-1, and ccp-2 plants at 0 and 48 hpi. (E and F) Relative ratio of the PR1 mRNA versus the endogenous actin mRNA in Col-0, npr1-1, CCPOE-3 (E), or ccp-1 (F) leaves infiltrated with Pst DC3000 (OD600=0.0002) at 0 and 24 hpi. In (C–F), scale bars represent means ±SE (n=3). Different letters indicate significant differences (P<0.05, n=3) determined by ANOVA by Turkey’s multiple comparison test analysis. (This figure is available in color at JXB online.)
Interaction of CCP with the N-terminal repression domain of TGA2
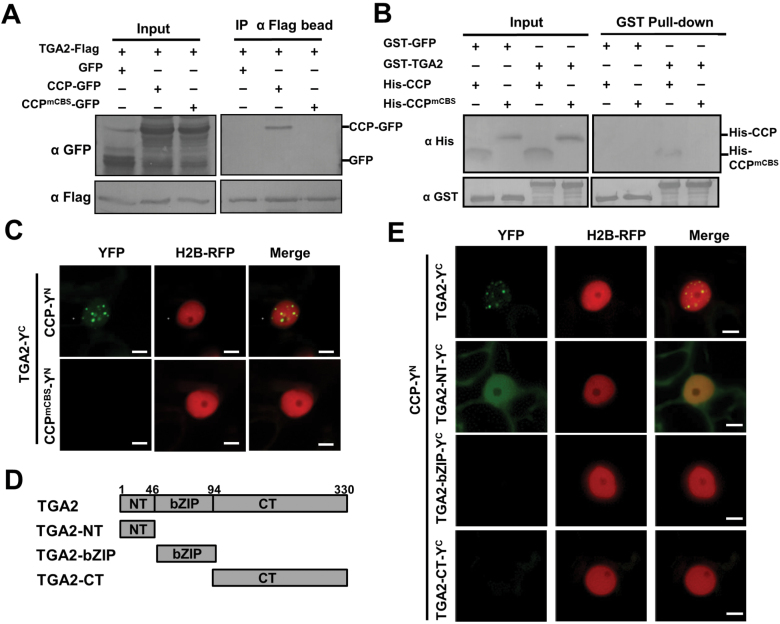
Upon plant pathogen infections and SA treatment, CCP expression is induced and undergoes nuclear translocation mediated by CBS and NLS motifs. Moreover, CCP is required for plant resistance to bacterial pathogens. Given that the TGA transcription factors are important regulators in the SA signaling pathway, we hypothesized that TGA transcription factors might be targeted by CCP in the nucleus. To verify this hypothesis, we performed Co-IP assays to examine the potential interaction of CCP and TGA2. The TGA2-Flag protein was co-expressed with CCP–GFP or CCPmCBS–GFP in N. benthamiana leaves by agroinfiltration. At 2 dpi, the TGA2-Flag protein and associated proteins were precipitated with anti-Flag affinity beads. Western blotting analyses revealed that TGA2-Flag was precipitated with CCP–GFP, but not with CCPmCBS–GFP or free GFP (Fig. 5A). In vitro pull-down assays were further carried out to determine the direct interaction of CCP and TGA2 using CCP-His, CCPmCBS-His, GST–GFP, and GST–TGA2. Pull-down assays revealed that CCP-His, but not CCPmCBS-His, was pulled down with GST–TGA2 (Fig. 5B). On the other hand, the GST–GFP control did not interact with CCP-His or CCPmCBS-His (Fig. 5B).
Fig. 5.
Interaction of CCP with TGA2 in vivo and in vitro. (A) Co-IP assays showing the interaction of TGA2 and CCP in vivo. Nicotiana benthamiana leaves were agroinfiltrated with constructs for co-expressing TGA2-Flag with CCP–GFP, CCPmCBS–GFP, or GFP. At 2 dpi, total proteins isolated from the agroinfiltrated N. benthamiana leaves were immunoprecipitated with anti-Flag beads. Total proteins (Input) and immunoprecipitated proteins (IP) were analyzed by western blotting analyses with anti-Flag and anti-GFP antibodies. (B) GST pull-down analysis of the interaction between TGA2 and CCP in vitro. GST–GFP or GST–TGA2 were incubated with His-CCP or His-CCPmCBS, and then immunoprecipitated with glutathione–Sepharose beads in vitro. The input and pull-down products were detected by western blotting analysis with anti-GST or anti-His antibodies. (C) BiFC analysis of the interaction of TGA2 with CCP or CCPmCBS in H2B transgenic N. benthamiana leaves. Agrobacterium strains harboring plasmids expressing YN- or YC-tagged tested proteins were infiltrated in H2B transgenic N. benthamiana leaves. At 2 dpi, BiFC fluorescence was monitored by confocal scanning laser microscopy. Scale bar=5 μm. (D) Schematic diagrams of the TGA2 domains. (E) BiFC analysis of the interaction between the three main domains of TGA2 and CCP in H2B transgenic N. benthamiana leaves. Scale bar=5 μm. (This figure is available in color at JXB online.)
We next determined whether CCP associates with TGA2 in vivo using a BiFC assay. To this end, the TGA2 coding sequence was fused with the C-terminal half of YFP (TGA2-YC), and coding sequences of CCP or CCPmCBS were fused with the N-terminal half of YFP (CCP-YN or CCPmCBS-YN). Co-expression of TGA2-YC and CCP-YN resulted in YFP fluorescent speckles in the nuclei of infiltrated cells (Fig. 5C). Interestingly, the SA treatment increase the YFP intensity of TGA2-YC and CCP-YN in the nucleus compared with mock treatment (Supplementary Fig. S11). In contrast, leaf cells co-expressing TGA2-YC with CCPmCBS-YN or negative control HY5-YN failed to produce YFP signal (Fig. 5C; Supplementary Fig. S5B).
TGA2 contains an N-terminal domain (amino acids 1–46), a middle bZIP domain (amino acids 47–94), and a C-terminal domain (amino acids 95–330) (Fig. 5D) (Rochon et al., 2006; Boyle et al., 2009). To determine the CCP-binding domain in the TGA2 protein, the three domains were individually fused with YC and co-expressed with CCP-YN in N. benthamiana leaves. BiFC results revealed that the NT domain of TGA2 interacted with the CCP protein in vivo (Fig. 5E). It should be noted that the BiFC signal of CCP-YN and TGA2-NT-YC localized evenly in the cytoplasm and nucleus, which was different from the speckles formed by CCP-YN and TGA2-YC (Fig. 5E). To explain this different localization, GFP–TGA2, GFP–NT, GFP–bZIP, and GFP–CT were expressed in H2B–RFP transgenic N. benthamiana plants through agroinfiltration. The results showed that GFP signals were observed in speckles mainly in the nuclei as GFP–TGA2, whereas GFP signals from GFP–NT and GFP–CT localized evenly in the nuclei and cytoplasm. On the other hand, GFP signals from GFP–bZIP were mainly present in the nucleolus (Supplementary Fig. S12). Collectively, these results indicate that CCP interacts directly with TGA2, and that the CCP CBS motif and the TGA2 NT domain are essential for their interactions.
CCP functions together with TGA2 to activate the expression of PR1
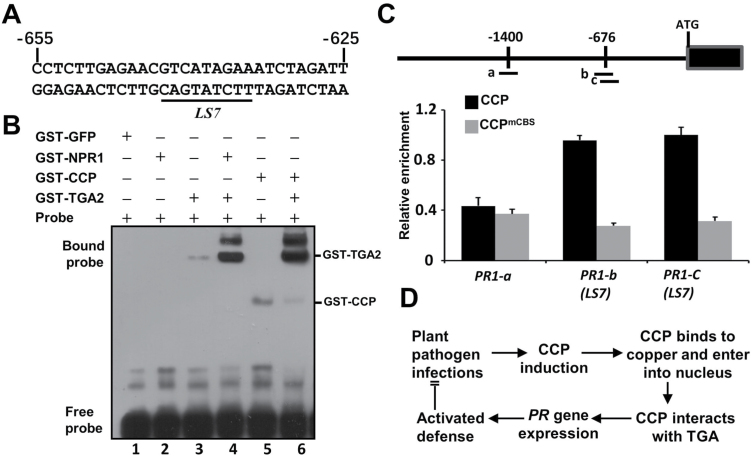
Previous studies have shown that TGA2 binds to the linker scan 7 element (LS7; Fig. 6A) of the PR1 promoter to positively regulate PR1 expression (Zhang et al., 1999). The NPR1 protein enhances the DNA binding affinity of TGA2 and then activates defense gene expression (Després et al., 2000). Since the N-terminus of TGA2 has been shown to be a repression domain (Boyle et al., 2009), the interaction of CCP and the N-terminal domain of TGA2 may enhance the binding of TGA2 to the LS7 element of the PR1 promoter and then activates pathogenesis-related gene expression. To test the hypothesis, the LS7 element and surrounding region of the PR1 promoter was labeled by biotin for EMSAs (Fig. 6A). The LS7 probe was incubated with different combinations of GST-tagged GFP, TGA2, NPR1, CCP, and TGA2. No retarded bands were observed on incubation of the LS7 probe with GST–GFP or GST–NPR1 (Fig. 6B), indicating that GST, GFP, or NPR1 do not bind to the LS7 probe. In contrast, incubation with GST–TGA2 or GST–CCP resulted in retarded bands of the LS7 probe, indicating that TGA2 and CCP have faint binding activity of the LS7 probe (Fig. 6B, lanes 3 and 5). Furthermore, GST–TGA2 incubated with GST–NPR1 or GST–CCP binds to a substantially increased amount of the LS7 probe compared with GST–TGA2 alone (Fig. 6B, compare lanes 4 and 6 with 3). Therefore, these results suggest that CCP and NPR1 enhance the binding of TGA2 with the positive regulatory element (LS7) of the PR1 promoter.
Fig. 6.
CCP enhances the binding activity of TGA2 to the LS7 element of the PR1 promoter in vivo. (A) The nucleotide sequence of the LS7 element (underlined) and surrounding region in the PR1 promoter. The numbers on the sequence indicate the position of the nucleotide relative to the transcription start site of the PR1 mRNA. The sequence was used as a probe in EMSAs. (B) EMSA showing the binding of TGA2 with the LS7 element of the PR1 promoter. GST-tagged GFP, TGA2, NPR1, and CCP proteins were purified from E. coli and mixed in different combinations. Equal amounts of the biotin-labeled LS7 element were incubated with different protein combinations, and the DNA binding was analyzed by EMSAs. (C) ChIP-qPCR assays showing binding of CCP to fragments of the PR1 promoter in vivo. Chromatins from 35S:CCP and 35S:CCPmCBS transgenic Arabidopsis seedlings with anti-Flag beads were collected for DNA extraction and quantitative PCR assays. Non-transgenic Col-0 plants acted as a negative control. Mean ±SD, n=3. Top, diagram of the PR1 promoter and the fragments for Chip-qPCR. (D) A proposed working model of CCP-mediated defense activation. Upon plant pathogen challenges, CCP is induced and binds to copper. CCP is translocated into the nucleus and interacts with TGA2, resulting in induction of defense response genes and improved immunity against plant pathogens.
To examine whether CCP is associated with the PR1 promoter in vivo, we performed ChIP-PCR assays using 4-week-old seedlings of Col-0, CCPOE-3, and CCPmCBS/OE infected with Pst DC3000. CCPmCBS/OE-1 was generated by transforming Col-0 with CCPmCBS under the control of the CaMV 35S promoter. The qRCR results revealed that ‘b’ and ‘c’ amplicons containing the LS7 element of the PR1 promoter were enriched in the CCPOE-3 immunoprecipitated samples compared with CCPmCBS/OE (Fig. 6C). In contrast, the ‘a’ amplicon located 1400 bp upstream of the PR1 start codon was not enriched in the CCPOE-3 samples (Fig. 6C). These results indicate that CCP is preferentially associated with the TGA2-targeting LS7 element of the PR1 promoter.
Collectively, CCP is induced by plant pathogen infections and SA treatment. The CBS and NLS motif mediate the nuclear translocation, and then physically bind TGA2 in vivo at the LS7 element of the PR1 promoter to induce expression of defense genes (Fig. 6D).
Discussion
Copper is an essential nutrient in various biological processes, but highly concentrated free copper is extremely toxic to cells. Thus, intracellular copper must be chelated by copper chaperones to avoid copped-induced detrimental effects. In addition, copper chaperones are responsible for copper delivery to specific copper proteins and compartments. In previous studies, three copper chaperone classes were named based on their interaction proteins and functions: interaction with SODs (CCS), interaction with copper-transporting ATPases (ATX and CCH), and function in mitochondrial respiration (COX17, COX11, HCC1, and HCC2) (Burkhead et al., 2009; Attallah et al., 2011). Here, we have identified a novel class of putative copper chaperone (CCP) that was induced by pathogen infections and SA treatment. In healthy plants, CCP was mainly expressed in roots and inflorescence tissues, and rapidly up-regulated upon infections with CMV and Pst DC3000, as well as SA treatment (Fig. 3). These results suggest that CCP has important roles in defense responses to pathogen infections. Blast search revealed that many monocot and dicot plants have a single CCP ortholog (Fig. 1F), indicating that the conserved CCP orthologs provide a common strategy for regulation of defense response in plants.
In plants, the copper-dependent biological processes have been extensively elucidated in the cytoplasm (Pilon et al., 2006; Robinson and Winge, 2010). However, whether copper is transferred into the nucleus and mediates plant defense responses has not yet been investigated (Pilon et al., 2006; Robinson and Winge, 2010). Intriguingly, we found that CCP predominantly formed nuclear speckles (Fig. 2). The mammalian ATOX1 has been shown to be a copper-dependent transcription regulator in the nucleus (Itoh et al., 2008; Muller and Klomp, 2009; Kamiya et al., 2018). A lysine-rich region (KKTGK) in the C-terminus of ATOX1 is an NLS (Hamza et al., 2001; Itoh et al., 2008; Muller and Klomp, 2009). The alignment of CCP with ATOX1 revealed that a KKVGF at the C-terminus of CCP was an NLS, which was supported by point mutation analyses (Fig. 2). In contrast, ATX1 and CCH do not localize to the nucleus (Supplementary Fig. S12). Thus, CCP represents a putative plant copper chaperone functioning in the nucleus.
The mammalian ATOX1 protein requires copper for nuclear translocation and transcriptional activation in the nucleus (Itoh et al., 2008; Muller and Klomp, 2009). Furthermore, copper-dependent dimerization of ATOX1 was revealed by X-ray crystallography and biochemical analyses (Wernimont et al., 2000). Thus, it has been proposed that ATOX1 homodimerization might block interaction with its cytoplasmic interactors, ATP7A and ATP7B of P-type ATPases, and trigger ATOX1 to translocate into the nucleus to regulate transcription (Muller and Klomp, 2009). In agreement with mammalian ATOX1, plant CCP also requires copper binding for its nuclear translocation, self-interaction activity, and interaction with TGA2 (Figs 2, 6). In addition, it should be noted that CCP might also target some components for copper delivery and mediate unknown biological processes in the cytoplasm.
Mammalian ATOX1 has been shown to be a copper-dependent transcription regulator; however, details of the functions of ATOX1 in these processes remain elusive (Itoh et al., 2008; Muller and Klomp, 2009; Kahra et al., 2015; Kamiya et al., 2018; Matson Dzebo et al., 2018). Previous studies found that ATOX1 could directly bind the GAAAGA promoter sequence as a transcription factor (Itoh et al., 2008; Muller and Klomp, 2009; Kamiya et al., 2018). However, other studies found that ATOX1 had no DNA binding activities in vitro, and regulated gene transcription via DNA-binding transcription regulators. Here, CCP moves into the nucleus and activates expression of defense genes. We detected slight binding activity of CCP with the elements in the promoter of PR1 (Fig. 6A). Moreover, CCP may regulate PR1 gene expression by interacting with the TGA2 transcription factor that binds to the as-1 elements of the PR gene promoters (Zhang, 1999; Zhou, 2000; Johnson et al., 2003). In normal conditions, TGA2 is a transcriptional repressor that requires its N-terminus as a repression domain (Zhang et al., 2003; Rochon et al., 2006; Boyle et al., 2009). Upon pathogen infection or SA treatment, the N-terminal BTB/POZ domain of NPR1 interacts with TGA2, masking its repressor domain and forming an enhancesome with TGA2 to activate PR1 transcription (Boyle et al., 2009). Similarly, we found that CCP also interacted with the N-terminal repression domain of TGA2 (Fig. 5E), which induced PR1 expression probably by inactivating the repression activity of TGA2. In agreement with this hypothesis, the CCP protein can induce PR1 expression and enhance immunity against Pst DC3000 (Fig. 4). Given that TGA2, TGA5, and TGA6 act as redundant transcription factors in the SA signaling pathway (Zhang et al., 2003; Dröge-Laser et al., 2018), whether CCP interacts with TGA5 and TGA6 needs to be established in future studies. More interestingly, we found that GST–NPR1, rather than the negative control GST–GFP, could pull-down His-CCP and His-TGA2 together in the presence of Cu2+ (Supplementary Fig. S13), indicating that NPR1, CCP, and TGA2 are probably in a complex.
Taken together, our results provide insights that CCP is a putative copper chaperone family inducing defense against plant pathogens in the nucleus. As summarized in Fig. 6D, upon pathogen infections, the up-regulated CCP forms copper-dependent homodimers, and then undergoes nuclear translocation. Within the nucleus, the CCP homodimer binds the N-terminus of TGA2 that has been shown to be a repressor of transcription of PR1 without SA induction (Zhang et al., 2003; Mosher et al., 2006; Rochon et al., 2006; Boyle et al., 2009). The CCP homodimer binds to and masks the N-terminal repressive domain of TGA2, which probably results in negating the repression activity of TGA2 and positively regulating defense response genes to improve immunity against plant pathogens. There are still some difference between CCP and NPR1 regulatory mechanisms in plant immune responses. For example, since NPR1 transcription is not dramatically up-regulated in response to plant pathogen infections (Wang et al., 2006), post-translational modifications of NPR1 regulate its localization, turnover, and functions in plant immunity (reviewed in Withers and Dong, 2016). in comparison, CCP transcription is dramatically induced upon pathogen challenge. Future studies on the CCP-binding transcription factors as well as the mechanism of their genetic metabolic regulation in various plants will provide evidence for copper-induced defense in plants.
Supplementary data
The following supplementary data are available at JXB online.
Fig. S1. Identification of a CMV-induced copper chaperone in Arabidopsis.
Fig. S2. RT–PCR confirming the deletion of Lys7 in the lys7 mutant.
Fig. S3. Western blotting showing accumulation of GFP, GFP–CCP, GFP–CCPmCBS, and GFP–CCPmNLS.
Fig. S4. Confocal micrographs showing the subcellular localization of GUS–GFP–CCP, GUS–GFP–CCPmCBS, and GUS–GFP–CCPmNLS.
Fig. S5. Self-interactions of CCP derivatives and the negative controls of BiFC in N. benthamiana leaves.
Fig. S6. Subcellular localization of CCP in H2B–RFP transgenic N. benthamiana leaves treated with buffer or BCS.
Fig. S7. Subcellular localization of NbCCP, AtATX1, and AtCCH in H2B–RFP transgenic N. benthamiana leaves.
Fig. S8. Accumulation of CCP under excess copper.
Fig. S9. Accumulation of the CCP mRNA and protein in CCP overexpression lines.
Fig. S10. Identification of ccp-1 and ccp-2 mutant lines.
Fig. S11. BiFC analyzing the interactions of CCP and TGA2 treated with buffer or SA.
Fig. S12. Subcellular localization of TGA2 and mutants GFP, GFP–TGA2, GFP–TGA2–NT, GFP–TGA2–bZIP, and GFP–CT in H2B–RFP transgenic N. benthamiana leaves.
Fig. S13. GST pull-down analysis of the complex NPR1, TGA2, and CCP in vitro.
Table S1. Primers used in this study
Acknowledgements
We thank Jialin Yu, Dawei Li, Chenggui Han, Ying Wang, and Yongliang Zhang for their helpful suggestions and constructive criticism. We thank Professor Jun Fan at College of Plant Protection of China Agricultural University for his gift of the bacterial strain. We would also like to thank Professor Michael M. Goodin (University of Kentucky, Lexington, KY, USA) for H2B–RFP transgenic N. benthamiana seeds. This work was supported by grants from the Natural Science Foundation of China (31872920, 31571978) and the Fundamental Research Funds for the Central Universities (CAU, 2020TC163).
Glossary
Abbreviations:
- CBS
copper-binding site
- CCP
copper chaperone induced by pathogens
- CMV
Cucumber mosaic virus
- NLS
nuclear localization signal
Data availability
Accession numbers of genes used in this study are: Arabidopsis CCP (MN180192), ATX1 (NM_105295), CCH (NM_001339780), CCS (NM_101123); N. benthamiana CCP (NbCCP, XM_00959332); S. cerevisiae Lys7 (CCS, CP036475); and mammalian antioxidant-1 (ATOX1) (XM_004042855). The accession number of the RNA-seq data in this paper is PRJNA593673.
Author contributions
XBW and LXC designed the research; LXC, KD, ZZ, XPZ, XT, FFZ, and JZZ performed the experiments; XBW, XDF, TY, and QG analyzed the data and drafted the manuscript. XBW and LXC proofread and finalized the manuscript.
References
- Abdel-Ghany SE, Burkhead JL, Gogolin KA, Andrés-Colás N, Bodecker JR, Puig S, Peñarrubia L, Pilon M. 2005. AtCCS is a functional homolog of the yeast copper chaperone Ccs1/Lys7. FEBS Letters 579, 2307–2312. [DOI] [PubMed] [Google Scholar]
- Attallah CV, Welchen E, Martin AP, Spinelli SV, Bonnard G, Palatnik JF, Gonzalez DH. 2011. Plants contain two SCO proteins that are differentially involved in cytochrome c oxidase function and copper and redox homeostasis. Journal of Experimental Botany 62, 4281–4294. [DOI] [PubMed] [Google Scholar]
- Balandin T, Castresana C. 2002. AtCOX17, an Arabidopsis homolog of the yeast copper chaperone COX17. Plant Physiology 129, 1852–1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle P, Le Su E, Rochon A, Shearer HL, Murmu J, Chu JY, Fobert PR, Després C. 2009. The BTB/POZ domain of the Arabidopsis disease resistance protein NPR1 interacts with the repression domain of TGA2 to negate its function. The Plant Cell 21, 3700–3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkhead JL, Reynolds KA, Abdel-Ghany SE, Cohu CM, Pilon M. 2009. Copper homeostasis. New Phytologist 182, 799–816. [DOI] [PubMed] [Google Scholar]
- Cao H, Bowling SA, Gordon AS, Dong X. 1994. Characterization of an Arabidopsis mutant that is nonresponsive to inducers of systemic acquired resistance. The Plant Cell 6, 1583–1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu CC, Lee WC, Guo WY, Pan SM, Chen LJ, Li HM, Jinn TL. 2005. A copper chaperone for superoxide dismutase that confers three types of copper/zinc superoxide dismutase activity in Arabidopsis. Plant Physiology 139, 425–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis MD, Grossniklaus U. 2003. A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiology 133, 462–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Després C, DeLong C, Glaze S, Liu E, Fobert PR. 2000. The Arabidopsis NPR1/NIM1 protein enhances the DNA binding activity of a subgroup of the TGA family of bZIP transcription factors. The Plant Cell 12, 279–290. [PMC free article] [PubMed] [Google Scholar]
- Dröge-Laser W, Snoek BL, Snel B, Weiste C. 2018. The Arabidopsis bZIP transcription factor family—an update. Current Opinion in Plant Biology 45, 36–49. [DOI] [PubMed] [Google Scholar]
- Fang XD, Yan T, Gao Q, Cao Q, Gao DM, Xu WY, Zhang ZJ, Ding ZH, Wang XB. 2019. A cytorhabdovirus phosphoprotein forms mobile inclusions trafficked on the actin/ER network for viral RNA synthesis. Journal of Experimental Botany 70, 4049–4062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu ZQ, Dong X. 2013. Systemic acquired resistance: turning local infection into global defense. Annual Review of Plant Biology 64, 839–863. [DOI] [PubMed] [Google Scholar]
- Goodin MM, Dietzgen RG, Schichnes D, Ruzin S, Jackson AO. 2002. pGD vectors: versatile tools for the expression of green and red fluorescent protein fusions in agroinfiltrated plant leaves. The Plant Journal 31, 375–383. [DOI] [PubMed] [Google Scholar]
- Guo Z, Wang XB, Wang Y, Li WX, Gal-On A, Ding SW. 2018. Identification of a new host factor required for antiviral RNAi and amplification of viral siRNAs. Plant Physiology 176, 1587–1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamza I, Faisst A, Prohaska J, Chen J, Gruss P, Gitlin JD. 2001. The metallochaperone Atox1 plays a critical role in perinatal copper homeostasis. Proceedings of the National Academy of Sciences, USA 98, 6848–6852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison MD, Jones CE, Dameron CT. 1999. Copper chaperones: function, structure and copper-binding properties. Journal of Biological Inorganic Chemistry 4, 145–153. [DOI] [PubMed] [Google Scholar]
- Himelblau E, Mira H, Lin SJ, Culotta VC, Peñarrubia L, Amasino RM. 1998. Identification of a functional homolog of the yeast copper homeostasis gene ATX1 from Arabidopsis. Plant Physiology 117, 1227–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh S, Kim HW, Nakagawa O, Ozumi K, Lessner SM, Aoki H, Akram K, McKinney RD, Ushio-Fukai M, Fukai T. 2008. Novel role of antioxidant-1 (Atox1) as a copper-dependent transcription factor involved in cell proliferation. Journal of Biological Chemistry 283, 9157–9167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson C, Boden E, Arias J. 2003. Salicylic acid and NPR1 induce the recruitment of trans-activating TGA factors to a defense gene promoter in Arabidopsis. The Plant Cell 15, 1846–1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahra D, Mondol T, Niemiec MS, Wittung-Stafshede P. 2015. Human copper chaperone Atox1 translocates to the nucleus but does not bind DNA in vitro. Protein and Peptide Letters 22, 532–538. [DOI] [PubMed] [Google Scholar]
- Kamiya T, Takeuchi K, Fukudome S, Hara H, Adachi T. 2018. Copper chaperone antioxidant-1, Atox-1, is involved in the induction of SOD3 in THP-1 cells. Biometals 31, 61–68. [DOI] [PubMed] [Google Scholar]
- Kinkema M, Fan W, Dong X. 2000. Nuclear localization of NPR1 is required for activation of PR gene expression. The Plant Cell 12, 2339–2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliebenstein DJ, Monde RA, Last RL. 1998. Superoxide dismutase in Arabidopsis: an eclectic enzyme family with disparate regulation and protein localization. Plant Physiology 118, 637–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JZ, Zhou LY, Peng YL, Fan J. 2019. Pseudomonas bacteriocin syringacin M released upon desiccation suppresses the growth of sensitive bacteria in plant necrotic lesions. Microbial Biotechnology 13, 134–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin K, Kopperud K, Chakrabarty R, Banerjee R, Brooks R, Goodin MM. 2009. Transient expression in Nicotiana benthamiana fluorescent marker lines provides enhanced definition of protein localization, movement and interactions in planta. The Plant Journal 59, 150–162. [DOI] [PubMed] [Google Scholar]
- Matson Dzebo M, Blockhuys S, Valenzuela S, Celauro E, Esbjörner EK, Wittung-Stafshede P. 2018. Copper chaperone Atox1 interacts with cell cycle proteins. Computational and Structural Biotechnology Journal 16, 443–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosher RA, Durrant WE, Wang D, Song J, Dong X. 2006. A comprehensive structure–function analysis of Arabidopsis SNI1 defines essential regions and transcriptional repressor activity. The Plant Cell 18, 1750–1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mou Z, Fan W, Dong X. 2003. Inducers of plant systemic acquired resistance regulate NPR1 function through redox changes. Cell 113, 935–944. [DOI] [PubMed] [Google Scholar]
- Muller PA, Klomp LW. 2009. ATOX1: a novel copper-responsive transcription factor in mammals? International Journal of Biochemistry & Cell Biology 41, 1233–1236. [DOI] [PubMed] [Google Scholar]
- Oyama T, Shimura Y, Okada K. 1997. The Arabidopsis HY5 gene encodes a bZIP protein that regulates stimulus-induced development of root and hypocotyl. Genes & Development 11, 2983–2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilon M, Abdel-Ghany SE, Cohu CM, Gogolin KA, Ye H. 2006. Copper cofactor delivery in plant cells. Current Opinion in Plant Biology 9, 256–263. [DOI] [PubMed] [Google Scholar]
- Puig S, Mira H, Dorcey E, et al. 2007. Higher plants possess two different types of ATX1-like copper chaperones. Biochemical and Biophysical Research Communications 354, 385–390. [DOI] [PubMed] [Google Scholar]
- Qin XF, Holuigue L, Horvath DM, Chua NH. 1994. Immediate early transcription activation by salicylic acid via the cauliflower mosaic virus as-1 element. The Plant Cell 6, 863–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson NJ, Winge DR. 2010. Copper metallochaperones. Annual Review of Biochemistry 79, 537–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochon A, Boyle P, Wignes T, Fobert PR, Després C. 2006. The coactivator function of Arabidopsis NPR1 requires the core of its BTB/POZ domain and the oxidation of C-terminal cysteines. The Plant Cell 18, 3670–3685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seyfferth C, Tsuda K. 2014. Salicylic acid signal transduction: the initiation of biosynthesis, perception and transcriptional reprogramming. Frontiers in Plant Science 5, 697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin LJ, Lo JC, Yeh KC. 2012. Copper chaperone antioxidant protein1 is essential for copper homeostasis. Plant Physiology 159, 1099–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin LJ, Yeh KC. 2012. Overexpression of Arabidopsis ATX1 retards plant growth under severe copper deficiency. Plant Signaling & Behavior 7, 1082–1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uknes S, Mauch-Mani B, Moyer M, Potter S, Williams S, Dincher S, Chandler D, Slusarenko A, Ward E, Ryals J. 1992. Acquired resistance in Arabidopsis. The Plant Cell 4, 645–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter M, Chaban C, Schütze K, et al. 2004. Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. The Plant Journal 40, 428–438. [DOI] [PubMed] [Google Scholar]
- Wang D, Amornsiripanitch N, Dong X. 2006. A genomic approach to identify regulatory nodes in the transcriptional network of systemic acquired resistance in plants. PLoS Pathogens 2, e123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Guo C, Peng J, et al. 2019. ABRE-BINDING FACTORS play a role in the feedback regulation of ABA signaling by mediating rapid ABA induction of ABA co-receptor genes. New Phytologist 221, 341–355. [DOI] [PubMed] [Google Scholar]
- Weigel RR, Bäuscher C, Pfitzner AJ, Pfitzner UM. 2001. NIMIN-1, NIMIN-2 and NIMIN-3, members of a novel family of proteins from Arabidopsis that interact with NPR1/NIM1, a key regulator of systemic acquired resistance in plants. Plant Molecular Biology 46, 143–160. [DOI] [PubMed] [Google Scholar]
- Wernimont AK, Huffman DL, Lamb AL, O’Halloran TV, Rosenzweig AC. 2000. Structural basis for copper transfer by the metallochaperone for the Menkes/Wilson disease proteins. Nature Structural Biology 7, 766–771. [DOI] [PubMed] [Google Scholar]
- Withers J, Dong X. 2016. Posttranslational modifications of NPR1: a single protein playing multiple roles in plant immunity and physiology. PLoS Pathogens 12, e1005707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing HL, Dong L, Wang ZP, Zhang HY, Han CY, Liu B, Wang XC, Chen QJ. 2014. A CRISPR/Cas9 toolkit for multiplex genome editing in plants. BMC Plant Biology 14, 327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan S, Dong X. 2014. Perception of the plant immune signal salicylic acid. Current Opinion in Plant Biology 20, 64–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan T, Zhu JR, Di D, Gao Q, Zhang Y, Zhang A, Yan C, Miao H, Wang XB. 2015. Characterization of the complete genome of Barley yellow striate mosaic virus reveals a nested gene encoding a small hydrophobic protein. Virology 478, 112–122. [DOI] [PubMed] [Google Scholar]
- Yoo SD, Cho YH, Sheen J. 2007. Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nature Protocols 2, 1565–1572. [DOI] [PubMed] [Google Scholar]
- Zhang XP, Liu DS, Yan T, Fang XD, Dong K, Xu J, Wang Y, Yu JL, Wang XB. 2017. Cucumber mosaic virus coat protein modulates the accumulation of 2b protein and antiviral silencing that causes symptom recovery in planta. PLoS Pathogens 13, e1006522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Tessaro MJ, Lassner M, Li X. 2003. Knockout analysis of Arabidopsis transcription factors TGA2, TGA5, and TGA6 reveals their redundant and essential roles in systemic acquired resistance. The Plant Cell 15, 2647–2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YL, Fan WH, Kinkema M, Li X, Dong X. 1999. Interaction of NPR1 with basic leucine zipper protein transcription factors that bind sequences required for salicylic acid induction of the PR gene. Proceedings of the National Academy of Sciences, USA 96, 6523–6528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Tong X, Liu SY, Chai LX, Zhu FF, Zhang XP, Zou JZ, Wang XB. 2019. Genetic analysis of a Piezo-like protein suppressing systemic movement of plant viruses in Arabidopsis thaliana. Scientific Reports 9, 3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou JM, Trifa Y, Silva H, Pontier D, Lam E, Shah J, Klessig DF. 2000. NPR1 differentially interacts with members of the TGA/OBF family of transcription factors that bind an element of the PR-1 gene required for induction by salicylic acid. Molecular Plant-Microbe Interactions 13, 191–202. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Accession numbers of genes used in this study are: Arabidopsis CCP (MN180192), ATX1 (NM_105295), CCH (NM_001339780), CCS (NM_101123); N. benthamiana CCP (NbCCP, XM_00959332); S. cerevisiae Lys7 (CCS, CP036475); and mammalian antioxidant-1 (ATOX1) (XM_004042855). The accession number of the RNA-seq data in this paper is PRJNA593673.