Abstract
Using an unbiased high‐throughput microRNA (miRNA)‐silencing screen combined with functional readouts for mitochondrial oxidative capacity in C2C12 myocytes, we previously identified 19 miRNAs as putative regulators of skeletal muscle mitochondrial metabolism. In the current study, we highlight miRNA‐204‐5p, identified from this screen, and further studied its role in the regulation of skeletal muscle mitochondrial function. Following silencing of miRNA‐204‐5p in C2C12 myotubes, gene and protein expression were assessed using quantitative polymerase chain reaction, microarray analysis, and western blot analysis, while morphological changes were studied by confocal microscopy. In addition, miRNA‐204‐5p expression was quantified in human skeletal muscle biopsies and associated with in vivo mitochondrial oxidative capacity. Transcript levels of PGC‐1α (3.71‐fold; p < .01), predicted as an miR‐204‐5p target, as well as mitochondrial DNA copy number (p < .05) and citrate synthase activity (p = .06) were increased upon miRNA‐204‐5p silencing in C2C12 myotubes. Silencing of miRNA‐204‐5p further resulted in morphological changes, induced gene expression of autophagy marker light chain 3 protein b (LC3B; q = .05), and reduced expression of the mitophagy marker FUNDC1 (q = .01). Confocal imaging revealed colocalization between the autophagosome marker LC3B and the mitochondrial marker OxPhos upon miRNA‐204‐5p silencing. Finally, miRNA‐204‐5p was differentially expressed in human subjects displaying large variation in oxidative capacity and its expression levels associated with in vivo measures of skeletal muscle mitochondrial function. In summary, silencing of miRNA‐204‐5p in C2C12 myotubes stimulated mitochondrial biogenesis, impacted on cellular morphology, and altered expression of markers related to autophagy and mitophagy. The association between miRNA‐204‐5p and in vivo mitochondrial function in human skeletal muscle further identifies miRNA‐204‐5p as an interesting modulator of skeletal muscle mitochondrial metabolism.
Keywords: C2C12, microRNA, mitochondria, mitophagy, skeletal muscle
Silencing of microRNA‐204‐5p (miRNA‐204‐5p) in C2C12 myotubes enhanced mitochondrial biogenesis, via regulation of PGC‐1α. In humans, low expression of miRNA‐204‐5p in skeletal muscle was associated high oxidative capacity. These findings identify miRNA‐204‐5p as an interesting modulator of mitochondrial function in human skeletal muscle.
1. INTRODUCTION
Mitochondria play critical roles in the regulation of important biological processes such as energy production (Kasahara & Scorrano, 2014), reactive oxygen species generation (Di Meo, Iossa, & Venditti, 2017), cell growth (Di Meo et al., 2017), and insulin response (Di Meo et al., 2017). Consequently, the mitochondrial dysfunction is associated with several disorders, such as cardiovascular disease (el Azzouzi et al., 2013), cancer (Chourasia, Boland, & Macleod, 2015), neurodegenerative disorders (Pickrell & Youle, 2015), and metabolic abnormalities such as Type 2 diabetes (Bollinger, Powell, Houmard, Witczak, & Brault, 2015; Munasinghe et al., 2016). Even in the absence of disease, maintaining a healthy functioning pool of mitochondria is essential for cellular/tissue/organismal homeostasis (Price et al., 2012; Russell, Foletta, Snow, & Wadley, 2014). Sustained physical activity is considered as one of the most potent interventions to maintain a healthy mitochondrial function and is associated with several cardiovascular and metabolic health effects. Interestingly, physical activity stimulates mitochondrial function by promoting mitochondrial biogenesis (Montero & Lundby, 2017), but also by actively inducing the degradation of damaged dysfunctional mitochondria through mitophagy, the selective degradation of mitochondria by the autophagic machinery, a process recognized as essential to maintain mitochondrial function (Lira et al., 2013). However, the exact molecular mechanisms linking physical exercise and mitochondrial adaptations in skeletal muscle are incompletely understood.
In this context, increasing amounts of evidence indicate that microRNAs (miRNAs) can modulate different aspects of the regulation of mitochondrial function (M. Cheng et al., 2016; Mohamed, Hajira, Pardo, & Boriek, 2014). MiRNAs are short oligonucleotides of 18–20 nucleotides in length that posttranscriptionally regulate many biological processes by binding protein‐coding transcripts (Bartel, 2004), adding a new player on an already complex network of gene regulation. It has been demonstrated that most of the transcriptome is in fact regulated by miRNAs, including both nuclear‐ and mitochondria‐encoded messenger RNAs (mRNAs) coding for mitochondrial proteins (Duarte, Palmeira, & Rolo, 2014). Moreover, miRNAs have been shown to regulate proteins involved in mitochondrial maintenance, such as mitochondrial biogenesis (Mohamed et al., 2014), protein quality control mechanisms (Dahlmans, Houzelle, Schrauwen, & Hoeks, 2016), mitochondrial dynamics (Dahlmans et al., 2016), and autophagy (M. Cheng et al., 2016).
Previously, we conducted unbiased hypothesis‐free miRNA‐silencing screens in C2C12 muscle cells and identified 19 miRNAs as regulators of skeletal muscle mitochondrial function (Dahlmans et al., 2017). In a subsequent experiment, we individually silenced these 19 candidate miRNAs in differentiated C2C12 myotubes and quantified the transcripts of 27 genes involved in different aspects of mitochondrial function (Dahlmans et al., 2019). This series of experiments identified several interesting candidate miRNAs, but here we focus on miRNA‐204‐5p since silencing of this miRNA clearly increased the expression of the mRNA encoding the peroxisome proliferator‐activated receptor‐gamma coactivator‐1α (PGC‐1α) in C2C12 myotubes, both 24 and 48 hr posttransfection. PGC‐1α is a major regulator for mitochondrial biogenesis and oxidative metabolism, plays an important role in exercise adaptation in muscle, and has been reported to be downregulated in skeletal muscle of insulin‐resistant and Type 2 diabetic subjects (Patti et al., 2003), whereas its overexpression in mice resulted in an endurance‐trained phenotype (Lin et al., 2002).
On the basis of these earlier data, the aim of the present study is to test the hypothesis that miRNA‐204‐5p is involved in the regulation of skeletal muscle mitochondrial biogenesis and to study its role in skeletal muscle mitochondrial metabolism in more detail.
2. MATERIALS AND METHODS
2.1. Cell culture, reagents, and antibodies
Mouse skeletal muscle C2C12 myoblasts were cultivated in growth medium consisting of high‐glucose Dulbecco's modified Eagle's medium (DMEM; Gibco, Carlsbad, CA) supplemented with 10% fetal bovine serum (Batch Number 1693362), 1% non‐essential amino acids (Gibco), and 20 mM n‐2‐hydroxyethylpiperazine‐n‐2‐ethane sulfonic acid (HEPES; Gibco). Upon reaching 80–90% confluence, cells were seeded in 12‐well CellBIND plates (Coring Life Science, Lowell, CA) or staining slide flasks (Nunc Lab‐Tek Flask on Slide; Thermo Fisher Scientific, Waltham, MA) at a density of 100,000 cells/well or 240,000 cells/slide flasks, respectively. Following seeding, cells were allowed to grow for 24 hr in growth medium. Then, C2C12 myoblasts were differentiated into myotubes for 5 days in high‐glucose DMEM supplemented with 2% horse serum (Gibco), 1% non‐essential amino acids (Gibco), and 20 mM HEPES (Gibco).
2.2. In silico analysis
TargetScanMouse, miRanda, and microT‐CDS algorithms were used to predict the targets of miRNA‐204‐5p. Predicted targets were only considered when they were predicted by at least two of the databases. In addition, all predictions present at least a 7mer site‐matching, meaning that the entire seed sequence of miRNA‐204‐5p matches the 3′‐ untranslated region (3′‐UTR) sequence of the gene of interest.
2.3. C2C12 transfection
At Day 5 of differentiation, C2C12 myotubes were transfected with either a specific miRCURY LNA™ miRNA inhibitor targeting miRNA‐204‐5p (Exiqon A/S, Vedbæk, Denmark), or a specific locked nucleic acid (LNA) inhibitor, was used as a negative control. This control LNA (sequence: TAACACGTCTATACGCCCA) does not target any known mature miRNA.
Transfection was performed using lipofectamine RNAiMAX (Invitrogen, Carlsbad), according to the manufacturer's instructions. Shortly, lipofectamine complexes were stabilized in OptiMEM (Gibco) for 5 min and incubated with the LNAs, diluted to 500 nM in OptiMEM, for 20 min. Finally, the lipofectamine/LNA complexes were diluted 1:5 in differentiation media (Dahlmans et al., 2017).
2.4. RNA preparation and gene expression analysis in C2C12 cells
Cells were washed with 1 ml cold 1X phosphate‐buffered saline (PBS) and harvested in 700 µl TRIzol reagent (Invitrogen) and kept at −80oC for subsequent analyses. RNeasy Mini Kits (Qiagen, Venlo, The Netherlands) were used to extract RNA. RNA quantity and quality were measured using a nanodrop microvolume spectrophotometer (ND‐1000; NanoDrop Technologies, Wilmington, DE) and complementary DNA (cDNA) was synthesized using the High‐Capacity RNA‐to‐cDNA Kit (Applied Biosystems) according to manufacturer's guidelines. Quantitative polymerase chain reaction (qPCR) was performed using the SensiMix SYBR Hi‐ROX Kit (Bioline, London, UK) and specific primer sets (Sigma‐Aldrich, St. Louis, MI) to quantify the abundance of mRNAs of interest. The qPCR analyses were performed on the CFX384 Touch™ Real‐Time PCR Detection System (Bio‐Rad, Hercules, CA).
2.5. Mitochondrial DNA copy number quantification
For the determination of mitochondrial DNA (mtDNA) copy number, DNA was isolated at 24, 36, and 48 hr posttransfection using the DNeasy Blood & Tissue Kit (Qiagen). DNA was quantified and the integrity was checked by spectrophotometry using the Nanodrop. Relative amounts of nuclear and mtDNA were quantified by qPCR, in which nuclear DNA was represented by the UCP2 gene and mtDNA by the COX2 gene. qPCR was performed on the ABI Prism 7900HT Real‐Time PCR System (Applied Biosystems, Foster City, CA). MtDNA copy number was calculated using the ΔC t method.
2.6. Luciferase reporter assay
C2C12 cells were cultured in DMEM (4.5 g/L d‐glucose) supplemented with 10% fetal bovine serum, 2% HEPES, and 1% minimum essential medium non‐essential amino acids solution and seeded in 24‐well plates at a density of 50,000 cells/well. The next day, cells were transfected with hsa‐miRNA‐204‐5p or Neg control #1 (Ambion; Life Technologies, Waltham) precursor molecules and cotransfected with the LightSwitch_PPARGC1A_3′UTR vector (Active Motif, Carlsbad) using Lipofectamine 2000 (Invitrogen, Waltham). After 24 hr, the luciferase activity was determined using the LightSwitch Luciferase Assay Reagent (SwitchGear Genomics, Carlsbad) according to the manufacturer's instructions using a GLOMAX Microplate Luminometer (Promega Corporation, Madison, WI).
2.7. Microarray analysis
Purified RNA was labeled with the Ambion WT Expression Kit (Carlsbad) and hybridized to an Affymetrix Mouse Gene 1.1 ST array plate (Affymetrix, Santa Clara, CA). Hybridization, washing, and scanning were carried out on an Affymetrix GeneTitan platform according to the manufacturer's instructions. Normalized expression estimates were obtained from the raw intensity values applying the robust multiarray analysis preprocessing algorithm available in the Bioconductor library affyPLM with default settings (Bolstad, Irizarry, Astrand, & Speed, 2003; Irizarry et al., 2003). Probe sets were defined according to Dai et al. (2005). In this method, probes are assigned to Entrez IDs as a unique gene identifier. In this study, probes were reorganized based on the Entrez Gene database, build 37, Version 1 (remapped CDF v22). q values were calculated using an intensity‐based moderated T‐statistic (Sartor et al., 2006). Genes were defined as significantly changed when q < .05.
2.8. Protein extraction and western blot analysis in C2C12 muscle cells
Cells were washed with 1 ml cold 1X PBS and scraped in 150 µl Bio‐Plex cell lysis buffer (Bio‐Rad), and processed according to the manufacturer's instruction. Protein loading for each sample was first determined using InstantBlue Coomassie (Expedeon, San Diego, CA). Equal amounts of protein were loaded in precast sodium dodecyl sulfate‐polyacrylamide gel electrophoresis (Mini TGX AnyKD or Criterion TGX AnykD; Bio‐Rad). Next, LC3B‐I and LC3B‐II were detected using an antibody directed against LC3B, diluted 1:1000 (L7543; Sigma‐Aldrich). Furthermore, an antibody directed against β‐actin was diluted 1:25,000 (A5316; Sigma‐Aldrich), and was used as a loading control. Finally, membranes were quantified at the appropriate wavelengths using an Odyssey CLx Imaging System (Li‐COR, Lincoln, NE).
2.9. Live cell imaging in C2C12 cells
C2C12 myoblasts were seeded in 12‐well CellBIND plates (Corning) at 100,000 cells/well and allowed to settle overnight in the growth medium. Then, the medium was replaced to differentiation medium and cells were transfected as previously described. Continuous cell monitoring was performed for 10 days using the IncuCyte ZOOM (Essen Bioscience, Ltd., Hertfordshire, UK). The differentiation medium was replaced every 48 hr to maintain the cells for the duration of the experiments.
2.10. Immunocytochemistry in C2C12 cells
C2C12 myoblasts were seeded in slide flasks (Nunc Lab‐Tek Flask on Slide; Thermo Fisher Scientific) at 240,000 cells/flask and differentiated for 5 days. Then, myotubes were transfected and fixated 48 hr posttransfection. To fix the cells, myotubes were washed three times in PBS and incubated for 1 hr in 3.7% formaldehyde at room temperature. Finally, fixed myotubes were washed three times in PBS and stored in PBS at 4°C until immunocytochemistry was performed. Before staining, slides were rinsed for 5 min in PBS and directly incubated with primary antibodies diluted in 0.05% Tween‐20 in PBS. Mitochondria were stained by an anti‐OxPhos antibody cocktail (MS604; MitoSciences, Eugene, OR), targeting structural subunits of all OxPhos complexes. Autophagosomes were visualized by using an anti‐LC3B antibody (L7543; Sigma‐Aldrich). Image acquisition was performed using a Nikon Eclipse E800 fluorescent microscope equipped with ×40 oil immersion objective (Nikon Instruments, Amsterdam, The Netherlands) or a Leica TCS SP8 confocal microscope equipped with a ×63 1.40NA oil immersion objective (Leica, Amsterdam). The confocal images were processed using deconvolution software (Huygens Professional; Scientific Volume Imaging B.V., Hilversum, The Netherlands).
2.11. Subject selection and human muscle biopsies collection
The subjects included in this study participated in previous studies (Phielix, Meex, Moonen‐Kornips, Hesselink, & Schrauwen, 2010; Phielix et al., 2012; Vosselman et al., 2015; van de Weijer et al., 2015) that were approved by the institutional medical ethics committee, registered at ClinicalTrials.gov (NCT00943059 and NCT01298375) and in the Dutch trial register (NL1888) and for which all participants provided their written informed consent in accordance with the declaration of Helsinki. From these studies, we selected four groups of male, weight‐stable individuals for the analysis of miRNA‐204‐5p expression in skeletal muscle and its relation to parameters for in vivo oxidative capacity. These four groups included: (a) Type 2 diabetic patients (n = 12); (b) overweight/obese nondiabetic subjects (n = 12); (c) young lean individuals (n = 12), and (d) young endurance‐trained athletes (n = 12). The primary selection was based on the availability of muscle material and the presence of reliable data for in vivo oxidative capacity (maximal aerobic capacity [VO2 max] and phosphocreatine [PCr] recovery rate). Second, subjects were also selected with the intention to create age‐ and body mass index‐matched groups (i.e., Group 1 vs. Group 2 and Group 3 vs. Group 4).
All Type 2 diabetes patients were on metformin treatment and the use of statins was permitted. Obese nondiabetic individuals were considered healthy, that is, they did not use medication, did not present any pathology (hypertension, cardiovascular disorders, and liver dysfunction), and did not have any first‐degree relatives with Type 2 diabetes.
Lean sedentary (LS) subjects were included if their VO2 max was below 45 ml−1·min−1·kg and did not participate in more than 1 hr of organized exercise per week for the previous 2 years. Endurance‐trained athletes were enrolled in the study if their VO2 max was higher than 55 ml−1·min−1·kg and if they participated in endurance training at least three times per week for the last 2 years.
All biopsies were collected after an overnight (≥10 hr) fast and before any intervention. Subjects were instructed to avoid (strenuous) physical exercise in the 3‐day period preceding the muscle biopsy.
2.12. Subject characterization
All subjects included in this study were thoroughly phenotyped in term of body composition and aerobic capacity. Body composition was determined either via dual X‐ray absorptiometry (Discovery A; Hologic, Bedford, MA) or hydrostatic weighing (Siri, 1993). Maximal aerobic capacity was determined through an incremental cycling test until exhaustion, as described previously (Kuipers, Verstappen, Keizer, Geurten, & van Kranenburg, 1985). In vivo mitochondrial function was assessed via quantification of PCr resynthesis rates using a 31P magnetic resonance spectrometry approach on a 3 T whole‐body scanner (Achieve 3T‐X; Philips Healthcare, The Netherlands), as previously described (Lindeboom et al., 2014).
2.13. MiRNA quantitative PCR in muscle biopsies
A 10–15 mg piece of frozen muscle tissue was homogenized in 700 µl of TRIzol with an Ultra‐Turrax (IKA, Staufen, Germany) for 1 min at 17,000 rpm. RNA isolation was then performed using a miRNeasy Mini Kit (Qiagen) according to the manufacturer's instructions. The yield was improved by reloading the final eluate onto the column membrane twice. RNA concentrations were measured using a microvolume spectrophotometer (Nanodrop), whereas RNA quality was determined using a Bioanalyzer System (Agilent Technologies, Santa Clara, CA). Then, cDNA synthesis was performed using the Universal cDNA Synthesis Kit II (Exiqon) according to the manufacturer's protocol. Finally, cDNA samples were analyzed using predesigned 384‐well Pick‐&‐Mix microRNA PCR Panel plates (Exiqon), according to the manufacturer's guidelines using a CFX384 Touch Real‐time PCR Detection System (Bio‐Rad). Relative miRNA expression levels were determined using the ΔΔC t method. Synthetic UniSP6 spike‐in control was used to monitor the cDNA synthesis reaction, that is, as a positive control of the reverse transcription. Furthermore, an interpolate calibrator was added to all qPCR plates to uniformly normalize C t values across all the experiments.
2.14. Statistics analysis
All results are expressed as means ± standard deviation. Each set of in vitro data presented in this study is the result of three independently executed experiments, each of which includes three technical replicates (unless indicated otherwise in the figure legend). Comparisons between in vitro conditions were performed by unpaired two‐tailed Student's t test. Comparisons over time were performed by two‐way analysis of variance (ANOVA). p < .05 was considered statistically significant.
Differences between the various human subject groups were assessed via one‐way ANOVA with Tukey's multiple comparisons test. Pearson correlation analysis was used to correlate miRNA‐204‐5p expression in skeletal muscle to in vivo parameters of oxidative capacity.
3. RESULTS
3.1. miRNA‐204‐5p silencing in C2C12 myotubes increases PGC‐1α gene expression
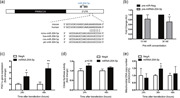
We previously silenced 19 individual candidate miRNAs, identified in a large miRNA‐silencing screen (Dahlmans et al., 2017), and subsequently measured 27 genes involved in mitochondrial function (Dahlmans et al., 2019). Interestingly, the silencing of miRNA‐204‐5p showed a pronounced induction of the expression of PGC‐1α, a well‐known regulator of mitochondrial biogenesis, both when silenced for 24 (2.43‐fold; p < .05) and 48 hr (3.71‐fold; p < .01; Figure 1). Subsequent in silico analysis by TargetScan and miRanda revealed a specific sequence in the 3′‐UTR of PGC‐1α as a predicted target to the seed region of miRNA‐204‐5p (Figure 1a). In addition, both miRNA‐204‐5p and the binding site in the 3′‐UTR of PGC‐1α were conserved over many different invertebrates as well as vertebrate species including humans, but not worm (Figure 1a). Moreover, activity assays using a luciferase reporter construct harboring the PGC‐1α 3′‐UTR revealed that increasing concentrations of pre‐miRNA‐204‐5p significantly and dose‐dependently reduced the luciferase reporter signal, indicating a functional interaction between miRNA‐204‐5p and the 3′‐UTR of PGC‐1α (Figure 1b).
Figure 1.
miRNA‐204‐5p silencing induces mitochondrial biogenesis through PGC‐1α induction. (a) Representation of the binding between the 3′‐UTR of PGC‐1α mRNA and miRNA‐204‐5p in different species. (b) Activity assay of a luciferase reporter constructs harboring the PGC‐1α 3′‐UTR after transfection with a synthetic precursor for miRNA‐204‐5p. Data shown are the result of three independently executed duplicate experiments. (c) Relative PGC‐1α gene expression upon miRNA‐204‐5p silencing compared to Negative Control A (NegA), 24 and 48 hr posttransfection in C2C12 myotubes. (d) Citrate synthase activity following 24 and 48 hr transfection of anti‐miRNA‐204‐5p versus NegA in C2C12 myotubes. (d) Mitochondrial DNA copy number and nuclear DNA upon miRNA‐204‐5p silencing versus NegA. *p < .05 and **p < .01 (n = 3). 3′‐UTR, 3′‐untranslated region; miRNA, microRNA; mRNA, messenger RNA; PGC‐1α, peroxisome proliferator‐activated receptor‐gamma coactivator‐1α
To address the consequences of the increase in PGC‐1α expression (Figure 1c), we next assessed citrate synthase activity as well as mtDNA copy number as markers for mitochondrial density. In line with the elevated PGC‐1α expression, citrate synthase activity (Figure 1d) showed a strong tendency to be increased after 24 hr miRNA‐204‐5p silencing (25% increase; p = .06), whereas the difference did not reach statistical significance at 48 hr. MtDNA copy number was significantly increased (46% increase; p < .05) 36 hr posttransfection, but not after 24 or 48 hr miRNA‐204‐5p silencing (Figure 1e). Together, these results indicate a (transient) increase in mitochondrial content upon miRNA‐204‐5p silencing, which prompted us to further analyze the role of miRNA‐204‐5p in regulating mitochondrial metabolism.
3.2. miRNA‐204‐5p silencing induces morphological aberrations in C2C12 myotubes
To investigate if the silencing of miRNA‐204‐5p also resulted in morphological changes that could reveal enhanced mitochondrial density, we next studied the effect of miRNA‐204‐5p silencing on C2C12 myotube development and continuously monitored the transfected C2C12 myotubes for 72 hr after transfection. Interestingly, myotubes transfected with anti‐miRNA‐204‐5p displayed morphological changes that were most apparent 48 hr posttransfection. Thus, whereas nontransfected and Negative Control A transfected C2C12 myotubes displayed a normal morphology and similar appearance as control cells (Figure 2a–e) 48 hr posttransfection, (i.e., at Day 7 of differentiation) myotubes transfected with anti‐miRNA‐204‐5p showed large round anomalous structures within the myotubes (Figure 2f–j).
Figure 2.
Silencing miRNA‐204‐5p induces the development of an aberrant morphology in C2C12 myotubes. C2C12 myoblasts were differentiated for 5 days prior transfection with either Negative Control A or an LNA targeting miRNA‐204‐5p, and were subsequently observed for 72 hr. The development of the aberrant phenotype was consistently observed 48 hr posttransfection. (a–e) C2C12 myotubes transfected with of Negative Control A. (f–j) C2C12 myotubes transfected with an LNA directed against miRNA‐204‐5p. Panels are bright‐field micrographs. Red arrows indicate C2C12 myotubes with atypical morphology. Scale bar = 400 μm (n = 3). LNA, locked nucleic acid; miRNA, microRNA
Strikingly, when undifferentiated C2C12 muscle cells were transfected with anti‐miRNA‐204‐5p and subsequently underwent differentiation, the aberrant structures did not appear before Day 5 of differentiation (Figure 3). In other words, C2C12 muscle cells transfected with Negative Control A or anti‐miRNA‐204‐5p in the myoblasts stage, displayed similar and normal morphology during the first 4 days of subsequent differentiation (Figure 3). Again, although the morphological abnormalities started to appear at Day 5 of differentiation, the irregular cellular morphology was most apparent at Day 7 of differentiation (Figure 3h), comparable to the changes seen after silencing miRNA‐204‐5p in already differentiated C2C12 myotubes.
Figure 3.
miRNA‐204‐5p silencing only affects fully differentiated C2C12 myotubes. Undifferentiated C2C12 myoblasts were transfected with either Negative Control A or an LNA targeting miRNA‐204‐5p, and were subsequently observed for 7 days. (a–d) Negative Control A. (e–h) Transfection of an LNA against miRNA‐204‐5p. Red arrows indicate C2C12 myotubes with atypical morphology. Panels are bright‐field micrographs (n = 2). Scale bar = 400 μm. LNA, locked nucleic acid; miRNA, microRNA
3.3. Silencing of miRNA‐204‐5p affects markers for autophagy/mitophagy
We next hypothesized that the abnormal phenotype of the C2C12 myotubes upon miRNA‐204‐5p silencing could possibly be explained by increased autophagic activity, especially since microtubule‐associated protein 1A/1B‐LC3B, a major player in autophagosome formation, is another predicted target (Figure 4a) of miRNA‐204‐5p as shown previously in renal cell carcinoma (Mikhaylova et al., 2012). To study whether the morphological abnormalities that we observed in the C2C12 myotubes could be related to autophagy, we first performed a microarray analysis 24 hr posttransfection and focused on the expression of several autophagy‐related genes. Indeed, the miRNA‐204‐5p predicted target LC3B, as well as autophagosome and lysosome fuser VAMP8 and lysosomal cysteine protease inhibitor cystatin B (CSTB), were significantly increased following anti‐miRNA‐204‐5p treatment (Figure 4b). A similar expression pattern was observed for the autophagy markers GABARAPL1 and SQSTM1 (Figure 4b), although these changes did not reach statistical significance. Interestingly, however, mitophagy markers PARK2, BNIP3, BNIP3L, and FUNDC1 appeared to be decreased after miRNA‐204‐5p lowering, although this was only statistically significant for FUNDC1 (Figure 4b).
Figure 4.
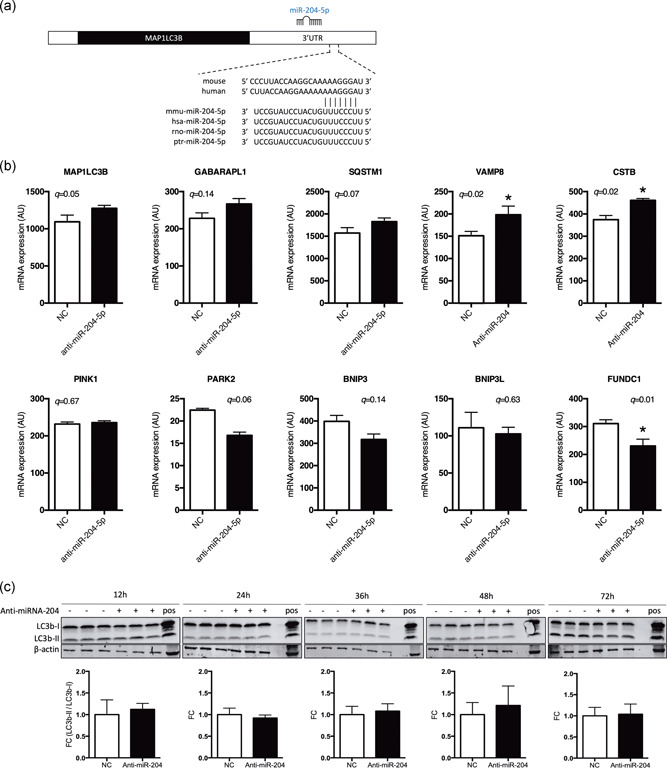
miRNA‐204‐5p silencing does not increase LC3B‐I or LC3B‐II protein abundance. (a) Representation of the binding between the 3′‐UTR of LC3B (MAP1LC3B) mRNA and miRNA‐204‐5p across different species. (b) Gene expression analysis (microarray analysis) following anti‐miRNA‐204‐5p (24 hr) in differentiated C2C12 myotubes. *q < .05 (FDR adjusted q‐value intensity‐based moderated T‐statistic, n = 3). (c) LC3B‐I and LC3B‐II protein levels were quantified using western blot analysis 12, 24, 36, 48, and 72 hr posttransfection with either Negative Control A or anti‐miRNA‐204‐5p. In addition, transfected cells were compared to chloroquine treated C2C12 myotubes as a positive control (n = 3). 3′‐UTR, 3′‐untranslated region; FDR, false discovery rate; LC3B, light chain 3 protein b; miRNA, microRNA; mRNA, messenger RNA; NC, negative control
Next, to investigate if the induction of autophagy markers was translated to the protein level, we quantified the abundance of LC3B‐I and LC3B‐II protein 12, 24, 36, 48, and 72 hr posttransfection. Despite the apparent induction of several autophagy‐related genes and the aberrant phenotype of the cells, 48 hr posttransfection with anti‐miRNA‐204‐5p (as displayed in Figure 2), we could not detect changes in protein abundance of LC3B‐I or LC3B‐II after silencing miRNA‐204‐5p (Figure 4c). However, since we previously demonstrated that the morphological phenotype only occurs in fully differentiated C2C12 myotubes, and not myoblasts, changes in LC3B abundance might have been masked by the presence of undifferentiated myoblasts in our cell cultures. To circumvent this problem, we finally performed immunocytochemistry with an antibody against LC3B and investigated LC3B in miRNA‐204‐5p silenced C2C12 myotubes. Indeed, while C2C12 myotubes transfected with Negative Control A displayed a diffuse, ubiquitous LC3B expression (Figure 5a), miRNA‐204‐5p silencing induced a typical punctuated staining of LC3B, 48 hr posttransfection (Figure 5b). The observed LC3B staining was similar to the pattern observed in C2C12 myotubes transfected with Negative Control A and subsequently treated with chloroquine, which is known to induce the accumulation of LC3B‐positive autophagosomes by inhibiting their clearance (Figure 5c). Finally, we performed double staining for LC3B and OxPhos, as a marker for mitochondria, in C2C12 myotubes, 48 hr posttransfection with either Negative Control A or anti‐miRNA‐204‐5p. Interestingly, whereas OxPhos displayed a diffuse homogenous staining in the Negative Control A transfected cells, we observed a punctuated OxPhos localization in the anti‐miRNA‐204‐5p transfected cells that were similar to the LC3B staining pattern. Furthermore, the merged images of OxPhos and LC3B demonstrated the colocalization of both mitochondria and the autophagosomal marker LC3B (Figure 6).
Figure 5.
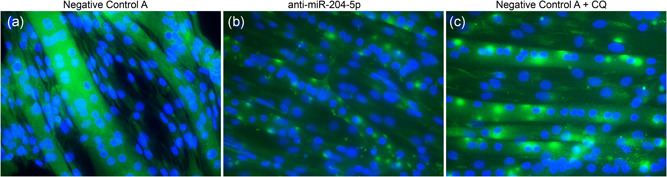
miRNA‐204‐5p silencing leads to the activation of LC3B and the accumulation of autophagosomes in C2C12 myotubes. C2C12 myoblasts were differentiated for 5 days and transfection with either Negative Control A (a) or an LNA targeting miRNA‐204‐5p (b) for 48 hr. Immunocytochemistry targeting the autophagosome marker LC3B (green) or nuclei (blue) was performed. (c) Twenty‐four hours chloroquine incubation (1 μg/ml) was used as a positive control for autophagosome accumulation (n = 4). LC3B, light chain 3 protein b; LNA, locked nucleic acid; miRNA, microRNA
Figure 6.
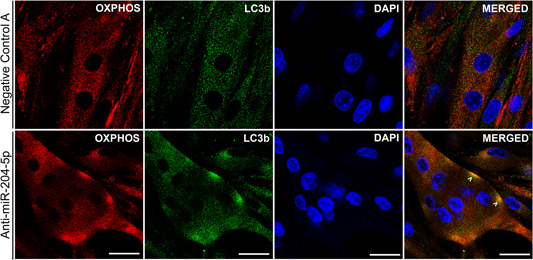
miRNA‐204‐5p silencing targets mitochondria for autophagy. C2C12 myotubes silenced with an anti‐miRNA‐204‐5p displayed colocalization of the autophagosome marker LC3B (green) and the mitochondrial marker OxPhos (red), indicating that mitochondria are targeted for autophagy (mitophagy). Scale bar = 25 μm (n = 3). DAPI, 4′,6‐diamidino‐2‐phenylindole; LC3B, light chain 3 protein b; miRNA, microRNA
3.4. miRNA‐204‐5p expression correlates to in vivo mitochondrial function in human skeletal muscle
To test whether these findings are relevant in human physiology, we then selected (from previous human clinical trials performed in our facilities [Phielix et al., 2010; Phielix et al., 2012; van de Weijer et al., 2015; Vosselman et al., 2015]) four subject groups known to be characterized by different mitochondrial abundance and oxidative capacity, including Type 2 diabetes patients (Type 2 diabetes mellitus [T2DM]), obese nondiabetic subjects (O), LS individuals, and endurance‐trained athletes (A) and investigated miRNA‐204‐5p expression in skeletal muscle biopsies. Also, VO2 max and PCr recovery rates were determined, although the latter could not be assessed in the entire cohort (n = 38). The characteristics of the subjects included for the current study are displayed in Table 1. VO2 max and PCr recovery were highest in endurance‐trained athletes and lowest in patients with T2DM (Table 1).
Table 1.
Subject characteristics
| T2DM# | Obese$ | Lean Sedentary* | Athletes& | |
|---|---|---|---|---|
| n | 11 | 12 | 11 | 11 |
| Age, years | 58.6 ± 4.0 | 56.7 ± 7.2 | 22.2 ± 2.7 | 25.3 ± 4.5 |
| ***, &&& | ***, &&& | ###, $$$ | ###, $$$ | |
| Weight, kg | 100.2 ± 12.6 | 94.1 ± 13.8 | 73.7 ± 7.1 | 70.6 ± 7.7 |
| ***, &&& | ***, &&& | ###, $$$ | ###, $$$ | |
| Height, m | 1.77 ± 0.08 | 1.74 ± 0.07 | 1.83 ± 0.05 | 1.83 ± 0.07 |
| *, & | $ | $ | ||
| BMI, kg/m2 | 32.4 ± 3.7 | 31.0 ± 3.7 | 22.1 ± 1.8 | 21.0 ± 1.5 |
| ***, &&& | ***, &&& | ###, $$$ | ###, $$$ | |
| Fat (%) | 33.8 ± 4.9 | 34.7 ± 7.0 | 18.1 ± 3.7 | 12.7 ± 2.1 |
| ***, &&& | ***, &&& | ###, $$$ | ###, $$$ | |
| VO2 max, ml−1·min−1·kg (lean mass) | 37.6 ± 4.5 | 42.8 ± 6.9 | 50.4 ± 3.4 | 68.9 ± 4.6 |
| ***, &&& | **, &&& | ###, $$, &&& | ###, $$$, *** | |
| PCr recovery, s a | 26.7 ± 6.1 | 21.3 ± 4.4 | 20.8 ± 3.9 | 15.9 ± 5.1 |
| &&& | ### |
Note: Age, weight, height, body mass index (BMI), body composition, and in vivo (mitochondrial) oxidative capacity in Type 2 diabetic patients (T2DM), nondiabetic overweight/obese individuals, lean sedentary individuals, and endurance‐trained athletes. Values presented are mean ± SD. Significance is indicated with #, $, *, and & representing changes compared to T2DM, obese, lean sedentary, and athletic subjects, respectively, and with 1, 2, and 3 symbols representing p < .05, p < .01, and p < .001, respectively. Statistical significances are based on one‐way ANOVA with Tukey's post hoc test.
Abbreviations: ANOVA, analysis of variance; PCr, phosphocreatine; SD, standard deviation; T2DM, Type 2 diabetes mellitus; VO2 max, maximal aerobic capacity.
For PCr recovery, group size for the lean sedentary subjects and the endurance‐trained athletes are n = 6 and n = 9, respectively.
Interestingly, the expression of miRNA‐204‐5p in skeletal muscle was different between the Type 2 diabetic individuals and endurance‐trained athletes as assessed by one‐way ANOVA with Tukey's post hoc correction for multiple testing. Thus, the expression of miRNA‐204‐5p was 1.85‐fold (p = .008) higher in Type 2 diabetes patients with a low mitochondrial oxidative capacity when compared to the endurance‐trained athletes with an optimal mitochondrial oxidative capacity (Figure 7a). In addition, miRNA‐204‐5p expression negatively correlated with VO2 max (r = −.38, p = .011; Figure 7b) and positively correlated with PCr recovery rates (r = .37, p = .023; Figure 7c). Collectively, these collective results indicate that low miRNA‐204‐5p expression is associated with high in vivo mitochondrial oxidative capacity in humans, that is, a high VO2 max and a fast PCr recovery.
Figure 7.
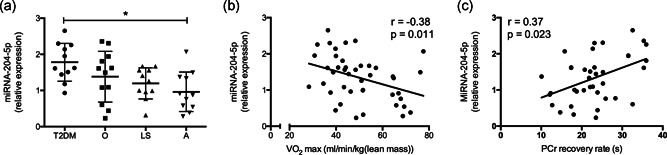
miRNA‐204‐5p levels in human biopsies correlates with oxidative capacity in vivo. (a) miRNA‐204‐5p expression was assessed in skeletal muscle (vastus lateralis) biopsies from Type 2 diabetic (T2DM), obese (O), lean sedentary (LS), and athletic (A) subjects. *p < .05 one‐way ANOVA with Tukey's post hoc testing for multiple comparisons. Pearson's correlation between miRNA‐204‐5p expression and (b) VO2 max (normalized for lean body mass) and between miRNA‐204‐5p expression and (c) PCr recovery rate, a measure for in vivo mitochondrial capacity. ANOVA, analysis of variance; miRNA, microRNA; PCr, phosphocreatine; T2DM, Type 2 diabetes mellitu; VO2 max, maximal aerobic capacity
4. DISCUSSION
We previously conducted a high‐throughput hypothesis‐free miRNA‐silencing screen in C2C12 muscle cells and identified several miRNAs as potential regulators of mitochondrial function (Dahlmans et al., 2017). Here, we further investigated one of these miRNAs and observed that miRNA‐204‐5p silencing in C2C12 myotubes resulted in the induction of PGC‐1α gene expression and mitochondrial biogenesis and that miRNA‐204‐5p expression in human muscle negatively correlated to oxidative capacity.
In the present study, miRNA‐204‐5p silencing induced PGC‐1α, a transcriptional cofactor pivotal in the regulation of mitochondrial biogenesis, which prompted us to study the role of miRNA‐204‐5p in the regulation of mitochondrial metabolism in more detail. Interestingly, the increase in PGC‐1α gene expression was accompanied by an increase in citrate synthase activity and mtDNA copy number, known markers of mitochondrial content. We could also show a direct interaction between miRNA‐204‐5p and the 3′‐UTR of PGC‐1α. To the best of our knowledge, this is the first study to directly link miRNA‐204‐5p to mitochondrial biogenesis in skeletal muscle, whereas most studies regarding miRNA‐204‐5p have been focusing on its roles in cancer. In the context of gastric cancer, however, miRNA‐204‐5p has also been reported to regulate SIRT1 expression, a known metabolic regulator, and upstream target of PGC‐1α (Canto & Auwerx, 2009), indicating that miRNA‐204‐5p may also regulate mitochondrial function in an indirect fashion.
Intriguingly, the silencing of miRNA‐204‐5p also led to the consistent development of an aberrant morphological phenotype. In this context, previous studies in renal carcinoma cells have demonstrated that miRNA‐204‐5p is able to regulate autophagy by directly targeting LC3B's broadly conserved 3′‐UTR (Mikhaylova et al., 2012; Xiao et al., 2011). Therefore, we studied several markers of autophagy and found that miRNA‐204‐5p silencing increased LC3B gene—but not protein—expression levels and resulted in an LC3B staining pattern suggestive for autophagosome formation. Since the autophagosomal marker LC3B also colocalized with mitochondria upon miRNA‐204‐5p silencing, this possibly reflects the autophagic clearance of mitochondria, a process called mitophagy. Arguing against this notion, gene expression of the mitophagy marker FUNDC1 was significantly reduced. Furthermore, since we did not directly assess autophagic flux, our data do not exclude the possibility that anti‐miRNA‐204‐5p treatment may inhibit—rather than activate—autophagy by blocking lysosome and autophagosome fusion, ultimately resulting in failure to form autophagosomes. In fact, we found some support for this possibility since gene expression of autophagosome and lysosome fuser VAMP8 and proteinases release protector from lysosomes CSTB appeared to be increased in anti‐miRNA‐204‐5p treated C2C12 myotubes. Thus, we found support for the involvement of miRNA‐204‐5p in autophagy although its regulatory function in this process requires further study.
In addition, the phenotype of the cells may have been affected by changes in the differentiation process, as miRNA‐204‐5p has been shown to inhibit C2C12 myoblast differentiation, most likely through inhibition of MEFC2 an ERRγ (X. Cheng et al., 2018). Interestingly, miRNA‐204‐5p expression was also decreased during C2C12 myotube differentiation (X. Cheng et al., 2018), whereas the oxidative phenotype of C2C12 myotubes has been shown to increase during myotube differentiation (Remels et al., 2010). This enhanced capacity for oxidative phosphorylation requires extensive remodeling of the mitochondrial network and it has been shown that autophagy/mitophagy is required for this remodeling, which is also integratively linked to C2C12 myotube differentiation (Sin et al., 2016). Taken together, these findings indicate that miRNA‐204‐5p may play a role in the interplay of mitochondrial biogenesis, autophagy/mitophagy, and myogenic differentiation.
Interestingly, clearing dysfunctional mitochondria through mitophagy appears important for maintaining metabolic health, as this process has been shown to be dysregulated in obese (Bollinger et al., 2015) and Type 2 diabetic subjects (Munasinghe et al., 2016), and has shown to be important for exercise‐induced adaptations (He et al., 2012; Lira et al., 2013; Moller et al., 2015; Saleem, Carter, & Hood, 2014; Vainshtein, Tryon, Pauly, & Hood, 2015). Furthermore, it has been demonstrated that PGC‐1α also plays a role in the regulation of mitophagy (Vainshtein et al., 2015). Thus, acute exercise increased the expression of mitophagy‐related genes in wild‐type, but not in PGC‐1α knock‐out mice. In addition, mitochondria were targeted for mitophagy post‐exercise in wild‐type mice, whereas this response was diminished in PGC‐1α knock‐out mice (Vainshtein et al., 2015), indicating a dual role for PGC‐1α in both mitochondrial biogenesis and mitophagy. The observed increases in PGC‐1α gene expression, in combination with the observed colocalization of mitochondria with autophagosomes, could be suggestive of increased mitochondrial turnover, although this could not be directly tested in the current study.
To address the human relevance of our findings, we also determined the expression of miRNA‐204‐5p in human skeletal muscle biopsies from individuals ranging widely in mitochondrial oxidative capacity. We observed that individuals with a high mitochondrial oxidative capacity are characterized by a low expression of miRNA‐204‐5p. It should be noted though that age may be a confounding factor in this correlation given the significant age difference between certain subject groups (Table 1). Nonetheless, exercise training has been shown to induce PGC‐1α and mitochondrial biogenesis as well as markers for autophagy and mitophagy in human skeletal muscle (Balan et al., 2019; Brandt, Gunnarsson, Bangsbo, & Pilegaard, 2018; Egan, O'Connor, Zierath, & O'Gorman, 2013; Popov, Lysenko, Makhnovskii, Kurochkina, & Vinogradova, 2017). Together with our results in C2C12 cells upon silencing of miRNA‐204‐5p, one may postulate that low levels of miRNA‐204‐5p simultaneously induce mitochondrial biogenesis as well as mitochondrial degradation and that this enhanced mitochondrial “turnover” is linked to oxidative capacity in human skeletal muscle.
In summary, we here demonstrated that the silencing of miRNA‐204‐5p in C2C12 myotubes enhanced mitochondrial biogenesis, via regulation of PGC‐1α. In addition, the anti‐miRNA‐204‐5p treatment also substantially affected the morphology of differentiated C2C12 myotubes and altered several markers related to autophagy and mitophagy. Finally, in humans, low expression of miRNA‐204‐5p in skeletal muscle was associated with high oxidative capacity. In conclusion, this study identifies miRNA‐204‐5p as an interesting modulator of mitochondrial function in human skeletal muscle.
CONFLICT OF INTERESTS
The authors declare that there are no conflict of interests.
AUTHOR CONTRIBUTIONS
A. H. designed and performed the experiments, analyzed data, and wrote the manuscript. D. D. designed and performed the experiments and analyzed data. J. A. J., E. M.‐K., G. S., E. B. M. N., S. K., and X. W. assisted during the experiments and data analysis. J. H. contributed to the design of the study, analyzed and interpreted the data, and reviewed and edited the manuscript. All authors reviewed and approved the final version of the manuscript.
ACKNOWLEDGMENTS
The work of Joris Hoeks was supported by a Vidi (Grant Number 917.14.358) for innovative research from the Netherlands Organization for Scientific Research and a Senior Fellowship from the Dutch Diabetes Research Foundation (Grant Number 2013.82.1639).
Houzelle A, Dahlmans D, Nascimento EBM, et al. MicroRNA‐204‐5p modulates mitochondrial biogenesis in C2C12 myotubes and associates with oxidative capacity in humans. J Cell Physiol. 2020;235:9851–9863. 10.1002/jcp.29797
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- el Azzouzi, H. , Leptidis, S. , Dirkx, E. , Hoeks, J. , van Bree, B. , Brand, K. , … De Windt, L. J. (2013). The hypoxia‐inducible microRNA cluster miR‐199a approximately 214 targets myocardial PPARδ and impairs mitochondrial fatty acid oxidation. Cell Metabolism, 18(3), 341–354. [DOI] [PubMed] [Google Scholar]
- Balan, E. , Schwalm, C. , Naslain, D. , Nielens, H. , Francaux, M. , & Deldicque, L. (2019). Regular endurance exercise promotes fission, mitophagy, and oxidative phosphorylation in human skeletal muscle independently of age. Frontiers in Physiology, 10, 1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel, D. P. (2004). MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell, 116(2), 281–297. [DOI] [PubMed] [Google Scholar]
- Bollinger, L. M. , Powell, J. J. , Houmard, J. A. , Witczak, C. A. , & Brault, J. J. (2015). Skeletal muscle myotubes in severe obesity exhibit altered ubiquitin‐proteasome and autophagic/lysosomal proteolytic flux. Obesity (Silver Spring, MD), 23(6), 1185–1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolstad, B. M. , Irizarry, R. A. , Astrand, M. , & Speed, T. P. (2003). A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics, 19(2), 185–193. [DOI] [PubMed] [Google Scholar]
- Brandt, N. , Gunnarsson, T. P. , Bangsbo, J. , & Pilegaard, H. (2018). Exercise and exercise training‐induced increase in autophagy markers in human skeletal muscle. Physiological Reports, 6(7), e13651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canto, C. , & Auwerx, J. (2009). PGC‐1alpha, SIRT1 and AMPK, an energy sensing network that controls energy expenditure. Current Opinion in Lipidology, 20(2), 98–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, X. , Du, J. , Shen, L. , Tan, Z. , Jiang, D. , Jiang, A. , … Zhu, L. (2018). MiR‐204‐5p regulates C2C12 myoblast differentiation by targeting MEF2C and ERRγ. Biomedicine & Pharmacotherapy, 101, 528–535. [DOI] [PubMed] [Google Scholar]
- Cheng, M. , Liu, L. , Lao, Y. , Liao, W. , Liao, M. , Luo, X. , … Xu, N. (2016). MicroRNA‐181a suppresses parkin‐mediated mitophagy and sensitizes neuroblastoma cells to mitochondrial uncoupler‐induced apoptosis. Oncotarget, 7(27), 42274–42287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chourasia, A. H. , Boland, M. L. , & Macleod, K. F. (2015). Mitophagy and cancer. Cancer & Metabolism, 3, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlmans, D. , Houzelle, A. , Andreux, P. , Jorgensen, J. A. , Wang, X. , de Windt, L. J. , … Hoeks, J. (2017). An unbiased silencing screen in muscle cells identifies miR‐320a, miR‐150, miR‐196b, and miR‐34c as regulators of skeletal muscle mitochondrial metabolism. Molecular Metabolism, 6(11), 1429–1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlmans, D. , Houzelle, A. , Andreux, P. , Wang, X. , Jorgensen, J. A. , Moullan, N. , … Hoeks, J. (2019). MicroRNA‐382 silencing induces a mitonuclear protein imbalance and activates the mitochondrial unfolded protein response in muscle cells. Journal of Cellular Physiology, 234(5), 6601–6610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlmans, D. , Houzelle, A. , Schrauwen, P. , & Hoeks, J. (2016). Mitochondrial dynamics, quality control and miRNA regulation in skeletal muscle: Implications for obesity and related metabolic disease. Clinical Science (London), 130(11), 843–852. [DOI] [PubMed] [Google Scholar]
- Dai, M. , Wang, P. , Boyd, A. D. , Kostov, G. , Athey, B. , Jones, E. G. , … Meng, F. (2005). Evolving gene/transcript definitions significantly alter the interpretation of GeneChip data. Nucleic Acids Research, 33(20), e175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Meo, S. , Iossa, S. , & Venditti, P. (2017). Skeletal muscle insulin resistance: Role of mitochondria and other ROS sources. Journal of Endocrinology, 233(1), R15–R42. [DOI] [PubMed] [Google Scholar]
- Duarte, F. V. , Palmeira, C. M. , & Rolo, A. P. (2014). The role of microRNAs in mitochondria: Small players acting wide. Genes (Basel), 5(4), 865–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan, B. , O'Connor, P. L. , Zierath, J. R. , & O'Gorman, D. J. (2013). Time course analysis reveals gene‐specific transcript and protein kinetics of adaptation to short‐term aerobic exercise training in human skeletal muscle. PLoS One, 8(9), e74098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, C. , Bassik, M. C. , Moresi, V. , Sun, K. , Wei, Y. , Zou, Z. , … Levine, B. (2012). Exercise‐induced BCL2‐regulated autophagy is required for muscle glucose homeostasis. Nature, 481(7382), 511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry, R. A. , Bolstad, B. M. , Collin, F. , Cope, L. M. , Hobbs, B. , & Speed, T. P. (2003). Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Research, 31(4), e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasahara, A. , & Scorrano, L. (2014). Mitochondria: From cell death executioners to regulators of cell differentiation. Trends in Cell Biology, 24(12), 761–770. [DOI] [PubMed] [Google Scholar]
- Kuipers, H. , Verstappen, F. T. , Keizer, H. A. , Geurten, P. , & van Kranenburg, G. (1985). Variability of aerobic performance in the laboratory and its physiologic correlates. International Journal of Sports Medicine, 6(4), 197–201. [DOI] [PubMed] [Google Scholar]
- Lin, J. , Wu, H. , Tarr, P. T. , Zhang, C. Y. , Wu, Z. , Boss, O. , … Spiegelman, B. M. (2002). Transcriptional co‐activator PGC‐1 alpha drives the formation of slow‐twitch muscle fibres. Nature, 418(6899), 797–801. [DOI] [PubMed] [Google Scholar]
- Lindeboom, L. , Nabuurs, C. I. , Hoeks, J. , Brouwers, B. , Phielix, E. , Kooi, M. E. , … Schrauwen‐Hinderling, V. B. (2014). Long‐echo time MR spectroscopy for skeletal muscle acetylcarnitine detection. Journal of Clinical Investigation, 124(11), 4915–4925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lira, V. A. , Okutsu, M. , Zhang, M. , Greene, N. P. , Laker, R. C. , Breen, D. S. , … Yan, Z. (2013). Autophagy is required for exercise training‐induced skeletal muscle adaptation and improvement of physical performance. FASEB Journal, 27(10), 4184–4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikhaylova, O. , Stratton, Y. , Hall, D. , Kellner, E. , Ehmer, B. , Drew, A. F. , … Czyzyk‐Krzeska, M. F. (2012). VHL‐regulated MiR‐204 suppresses tumor growth through inhibition of LC3B‐mediated autophagy in renal clear cell carcinoma. Cancer Cell, 21(4), 532–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohamed, J. S. , Hajira, A. , Pardo, P. S. , & Boriek, A. M. (2014). MicroRNA‐149 inhibits PARP‐2 and promotes mitochondrial biogenesis via SIRT‐1/PGC‐1alpha network in skeletal muscle. Diabetes, 63(5), 1546–1559. [DOI] [PubMed] [Google Scholar]
- Moller, A. B. , Vendelbo, M. H. , Christensen, B. , Clasen, B. F. , Bak, A. M. , Jorgensen, J. O. , … Jessen, N. (2015). Physical exercise increases autophagic signaling through ULK1 in human skeletal muscle. Journal of Applied Physiology, 118(8), 971–979. [DOI] [PubMed] [Google Scholar]
- Montero, D. , & Lundby, C. (2017). Refuting the myth of non‐response to exercise training: 'non‐responders' do respond to higher dose of training. Journal of Physiology, 595(11), 3377–3387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munasinghe, P. E. , Riu, F. , Dixit, P. , Edamatsu, M. , Saxena, P. , Hamer, N. S. , … Katare, R. (2016). Type‐2 diabetes increases autophagy in the human heart through promotion of Beclin‐1 mediated pathway. International Journal of Cardiology, 202, 13–20. [DOI] [PubMed] [Google Scholar]
- Patti, M. E. , Butte, A. J. , Crunkhorn, S. , Cusi, K. , Berria, R. , Kashyap, S. , … Mandarino, L. J. (2003). Coordinated reduction of genes of oxidative metabolism in humans with insulin resistance and diabetes: Potential role of PGC1 and NRF1. Proceedings of the National Academy of Sciences of the United States of America, 100(14), 8466–8471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phielix, E. , Meex, R. , Moonen‐Kornips, E. , Hesselink, M. K. , & Schrauwen, P. (2010). Exercise training increases mitochondrial content and ex vivo mitochondrial function similarly in patients with type 2 diabetes and in control individuals. Diabetologia, 53(8), 1714–1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phielix, E. , Meex, R. , Ouwens, D. M. , Sparks, L. , Hoeks, J. , Schaart, G. , … Schrauwen, P. (2012). High oxidative capacity due to chronic exercise training attenuates lipid‐induced insulin resistance. Diabetes, 61(10), 2472–2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickrell, A. M. , & Youle, R. J. (2015). The roles of PINK1, parkin, and mitochondrial fidelity in Parkinson's disease. Neuron, 85(2), 257–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popov, D. V. , Lysenko, E. A. , Makhnovskii, P. A. , Kurochkina, N. S. , & Vinogradova, O. L. (2017). Regulation of PPARGC1A gene expression in trained and untrained human skeletal muscle. Physiological Reports, 5(23), e13543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price, N. L. , Gomes, A. P. , Ling, A. J. , Duarte, F. V. , Martin‐Montalvo, A. , North, B. J. , … Sinclair, D. A. (2012). SIRT1 is required for AMPK activation and the beneficial effects of resveratrol on mitochondrial function. Cell Metabolism, 15(5), 675–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remels, A. H. , Langen, R. C. , Schrauwen, P. , Schaart, G. , Schols, A. M. , & Gosker, H. R. (2010). Regulation of mitochondrial biogenesis during myogenesis. Molecular and Cellular Endocrinology, 315(1–2), 113–120. [DOI] [PubMed] [Google Scholar]
- Russell, A. P. , Foletta, V. C. , Snow, R. J. , & Wadley, G. D. (2014). Skeletal muscle mitochondria: A major player in exercise, health and disease. Biochimica et Biophysica Acta/General Subjects, 1840(4), 1276–1284. [DOI] [PubMed] [Google Scholar]
- Saleem, A. , Carter, H. N. , & Hood, D. A. (2014). p53 is necessary for the adaptive changes in cellular milieu subsequent to an acute bout of endurance exercise. American Journal of Physiology: Cell Physiology, 306(3), C241–C249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartor, M. A. , Tomlinson, C. R. , Wesselkamper, S. C. , Sivaganesan, S. , Leikauf, G. D. , & Medvedovic, M. (2006). Intensity‐based hierarchical Bayes method improves testing for differentially expressed genes in microarray experiments. BMC Bioinformatics, 7, 538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sin, J. , Andres, A. M. , Taylor, D. J. , Weston, T. , Hiraumi, Y. , Stotland, A. , … Gottlieb, R. A. (2016). Mitophagy is required for mitochondrial biogenesis and myogenic differentiation of C2C12 myoblasts. Autophagy, 12(2), 369–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siri, W. E. (1993). Body composition from fluid spaces and density: Analysis of methods. 1961. Nutrition, 9(5), 480–491. [PubMed] [Google Scholar]
- Vainshtein, A. , Tryon, L. D. , Pauly, M. , & Hood, D. A. (2015). Role of PGC‐1alpha during acute exercise‐induced autophagy and mitophagy in skeletal muscle. American Journal of Physiology: Cell Physiology, 308(9), C710–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vosselman, M. J. , Hoeks, J. , Brans, B. , Pallubinsky, H. , Nascimento, E. B. , van der Lans, A. A. , … Lichtenbelt, W. D. (2015). Low brown adipose tissue activity in endurance‐trained compared with lean sedentary men. International Journal of Obesity (2005), 39(12), 1696–1702. [DOI] [PubMed] [Google Scholar]
- van de Weijer, T. , Phielix, E. , Bilet, L. , Williams, E. G. , Ropelle, E. R. , Bierwagen, A. , … Schrauwen, P. (2015). Evidence for a direct effect of the NAD+ precursor acipimox on muscle mitochondrial function in humans. Diabetes, 64(4), 1193–1201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao, J. , Zhu, X. , He, B. , Zhang, Y. , Kang, B. , Wang, Z. , & Ni, X. (2011). MiR‐204 regulates cardiomyocyte autophagy induced by ischemia‐reperfusion through LC3‐II. Journal of Biomedical Science, 18, 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


