We discuss the contribution of the versatile and atypical plant histone deacetylase, HDA9, to plant growth, development, and acclimation to the environment, and propose novel leads for future research.
Keywords: ABA INSENSITIVE 4 (ABI4), Arabidopsis, EARLY FLOWERING 3 (ELF3), gene regulation, HDA9, histone deacetylase 9, HIGH EXPRESSION OF OSMOTICALLY RESPONSIVE GENES 15 (HOS15), ELONGATED HYPOCOTYL 5 (HY5), POWERDRESS (PWR), WRKY53
Abstract
Plants tightly control gene transcription to adapt to environmental conditions and steer growth and development. Different types of epigenetic modifications are instrumental in these processes. In recent years, an important role for the chromatin-modifying RPD3/HDA1 class I HDAC HISTONE DEACETYLASE 9 (HDA9) emerged in the regulation of a multitude of plant traits and responses. HDACs are widely considered transcriptional repressors and are typically part of multiprotein complexes containing co-repressors, DNA, and histone-binding proteins. By catalyzing the removal of acetyl groups from lysine residues of histone protein tails, HDA9 negatively controls gene expression in many cases, in concert with interacting proteins such as POWERDRESS (PWR), HIGH EXPRESSION OF OSMOTICALLY RESPONSIVE GENES 15 (HOS15), WRKY53, ELONGATED HYPOCOTYL 5 (HY5), ABA INSENSITIVE 4 (ABI4), and EARLY FLOWERING 3 (ELF3). However, HDA9 activity has also been directly linked to transcriptional activation. In addition, following the recent breakthrough discovery of mutual negative feedback regulation between HDA9 and its interacting WRKY-domain transcription factor WRKY53, swift progress in gaining understanding of the biology of HDA9 is expected. In this review, we summarize knowledge on this intriguing versatile—and long under-rated—protein and propose novel leads to further unravel HDA9-governed molecular networks underlying plant development and environmental biology.
Introduction
Eukaryotic DNA is orderly and densely packed into higher order structures, called chromatin. The first level of chromatin compaction comprises a histone protein octamer that wraps ~147 bp (Luger et al., 1997; Rosa and Shaw, 2013). This basal protein–DNA unit, called a nucleosome, contains a tetramer of two dimers consisting of four core histone (H) proteins each: H2A/H2B and H3/H4. Besides the four canonical histones, various histone variants exist with different physical properties and biological functions (Henikoff and Smith, 2015; Talbert and Henikoff, 2017). Histone proteins contain unstructured N-terminal tails that extrude from the nucleosomes and are prone to post-translational epigenetic modifications such as acetylation, methylation, SUMOylation, ubiquitination, and phosphorylation (Berger, 2007; Rosa and Shaw, 2013; Liu et al., 2014). Such epigenetic modifications regulate the accessibility of DNA to binding proteins, such as transcription factors and DNA polymerases, by modulating the electrostatic interactions between the histones and DNA molecule (Bowman and Poirier, 2015).
Histone acetylation is a dynamic and versatile epigenetic mark that occurs at lysine (K) residues on the histone tails and causes histones to shift from a positive to a neutral charge, thereby typically allowing for a transcriptionally prone, decondensed chromatin environment. Histone acetyltransferases (HATs) catalyze the deposition of acetyl groups, whereas histone deacetylases (HDACs) remove these marks (Pandey et al., 2002; Liu et al., 2014; Chen et al., 2020). Hence, HDACs are associated with SWI-INDEPENDENT3 (SIN3)-like co-repressors and are often—but not exclusively—associated with silenced genes (Li et al., 2002; Tian et al., 2005; Alinsug et al., 2009). Other factors in HDAC multiprotein co-repressor complexes typically are DNA-binding factors, chromatin-modifying enzymes, and several other structural and regulatory proteins (Grzenda et al., 2009; Perrella et al., 2013, 2016; Liu et al., 2014). Together, HDAC multiprotein complexes orchestrate enzymatic activity, cofactor recruitment, substrate binding, and genomic targeting.
In Arabidopsis thaliana, there are 18 proteins recognized as HDACs that are categorized into three families: the Reduced Potassium Dependence3 (RPD3/HDA1-like) family, the plant-specific HD2-type family, and the NAD-dependent Silent Information Regulator (SIR) family. These families contain twelve, four, and two members, respectively. The RPD3/HDA1-like family is subdivided into three classes (I–III) based on sequence similarity (Pandey et al., 2002; Hollender and Liu, 2008; Alinsug et al., 2009). HDACs exert diverse functions in plants. For a detailed overview of HDACs, we refer the reader to Liu et al. (2014) and Chen et al. (2020).
In recent years, the RPD3/HDA1 class I HDAC HISTONE DEACETYLASE 9 (HDA9) has gained increasing attention. Phylogenetic analyses indicate that HDA9 is homologous to the functional HDACs: HDA6, HDA7, and HDA19 (Pandey et al., 2002; Hollender and Liu, 2008; Alinsug et al., 2009). In addition, HDA9 is closely related to HDA10 and HDA17, which are physically located next to HDA9 on the genome. These pseudogenes lack a catalytic HDAC domain and probably originated from a HDA9 duplication and genomic rearrangement event (Pandey et al., 2002; Alinsug et al., 2009).
Unlike other functional plant HDACs, HDA9 contains a BH3-only pro-apoptotic (BAD) domain (Alinsug et al., 2009), that allows for interaction with 14-3-3 proteins that are associated with a multitude of signaling proteins and have a role in hormone, kinase, phosphatase, and transmembrane receptor signaling pathways (Jaspert et al., 2011; Camoni et al., 2018).
HDA9 expression is observed in several Arabidopsis organs and tissues across developmental stages, which suggests that HDA9 functions throughout the plant’s life cycle (Van Zanten et al., 2014; Kang et al., 2015; Suzuki et al., 2018; Mayer et al., 2019; van der Woude et al., 2019) (Table 1). In germinating seedlings, HDA9 is mainly present in below-ground parts and the root–hypocotyl junction (Van Zanten et al., 2014; van der Woude et al., 2019), and the gene becomes more ubiquitously expressed later in development (Hollender and Liu, 2008; Kang et al., 2015; Mayer et al., 2019). Accordingly, the Brassica juncea HDA9 homolog (BjuHDA9) is ubiquitously detected throughout the plant and particularly in floral tissues (Yan et al., 2018).
Table 1.
Confirmed HDA9 expression domains across plant developmental stages and their corresponding literature references
HDA9 substrates include H3K9Ac, H3K14Ac, H3K18Ac, H3K27Ac, H3K36Ac, and H3K56Ac (Kim et al., 2013; Van Zanten et al., 2014; Kang et al., 2015; Chen et al., 2016; Kim et al., 2016; Mayer et al., 2019; Park et al., 2019; van der Woude et al., 2019; Yang et al., 2020; Zeng et al., 2020; Zheng et al., 2020), but not H4 or H2A lysines (Kim et al., 2016; Mayer et al., 2019). In addition, hda9 mutants display altered H3K9Me1, H3K9Me2, H3K27Me1, H3K27Me2, and H3K36Me2 levels (Chen et al., 2016; Kim et al., 2016; Mayer et al., 2019; Zeng et al., 2020). How HDA9 affects histone methylation is unknown, but HDA9 is likely to play a facilitating role, as HDA9-mediated H3K27 deacetylation is required for Polycomb Repressive Complex 2 (PRC2)-mediated H3K27me3 (Zeng et al., 2020). Furthermore, accumulation of miRNAs (miR157, miR162, and miR172) was impaired in the hda9 mutant background, suggesting a possible role for HDA9 in the regulation of miRNA production (Kim et al., 2009).
In general, HDA9 targets histones positioned close to transcriptional start sites of actively transcribed genes at euchromatic regions (Fig. 1) (Chen et al., 2016; Kim et al., 2016; Mayer et al., 2019; Yang et al., 2019). Consistent with its role in transcriptional regulation, the association of HDA9 with genomic targets correlates well with mRNA expression levels (Chen et al., 2016; Kim et al., 2016; Mayer et al., 2019; Yang et al., 2019; Baek et al., 2020).
Fig. 1.
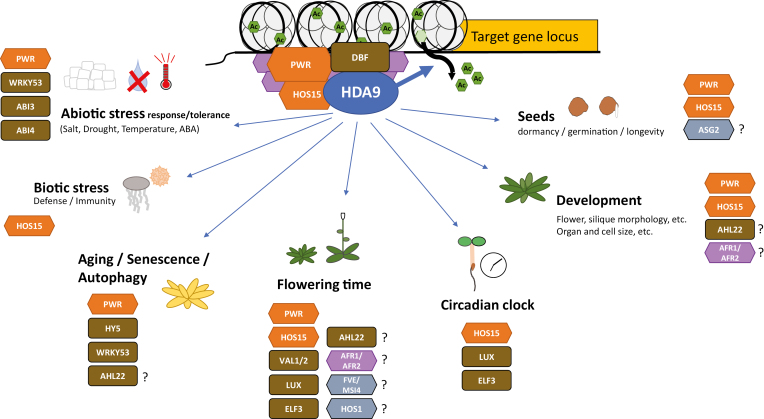
Schematic representation of the HDA9–PWR–HOS15 core histone deacetylase complex and their roles in plant development and responses to the environment. The catalytic HDAC HDA9 (blue oval), together with its core complex components PWR and HOS15 (orange elongated hexagons) and other structural components (purple hexagon), such as AFR1/AFR2, facilitate the de-acetylation (green hexagons) of histones in nucleosome complexes (gray circles), around which two turns of DNA are wrapped (black lines). This affects chromatin accessibility for regulatory proteins and the transcription machinery, and thereby controls the expression of its target genes (yellow box). The HDA9–core histone deacetylase complex is targeted to DNA promoter elements by DNA-binding factors (DBFs; brown boxes), that includes transcription factors such as WRKY53, HY5, ELF3, ABI3, and ABI4. Other known HDA9 partners are the DNA-binding proteins AHL22, VAL1, and VAL2, as well as ASG2, FVE/MSI4, and HOS1 (gray hexagons). The HDA9–PWR–HOS15 complex regulates diverse processes throughout the plant’s life cycle as well as responses and tolerance to the indicated biotic and abiotic stresses. The diverse HDA9-mediated processes and responses rely on different DNA-binding and other proteins (known factors are depicted in association with the mentioned process/response).
Despite the fact that some HDACs target non-histone protein substrates (Hartl et al., 2017; Zheng et al., 2020), for a long time there was no evidence suggesting that HDA9 can deacetylate proteins other than histone H3, even though HDA9 has been detected in both the cytoplasm and the nucleus (Kang et al., 2015; Ducos et al., 2017; Suzuki et al., 2018; Mayer et al., 2019; Yang et al., 2019). A recent study, however, demonstrated that HDA9 can remove acetyl groups and thereby negatively regulates the transcriptional activity of its interacting transcription factor protein WRKY53 (Zheng et al., 2020). Pharmacological evidence showed that HDA9 is prone to proteasomal regulation (Mayer et al., 2019) and it has been suggested that HDA9 may be associated with a CUL4-based E3 ligase (Park et al., 2019).
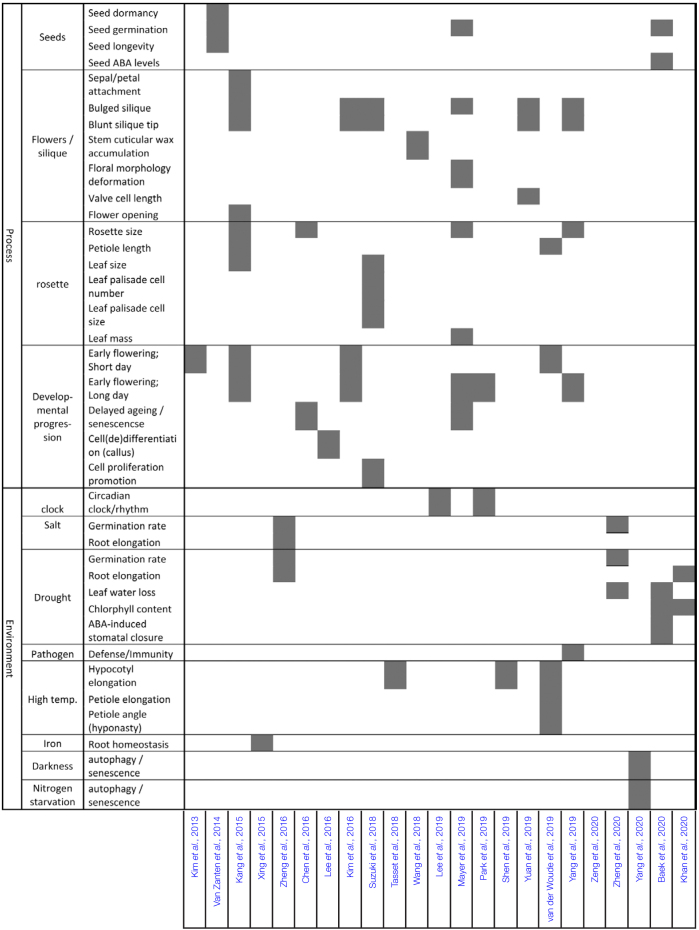
On the phenotypic level (Fig. 1), HDA9 regulates diverse traits including seed dormancy (Van Zanten et al., 2014; Baek et al., 2020), flowering time (Kim et al., 2013; Kang et al., 2015; Kim et al., 2016; Mayer et al., 2019; Park et al., 2019; van der Woude et al., 2019; Zeng et al., 2020), leaf senescence (Chen et al., 2016; Mayer et al., 2019; Yang et al., 2020), cellular differentiation (Lee et al., 2016), cell proliferation (Suzuki et al., 2018), suppression of stem cuticular wax crystal accumulation (Wang et al., 2018), flower opening, petal and sepal attachment to the receptacles (Kang et al., 2015), and several other developmental and physiological phenotypes (Fig. 1; Table 2). Moreover, HDA9 mediates responses to environmental signals such as salt, drought (Zheng et al., 2016; Baek et al., 2020; Khan et al., 2020; Zheng et al., 2020), and warm temperatures (Tasset et al., 2018; Shen et al., 2019; van der Woude et al., 2019).
Table 2.
Confirmed HDA9-mediated processes and responses to environmental stresses and their corresponding literature references
In this review, we report in detail the intriguing findings on the versatile role of the pleiotropic HDA9 chromatin-modifying protein (Fig. 1) and discuss possible future directions required to further unravel the function and regulation of HDA9-governed molecular networks.
HDA9-interacting proteins; the HDA9–HOS15–PWR core HDAC complex
The SANT (Swi3, Ada2, N-Cor, TFIIIB) domain-containing protein POWERDRESS (PWR) was identified by an immuno-purification approach as a high-confident HDA9-interacting protein (Chen et al., 2016) (Fig. 1; Table 3). In addition, HDA9 was identified in a screen for hdac mutants with early flowering and bulged silique phenotypes similar to pwr mutants (Yumul et al., 2013; Kim et al., 2016). Consistent with the proposed role for PWR in HDAC multiprotein complexes, a histone H3 hyperacetylation phenotype was observed in pwr mutants, and pwr-hyperacetylated sites significantly overlapped with those found in the hda9 mutant background (Chen et al., 2016; Kim et al., 2016; Mayer et al., 2019). Furthermore, the WD40-repeat protein HIGH EXPRESSION OF OSMOTICALLY RESPONSIVE GENE 15 (HOS15) was shown to interact with both HDA9 and PWR (Fig. 1; Table 3) (Park et al., 2018a, b; Suzuki et al., 2018; Mayer et al., 2019; Park et al., 2019; Yang et al., 2019), and the hos15 mutant displayed histone hyperacetylation and methylation changes similar to hda9 and pwr mutants (Suzuki et al., 2018; Mayer et al., 2019; Yang et al., 2019). Moreover, HDA9 chromatin binding was reduced in hos15 (Chen et al., 2016) and pwr mutants (Kim et al., 2016), suggesting that PWR and HOS15 are required for HDA9 genome targeting.
Table 3.
Confirmed HDA9-interacting proteins
| Interacting protein | Reference | Technique(s) used for interaction study | Target gene identification/ confirmation method(s)a |
|---|---|---|---|
| WRKY53 | Zheng, 2020 | Y2H, Co-IP, BiFC | qRT-PCR, ChIP-PCR, transient expression assays |
| PWR, WRKY53, AHL22 | Chen, 2016 | Co-IP, in vitro IP | qRT-PCR, RNA-seq, ChIP-PCR, ChIP-seq |
| PWR | Kim, 2016 | Co-IP, Y2H | qRT-PCR, RNA-seq, ChIP-PCR, ChIP-seq |
| HOS15, PWR | Suzuki, 2018 | Y2H | qRT-PCR |
| HOS15, PWR | Mayer 2019 | IP-MS, IP, BiFC | qRT-PCR, RNA-seq, ChIP-PCR, ChIP-seq |
| HOS15 | Yang, 2019 | Co-IP | qRT-PCR, RNA-seq, ChIP-PCR |
| HOS15 | Park 2018a | Split-LUC | qRT-PCR, ChIP-PCR |
| HOS15 | Park 2018b | IP-MS, Co-IP, Y2H, LCI | NA |
| HOS15, ELF3, LUX | Park 2019 | Co-IPb | qRT-PCR, RNA-seq, ChIP-PCR |
| ELF3 | Lee, 2019 | Y2H, Co-IP, BiFC | qRT-PCR, ChIP-PCR, transient expression assays. |
| AHL22 | Yun, 2012 | BiFC, in vitro pulldown | qRT-PCR, ChIP-PCR, EMSA, MAR binding assay |
| AFR1, AFR2 | Gu, 2013 | Y2H | qRT-PCR, ChIP-PCR |
| HOS1, FVE/MSI4 | Jung, 2013 | Y2H | qRT-PCR, ChIP-PCR |
| CYP1-1. HDA6, HDA19 c | Zheng, 2016 | Y2H | qRT-PCR, RNA-seq, ChIP-PCR |
| ASG2 | Ducos, 2017 | BiFC | NA |
| VAL1, VAL2 | Zeng, 2020 | Co-IP, Y2H | qRT-PCR, ChIP-PCR |
| HY5 | Yang, 2020 | Co-IP, BiFC | qRT-PCR, ChIP-PCR, dual-luciferase reporter assay |
| ABI4 | Baek, 2020 | Co-IP, Y2H | qRT-PCR, ChIP-PCR |
| ABI4, ABI3 | Khan, 2020 | Co-IP, Y2H | qRT-PCR, ChIP-PCR |
a The indicated techniques were used to identify target genes of either HDA9 and/or of the specified HDA9-interacting protein.
b Co-IP data by Park et al. (2019) suggest HDA9–LUX interaction, but a yeast two-hybrid assay did not confirm this interaction (Lee et al., 2019).
c These proposed interactions (yeast two-hybrid-based) should be considered with care, as Yuan et al. (2019) did not detect interaction between HDA9 and HDA6.
Abbreviations; Y2H, yeast two-hybrid; Co-IP, co-immunoprecipitation; IP-MS, immunoprecipitation followed by MS; BiFC, bimolecular fluorescence complementation; LCI, luciferase complementation imaging, qRT-PCR, quantitative real-time PCR; ChIP-PCR, chromatin immunoprecipitation-PCR; ChIP-seq, ChIP sequencing; RNA-seq, RNA sequencing (whole-transcriptome sequencing); MAR, matrix-attachment region; NA, not applicable.
Several hda9 mutant phenotypes, including altered leaf size, leaf palisade cell number and palisade cell size (Suzuki et al., 2018), and other traits further discussed below, are equally affected in hos15 (and pwr) single mutants and, to the best of our knowledge, no clear additive effects were observed in higher order mutants for any of the tested phenotypes. Moreover, HDA9, PWR, and HOS15 are co-expressed in different tissues (Mayer et al., 2019) and hda9, hos15, and pwr mutant transcriptomes exhibit a large overlap (Chen et al., 2016; Mayer et al., 2019). In fact, nucleocytoplasmic fractionation assays demonstrated that PWR and HOS15 are required for HDA9 accumulation in the nucleus, and pwr and hos15 mutants show significantly reduced nuclear HDA9 levels (Chen et al., 2016; Mayer et al., 2019). However, the mutant transcriptomes of pwr and hos15 suggest that both display HDA9-independent effects on gene regulation, possibly by interacting with other HDAC transcriptional co-repressors. Indeed, unlike HDA9 and PWR, HOS15 also targets acetylated H4 (Zhu et al., 2008).
Taken together, HDA9–PWR–HOS15 form a core HDAC complex to control gene transcription (Fig. 1). In addition, HDA9 physically interacts with the DNA-binding AT-HOOK MOTIF-CONTAINING 22 (AHL22) protein (Yun et al., 2012; Chen et al., 2016) and with AP30 FUNCTION-RELATED 1 (AFR1) and AFR2, being the plant relatives of yeast SAP30 FUNCTION-RELATED 1, a Sin3-associated structural component of HDAC complexes (Gu et al., 2013) (Table 3). Up to now, the contribution of AHL22 and AFR1/AFR2 to HDA9-mediated phenotypes is poorly understood. However, AHL22 overexpression results in short and stunted siliques and compact plants (Yun et al., 2012), similar to pwr and hda9 mutants. However, unlike pwr and hda9 (Kim et al., 2013; Yumul et al., 2013; Kang et al., 2015; Kim et al., 2016; Mayer et al., 2019; Park et al., 2019; van der Woude et al., 2019; Yang et al., 2019; Zeng et al., 2020), AHL22 overexpression leads to delayed flowering (Yun et al., 2012). Furthermore, afr1 and afr2 mutants exhibit elongated petioles and an open rosette structure, which is in contrast to the stunted hda9 mutant phenotype (Gu et al., 2013), whereas similarly to hda9 and pwr, the afr mutants exhibit early flowering (Gu et al., 2013). Additional proteins shown to interact with HDA9 include ASG2 (Ducos et al., 2017), EARLY FLOWERING3 (ELF3), and possibly LUX ARRHYTHMO (LUX) (Lee et al., 2019; Park et al., 2019), VP1/ABI3-LIKE 1 (VAL1) and VAL2 (Zeng et al., 2020), ELONGATED HYPOCOTYL 5 (HY5) (Yang et al., 2020), and ABA INSENSITIVE4 (ABI4) and ABI3 (Baek et al., 2020, Khan et al., 2020). These interactions are discussed below (Fig. 1; Table 3). The biological function of the indicated interaction between HDA9, HOS1, and FVE/ MULTICOPY SUPPRESSOR OF IRA1 4 (MSI) (Jung et al., 2013) requires further investigation (Table 3).
Interestingly, similar to the hda9 mutant phenotypes, mutants in histone deacetylase complex 1 (HDC1), a factor that interacts with HDACs and quantitatively determines histone acetylation levels, exhibited short petioles and a compact stature (Perrella et al., 2013, 2016). This suggests that HDC1 may also be part of the HDA9–PWR–HOS15 multiprotein complex. However, a possible direct interaction between HDC1 and HDA9 remains to be established. Notably, to the best of our knowledge, the HDA9-interacting proteins so far identified are fundamentally different from other HDACs studied. In particular, co-immunoprecipitation (Co-IP) using HDA6 or HDA19 as baits revealed mainly interactions with the conserved subunits of the RPD3-containing HDAC complex, including SIN3-like co-repressor proteins (SNL1–SNL6) and MSI1, and with each other (Perrella et al., 2013; Mehdi et al., 2016; Ning et al., 2019). This could indicate that the HDAC complex containing HDA9 may be fundamentally divergent from related canonical HDACs.
The transcription factors WRKY53 (Chen et al., 2016; Zheng et al., 2020), ABI4, ABI3 (Baek et al., 2020, Khan et al., 2020), and HY5 (Yang et al., 2020), the epigenome readers VP1/ABI3-LIKE 1 (VAL1) and VAL2 (Zeng et al., 2020), the circadian clock Evening Complex (EC) transcription factor(s) ELF3 and possibly LUX (Lee et al., 2019; Park et al., 2019), and AT-hook motif-containing protein AHL22 (Yun et al., 2012) are currently the only confirmed HDA9-interacting proteins with DNA binding capacity (Fig. 1). In particular, the molecular mechanism of the HDA9–WRKY53 interaction is now understood in detail (Zheng et al., 2020). Despite the limited number of confirmed HDA9 interactors, it is likely that HDA9 associates directly—or as part of a bigger HDAC multiprotein complex—with many more yet to be discovered DNA-binding factors.
The role of HDA9 in circadian clock regulation
Coordinated plant growth and development depend on tight regulation by the circadian clock. Circadian rhythms are entrained by environmental cues such as daylength and ambient temperature, and regulate vital processes such as metabolism, energy homeostasis, plant growth, stomatal closure, positional movement of leaves, and flowering initiation (Jouve et al., 1998; Dowson-Day and Millar, 1999; McClung, 2006; Park et al., 2019). At the core of the complex circadian clock regulation are multiple interlocking transcriptional feedback loops that regulate the clock’s output across a day/night cycle. The so-called central oscillator consists, among other factors, of two morning-expressed MYB transcription factors, LATE ELONGATED HYPOCOTYL (LHY) and CIRCADIAN CLOCK-ASSOCIATED 1 (CCA1), and the evening-expressed TIMING OF CAB EXPRESSION 1 (TOC1) (Gendron et al., 2012) (also referred to as PSEUDO RESPONSE REGULATOR 1 or PRR1), as well as other PRR family members such as PPR5, PPR7, and PPR9 (Nakamichi et al., 2010; Chow et al., 2012), GIGANTEA (GI), and the EC factors LUX, ELF3, and ELF4 (Ezer et al., 2017; McClung, 2019).
Over a third of Arabidopsis gene transcripts are controlled by the circadian clock (Michael and McClung, 2003; Kim et al., 2017), and rhythmic chromatin modifications have been associated with Arabidopsis circadian clock regulation (Farinas and Mas, 2011; Malapeira et al., 2012; Hung et al., 2018). The activity of CCA1, together with the MYB transcription factor REVEILLE8 (RVE8) for instance, causes differential H3 acetylated states at the TOC1 promoter region. At dawn, CCA1 represses chromatin accessibility via the recruitment of HDACs or repression of HATs (Perales and Más, 2007). During the daytime, CCA1 is antagonized by RVE8, correlating with H3 acetylation (Farinas and Mas, 2011; Malapeira et al., 2012; Hung et al., 2018), and rhythmic changes in histone marks are closely associated with clock activity (Perales and Más, 2007; Farinas and Mas, 2011; Lee et al., 2019).
A recent study demonstrated that expression of the circadian clock genes CCR2, CAB2, CCA1, and TOC1 displays signs of period shortening and advanced rhythmic phase in the hda9 mutant background (Lee et al., 2019). However, HDA9 expression itself did not show a significant circadian oscillation in wild-type plants. Subsequent analysis demonstrated that HDA9 is recruited to the TOC1 promoter region, thereby promoting H3 deacetylation. This resulted in TOC1 repression after its peak expression during the night (Lee et al., 2019).
Furthermore, it was recently found that HDA9 interacts with ELF3 when in complex with LUX (Lee et al., 2019; Park et al., 2019) (Table 3). Whether HDA9 directly interacts with LUX is not yet clear. Co-IP data by Park et al. (2019) would suggest so. Yet, a yeast two-hybrid assay did not confirm a direct interaction between the two proteins (Lee et al., 2019). Nevertheless, TOC1 repression is mediated by HDA9 via a direct and rhythmic interaction with ELF3 (Lee et al., 2019). Indeed, HDA9-dependent deacetylation and HDA9 association with the TOC1 promoter was impaired in elf3 mutants, comparable with hda9 mutants. Similarly, the HOS15–HDA9–EC complex dampens the rhythmic expression of GI, by mediating the deacetylation of GI-associated histone proteins, mainly in the late afternoon (Park et al., 2019). Moreover, it was shown that HOS15–HDA9 is targeted to the GI locus by LUX and ELF3 and that this is necessary for the deacetylation of H3 at the GI promoter to repress flowering (Park et al., 2019).
HDA9 control of flowering time
Flowering time is tightly regulated by several endogenous developmental cues and environmental variables such as temperature and photoperiod (Cho et al., 2017). Several studies have reported an intrinsic role for HDA9 in flowering time control.
Mutations in HDA9 lead to a mild early flowering phenotype under otherwise non-inductive short-day (SD) photoperiod conditions, seemingly independent of the CONSTANS/SUPPRESSOR OF OVEREXPRESSION OF CO1 (CO/SOC1) pathway (Kim et al., 2013; Kang et al., 2015; Kim et al., 2016; van der Woude et al., 2019). Subsequent analysis revealed that hda9 mutants show increased expression levels of the floral activator AGAMOUS-LIKE19 (AGL19) in SDs, which is accompanied by increased H3K9Ac and H3K27Ac levels at the AGL19 chromatin. Subsequent ChIP experiments indicated that HDA9 is indeed capable of binding to the AGL19 locus and directly affects AGL19 transcription by mediating deacetylation, thereby repressing flowering (Kim et al., 2013; Kang et al., 2015). Similar results were found in the hos15 mutant in inductive long-day (LD) conditions, where AGAMOUS-LIKE 19 (AGL19) and AGL24 as well as CO and SOC1 in these conditions (Park et al., 2019) were up-regulated.
Kim et al. (2013) did not observe altered expression or differential H3K9Ac or H3K27Ac levels of the flowering time regulator FLOWERING LOCUS C (FLC) under SD or LD conditions in hda9 mutants. Also, Park et al. (2019) reported that levels of the floral repressor FLC were unchanged in the hos15 mutant background in LD conditions. Kang et al. (2015), however, demonstrated that loss of HDA9 led to a slight reduction in FLC, as well as MAF4 and MAF5, mRNA levels in both LD and SD conditions. Yet, their genetic analyses suggested that HDA9 mediates flowering time largely independently of FLC (Kang et al., 2015). However, a very recent report showed that HDA9 associates with the CURLY FLOWER (CLF)–PRC2 transcriptional repressor complex, to regulate FLC repression and thereby flowering time, based on a forward genetic approach (Zeng et al., 2020). The authors reported that FLC transcription was markedly up-regulated in the hda9 mutant background in LD conditions and accordingly, HDA9 associated with the FLC locus and directly mediated local histone deacetylation (Zeng et al., 2020). CLF–PRC2 recruitment and H3K27Me3 levels at the FLC locus were partly reduced in the hda9 mutant background. This suggests that HDA9-mediated H3K27 deacetylation is a prerequisite for CLF–PRC2-mediated repressive H3K27Me3 marker deposition and thereby FLC repression (Zeng et al., 2020). Interestingly, genome-wide analysis showed that this requirement applies across the genome and is not restricted to FLC alone. In addition, HDA9 was shown to physically interact with the CLF–PRC2-interacting proteins VP1/ABI3-LIKE 1 (VAL1) and VAL2, that possess a plant-specific B3 DNA-binding domain and recognize the CME element in the FLC promoter. Hence, HDA9 acts in concert with the CLF–PRC2 complex to suppress the expression of FLC and the floral integrator FLOWERING LOCUS T (FT), via mutual physical interactions with the epigenome readers VAL1 and VAL2 (Zeng et al., 2020).
Further evidence showed that a mutation in the FT locus suppressed the hda9 early flowering phenotype, and FT mRNA levels were increased in the hda9 mutant background (Kang et al., 2015; Park et al., 2019; Zeng et al., 2020). This suggests that HDA9 acts upstream of FT in flowering time regulation. This effect is likely to be a direct consequence of altered AGL19 transcription in the hda9 mutant, as H3Ac levels of the FT locus were unaltered in the hda9 mutant background, in contrast to the AGL19 locus (Kang et al., 2015). Genetic analyses further indicated that HDA9 negatively regulates the autonomous flowering pathway, as the late-flowering phenotype of a plant line carrying an active FRIGIDA allele was partially suppressed by the hda9 mutation (Kang et al., 2015). The photoperiodic pathway was similarly affected by HDA9, although to a lesser extent. In LD conditions, double mutants between hda9 and gigantea (gi-2) or constans (co-101) displayed a late flowering phenotype compared with the wild type; however, each double mutant flowered slightly earlier than the respective single mutants (Kang et al., 2015). Similar results were presented by Park et al. (2019), who demonstrated that HOS15 might function upstream of GI, CO, and FT, as the respective double mutant combinations with hos15 were late flowering in LD conditions, whereas the hos15 single mutants were early flowering. The latter effect is most probably due to the high levels of GI expression in hos15 mutants due to H3 hyperacetylation at the GI locus (Park et al., 2019). Furthermore, in the absence of hos15, the HDA9–HOS15–LUX/ELF3 complex cannot target the GI promoter for deacetylation. Notably, the early flowering of the hos15 mutant under SD conditions was independent of GI (Park et al., 2019).
Taken together, the role of HDA9 in flowering time control is highly complex as it depends on many environmental factors, including daylength, where HDA9 appears to modulate at the same time the expression of positive (e.g. AGL19, GI, and FT) and negative floral regulators (e.g. FLC). For example, the observation that hda9 mutants flower like the wild type in LD conditions (Kang et al., 2015; Zeng et al., 2020), despite markedly high FLC repression levels in this mutant (Zeng et al., 2020), can be possibly explained by the misexpression of other floral regulators such as FT.
The complex and sometimes contrasting findings in Arabidopsis prohibit drawing firm conclusions on the role of HDA9 in flowering as of yet. However, the role of HDA9 is at least partially conserved in different plant species, as the HDA9 homolog of the oil seed and vegetable crop Brassica juncea (BjuHDA9) was shown to interact with the promoters of BjuSOC1 and BjuAGL24 (Jiang et al., 2018). Interestingly, BjuHDA9 transcript levels were higher in an SD photoperiod than in LDs (Jiang et al., 2018). Moreover, overexpression of the floral regulator BjuAGL18 resulted in the transcriptional up-regulation of BjuHDA9 during flowering (Yan et al., 2018). Whether HDA9 is also transcriptionally regulated by the photoperiod and/or floral regulators in Arabidopsis remains to be investigated.
HDA9 controls leaf aging, senescence, autophagy, and cellular proliferation and de-differentiation
Despite the delayed flowering initiation observed in hda9 mutants, HDA9 is considered to play a generic role in promoting developmental progression (Suzuki et al., 2018). This was proposed based on quantification of leaf heteroblasty progression of hos15 mutants, which revealed a slightly delayed juvenile to adult phase transition, which probably also accounts for hda9 (Suzuki et al., 2018). In addition, HDA9 promotes cell proliferation in leaf primordia. Hence, hda9 mutants produce smaller leaves with a reduced number of palisade cells (Suzuki et al., 2018). In contrast, HDA9 also promotes cellular de-differentiation (Lee et al., 2016), as hda9 mutants displayed reduced ability of pluripotent callus formation, and several genes involved in the de-differentiation process were down-regulated in leaves and calli of the hda9 mutant. Moreover, HDA9 itself is transcriptionally up-regulated in callus tissues (Lee et al., 2016).
Compelling evidence for a role for HDA9 in developmental progression was provided by Chen et al. (2016), who demonstrated that HDA9 stimulates leaf aging and senescence by targeting multiple senescence-regulating pathways simultaneously. In a search for PWR-interacting proteins by immunoaffinity purification followed by MS, HDA9, WRKY53, and AHL22 were identified as the most abundant peptides co-purifying with PWR (Table 3). Subsequent analysis indicated that age-related and dark-induced leaf senescence was delayed in hda9 and pwr single mutants and their hda9 pwr double mutant combination (Chen et al., 2016; Yang et al., 2020). Transcription of various positive regulators of senescence, such as SENESCENCE 4 (SEN4), SENESCENCE ASSOCIATED GENE 12 (SAG12), and SAG113, was attenuated in the hda9 mutant background (Chen et al., 2016). Similarly, down-regulation of a significant fraction of genes known to be repressed during senescence was impaired in the hda9 mutant background (Chen et al., 2016). In agreement with the influence of HDA9 on the senescence transcriptome, the protein was mildly up-regulated in early-senescent leaves. Abscisic acid (ABA)-responsive genes were significantly down-regulated in hda9 mutants, suggesting that the ABA phytohormone signaling pathway, known to be involved in senescence (Jibran et al., 2013), is impaired in these mutants (Chen et al., 2016). Furthermore, among the senescence-associated genes differentially expressed in hda9 is WRKY57, encoding a transcription factor involved in the repression of jasmonic acid (JA) during leaf senescence that was demonstrated to be a direct target of HDA9 (Chen et al., 2016).
The observation that the W-box promoter element, recognized by WRKY transcription factors, was over-represented among HDA9 chromatin-binding targets also suggests a functional connection between HDA9 and WRK53 in senescence (Chen et al., 2016). However, the role of HDA9–WRKY53 interactions in regulating leaf senescence remains to be confirmed empirically.
Autophagy is one of the processes involved in leaf senescence (Hanaoka et al., 2002; Avila-Ospina et al., 2014). Autophagy is a metabolic process in which cytoplasmic components such as proteins and dysfunctional organelles are sequestered to the vacuole or lysosome for degradation and recycling, which is important for tolerance to adverse environmental conditions. The process of autophagy is regulated by the so-called autophagy-related genes (ATGs).
A recent study demonstrated the involvement of HDA9 in the transcriptional regulation of ATGs (Yang et al., 2020). The authors showed that nitrogen starvation and darkness induce autophagy and modulate ATG expression. Based on the premise of light-mediated transcriptional regulation of these ATGs, the versatile light signaling regulator bZIP transcription factor HY5 (Gangappa and Botto, 2016) was selected for further study. Indeed, HY5 negatively regulates autophagy in darkness and under nitrogen starvation conditions, and was shown to target the promoters of ATG5 and ATG8e (Yang et al., 2020). As a next step, HDA9 was identified in a screen for HDACs that interact with HY5 (Table 3), and mutants in hda9 are more tolerant of nitrogen starvation than the corresponding wild type and displayed more autophagosomes. Accordingly, ATG5 and ATG8e transcript and protein levels were enhanced in the hda9 mutant, and disruption of autophagy by mutating atg5 or atg7 abolished the enhanced nitrogen starvation tolerance phenotypes of hy5 and hda9 mutants. Accordingly, ChIP-PCR experiments indicated that HDA9 is targeted to the ATG5 and ATG8e genomic loci in a HY5-dependent manner. Double mutant analysis confirmed that HY5 and HDA9 synergistically regulate cell autophagy upstream of ATGs by H3K9 and H3K27 deacetylation of the ATG5 and ATG8e genomic loci, thereby regulating their expression (Yang et al., 2020). Interestingly, the HY5–HDA9 complex dissociated from the chromatin of ATG5 and ATG8e in response to darkness and nitrogen starvation, and the HY5–HDA9 protein–protein interaction was broken. In addition, darkness and nitrogen starvation conditions led to HY5, but not HDA9, 26S proteasomal degradation in a COP1-dependent manner (Yang et al., 2020).
Taken together, a model was proposed whereby, under light and high nitrogen conditions, HY5 recruits HDA9 to repress ATG expression by decreasing acetylation levels, thereby suppressing cell autophagy. In response to nitrogen starvation and darkness, HY5 is degraded in a COP1-dependent manner, leading to the dissociation of HDA9 and acetylation of ATGs, followed by their transcriptional induction and activation of cell autophagy, which ultimately results in enhanced tolerance to these environmental conditions (Yang et al., 2020).
The role of HDA9 in regulating seed dormancy and germination
Seed dormancy is defined as a state of quiescence in viable seeds, during which germination is prohibited, even if environmental conditions are favorable for germination (e.g. seasonal optimal temperature, moisture, and light conditions; Baskin and Baskin, 2004; Née et al., 2017). Treatment of dormant Arabidopsis wild-type Columbia-0 (Col-0) seeds with the HDAC inhibitors trichostatin-A (TSA) and butyric acid sodium salt released dormancy in a dose-dependent manner. Subsequent reverse genetic analysis revealed that mutants in hda9 displayed reduced dormancy (Van Zanten et al., 2014). Moreover, hda9 mutants germinated faster (Van Zanten et al., 2014; Baek et al., 2020) and exhibited improved seed longevity (storability) (Van Zanten et al., 2014). The role of different HDACs in seed biology, however, depends on the species studied (Van Zanten et al., 2013). For instance, TSA application leads to a delay in germination in maize (Zhang et al., 2011).
Germination and dormancy are tightly regulated by the balance between the phytohormones gibberellin (GA) and ABA, where GA typically stimulates germination and ABA is associated with the repression of germination and dormancy enhancement (Finkelstein et al., 2008). ABA levels were reduced in seeds of hda9 mutants and increased in heterotrophic seedlings (Baek et al. 2020). It remains an open question if and how the recently identified interaction between HDA9, ABI3, and ABI4 (Baek et al 2020; Khan et al., 2020) contributes to regulating seed dormancy and germination. However, pharmacological analysis indicated that ABA and GA sensitivity of seeds was unaltered in the hda9 mutant (Van Zanten et al., 2014), suggesting that HDA9 affects dormancy and germination largely independently of these phytohormones. Accordingly, meta-analysis of transcriptome data obtained from wild-type and hda9 mutant seeds, compared with published datasets, did not reveal a significant similarity that would suggest an involvement of GA and ABA (Van Zanten et al., 2014). However, unexpectedly, many of the differentially regulated genes in the hda9 mutant coded for factors involved in photosynthesis, the Calvin cycle, and secondary metabolism (Van Zanten et al., 2014). This included the 2B subunit of Rubisco and Rubisco activase (RCA). ChIP-PCR experiments confirmed that H3K9Ac levels on the loci of these genes were increased in hda9 compared with the wild type, especially in the 5' (+500 bp) region (Kim et al., 2013; Van Zanten et al., 2014). Moreover, Rubisco protein levels were enhanced in hda9 mutant dry seeds (Van Zanten et al., 2014). Taken together, HDA9 can be considered a positive regulator of seed dormancy and a repressor of germination and of vegetative properties in dry seeds. Interestingly, the opposite function was shown for the HDA9 homologs HDA6 and HDA19—these HDACs are involved in repression of embryonic properties in autotrophic seedlings (Tanaka et al., 2008).
ASG2 (ALTERED SEED GERMINATION 2) is a WD40 and Tetratrico Peptide Repeat (TPR) domain protein that is involved in ABA signaling. Mutant asg2 seeds exhibited increased weight, oil body density, and higher fatty acid contents that affected seed germination (Dutilleul et al., 2016; Ducos et al., 2017). The farnesylated form of ASG2 was shown to interact with HDA9 in the cytosol, but not in the nucleus (Ducos et al., 2017) (Fig. 1; Table 3). Future work should address the biological function of this interaction, especially whether HDA9 affects seed fatty acid content and how it is connected through ASG2 to the diverse roles of HDA9 in seed dormancy, germination, repression of vegetative properties, and possibly other biological processes.
Involvement of HDA9 in regulating responses to environmental signals: drought and salt stress
Plants have to deal with a large number of biotic and abiotic cues (Zhu, 2016), and HDA9 has been reported to play a role in orchestrating the responses to various environmental conditions (Zheng et al., 2016; Tasset et al., 2018; Shen et al., 2019; van der Woude et al., 2019; Yang et al., 2019; Zheng et al., 2020) (Fig. 1; Table 2). For instance, hda9 mutants accumulate high levels of iron in their roots, suggesting a role in iron homeostasis (Xing et al., 2015) and, as described above, HDA9 contributes to regulating darkness- and nitrogen starvation-mediated autophagy/leaf senescence (Yang et al., 2020). In addition, HDA9 is reported to function as a negative regulator of salt and drought stress tolerance, due to its repressive effect on stress-responsive genes in Arabidopsis (Zheng et al., 2016, 2020). Observations in broccoli (Brassica oleracea) suggest that salt-mediated regulation of HDA9 transcript levels may have a role in bud senescence (Yan, et al., 2020). Arabidopsis hda9 mutants displayed a decrease in the inhibition of seed germination and root growth, and thus an increase in tolerance to high NaCl concentration and simulated drought stress (PEG; polyethene glycol) conditions compared with the wild type (Zheng et al., 2016, 2020). In two recent studies, Baek et al. (2020) and Khan et al. (2020), however, proposed that HDA9 and PWR are positive regulators of physiological drought stress tolerance (i.e. progressive drought by withholding watering). Mutants in pwr (Khan et al., 2020) and hda9 (Baek et al., 2020) displayed reduced sensitivity to ABA regarding stomatal closure, and HDA9 was transcriptionally induced under drought conditions (Baek et al., 2020). Interestingly, yeast two-hybrid and Co-IP analyses demonstrated that HDA9 physically interacts with the transcription factors ABI4 (Baek et al 2020; Khan et al., 2020) and ABI3 (Khan et al., 2020) (Fig. 1; Table 3). Combined, the data support a model in which a PWR–HDA9–ABI4 complex targets the loci of ABA catabolism and ABA signaling genes and regulates their histone acetylation status and transcription (Baek et al., 2020; Khan et al., 2020). Transcript levels of the ABA catabolism genes CYP707A1 (hda9 and pwr) and CYP707A2 (hda9) were indeed enhanced, whereas ABA phytohormone levels were reduced in hda9 and abi4 mutant plants under drought stress. Moreover, H3 acetylation levels were enhanced at the CYP707A1 locus in the hda9 and pwr mutant backgrounds (Khan et al., 2020). Stomatal aperture and water loss were accordingly increased in these mutant backgrounds, resulting ultimately in dehydration hypersensitivity (Baek et al., 2020; Khan et al., 2020).
Similar to CYP707A1 and CYP707A2 (Baek et al., 2020; Khan et al., 2020), Zheng et al. (2016, 2020) found that several drought stress-related genes were highly induced in the hda9 mutant background upon drought/salt stress application, which correlated with enhanced H3K9Ac levels in promoter regions of a selection of these genes. Furthermore, yeast two-hybrid analysis indicated an interaction of HDA9 with HDA6, HDA19, and AtCYP1-1 (cyclophilin-like peptidyl-prolyl cis–trans isomerase family protein) (Table 3), all of which have been associated with salt and/or drought stress before (Zheng et al., 2016). However, later work from Yuan et al. (2019) did not confirm an interaction between HDA6, or HDA19, and HDA9, and the possible association between HDA9 and HDA6, HDA19 and AtCYP1-1 in drought and/or salt stress responsiveness was not functionally validated in planta (Zheng et al., 2016).
The interaction between WRK53 and HDA9, that was previously described in the context of leaf senescence (Chen et al., 2016), was confirmed (Zheng et al., 2020). In contrast to HDA9, in the latter study WRKY53 was shown to act as a positive regulator of salt and drought stress responses, and the mutual and antagonistic roles of HDA9 and WRKY53 have now been elucidated in great molecular depth (Zheng et al., 2020). In detail, the authors showed that HDA9 repressed WRKY53 transcription—and therewith several WRKY53 target genes—under non-stressed conditions and thereby prevented WRKY53 gene induction under salt stress. Unexpectedly, HDA9 did not, however, associate with the chromatin of WRKY53 target genes, nor were histone acetylation levels at the WRKY53 locus affected in the hda9 mutant. However, H3K4Me2/Me3 levels were enhanced, correlating with the increased WRKY53 expression in the hda9 mutant background under salt stress (Zheng et al., 2020). These observations prompted the authors to test whether HDA9 could target the WRKY53 protein directly. Indeed, post-translational K12Ac, K26Ac, K27AC, K58Ac, K169Ac, K175Ac, and K268Ac modification levels of the WRKY53 protein were higher in the hda9 mutant and lower in a HDA9 overexpression line, which was confirmed by several biochemical validations (Zheng et al., 2020). The authors thus revealed that HDA9 is able to modify the acetylation status of a non-histone protein.
Additional studies indicated that HDA9 represses WRKY53 cis transcriptional activity by preventing the transcription factor from binding to its own promoter (Zheng et al., 2020). Accordingly, the central deacetylase domain of HDA9 interacts directly with the WRKY53 DNA-binding domain. HDAC inhibition with TSA did not interfere with the negative effect of HDA9 on WRKY53 DNA binding capacity, suggesting that this probably occurs independently of WRKY53 lysine deacetylation. WRKY53 lysine acetylation is, however, important for WRKY53 transcriptional activity in trans (Zheng et al., 2020).
Interestingly, H3K9Ac/H3K27Ac levels were increased and decreased, respectively, in WRKY53 overexpression and wrky53 mutant lines, suggesting that WRKY53 in turn regulates HDA9 activity. This was confirmed by direct HDAC activity assays using purified HDA9 protein, derived from the WRKY53 overexpression and wrky53 mutant line, and by experiments with recombinant WRKY53 protein. The repression of HDA9 activity required the WRKY53 DNA-binding domain, which probably masks the HDAC catalytic domain (Zheng et al., 2020).
In conclusion, HDA9 modulates salt and drought stress tolerance responses by directly targeting and repressing the DNA binding and transcriptional activity of the high hierarchical positive regulator of stress responses; WRKY53 (Zheng et al., 2020).
Involvement of HDA9 in regulating responses to environmental signals: thermomorphogenesis
While HDA9 is considered to function mainly as a negative regulator of salt and drought stress responsiveness, the protein was identified as a positive regulator of plant thermomorphogenesis—a suite of architectural traits induced by plants to mitigate negative effects of mildly increased temperatures—by improving cooling capacity (Quint et al., 2016; Casal and Balasubramanian, 2019). Thermomorphogenesis is mediated by the high temperature-induced transcription factor PHYTOCHROME INTERACTING FACTOR4 (PIF4) (Koini et al., 2009; Sun et al., 2012) and regulated by the EC component ELF3 (Box et al., 2015; Raschke et al., 2015). PIF4 activates the expression of auxin biosynthesis genes, including that encoding the rate-limiting enzyme YUCCA8 (YUC8), that subsequently stimulates auxin accumulation required for inducing thermomorphogenesis (Franklin et al., 2011; Sun et al., 2012), in concert with the brassinosteroid phytohormones (Ibañez et al., 2018). Furthermore, high temperatures lead to the eviction of the histone variant H2A.Z-containing nucleosomes from promoters of thermo-responsive genes, which then allows for the binding of transcriptional regulators, including PIF4, to the DNA (Kumar and Wigge, 2010; Cortijo et al., 2017).
Mutants in HDA9 and PWR are impaired in thermomorphogenesis, as exhibited by traits such as reduced hypocotyl elongation and maintenance of a compact rosette (Tasset et al., 2018; Shen et al., 2019; van der Woude et al., 2019). Some warm temperature-mediated features were, however, retained in hda9. For instance, the expression of HEAT SHOCK PROTEIN 70 (HSP70), a warm temperature-induced marker gene (Kumar and Wigge, 2010), and high temperature-induced flowering were comparable between hda9 and wild-type plants. This contrasted with pwr mutants that displayed reduced HSP70 expression and reduced sensitivity of thermal floral induction (Tasset et al., 2018). Moreover, opposite to PWR (Tasset et al., 2018), HDA9 is not involved in regulating PIF4 at the transcriptional level under warm temperatures (van der Woude et al., 2019). Interestingly, unlike pif4 mutants, hda9 loss-of-function alleles retain their responsiveness to light signals that induce the shade avoidance response that resembles thermomorphogenesis and is considered to be a competitive response to outgrow shading in dense canopies (Ballaré and Pierik, 2017). Furthermore, the effects of HDA9 on thermomorphogenesis occurred independent of the light and temperature sensor phytochrome B (phyB) (van der Woude et al., 2019). Together, this suggests that HDA9 is part of a thermosignaling pathway that operates independently of shade avoidance and temperature-induced flowering regulation.
At the protein level, HDA9 accumulates at dawn and becomes less abundant over the photoperiod in response to high temperature (27 °C), whereas no marked (diurnal/circadian) changes in HDA9 protein contents were observed at control temperatures. HDA9 mRNA and protein were mainly detected in young seedlings shortly after germination and declined during seedling establishment. Together, this suggests that HDA9 protein is regulated by temperature cues and can be considered as an early regulator of thermomorphogenesis (van der Woude et al., 2019).
Gene Ontology enrichment analysis revealed that high temperature-induced up-regulation of auxin-related genes was impaired in hda9 mutants (van der Woude et al., 2019), and subsequent analysis confirmed that this included YUC8 (Sun et al., 2012). In agreement, warm temperature-induced YUC8 induction was impaired in pwr mutants as well (Tasset et al., 2018). In line with reduced YUC8 expression, bioactive auxin (indole-3-acetic acid; IAA) levels were low in the hda9 mutant under warm temperature conditions, whilst the YUC8 enzyme substrate indole-3-pyruvic acid (IPyA) accumulated to high levels (van der Woude et al., 2019). ChIP-PCR analyses revealed hyperacetylation of the YUC8 promoter in the hda9 and pwr mutant backgrounds under high temperature and also in control temperature conditions for pwr, suggesting that histone deacetylation is required for YUC8 expression. Interestingly, HDA9-mediated H3K9K14 deacetylation of nucleosomes was associated with low H2A.Z levels at warm temperatures at the YUC8 locus, whereas hda9 mutants displayed high H2A.Z levels. These high H2A.Z levels consequently led to reduced PIF4 binding capacity to the G-box promoter element, which explains attenuated YUC8 transcriptional induction, prohibition of auxin biosynthesis, and suppression of thermomorphogenesis, in the hda9 mutant background. Probably, PWR is involved in this as well, as genes misregulated in pwr mutants exhibited significant overlap with known H2A.Z-enriched genes, and with differentially expressed genes in mutants disturbed in H2A.Z deposition (Tasset et al., 2018).
Altogether, HDA9–PWR-mediated deacetylation is associated with thermomorphogenesis via an induction of gene transcription (of YUC8), by promoting net depletion of the repressive histone variant H2A.Z. The role of PWR in thermomorphogenesis regulation appears broader than that of HDA9, given the more pleiotropic phenotypes of pwr compared with hda9. Whether HOS15 plays an active role in regulating thermomorphogenesis as well could be addressed in future studies.
It is worth mentioning that the notable role of HDA9 in activating gene expression is atypical, since HDACs are generally considered to act as transcriptional co-repressors (Li et al., 2002; Tian et al., 2005; Perrella et al., 2013). Further studies are required to reveal if HDA9 has a similar transcriptional activating role in other HDA9-mediated processes. Recent reports using genome-wide HDA9 ChIP-sequencing surveys showed that HDA9 indeed associates mainly with actively transcribed genes and that HDA9 binding positively correlates with gene expression (Kang et al., 2015; Chen et al., 2016; Kim et al., 2016; Mayer et al., 2019).
Involvement of HDA9 in plant immunity
In general, plants display two distinct types of immunity: pattern-triggered immunity (PTI) and effector-triggered immunity (ETI) to defend against microbial pathogens. PTI is based on recognition of conserved microbial or pathogen-associated molecular patterns (MAMPs and PAMPs), whereas ETI is based on recognition of pathogen-associated effectors or toxins (Jones and Dangl, 2006; Miller et al., 2017). Many of these pathogen-associated effectors are recognized by nucleotide-binding leucine-rich repeat/NOD-like intracellular immune receptor (NB-LRR or NLR) proteins (Meyers et al., 2003). Tight regulation of NLR genes is vital to balanced plant growth and defense. Constitutive expression of NLR genes suppresses plant growth and causes autoimmunity, whereas, on the other hand, adequate induction of NLR gene expression is crucial for timely recognition of pathogens and effective defense initiation.
Recent evidence indicated important roles for HDA9 and HOS15 in NLR transcriptional regulation (Yang et al., 2019). Arabidopsis plants defective in HDA9 and HOS15 show enhanced resistance against Pseudomonas syringae pv. tomato DC3000 (Yang et al., 2019). However, neither HOS15, HDA9 transcript, nor protein levels were altered in response to pathogen infection. Similarly, neither HDA9 nuclear–cytoplasmic transport nor HOS15–HDA9 protein–protein interaction was affected.
Nevertheless, hda9 and hos15 mutants together regulate a large fraction (approximately one-third) of known NLR genes in the genome (Yang et al., 2019). ChiP-seq experiments indicated that HDA9 and HOS15 target largely the same subset of NLR genes, and mainly those that are differentially regulated at the transcriptional level in the hos15 mutant background compared with the wild type. However, unlike in the hos15 mutant, not many defense response genes were differentially regulated in the hda9 mutant in the absence of infection. This indicates that HDA9 requires a pathogenic trigger for its involvement in defense regulation. Indeed, H3K9Ac status of a selection of NLR genes was only enhanced in the hda9 mutant background upon pathogen infection, whereas acetylation levels of these loci were constitutively high in the hos15 mutant background (Yang et al., 2019).
How infection is able to activate HDA9-mediated defense remains unknown. Post-translational modifications triggered by infection of inactive HDA9 that is potentially already bound to its target loci may play a role. In addition, WRKY DNA-binding proteins might be responsible for recruiting HDA9 (and possibly HOS15) to its tailLR gene target loci once plants are infected, as W-boxes are the only known cis-elements that are present in NLR promoter regions. This could point to a possible role for WRKY53 in HDA9-mediated NLR expression regulation (Chen et al., 2016; Yang et al., 2019; Zheng et al., 2020). Testing this hypothesis would require further studies.
Taken together, HDA9 and HOS15 function in the same pathway to suppress immunity. Given the constitutively enhanced immunity status of hos15 mutants, the typical stunted rosette phenotype of hos15 and possibly also hda9 mutants could be interpreted as a mild autoimmune phenotype; that is, the growth–immunity trade-off in these mutants has possibly shifted towards immunity at the expense of growth, despite NLR genes not being induced in hda9 in non-infected conditions.
HDA9 in the larger HDAC context
Despite the fact that HDA9 directly controls many physiological and molecular traits governing plant development, growth, and responses to a changing environment (Fig. 1), it is unlikely that HDA9 operates in isolation independent of other HDAC proteins. Evidence suggests that HDA9 can act in parallel, redundantly, synergistically, or antagonistically to other members of the HDAC family. For instance, hda9 mutants display typical blunt and bulged siliques (tips) attributed to enhanced valve cell elongation (Yuan et al., 2019). This phenotype was not observed in hda6 single mutants. However, the hda9 hda6 double mutant showed additively exaggerated bulged silique and valve cell elongation phenotypes, suggesting that HDA6 and HDA9 redundantly control silique morphology (Yuan et al., 2019). These phenotypes emerge through the coordinated regulation of auxin signaling genes by HDA6 and HDA9, as many auxin-related genes and auxin signaling are additively affected in the single and double mutants (Yuan et al., 2019).
On the contrary, different HDACs may also act independently by targeting specific branches of regulatory molecular networks that either translate the same input to diverse phenotypic outcomes or translate different input to the same phenotypic outcomes. For instance, in the context of thermomorphogenesis, HDA9 has distinct and overlapping functions with HDA15 and HDA19. Mutants of HDA9 and HDA19 showed impaired warm temperature-induced hypocotyl elongation, while, on the contrary, a mutant of HDA15 exhibited a constitutive enhanced thermomorphogenesis response (Shen et al., 2019). This was reflected at the molecular level, as in hda9 regulation of many warm temperature-regulated genes was impaired, while in hda15 many warm temperature response genes are differentially regulated already at control temperature conditions. In the hda19 mutant, mostly stress-regulated genes were affected, at both control and high temperature conditions (Shen et al., 2019). Thus, these HDACs target distinct sets of genes and have distinct functions in the regulation of plant thermomorphogenesis. At the same time, a large fraction of misregulated genes involved in metabolism were shared between hda9 and hda15, suggesting that these HDACs may control the same metabolic pathways, but diverge in the regulation of thermomorphogenesis (Shen et al., 2019). Yet, HDA9 may have antagonistic roles with respect to other HDACs. An example of this is the aforementioned role of HDA9 in repressing vegetative traits in seeds (Van Zanten et al., 2014), whereas HDA6 and HDA19 together repress embryonic properties in autotrophic young seedlings (Tanaka et al., 2008).
Concluding remarks
Diverse roles of HDA9 in the regulation of a multitude of plant traits and responses to the environment have been described in recent years (Fig. 1; Table 2) in concert with few established direct interacting proteins (Fig. 1; Table 3). Nevertheless, several important questions remain to be answered. For instance, it is currently unclear if the cytosolic HDA9 population functions in the deacetylation of non-histone proteins other than WRKY53 (Zheng et al., 2020) and whether the atypical role of HDA9 as a conditional activator of gene transcription extends beyond YUC8 (Van der Woude et al., 2019). Another intriguing question is why hda9 mutants were hardly identified in reverse genetic mutant screens, despite its pleiotropic roles in diverse plant processes. Furthermore, future efforts could address how knowledge on Arabidopsis HDA9 can be utilized and translated to improve crop performance and yield in response to climate change.
As described in detail in this review, HDA9 has many faces, as its mode of action is tailored to specific trait/response and sometimes has apparent opposite effects (as seen for drought stress tolerance for instance) (Table 2; Fig. 1). Nevertheless, the involvement of HDA9 in regulating responsiveness to diverse environmental stimuli (e.g. pathogens, salt, drought, high temperature, darkness, and iron) on one hand, and diverse plant responses to these stimuli (e.g. growth acclimation, autophagy, senescence, aging, dormancy, and germination) on the other, suggests that HDA9 is an essential player in the molecular networks mediating optimal plant performance under suboptimal environmental conditions.
It is likely that many more unidentified HDA9-mediated phenotypes and interacting proteins remain to be discovered. HDA9 is able to physically interact with several transcription factors (e.g. WRKY53, HY5, ABI3, and ABI4) (Fig. 1; Table 3), which might contribute to establishment of HDA9-dependent epigenetic states, particularly in response to environmental stimuli. In this regard, it has been extensively demonstrated that transcription factors can directly recruit histone modifiers to their DNA targets to reinforce the local epigenetic landscape (Bonasio et al., 2010). Given the substantial difference in HDA9-interacting proteins compared with those identified for HDA6 and HDA19 on one hand (Perrella et al., 2013; Mehdi et al., 2016; Ning et al., 2019), and the positive correlation of HDA9 presence with gene expression (Kang et al., 2015; Chen et al., 2016; Kim et al., 2016; Mayer et al., 2019; van der Woude et al., 2019) on the other, we speculate that HDA9 may be fundamentally divergent from related HDACs.
Intriguingly, hda9 mutants also display impaired histone methylation and miRNA levels (Kim et al., 2009), suggesting a possible crosstalk with other epigenetic modifications. Similar mechanisms have been shown for other HDACs, including HDA6 that regulates flowering time through the association with the histone demethylase FLOWERING LOCUS D (FLD) (Yu et al., 2011). Furthermore, HDA6 interacts with the DNA methyltransferase MET1, thereby regulating cytosine methylation and rDNA loci in heterochromatic regions (To et al., 2011; Liu et al., 2012). However, how HDA9 acts in concert with other HDACs to mediate PRC2-dependent histone trimethylation and whether such a mechanism can occur on other loci rather than FLC requires further investigation. Similarly, the involvement of HDA9 in regulating miRNA genesis is not yet fully understood.
To date, ‘HDA9’ as a search input in the NCBI PubMed database (2 July 2020; (https://www.ncbi.nlm.nih.gov/pubmed/?term=HDA9) recovered 25 papers out of which 22 were published after 2016 and no less than 15 in 2019/2020. Thus, our knowledge on this previous undercharacterized protein is currently accumulating rapidly, and integration and cross-validation of findings is needed to fully appreciate the impact that HDA9 has on plant growth and development, and environmental responses. This review discussing the multiple functions of HDA9 aims to help the growing HDA9 community in achieving this goal.
Author contributions
PGHdR, GP, EK, and MvZ wrote the article together. PGHdR and GP had an equal contribution. MvZ conceived the project, initiated, supervised, and completed the writing, and agrees to serve as the author responsible for contact and ensures communication. The authors declare no competing interest.
References
- Alinsug MV, Yu CW, Wu K. 2009. Phylogenetic analysis, subcellular localization, and expression patterns of RPD3/HDA1 family histone deacetylases in plants. BMC Plant Biology 9, 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avila-Ospina L, Moison M, Yoshimoto K, Masclaux-Daubresse C. 2014. Autophagy, plant senescence, and nutrient recycling. Journal of Experimental Botany 65, 3799–3811. [DOI] [PubMed] [Google Scholar]
- Baek D, Shin G, Kim MC, Shen M, Lee SY, Yun DJ. 2020. Histone deacetylase HDA9 with ABI4 contributes to abscisic acid homeostasis in drought stress response. Frontiers in Plant Science 11, 143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballaré CL, Pierik R. 2017. The shade-avoidance syndrome: multiple signals and ecological consequences. Plant, Cell & Environment 40, 2530–2543. [DOI] [PubMed] [Google Scholar]
- Baskin JM, Baskin CC. 2004. A classification system for seed dormancy. Seed Science Research 14, 1–16. [Google Scholar]
- Berger SL. 2007. The complex language of chromatin regulation during transcription. Nature 447, 407–412. [DOI] [PubMed] [Google Scholar]
- Bonasio R, Tu S, Reinberg D. 2010. Molecular signals of epigenetic states. Science 330, 612–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman GD, Poirier MG. 2015. Post-translational modifications of histones that influence nucleosome dynamics. Chemical Reviews 115, 2274–2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Box MS, Huang BE, Domijan M, et al. 2015. ELF3 controls thermoresponsive growth in Arabidopsis. Current Biology 25, 194–199. [DOI] [PubMed] [Google Scholar]
- Camoni L, Visconti S, Aducci P, Marra M. 2018. 14-3-3 Proteins in plant hormone signaling: doing several things at once. Frontiers in Plant Science 9, 297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casal JJ, Balasubramanian S. 2019. Thermomorphogenesis. Annual Review of Plant Biology 70, 321–346. [DOI] [PubMed] [Google Scholar]
- Chen X, Ding AB, Zhong X. 2020. Functions and mechanisms of plant histone deacetylases. Science China. Life Sciences 63, 206–216. [DOI] [PubMed] [Google Scholar]
- Chen X, Lu L, Mayer KS, Scalf M, Qian S, Lomax A, Smith LM, Zhong X. 2016. POWERDRESS interacts with HISTONE DEACETYLASE 9 to promote aging in Arabidopsis. eLife 5, e17214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho LH, Yoon J, An G. 2017. The control of flowering time by environmental factors. The Plant Journal 90, 708–719. [DOI] [PubMed] [Google Scholar]
- Chow BY, Helfer A, Nusinow DA, Kay SA. 2012. ELF3 recruitment to the PRR9 promoter requires other evening complex members in the Arabidopsis circadian clock. Plant Signaling & Behavior 7, 170–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortijo S, Charoensawan V, Brestovitsky A, Buning R, Ravarani C, Rhodes D, van Noort J, Jaeger KE, Wigge PA. 2017. Transcriptional regulation of the ambient temperature response by H2A.Z nucleosomes and HSF1 transcription factors in arabidopsis. Molecular Plant 10, 1258–1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowson-Day MJ, Millar AJ. 1999. Circadian dysfunction causes aberrant hypocotyl elongation patterns in Arabidopsis. The Plant Journal 17, 63–71. [DOI] [PubMed] [Google Scholar]
- Ducos E, Vergès V, Dugé de Bernonville T, Blanc N, Giglioli-Guivarc’h N, Dutilleul C. 2017. Remarkable evolutionary conservation of antiobesity ADIPOSE/WDTC1 homologs in animals and plants. Genetics 207, 153–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutilleul C, Ribeiro I, Blanc N, et al. 2016. ASG2 is a farnesylated DWD protein that acts as ABA negative regulator in Arabidopsis. Plant, Cell & Environment 39, 185–198. [DOI] [PubMed] [Google Scholar]
- Ezer D, Jung JH, Lan H, et al. 2017. The evening complex coordinates environmental and endogenous signals in Arabidopsis. Nature Plants 3, 17087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farinas B, Mas P. 2011. Functional implication of the MYB transcription factor RVE8/LCL5 in the circadian control of histone acetylation. The Plant Journal 66, 318–329. [DOI] [PubMed] [Google Scholar]
- Finkelstein R, Reeves W, Ariizumi T, Steber C. 2008. Molecular aspects of seed dormancy. Annual Review of Plant Biology 59, 387–415. [DOI] [PubMed] [Google Scholar]
- Franklin KA, Lee SH, Patel D, et al. 2011. Phytochrome-interacting factor 4 (PIF4) regulates auxin biosynthesis at high temperature. Proceedings of the National Academy of Sciences, USA 108, 20231–20235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gangappa SN, Botto JF. 2016. The multifaceted roles of HY5 in plant growth and development. Molecular Plant 9, 1353–1365. [DOI] [PubMed] [Google Scholar]
- Gendron JM, Pruneda-Paz JL, Doherty CJ, Gross AM, Kang SE, Kay SA. 2012. Arabidopsis circadian clock protein, TOC1, is a DNA-binding transcription factor. Proceedings of the National Academy of Sciences, USA 109, 3167–3172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grzenda A, Lomberk G, Zhang JS, Urrutia R. 2009. Sin3: master scaffold and transcriptional corepressor. Biochimica et Biophysica Acta 1789, 443–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu X, Wang Y, He Y. 2013. Photoperiodic regulation of flowering time through periodic histone deacetylation of the florigen gene FT. PLoS Biology 11, e1001649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanaoka H, Noda T, Shirano Y, Kato T, Hayashi H, Shibata D, Tabata S, Ohsumi Y. 2002. Leaf senescence and starvation-induced chlorosis are accelerated by the disruption of an Arabidopsis autophagy gene. Plant Physiology 129, 1181–1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl M, Füßl M, Boersema PJ, et al. 2017. Lysine acetylome profiling uncovers novel histone deacetylase substrate proteins in Arabidopsis. Molecular Systems Biology 13, 949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S, Smith MM. 2015. Histone variants and epigenetics. Cold Spring Harbor Perspectives in Biology 7, a019364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollender C, Liu Z. 2008. Histone deacetylase genes in Arabidopsis development. Journal of Integrative Plant Biology 50, 875–885. [DOI] [PubMed] [Google Scholar]
- Hung FY, Chen FF, Li C, Chen C, Lai YC, Chen JH, Cui Y, Wu K. 2018. The Arabidopsis LDL1/2–HDA6 histone modification complex is functionally associated with CCA1/LHY in regulation of circadian clock genes. Nucleic Acids Research 46, 10669–10681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibañez C, Delker C, Martinez C, et al. 2018. Brassinosteroids dominate hormonal regulation of plant thermomorphogenesis via BZR1. Current Biology 28, 303–310. [DOI] [PubMed] [Google Scholar]
- Jaspert N, Throm C, Oecking C. 2011. Arabidopsis 14-3-3 proteins: fascinating and less fascinating aspects. Frontiers in Plant Science 2, 96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W, Wei D, Zhou W, Wang Z, Tang Q. 2018. HDA9 interacts with the promoters of SOC1 and AGL24 involved in flowering time control in Brassica juncea. Biochemical and Biophysical Research Communications 499, 519–523. [DOI] [PubMed] [Google Scholar]
- Jibran R, A Hunter D, P Dijkwel P. 2013. Hormonal regulation of leaf senescence through integration of developmental and stress signals. Plant Molecular Biology 82, 547–561. [DOI] [PubMed] [Google Scholar]
- Jones JD, Dangl JL. 2006. The plant immune system. Nature 444, 323–329. [DOI] [PubMed] [Google Scholar]
- Jouve L, Greppin H, Degli Agosti R. 1998. Arabidopsis thaliana floral stem elongation: evidence for an endogenous circadian rhythm. Plant Physiology and Biochemistry 36, 469–472. [Google Scholar]
- Jung JH, Park JH, Lee S, To TK, Kim JM, Seki M, Park CM. 2013. The cold signaling attenuator HIGH EXPRESSION OF OSMOTICALLY RESPONSIVE GENE1 activates FLOWERING LOCUS C transcription via chromatin remodeling under short-term cold stress in Arabidopsis. The Plant Cell 25, 4378–4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang MJ, Jin HS, Noh YS, Noh B. 2015. Repression of flowering under a noninductive photoperiod by the HDA9–AGL19–FT module in Arabidopsis. New Phytologist 206, 281–294. [DOI] [PubMed] [Google Scholar]
- Khan IU, Ali A, Khan HA, et al. 2020. PWR/HDA9/ABI4 complex epigenetically regulates ABA dependent drought stress tolerance in arabidopsis. Frontiers in Plant Science 11, 623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim W, Benhamed M, Servet C, Latrasse D, Zhang W, Delarue M, Zhou DX. 2009. Histone acetyltransferase GCN5 interferes with the miRNA pathway in Arabidopsis. Cell Research 19, 899–909. [DOI] [PubMed] [Google Scholar]
- Kim JA, Kim HS, Choi SH, Jang JY, Jeong MJ, Lee SI. 2017. The importance of the circadian clock in regulating plant metabolism. International Journal of Molecular Sciences 18, 2680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim W, Latrasse D, Servet C, Zhou DX. 2013. Arabidopsis histone deacetylase HDA9 regulates flowering time through repression of AGL19. Biochemical and Biophysical Research Communications 432, 394–398. [DOI] [PubMed] [Google Scholar]
- Kim YJ, Wang R, Gao L, et al. 2016. POWERDRESS and HDA9 interact and promote histone H3 deacetylation at specific genomic sites in Arabidopsis. Proceedings of the National Academy of Sciences, USA 113, 14858–14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koini MA, Alvey L, Allen T, Tilley CA, Harberd NP, Whitelam GC, Franklin KA. 2009. High temperature-mediated adaptations in plant architecture require the bHLH transcription factor PIF4. Current Biology 19, 408–413. [DOI] [PubMed] [Google Scholar]
- Kumar SV, Wigge PA. 2010. H2A.Z-containing nucleosomes mediate the thermosensory response in Arabidopsis. Cell 140, 136–147. [DOI] [PubMed] [Google Scholar]
- Lee K, Mas P, Seo PJ. 2019. The EC–HDA9 complex rhythmically regulates histone acetylation at the TOC1 promoter in Arabidopsis. Communications Biology 2, 143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K, Park OS, Jung SJ, Seo PJ. 2016. Histone deacetylation-mediated cellular dedifferentiation in Arabidopsis. Journal of Plant Physiology 191, 95–100. [DOI] [PubMed] [Google Scholar]
- Li J, Lin Q, Wang W, Wade P, Wong J. 2002. Specific targeting and constitutive association of histone deacetylase complexes during transcriptional repression. Genes & Development 16, 687–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Yang S, Zhao M, Luo M, Yu CW, Chen CY, Tai R, Wu K. 2014. Transcriptional repression by histone deacetylases in plants. Molecular Plant 7, 764–772. [DOI] [PubMed] [Google Scholar]
- Liu X, Yu CW, Duan J, Luo M, Wang K, Tian G, Cui Y, Wu K. 2012. HDA6 directly interacts with DNA methyltransferase MET1 and maintains transposable element silencing in Arabidopsis. Plant Physiology 158, 119–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond TJ. 1997. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature 389, 251–260. [DOI] [PubMed] [Google Scholar]
- Malapeira J, Khaitova LC, Mas P. 2012. Ordered changes in histone modifications at the core of the Arabidopsis circadian clock. Proceedings of the National Academy of Sciences, USA 109, 21540–21545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer KS, Chen X, Sanders D, Chen J, Jiang J, Nguyen P, Scalf M, Smith LM, Zhong X. 2019. HDA9–PWR–HOS15 is a core histone deacetylase complex regulating transcription and development. Plant Physiology 180, 342–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClung CR. 2006. Plant circadian rhythms. The Plant Cell 18, 792–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClung CR. 2019. The plant circadian oscillator. Biology 8, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehdi S, Derkacheva M, Ramström M, Kralemann L, Bergquist J, Hennig L. 2016. The WD40 domain protein MSI1 functions in a histone deacetylase complex to fine-tune abscisic acid signaling. The Plant Cell 28, 42–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers BC, Kozik A, Griego A, Kuang H, Michelmore RW. 2003. Genome-wide analysis of NBS-LRR-encoding genes in Arabidopsis. The Plant Cell 15, 809–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael TP, McClung CR. 2003. Enhancer trapping reveals widespread circadian clock transcriptional control in Arabidopsis. Plant Physiology 132, 629–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller RN, Costa Alves GS, Van Sluys MA. 2017. Plant immunity: unravelling the complexity of plant responses to biotic stresses. Annals of Botany 119, 681–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamichi N, Kiba T, Henriques R, Mizuno T, Chua NH, Sakakibara H. 2010. PSEUDO-RESPONSE REGULATORS 9, 7, and 5 are transcriptional repressors in the Arabidopsis circadian clock. The Plant Cell 22, 594–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Née G, Xiang Y, Soppe WJ. 2017. The release of dormancy, a wake-up call for seeds to germinate. Current Opinion in Plant Biology 35, 8–14. [DOI] [PubMed] [Google Scholar]
- Ning YQ, Chen Q, Lin RN, Li YQ, Li L, Chen S, He XJ. 2019. The HDA19 histone deacetylase complex is involved in the regulation of flowering time in a photoperiod-dependent manner. The Plant Journal 98, 448–464. [DOI] [PubMed] [Google Scholar]
- Pandey R, Müller A, Napoli CA, Selinger DA, Pikaard CS, Richards EJ, Bender J, Mount DW, Jorgensen RA. 2002. Analysis of histone acetyltransferase and histone deacetylase families of Arabidopsis thaliana suggests functional diversification of chromatin modification among multicellular eukaryotes. Nucleic Acids Research 30, 5036–5055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park HJ, Baek D, Cha JY, Liao X, Kang SH, McClung CR, Lee SY, Yun DJ, Kim WY. 2019. HOS15 interacts with the histone deacetylase HDA9 and the evening complex to epigenetically regulate the floral activator GIGANTEA. The Plant Cell 31, 37–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J, Lim CJ, Khan IU, Jan M, Khan HA, Park HJ, Guo Y, Yun D-J. 2018a Identification and molecular characterization of HOS15-interacting proteins in Arabidopsis thaliana. Journal of Plant Biology 61, 336–345. [Google Scholar]
- Park J, Lim CJ, Shen M, et al. 2018. b Epigenetic switch from repressive to permissive chromatin in response to cold stress. Proceedings of the National Academy of Sciences, USA 115, E5400–E5409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perales M, Más P. 2007. A functional link between rhythmic changes in chromatin structure and the Arabidopsis biological clock. The Plant Cell 19, 2111–2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrella G, Carr C, Asensi-Fabado MA, Donald NA, Páldi K, Hannah MA, Amtmann A. 2016. The histone deacetylase complex 1 protein of arabidopsis has the capacity to interact with multiple proteins including histone 3-binding proteins and histone 1 variants. Plant Physiology 171, 62–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrella G, Lopez-Vernaza MA, Carr C, Sani E, Gosselé V, Verduyn C, Kellermeier F, Hannah MA, Amtmann A. 2013. Histone deacetylase complex1 expression level titrates plant growth and abscisic acid sensitivity in Arabidopsis. The Plant Cell 25, 3491–3505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quint M, Delker C, Franklin KA, Wigge PA, Halliday KJ, van Zanten M. 2016. Molecular and genetic control of plant thermomorphogenesis. Nature Plants 2, 15190. [DOI] [PubMed] [Google Scholar]
- Raschke A, Ibañez C, Ullrich KK, et al. 2015. Natural variants of ELF3 affect thermomorphogenesis by transcriptionally modulating PIF4-dependent auxin response genes. BMC Plant Biology 15, 197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosa S, Shaw P. 2013. Insights into chromatin structure and dynamics in plants. Biology 2, 1378–1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Lei T, Cui X, Liu X, Zhou S, Zheng Y, Guérard F, Issakidis-Bourguet E, Zhou DX. 2019. Arabidopsis histone deacetylase HDA15 directly represses plant response to elevated ambient temperature. The Plant Journal 100, 991–1006. [DOI] [PubMed] [Google Scholar]
- Sun J, Qi L, Li Y, Chu J, Li C. 2012. PIF4-mediated activation of YUCCA8 expression integrates temperature into the auxin pathway in regulating Arabidopsis hypocotyl growth. PLoS Genetics 8, e1002594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M, Shinozuka N, Hirakata T, Nakata MT, Demura T, Tsukaya H, Horiguchi G. 2018. OLIGOCELLULA1/HIGH EXPRESSION OF OSMOTICALLY RESPONSIVE GENES15 promotes cell proliferation with HISTONE DEACETYLASE9 and POWERDRESS during leaf development in Arabidopsis thaliana. Frontiers in Plant Science 9, 580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbert PB, Henikoff S. 2017. Histone variants on the move: substrates for chromatin dynamics. Nature Reviews. Molecular Cell Biology 18, 115–126. [DOI] [PubMed] [Google Scholar]
- Tanaka M, Kikuchi A, Kamada H. 2008. The Arabidopsis histone deacetylases HDA6 and HDA19 contribute to the repression of embryonic properties after germination. Plant Physiology 146, 149–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasset C, Singh Yadav A, Sureshkumar S, Singh R, van der Woude L, Nekrasov M, Tremethick D, van Zanten M, Balasubramanian S. 2018. POWERDRESS-mediated histone deacetylation is essential for thermomorphogenesis in Arabidopsis thaliana. PLoS Genetics 14, e1007280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian L, Fong MP, Wang JJ, Wei NE, Jiang H, Doerge RW, Chen ZJ. 2005. Reversible histone acetylation and deacetylation mediate genome-wide, promoter-dependent and locus-specific changes in gene expression during plant development. Genetics 169, 337–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- To TK, Kim JM, Matsui A, et al. 2011. Arabidopsis HDA6 regulates locus-directed heterochromatin silencing in cooperation with MET1. PLoS Genetics 7, e1002055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Woude LC, Perrella G, Snoek BL, et al. 2019. HISTONE DEACETYLASE 9 stimulates auxin-dependent thermomorphogenesis in Arabidopsis thaliana by mediating H2A.Z depletion. Proceedings of the National Academy of Sciences, USA 116, 25343–25354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Zanten M, Liu Y, Soppe W. 2013. Epigenetic signalling during the life of seeds. In: Grafi G, Ohad N, eds. Epigenetic memory and control in plants. Berlin Heidelberg: Springer, 127–153. [Google Scholar]
- Van Zanten M, Zöll C, Wang Z, Philipp C, Carles A, Li Y, Kornet NG, Liu Y, Soppe WJ. 2014. HISTONE DEACETYLASE 9 represses seedling traits in Arabidopsis thaliana dry seeds. The Plant Journal 80, 475–488. [DOI] [PubMed] [Google Scholar]
- Wang T, Xing J, Liu X, Yao Y, Hu Z, Peng H, Xin M, Zhou DX, Zhang Y, Ni Z. 2018. GCN5 contributes to stem cuticular wax biosynthesis by histone acetylation of CER3 in Arabidopsis. Journal of Experimental Botany 69, 2911–2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing J, Wang T, Liu Z, et al. 2015. GENERAL CONTROL NONREPRESSED PROTEIN5-mediated histone acetylation of FERRIC REDUCTASE DEFECTIVE3 contributes to iron homeostasis in arabidopsis. Plant Physiology 168, 1309–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan K, Li CC, Wang Y, Wang XQ, Wang ZM, Wei DY, Tang QL. 2018. AGL18-1 delays flowering time through affecting expression of flowering-related genes in Brassica juncea. Plant Biotechnology 35, 357–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan K, Ran M, Li S, Zhang J, Wang Y, Wang Z, Wei D, Tang Q. 2020. The delayed senescence of postharvest buds in salt ions was related to antioxidant activity, HDA9 and CCX1 in broccoli (Brassica oleracea L. var. Italic Planch.). Food Chemistry 324, 126887. [DOI] [PubMed] [Google Scholar]
- Yang L, Chen X, Wang Z. 2019. HOS15 and HDA9 negatively regulate immunity through histone deacetylation of intracellular immune receptor NLR genes in Arabidopsis. New Phytologist 226, 507–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Shen W, Yang L, et al. 2020. HY5–HDA9 module transcriptionally regulates plant autophagy in response to light-to-dark conversion and nitrogen starvation. Molecular Plant 13, 515–531. [DOI] [PubMed] [Google Scholar]
- Yu CW, Liu X, Luo M, Chen C, Lin X, Tian G, Lu Q, Cui Y, Wu K. 2011. HISTONE DEACETYLASE6 interacts with FLOWERING LOCUS D and regulates flowering in Arabidopsis. Plant Physiology 156, 173–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan L, Chen X, Chen H, Wu K, Huang S. 2019. Histone deacetylases HDA6 and HDA9 coordinately regulate valve cell elongation through affecting auxin signaling in Arabidopsis. Biochemical and Biophysical Research Communications 508, 695–700. [DOI] [PubMed] [Google Scholar]
- Yumul RE, Kim YJ, Liu X, Wang R, Ding J, Xiao L, Chen X. 2013. POWERDRESS and diversified expression of the MIR172 gene family bolster the floral stem cell network. PLoS Genetics 9, e1003218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun J, Kim YS, Jung JH, Seo PJ, Park CM. 2012. The AT-hook motif-containing protein AHL22 regulates flowering initiation by modifying FLOWERING LOCUS T chromatin in Arabidopsis. Journal of Biological Chemistry 287, 15307–15316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng X, Gao Z, Jiang C, Yang Y, Liu R, He Y. 2020. HISTONE DEACETYLASE 9 functions with polycomb silencing to repress FLOWERING LOCUS C expression. Plant Physiology 182, 555–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Qiu Z, Hu Y, et al. 2011. ABA treatment of germinating maize seeds induces VP1 gene expression and selective promoter-associated histone acetylation. Physiologia Plantarum 143, 287–296. [DOI] [PubMed] [Google Scholar]
- Zheng Y, Ding Y, Sun X, Xie S, Wang D, Liu X, Su L, Wei W, Pan L, Zhou DX. 2016. Histone deacetylase HDA9 negatively regulates salt and drought stress responsiveness in Arabidopsis. Journal of Experimental Botany 67, 1703–1713. [DOI] [PubMed] [Google Scholar]
- Zheng Y, Ge J, Bao C, Chang W, Liu J, Shao J, Liu X, Su L, Pan L, Zhou D-X. 2020. Histone deacetylase HDA9 and transcription factor WRKY53 are mutual antagonists in regulation of plant stress response. Molecular Plant 19, 30408–304013. [DOI] [PubMed] [Google Scholar]
- Zhu JK. 2016. Abiotic stress signaling and responses in plants. Cell 167, 313–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Jeong JC, Zhu Y, et al. 2008. Involvement of Arabidopsis HOS15 in histone deacetylation and cold tolerance. Proceedings of the National Academy of Sciences, USA 105, 4945–4950. [DOI] [PMC free article] [PubMed] [Google Scholar]