Abstract
The eukaryotic genome is packaged into transcriptionally active euchromatin and silent heterochromatin, with most studies focused on the former encompassing the majority of protein‐coding genes. The recent development of various sequencing techniques has refined this classic dichromatic partition and has better illuminated the composition, establishment, and evolution of this genomic and epigenomic “dark matter” in the context of topologically associated domains and phase‐separated droplets. Heterochromatin includes genomic regions that can be densely stained by chemical dyes, which have been shown to be enriched for repetitive elements and epigenetic marks, including H3K9me2/3 and H3K27me3. Heterochromatin is usually replicated late, concentrated at the nuclear periphery or around nucleoli, and usually lacks highly expressed genes; and now it is considered to be as neither genetically inert nor developmentally static. Heterochromatin guards genome integrity against transposon activities and exerts important regulatory functions by targeting beyond its contained genes. Both its nucleotide sequences and regulatory proteins exhibit rapid coevolution between species. In addition, there are dynamic transitions between euchromatin and heterochromatin during developmental and evolutionary processes. We summarize here the ever‐changing characteristics of heterochromatin and propose models and principles for the evolutionary transitions of heterochromatin that have been mainly learned from studies of Drosophila and yeast. Finally, we highlight the role of sex chromosomes in studying heterochromatin evolution.
Keywords: heterochromatin, histone modifications, chromatin conformation, sex chromosomes
In this review, we summarize the ever‐changing characteristics of heterochromatin and propose models and principles for the evolutionary transitions of heterochromatin that have been mainly learned from studies of Drosophila and yeast. We also highlight the role of sex chromosomes in studying heterochromatin evolution.

Heterochromatin: an evolving concept
The understanding of heterochromatin desmonstrates how the connotation of a biological paradigm can evolve with the development of research technology (Fig. 1). The term “heterochromatin” was first used by Emil Heitz in 1928,1 probably with reference to heterochromosomes (i.e., sex chromosomes),2, 3 to describe the chromosomal fragments that remain densely stained throughout the cell cycle, in contrast to the other fragments of euchromatin that become invisible after telophase. Heitz later extended the staining method that he developed in liverworts to over 100 plant species and several Drosophila species, including D. melanogaster. He established that heterochromatin/euchromatin comprise the fundamental architecture of eukaryotic chromosomes and hypothesized that euchromatin is genetically active and heterochromatin genetically passive.
Figure 1.
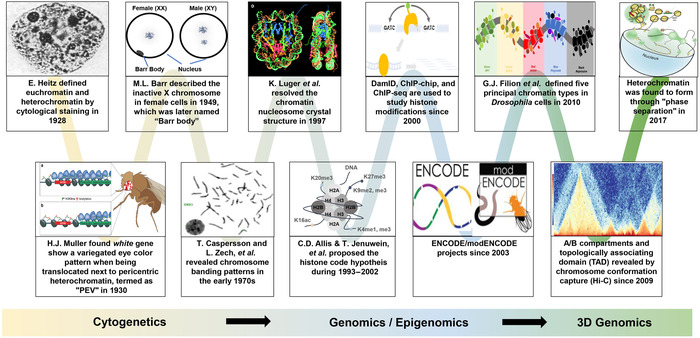
The concept of heterochromatin has been evolving since its first description in 1928 by Emil Heitz. Shown is an incomplete list of events that led to changes in the concept of heterochromatin during research developments from the cytogenetic era until very recently in the 3D genomics era. The characterization of proteins (e.g., HP1) regulating constitutive heterochromatin was largely attributed to the genetic screens in Drosophila for mutants affecting the position‐effect variegation (PEV) phenotype. The studies into facultative heterochromatin were first marked by the discovery of the Barr body, that is, the female‐specific inactivated X chromosome. After the discovery of nucleosome structure as the chromatin unit for gene regulation, many PEV‐related genes were found to be the reader or writer of histone modifications within the chromatin unit. With the development of chromatin immunoprecipitation (ChIP) technology targeting these histone modifications, combined with either microarray (ChIP‐chip) or sequencing analyses (ChIP‐seq), it became clear that heterochromatin is usually associated with highly repetitive regions and the histone methylation marks H3K9me2/3 and H3K27me3. This facilitated the definition of the heterochromatin region at the base pair resolution in the genomic and epigenomic era. Recently, it has been shown in Drosophila and humans that heterochromatin forms within phase‐separated droplets of HP1 protein and forms distinct topologically associated domains from those of euchromatin, as detected by Hi‐C technology.
Heterochromatic chromosomes or pieces of chromosomes contain no genes or somehow passive genes.1 It was later reported that heterochromatin has a heterogeneous distribution within and between chromosomes in Drosophila cells.4, 5, 6 In fact, Heitz noticed that heterochromatin is often associated with sex chromosomes, and some chromosomal regions are only stained in certain cell types. These were later recognized as facultative heterochromatin (fHet),7 compared with constitutive heterochromatin (cHet). From the work of Thomas Morgan, Heitz soon realized that the hypothesis of heterochromatin as being genetically inert was not entirely valid because, as Heitz noted, “genes which lie within heterochromatin do intervene in developmental process.”8, 9
More understanding of the composition and regulation of heterochromatin was initiated after Muller's seminal finding of the variegated eye color pattern of X‐ray–irradiated D. melanogaster in 1930.8 This phenotype, position‐effect variegation (PEV), was shown by subsequent examinations of polytene chromosomes to be associated with chromosomal rearrangements that positioned the white gene from euchromatin near the pericentric cHet, indicating the spreading effect of silencing heterochromatin. Interestingly, PEV has also been induced for some genes (e.g., light + and its nearby genes,10, 11 see below) that relocated from heterochromatin to distal euchromatin, suggesting that a proper dosage of heterochromatin‐enriched protein is required for the normal expression of these genes. PEV has become a very powerful tool for identifying the critical regulators of cHet (reviewed by Ref. 12) through genetic screens for secondary mutations modifying PEV phenotypes.
The discovery of cHet‐associated proteins has also been facilitated by localizing the candidate genes through immunostaining of Drosophila polytene chromosomes, where cHet regions are under‐replicated and concentrated at the chromocenter. Several identified proteins turned out to be histone lysine methyltransferases with evolutionarily conserved domains (e.g., chromodomain and SET domain) and to have an ortholog in yeast and humans that functions similarly in posttranslational modifications (PTMs) of histones.13, 14 While cHet is concentrated at telomeric and centromeric regions of all chromosomes across different cell types, in 1949 Barr and Bertram reported a female‐specific heterochromatic X chromosome in cat neuronal cells, later termed the Barr body.15 This special form of chromosome‐wide fHet was hypothesized to reflect the random inactivation of the X chromosome (XCI) to achieve dosage compensation.16 XCI is initiated by activity of the X‐linked long noncoding RNA Xist in eutherian mammals and is precisely controlled to be completed in the blastocyst stage, which gives rise to all somatic cells.17
Another classic paradigm of temporally regulated fHet was identified in the homeotic (Hox) genes. The repressed or activated expression state of Hox genes during embryonic development is maintained by Polycomb and Trithorax group proteins (PcG and TrxG).18 The first PcG gene, Polycomb (Pc), was discovered in the 1940s,19 and its mutation was later characterized to be responsible for transforming the anterior segments of Drosophila embryos to more posterior segments.18 The first identified TrxG gene, Trithorax (Trx), was identified as a regulator of Hox genes and to counteract PcG protein expression to produce more anterior embryonic segments.20, 21 Genetic screens producing a similar phenotype to that of Pc or Trx have identified many more genes that were later defined as PcG or TrxG genes. In particular, mutations in PcG genes result in the suppression of PEV associated with cHet, suggesting a distinct mechanism underlying the gene silencing effects of fHet and cHet.12, 18 A shared feature between the cHet‐ and fHet‐related genes is that many of them—for example, the cHet‐related Su(var)3‐9, the PcG gene E(z), and the TrxG genes dSet1 and Trx—contain the SET protein domain, suggesting that histone lysine methylation can either repress or activate gene expression. By the discovery of many more forms of histone modifications, along with the landmark visualization of nucleosome structure22, 23 as a chromatin unit for gene regulation, a histone code hypothesis was proposed.24 The hypothesis states that various PTMs of histone tails on specific residues, for example, acetylation or methylation, “extend” the genetic code by altering a heritable chromatin state and hence gene expression level through recruiting downstream effector proteins (so‐called “readers” and “writers” of PTMs, reviewed in Ref. 25) recognizing the histone modifications. It has become clear that cHet is usually associated with HP1 protein and di‐ and trimethylation of either histone H3 lysine 9 (H3K9me2/3) or histone H4 lysine 20 (H4K20me2/3, particularly at pericentric cHet). fHet is associated with PcG/TrxG proteins and trimethylation of lysine 27 (H3K27me3) alone as a repressed chromatin state but can form bivalent chromatin, together with H3K4me3, and switch to an activated chromatin state (i.e., euchromatin) in a spatiotemporal manner. For example, the Barr body is now known to be the result of H3K27me3 modification of the inactivated X chromosome in females by PcG genes directed by the Xist RNA. The inactivated X chromosome occupies a distinct region from its active homologous chromosome in the female nuclei, where the Barr body specifically overlaps with the silenced noncoding genomic regions.26 There are also established links between cHet and fHet, where one of the PcG proteins, EED, can bind H3K9me3,27 and loss of the H3K9 methyltransferase reduces the binding of PcG proteins to chromatin.28
The genome‐wide profiling of chromatin, a much higher resolution than chemical staining, became available after the development of microarray and Illumina sequencing, chromatin immunoprecipitation (ChIP‐chip or ChIP‐seq), and DNA adenine methyltransferase identification (DamID) experiments with antibodies targeting chromosomal proteins or various histone modifications. Such efforts have culminated in multiple international consortia that have collected massive ChIP‐seq datasets of various histone modifications from human tissues, for example, the Encyclopedia of DNA Elements (ENCODE) project29 and the Roadmap Epigenomics project,30 or from model organisms, such as Caenorhabditis elegans and D. melanogaster, that is, the modENCODE project.31, 32 A major aim of these consortia is to annotate the functions of noncoding genomes in the postgenomic era.
The modENCODE consortium first partitioned the Drosophila genome into nine chromatin states by combinatorial genome‐wide patterns of 18 histone modifications32 and, later, into 16 states on the basis of eight histone modifications.31 Before that, Filion et al. used the DamID technique and partitioned the genome into five color‐coded chromatin states, based on the binding maps of 53 chromatin proteins.33 The different chromatin state numbers between these works can be attributed to the differences in techniques (ChIP and DamID), antibodies (histone modifications versus chromatin proteins), and cell lines (S2 versus Kc167); the main differences are in the numbers of euchromatin states. Both identified genomic regions enriched for cHet‐associated epigenetic markers H3K9me2/3 and proteins HP1 and SU(VAR)3‐9 (termed “green” chromatin in Filion et al.) and fHet markers H3K27me3 and PcG proteins (“blue” chromatin). In addition, Filion et al. characterized a third type of heterochromatin, termed “black” chromatin (bHet), which covers nearly half of the Drosophila genome and corresponds to the modENCODE states without any enrichment of measured histone modifications. bHet is enriched in the binding of SuUR protein, whose mutations influence the binding of H3K9me3 and H3K27me3.34, 35 It shares many features with canonical cHet despite a lack of H3K9me2/3 enrichment, that is, it is late replicating, mainly contains low‐expressed or tissue‐specific genes, and transgenes inserted into bHet are much more likely to be silenced compared with those in the genomic background.33 Another bHet‐associated protein is lamin, a major component of nuclear lamina, where canonical cHet is also concentrated. Since large genomic regions lacking histone modifications have also been found in plants36 and mammals,37 it remains to be elucidated whether bHet‐associated proteins identified in Drosophila are also enriched in corresponding chromatin regions of other species. In summary, bHet together with cHet and fHet comprise the three major types of heterochromatin to our current knowledge.
Functions of heterochromatin
While fHet regions encompass genes and enhancers and can dynamically switch between the active and repressive states during development to regulate gene expression, cHet contains predominantly various types of repetitive sequences (satellite DNAs, ribosomal DNAs, and transposable elements (TEs)) and few functional genes that are usually silenced among most cell types. Such a genomic composition and the chromosomal distribution of cHet concentrated at centromeric and telomeric regions led to the assumption that the primary function of cHet is related to genome stability. Indeed, the disruption of cHet‐related genes, such as HP1 of Drosophila or Swi6 of fission yeast, produces mutant phenotypes of telomere fusions,38 aberrant subtelomeric recombination or dysregulation of telomere lengths.39, 40 Such mutations also result in the loss of pericentromeric cHet, which together with CENP‐A–containing (a histone H3 variant conserved across eukaryotes) centromeric chromatin comprise the functional centromeres (reviewed in Refs. 41 and 42). The affected cells of fission yeast, Drosophila, and mice tend to show chromosome segregation errors and a higher chance of chromosome loss.43, 44
In addition to acting as chromosome structural components, cHet also guards the genome integrity by suppressing the activities, including both recombination and transcription, of repetitive sequences.45 In species other than yeast, cHet is also distributed on chromosome arms in between the euchromatic regions (called intercalary heterochromatin in Drosophila). Repetitive sequences at different chromosomal regions are isolated from each other by cHet domains to prevent their ectopic recombination to avoid chromosomal rearrangements, such as large insertions, deletions, and translocations.46, 47 Another threat to genome integrity comes from DNA transposons and retrotransposons that can mobilize within the genome and disrupt gene functions by cut‐ or copy‐and‐paste mechanisms if not properly regulated (reviewed in Refs. 48 and 49). The suppression of these TEs is realized by various types of small RNAs and Argonaute family proteins that either cleave the TE transcripts (posttranscriptional gene silencing) or mediate cHet formation at the TE loci to directly suppress their transcription (transcriptional silencing). The latter has been extensively studied in model organisms because it directly informs the mechanisms of cHet establishment. In fission yeast, small interfering RNAs (siRNAs) transcribed by RNA polymerase II from centromeric repeats (outer repeats) flanking the CENP‐A/Cnp1–containing centromeric core regions form an RNAi machinery (RITS, including Argonaute protein AGO1) that recruits histone methyltransferase CLR4 to initiate H3K9m2/3, followed by the HP1 homolog Swi6 to establish and maintain cHet.41 The recruitment of HP1 to the H3K9me2/3 marks is realized by the characteristic chromodomain at the N‐terminus of the HP1 protein,50 while its chromoshadow domain (CSD)51 at the C‐terminus is responsible for the dimerization of HP1 proteins and further recruiting histone methyltransferase (i.e., spreading of heterochromatin). This classic paradigm of cHet assembly has been found in both plant52, 53 and animal53, 54 species with different types of small RNAs and proteins involved, associated with triggering DNA methylation or H3K9me2/3 modification.
The biogenesis pathway of the small RNAs related to cHet formation has been most extensively studied in Drosophila, where a large portion of small RNAs interacting with Argonaute protein PIWI (PIWI‐interacting RNAs, piRNAs) are transcribed from a few genomic clusters (piRNA clusters) that are predominantly composed of transposon relics.48 In D. melanogaster, transgenics carrying the 1360 DNA transposons or invader 4 retroposons demonstrate de novo heterochromatin formation at mobile element euchromatic insertion sites,55 while deletions of their encompassed piRNA sequences compromise ectopic assembly of heterochromatin. This supports a model in which piRNAs recognize the complementary sequences of nascent TE transcripts that initiate cHet formation at the TE loci (cotranscriptional silencing). More mechanistic details of this model have been recently uncovered; it is now known in D. melanogaster, for example, that an HP1 paralog gene Rhino 56, 57, 58 licenses the piRNA clusters to produce dual‐strand primary piRNA transcripts by RNA polymerase II in a cHet environment, facilitated by the gene Moonshiner,59 a paralog of transcription factor TFIIA. The PIWI downstream effector gene Panoramix then recruits histone demethylase LSD1 and methyltransferase EGG to remove the active chromatin mark H3K4me2 and establish H3K9me3 on the transposons targeted by the piRNAs.60, 61
The case of transcribing piRNA clusters indicates that cHet is not completely silenced. Hundreds of protein‐coding genes, in addition to the rRNA and small RNA loci, are also embedded in the cHet of diverse organisms,62 and active expression of many protein‐coding genes is in fact dependent on cHet acting in either a cis or a trans manner (reviewed by Ref. 63). In the 1930s, some essential genes (e.g., light +) in Drosophila originally located in pericentromeric cHet were found to show PEV after being displaced to a novel euchromatin and heterochromatin (eu‐het) boundary by chromosome rearrangements.10, 64 The gene variegated expression patterns suggested that certain proteins enriched in cHet are required for their normal expression. Insights into the actual mechanisms of how cHet paradoxically (i.e., the heterochromatin paradox62, 65) regulates expression of cHet‐encompassed genes were later gained from studies of the fourth chromosome pair of D. melanogaster.
This pair of unusual autosomes originated from a pair of ancestral heterochromatic sex chromosomes in Diptera species66 and still shares many features with canonical sex chromosomes after becoming autosomes (reviewed by Refs. 67 and 68). These chromosomes show many heterochromatic properties that were discovered as early as in the 1940s;69 they are small size and have a relatively low gene number (∼80 genes and are thus also called dot chromosomes), a very low recombination rate, and are enriched in repetitive elements compared with other autosomes. Transcribed genes are simultaneously coated by the cHet mark H3K9me3 and the active gene mark H3K36me3, and by the protein POF (painting of fourth) that specifically binds to the dot chromosome. POF's specific binding seems to derive from its ancestral function of specifically upregulating the hemizygous X chromosome in males of other Diptera species, similar to the MSL proteins of Drosophila species,70 the major protein responsible for dosage compensation. Depletion of either heterochromatin protein HP1a (an isoform of HP1 in Drosophila) or POF leads to a decrease in gene expression, and depletion of histone methyltransferase EGG leads to decreased binding of POF, HP1a, and H3K9me2/3 on the dot chromosomes except for the pericentromeric regions. These characteristics suggest that HP1 positively regulates the active expression of some genes, as opposed to its canonical role in gene silencing.
Such a function is not restricted to the dot chromosome genes already embedded in the cHet, but is also related to many euchromatin genes on the other autosomes of Drosophila. This was revealed by experiments disrupting or downregulating HP1 in Drosophila larvae or the Kc cell line in which many euchromatin genes, particularly cell‐cycle regulatory genes, are transcriptionally affected.71, 72 Although the actual mechanism of how HP1 positively regulates gene expression remains elusive, it has been reported that HP1 may facilitate transcript elongation by interacting with the RNA processing proteins hnRNPs.73 Specifically, for the dot chromosome, HP1 is recruited to be concentrated at the body of active genes marked by H3K9me3, but not H3K9me2, in a POF/EGG‐dependent manner, where POF probably binds to the nascent transcripts to upregulate gene expression.70, 74, 75 This contrasts with the repeat‐enriched cHet regions of the dot chromosome or of any other chromosomes where the recruitment of HP1 is independent of POF, and the H3K9me2 and H3K9me3 marks show a high correlation with each other.
These studies suggested that HP1 can participate in the active transcription of some genes and that different cHet regions can be assembled by distinct mechanisms. In parallel, a recent PEV study also suggested that the telomeric regions of the Drosophila Y chromosome probably have a distinct type of cHet.76
Finally, the trans‐acting regulatory function of cHet was originally indicated by the suppression of PEV found in flies carrying one extra Y chromosome (XYY) and the enhancement of PEV found in male flies without the Y chromosome in the 1980s.77, 78 Such a pattern is more likely to be caused by the different dosages of cHet rather than the very few functional genes harbored by the Y chromosome. Given a fixed supply of proteins (e.g., satellite‐binding factor D1 or HP1a) that are tightly regulated for packaging the cHet of the entire genome, it has been proposed that changes in the cHet content on one chromosome may cause redistribution of cHet and, hence, its enclosed or surrounding gene expression levels on other chromosomes (called the heterochromatin sink hypothesis).79, 80 Experimental support later came from studies of D. melanogaster strains that differ only in their origin of Y chromosomes.81, 82, 83 These studies found that hundreds of X‐linked and autosomal genes, particularly male‐biased expressed genes or genes already residing in repressive chromatin regions (e.g., bHet) of other chromosomes,84 are affected by their gene expression levels.81 Although the underlying mechanisms of such Y‐linked regulatory variations (YRV) remain unclear, it has been speculated that variations in repetitive sequences of the Y chromosome would either compete with other cHet regions for binding of the limited amount of heterochromatin‐targeted proteins under the heterochromatin sink hypothesis or produce various amounts of small RNAs that can affect the chromatin configuration elsewhere in the genome.85 Consistent with this, recent characterization of D. melanogaster strains with varying numbers of Y chromosomes has found genome‐wide changes in H3K9me2/3 binding, but not for the active histone mark H3K4me3,86 while Y‐linked noncoding RNAs have a regulatory role in autosomal gene expression in mice.87
Heterochromatin in space and time
The segregation and mutual exclusion between heterochromatin and euchromatin is one of the major mechanisms that drives the genome to spatially fold into separate regulatory domains. Such a self‐assembly model88 is supported in Drosophila 89, 90, 91 and plants,92, 93 where there is a strong correspondence between the chromatin state and the three‐dimensional (3D) folding of the genome. The latter is measured by the recently developed chromosome conformation capture methods, particularly the high‐throughput version called Hi‐C. Hi‐C captures genomic regions that are proximal to, and thus potentially interacting with each other, in 3D space in interphase nuclei and allows quantification of the frequency of such interactions by the numbers of normalized read pairs that span the regions in contact. At a megabase‐level scale, a given chromosome can be divided into two types of compartments that have preferential long‐range interactions within the regions of the same type, with the A compartment largely corresponding to euchromatin and the B compartment largely to heterochromatin.94, 95 Consistent with their chromatin types, B compartment regions have a higher frequency of interactions, that is, a more condensed chromatin configuration, than that of A compartment regions. At a finer scale, depending on the experimental and bioinformatic protocols and sequencing coverage, Hi‐C data can define topologically associated domains (TADs) spanning tens to hundreds of kilobase‐long genomic sequences or subTADs (several kb) within a TAD.96, 97, 98 In mammals, TAD boundaries are enriched for cohesion complex and insulator proteins, such as CTCF.98, 99 Removal of the CTCF/cohesion protein or CTCF binding sites leads to the disruption or shift in TAD boundaries.100, 101, 102, 103 This indicates that in addition to self‐assembly, a loop‐extrusion mechanism (i.e., chromatin being extruded by the cohesion complex until it encounters the TAD boundary elements) also plays a crucial role in TAD formation.104
Different parts of the folded genome occupy different territories within the nuclei; early electron microscopy studies found that the condensed heterochromatin is mostly concentrated near the nuclear periphery or around nucleoli.55, 105 Genome‐wide mapping using DamID or ChIP and targeting proteins of the nuclear lamina distributed throughout the nuclear inner membrane revealed lamina‐associated domains (LADs) that can comprise over 40% of the mammalian genome (reviewed in Ref. 106). Purification of nucleoli allowed the identification of nucleolus‐associated domains (NADs).107, 108 While TADs reflect the interactions within the genome, LADs and NADs reflect the architectural genome organization within the nuclei. Consistent with the microscopic observations, compartment B or large TADs of heterochromatin largely overlap with LADs and NADs.96, 109 Such a conventional nuclear architecture is inverted in the rod nuclei (inverted nuclei) of nocturnal mammals or mouse cells with disrupted lamina proteins (lamina B receptor), where heterochromatin is dissociated from the lamina and localized to the nuclear interior. Interestingly, in such inverted nuclei, genomic compartments and TADs are still preserved compared with the other cell types with conventional nuclear architecture of lamina–cHet associations. This suggests that the compartmentalization of the genome is not strictly dependent on the lamina. By further polymer simulations of chromatin models that fit the microscopic observation of heterochromatin distribution in the nuclei, a recent study further suggested that interactions between heterochromatic regions, but not euchromatic regions, are crucial for establishing the genomic compartments.110
This indicated that the nuclear position of cHet can vary between different cell types. Additionally, LAD‐ or NAD‐associated cHet can switch positions after mitosis.111 These dynamic features of cHet and the spatial formation of its membraneless domain cannot be explained solely by local compaction of chromatin facilitated by associated proteins. An alternative model of phase separation has been proposed from the characterization of HP1 in both Drosophila and humans.112, 113 Studies found that HP1 proteins form liquid droplets in vitro in a reversible manner dependent on either temperature or the phosphorylation state of the protein. Although it remains to be determined whether such phase‐separated HP1 droplets correspond to the actual genomic compartment B or to heterochromatic TADs identified by Hi‐C, they clearly share important features of heterochromatin domains, that is, nucleosomes and DNA, but not transcriptional factors preferentially partition into such droplets. Under the phase‐separation model, heterochromatin regulates gene expression by precluding transcription factors from its phase boundary, while still maintaining certain flexibilities of forming larger domains by dynamically fusing with other droplets or dissolving itself if HP1 is released from the chromatin.
Such dynamic changes in cHet domains can be observed during the establishment of cHet in early Drosophila embryos. Overall, epigenetic information—including histone modifications, DNA methylation, small RNAs, and 3D chromatin conformation—undergoes dramatic reprogramming to reset the embryo to acquire totipotency after fertilization (Fig. 2). Drosophila embryos initially remain in a naive state without zygotic transcription and chromatin architecture114, 115 and rely on maternally deposited transcripts and histone modifications that are required to activate the zygotic transcription at the later stage of mitotic cycle 14. Histone acetylation, but almost no methylation (including the canonical H3K27me3 and H3K9me2/3 marks), can be detected before zygotic activation, as early as mitotic cycle 8.116 The deposition of some acetylation marks (e.g., H3K18ac, H3K27ac, and H4K8ac) at TAD boundaries in mitotic cycle 12 suggests that they are associated with early establishment of chromatin conformation.115, 117 Demethylation of another active chromatin marker H3K4me2 is required for the proper establishment of H3K9me2 marks.118 From mitotic cycle 11 on, HP1 foci have been observed to grow, fuse, and dissolve according to the progression of cell cycles; the percentage of immobile HP1 without liquid properties gradually increases before the onset of zygotic expression. Such a maturation process of heterochromatic domains may correspond to either the inclusion of more DNA/nucleosomes or the formation of contacts with the lamina during early embryogenesis.112 Counterintuitively, the aggregation of HP1 droplets is probably realized by concentration of HP1 increasing rather than by decreasing of accessibility of buried histone residues within the octamer core through its CSD. The consequential change in the conformation of nucleosomes may further promote the interactions between nucleosomes to form phase‐separated liquid condensates.119 However, it remains largely unclear how heterochromatin is initially established in early embryos.
Figure 2.
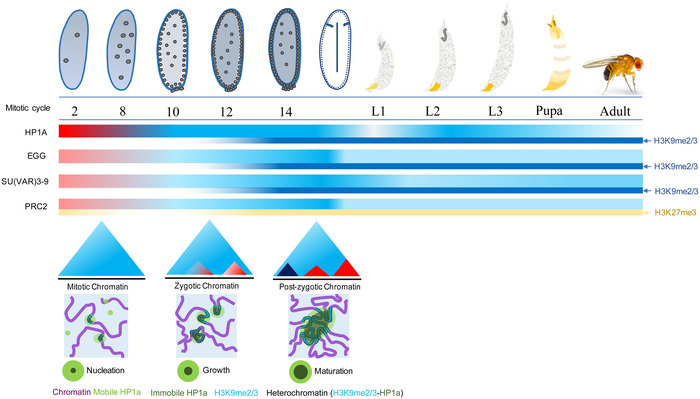
Establishment of heterochromatin during Drosophila embryogenesis. After fertilization, Drosophila embryos undergo 13 rapid cleavage divisions (cycles 2–13) with little zygotic transcription, as transcripts and proteins (e.g., EGG and PcG proteins) and histone modifications (e.g., H3K27me3) are mainly maternally inherited. During these early embryonic stages, both the state and topology of chromatin remain in a naive state, and there are more mobile HP1 proteins than immobile ones observed in the embryos. The establishment of the heterochromatin marker H3K9me2/3 has been recently shown to involve the histone methyltransferase EGG. At the onset of zygotic transcription at mitotic cycle 14, H3K9me2/3 can be detected by immunostaining, and there are significantly more structured TADs than in previous stages. Once the constitutive heterochromatin and chromatin topology are established, they remain largely stable across later developmental stages. The dynamics of mobile and immobile HP1a are adopted from Strom et al.110
Among the three H3K9 methyltransferases in the Drosophila genome, only EGG has recently been found to be required for the de novo establishment of cHet, while the other two (G9A and SU(VAR)3‐9) are probably required for the maintenance of cHet.120 PIWI also seems to play an important role, as maternal depletion of PIWI leads to the suppression of PEV in both somatic and gonadal tissues of later developmental stages, consistent with the piRNA‐guided model of heterochromatin establishment in early embryos. A small subset of piRNAs might participate in this process; live imaging shows that at the onset of zygotic transcription, some (e.g., 359 satellite sequences), but not all, repetitive sequences recruit HP1 to form cHet.121, 122 However, depletion of PIWI after the onset of zygotic expression has a much smaller impact on PEV,122 suggesting that the establishment and maintenance of cHet involve distinct pathways. Once heterochromatin is established, it seems to be generally conserved across different cell types with respect to its boundary with euchromatin.123
Evolution of heterochromatin and its associated proteins
Genomic sequences embedded in cHet, mainly TEs and satellite sequences, usually show rapid turnover among and within species. This can be partially explained by the mutagenic effect of DNA methylation in heterochromatic regions, where methylated CpG dinucleotides have a higher rate of forming TpG by oxidative deamination.124 Repetitive sequences can also readily have replication slippage and unequal crossovers, resulting in the expansion or contraction of copy numbers. In addition, a compact chromatin structure may restrict the accessibility of enclosed DNAs to repair complexes. Consistent with these explanations, a multivariate study of a cancer genome characterizing the association between the mutation rate and 46 genomic features, including various histone modifications, base compositions, and the recombination rate, found that the H3K9me3 binding level showed a positive correlation and alone could account for over 40% of the mutation rate variation.125 In addition to these mutational effects, purifying natural selection is expected to be low on the mostly silenced junk DNA in heterochromatin and to evolve mainly by genetic drift; however, emerging evidence supports a regulatory role of transposons and satellite DNAs in many cases.126, 127 Evidence for the great interspecific variation of heterochromatic sequences comes from extensive studies in closely related species of plants,128 Drosophila,129, 130 birds,131 and mammals.132 A recent characterization of six tunicate genomes revealed that TE divergence can contribute as much as a 12‐fold difference in genome size between related species.133
At the chromosome‐wide level, the mammalian Y chromosomes and Drosophila dot chromosomes afford good examples of the effects of turnover of heterochromatic sequences. While the dot chromosome is approximately 5 Mb long in D. melanogaster, it has expanded to over 18 Mb in Drosophila ananassae because of the massive accumulation of retroposons.134 By contrast, the dot chromosome of Drosophila willistoni has fused to an autosome and evolved a comparable level of recombination rate and codon usage as that of euchromatic autosomes, suggesting a possible transition from a heterochromatic ancestor to euchromatin.135 A parallel case is the fusion between the ancestral Y chromosome shared by all Drosophila species and the dot chromosome in Drosophila pseudoobscura,133, 136 with reduction in the size of intron and intergenic regions and some evidence of selective sweep on the Y chromosome after its transition to an autosome.137, 138 The ancestrally heterochromatic configuration changed on the Y chromosome after the fusion?
Mammalian Y chromosomes are more characterized than those of other species, with sequences of at least eight species having become available.139 The human Y chromosome contains 23 Mb of euchromatin and approximately 40 Mb of heterochromatin; the euchromatin contains a region that recently transposed from the X chromosome (X‐transposed) after the divergence of human and chimpanzee, a region with 20 single‐copy genes that are shared with the X chromosome (their common autosomal origin (X‐degenerate)) and ampliconic regions that contain massive palindrome sequences. The Y chromosome heterochromatin is shared between humans and gorillas on the Y long‐arms but absent on the Y chromosome of chimpanzees.139, 140, 141, 142 In contrast to the expectation that Y chromosomes are highly heterochromatic and gene poor owing to relatively low recombination, over 95% of the mouse Y chromosome contains euchromatic and ampliconic regions that contain 100–300 copies of three gene families with nearly identical sequences within each family and predominant expression in the male germline.143 Such interspecific heterochromatic changes occurred over 5 million years ago, thus affording few trackable clues indicating the actual mechanisms or functional consequences of heterochromatin turnovers.
There have generally been many more reported cases of euchromatin changing to heterochromatin (e.g., Ref. 144) than the opposite. Genetic manipulations in model organisms, such as Drosophila and yeast that cause euchromatin‐to‐heterochromatin transition, or comparative studies within populations or between closely related species, have provided important insights into the molecular mechanisms of heterochromatin evolution. Based on the previous work, we summarize three models of transition from euchromatin to heterochromatin (Fig. 3).
Figure 3.
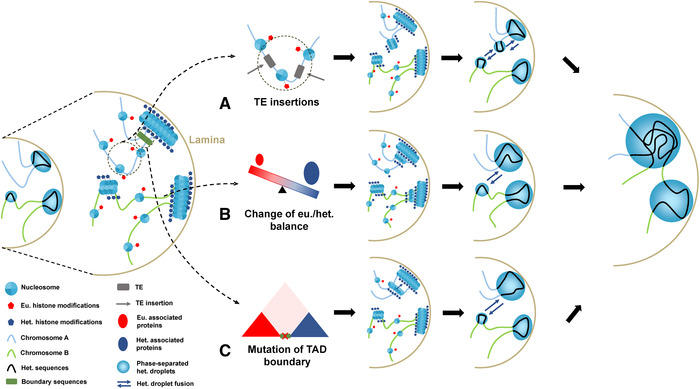
Transitions from euchromatin to heterochromatin. Three proposed models of euchromatin to heterochromatin transition. Heterochromatin is usually distributed close to the nuclear periphery and tethered to the lamina or around the nucleoli, while euchromatin is located in the nuclear interior. (A) The de novo formation of heterochromatin domains induced by TE insertions. A TE insertion into the euchromatic region may trigger heterochromatin formation mediated by small RNA pathways. This may further impact the expression of nearby genes by the spreading effect of newly formed heterochromatin domains. (B) Change in euchromatin/heterochromatin balance. The expansion of heterochromatin or ectopic formation of heterochromatin can be caused by upregulation of heterochromatin‐associated proteins (e.g., histone methyltransferase SU(VAR)3‐9, shown as dark blue circles) or downregulation of euchromatin‐associated proteins (red circles). This has been demonstrated in Drosophila and yeast. Such chromatin boundaries form without the participation of boundary elements, such as CTCF proteins, and are thus called negotiable borders. (C) Mutations of TAD boundary sequences (green bars between the two TADs) between euchromatin and heterochromatin domains. The TAD boundary sequences are usually CTCF binding sites or transcriptionally active genes or TEs. Removal or inversion of such boundary sequences may lead to the expansion of heterochromatin domains into euchromatin. The newly formed heterochromatin domains through the TE insertions, A, or expanded heterochromatin domains through B or C will convergently interact with other preexisting heterochromatin domains at the lamina through fusions of phase‐separated droplets. Such interactions may impact the nearby genes or genes on a different chromosome by reshaping the genome‐wide folding. Het, heterochromatin; Eu, euchromatin; TE, transposable element.
The first model (Fig. A) involves de novo establishment of the heterochromatin domain in a euchromatin background. This is demonstrated by the transgenic study of 1360 DNA transposons in Drosophila, which were targeted for heterochromatin formation after being inserted into euchromatin.55 A similar pattern has been observed for mouse embryonic stem cell lines carrying polymorphic retroposon insertions, where the H3K9me3 and H4K20me3 marks have been observed in some, but not all, types of retroposon insertions.145 In D. melanogaster populations, polymorphic TE insertions have been recently shown to cause epigenetic state changes in nearby genes due to the spreading effect of heterochromatin.146 Such a deleterious effect on gene expression seems to be strongly selected against by natural selection; thus, heterochromatin‐induced TEs (mainly long terminal repeat retroposons) are more likely to segregate at a low frequency in the population. Genomic regions with low levels of or no homologous recombination—for example, polymorphic inversions or sex chromosome regions—and insertion hotspots of TEs in euchromatin are vulnerable to transposon invasions and thus can readily form heterochromatin under this model.145, 147, 148
The second model (Fig. B) does not involve cis element–inducing heterochromatin, but rather changes in the balance between neighboring euchromatin and heterochromatin domains that shift the boundary between them. Such fluid borders between the two chromatin states form without the participation of boundary proteins, such as CTCF, and can change according to the dosage of heterochromatin‐ or euchromatin‐associated proteins92, 149 that act in trans. For example, in Drosophila, overexpression of cHet‐associated protein SU(VAR)3‐9 drives the expansion of cHet domains.149 Similarly, disruption of the euchromatin mark H4K16ac‐associated gene Sas2 or overexpression of the gene Sir3, which participates in the deacetylation in budding yeast,150, 151 leads to heterochromatin spreading. It has been demonstrated that the copy numbers and expression patterns of cHet‐associated genes dramatically vary between species,152, 153 probably in response to the rapid evolution of repetitive DNA sequences (see below). This may result in the shift in boundaries between euchromatin and heterochromatin, reshaping the chromatin architecture of the entire genome. Interestingly, different cHet‐ and euchromatin‐associated proteins may form a feedback loop to prevent promiscuous heterochromatin assembly. In budding yeast, disruption of histone demethylase EPE1 or histone acetyltransferase MST2 leads to heterochromatin spreading. EPE1/MST2 double mutant yeast, although initially deleteriously affected, quickly recovers through epigenetic mutations, which reduce the expression of another cHet‐associated gene, Clr4, (a homolog of the Drosophila Su(var)3‐9) to mitigate the negative effects of ectopic heterochromatin formation.154
The third model (Fig. C) involves the disruption of the original boundaries between euchromatin and heterochromatin through either mutations involving the TAD boundary sequences, for example, CTCF‐binding sites or transcriptionally active genes,96 or large genomic rearrangements, such as inversions (e.g., PEV), deletions, and duplications. Such changes may occur more frequently at a sub‐TAD level rather than involving two neighboring TADs. The removal or shift in TAD boundaries not only affects the local chromatin states near the boundary, but also erroneously creates contacts between enhancers and promoters of different TADs, leading to the misexpression of numerous genes. Consistent with this, comparison of chromatin architectures across several mammals has found that TAD boundaries are highly conserved in syntenic regions. Genomic rearrangements, if any, preferentially occur at the boundaries or within open chromatin regions, rather than disrupting the individual TAD structure as a regulatory module.96, 155 This model requires experimental test because the boundaries of TADs do not necessarily overlap with those between different types of chromatin. It is possible that in some cases, the euchromatin/heterochromatin boundary is regulated in trans and thus resilient to the change in TAD boundaries in cis.
In a broader sense, the impact of the evolutionary transition of euchromatin to heterochromatin is probably not restricted to their enclosed regions because of the fusion propensity of the phase‐separated heterochromatin droplets.112, 113, 119 The tethering of the newly evolved heterochromatin droplets formed through the first model or of the expanded droplets formed through the second or third model to the nuclear lamina and their subsequent fusions with other preexisting droplets, sometimes located on a different chromosome, can influence the expression of genes near these droplets in space if their spatial positions in the nucleus are altered. This may explain the cis‐spreading effect of heterochromatin and the trans‐regulatory function of the heterochromatic Y chromosomes or the YRV.81, 85 It has been recently shown that heterochromatin clustering is essential for the spatial compartmentalization of the entire genome.110 How the evolutionary transition of euchromatin to heterochromatin would influence heterochromatin clustering and what the consequences are for genome folding are intriguing questions that remain to be addressed.
Rapid turnover of heterochromatin does not simply reflect a passive evolutionary process driven by a faster mutation rate and genetic drift, but rather can have significant effects on the host genome. Species‐specific heterochromatin can function as a barrier for reproductive isolation. Taking the 359 satellite sequences mentioned above, a “pioneer” satellite recruits HP1 for establishing heterochromatin during early embryogenesis in D. melanogaster;121 such satellites are concentrated on the X chromosome of D. melanogaster but absent in Drosophila simulans. Female hybrid offspring between the two species die during embryogenesis because of lagging X‐linked chromatin derived from D. melanogaster and a resulting mitotic defect.156 This type of speciation process can be caused by the incompatibility between fast‐evolving heterochromatin and its regulatory proteins or RNAs in the hybrid genome. Consistent with this scenario, in hybrids between Drosophila mauritiana and D. simulans, the heterochromatin‐binding allele of OdsH from D. mauritiana erroneously decondenses Y chromosome heterochromatin of D. simulans and thereby causes male sterility.157 The rapid evolution of heterochromatin must be contained to avoid impairing its important structural and regulatory functions. This is manifested as an “arms race” of sorts between heterochromatin and its regulatory proteins and RNAs, including those involved in heterochromatin packaging,152, 153 telomere protection,158 and small RNA pathways.159, 160 These proteins either show an excess of amino acid change, that is, have the signature of positive selection,161, 162 or undergo rampant gene birth and death processes accompanied by newly evolved expression patterns that are usually restricted in the germline.152, 153 To date, a recent characterization of 64 Diptera genomes found a total of 121 HP1 duplications, but almost no duplications in SU(VAR)3‐9 and PcG proteins. Interestingly, the loss of a male‐specific HP1 family protein HP1E correlates with the transformation of the ancestral Y chromosome to an autosome in D. pseudoobscura, suggesting a relaxed selective constraint on HP1E due to the loss of Y‐linked heterochromatin.153
Sex chromosomes as a unique paradigm to study heterochromatin evolution
Sex chromosomes have been associated with heterochromatin ever since its discovery2, 3 (Fig. 1), as the Y or W chromosomes of most species are much more heterochromatic than other autosomes. This is because the evolution of sex necessitates the suppression of recombination between X and Y chromosomes (or Z and W chromosomes in species such as birds and butterflies), to prevent the male‐determining genes or male‐beneficial but female‐detrimental genes (so‐called sexually antagonistic genes) from appearing in females through recombination.163 The costs of sex on the Y chromosome include the massive accumulation of repetitive elements and loss of functional genes due to the reduced efficiency of natural selection in a nonrecombining environment. To balance the expression level resulting from Y gene loss, fHet is also involved in various dosage compensation mechanisms to downregulate X chromosome gene expression (e.g., C. elegans and eutherian mammals).17 The major difficulty in studying sex chromosomes is shared with that of studying heterochromatin: the highly repetitive sequence nature poses tremendous challenges for genome sequencing, assembly, and alignment, as well as gene mapping and manipulation. For example, the Y chromosome sequence of D. melanogaster is still not complete but received some significant improvement recently by PacBio sequencing,17, 164 nearly 20 years after the release of its first draft genome.165
Young sex chromosomes have initiated the heterochromatinization process very recently and still contain large portions of unique sequences, thus providing a paradigm to study the mechanisms and consequences of heterochromatin evolution. Such systems can include species that have evolved sex‐determining regions very recently or those that have recently undergone fusions between the ancestral sex chromosome and autosome (called neo‐sex chromosomes). The former are more concentrated in plant,166 fish,167 and amphibian168 species, while the latter have been extensively studied in Drosophila species,163 though similar systems have also been reported in birds169 and mammals.170
It seems that the heterochromatinization process can occur very quickly on the Y chromosome after recombination is suppressed. For example, the X and Y chromosomes of papaya have been estimated to diverge from each other 2–3 million years ago. The Y‐linked male‐specific region without recombination only accounts for 13% of the entire chromosome, but has already exhibited several heterochromatin knobs by cytogenetic staining and is also associated with an elevated level of DNA methylation.171 A more systematic study of heterochromatin evolution comes from Drosophila miranda, which formed a neo‐Y chromosome through a chromosome fusion between an autosome and an older Y chromosome that originated over 10 million years ago. The fused autosome is homologous to chr3 of D. pseudoobscura and only appears in males that do not have recombination, thus evolving like a true Y chromosome. The divergence time between D. miranda and D. pseudoobscura sets the maximum age of this neo‐Y to be within 1.5 million years.172 Previous cytogenetic studies showed that neo‐Y has already accumulated an excessive amount of retroposons relative to its homolog neo‐X.173, 174 This probably caused the chromosome‐wide increase in the H3K9me2 binding level of neo‐Y, indicated by both immunostaining and ChIP‐seq, relative to the neo‐X and autosomes. Indeed, the binding level of H3K9me2 shows a positive correlation with the copy numbers of TEs surrounding the neo‐Y linked genes. Interestingly, genes with a D. melanogaster ortholog located in black heterochromatin are much more likely to have become decorated by heterochromatin than any other genes on the neo‐Y. This suggests that the ancestral chromatin configuration affects the evolution propensity of heterochromatin.175
A similar state has been observed in another neo‐Y system of Drosophila busckii, where the homologous chromosome of the dot chromosome has fused to the ancestral Y chromosome within the last 1 million years. The active genes of the D. melanogaster dot chromosome are associated with the enrichment of H3K9me3 and the silencing of genes with H3K9me2, while the two heterochromatin marks coincide with each other in the rest of the genome.75 The neo‐Y in D. busckii seems to have adopted such a unique heterochromatin configuration and only has become more enriched for H3K9me2 on silent genes, but there was no difference for H3K9me3.176
Conclusions and perspective
A persistent interest in heterochromatin since its first description in the 1920s1 has been refueled by the development of sequencing techniques and new findings regarding developmental changes114, 121 and biophysical properties112, 113 of heterochromatin. Heterochromatic sequences had been previously assumed to be selfish and nonfunctional genomic parasites that only exist because of over‐replication. It is clear now that heterochromatin contributes a variety of important structural and regulatory functions to the host genome. Because of the formidable cost of acquiring the complete heterochromatic sequences and the technological limits of Illumina sequencing, the burst of genomic resources for various species in the past 10 years contributed little to advancing our understanding of the diversity and evolution of heterochromatin. Most sequencing projects tended to choose the homogametic sex (e.g., a female mammal or a male bird) and sometimes intentionally omitted the heterochromatin‐enriched Y or W chromosomes. High‐quality heterochromatic sequences were a privilege of a few model organisms or primate species.177, 178
The development of third‐generation sequencing, that is, PacBio or the nanopore platform, and its recent applications in Drosophila,173, 179 gorillas,180, 181 and humans179 have promised new discoveries regarding the diversity and functions of heterochromatin in the future. For example, centromere sequences of most species are not known except for yeast and humans, and they are epigenetically determined by CENP‐A chromatin without consensus genomic sequences across species and embedded in the heterochromatin.182 A recent study in D. melanogaster combined third‐generation sequencing and CENP‐A ChIP and discovered sequences that are not present in the published Drosophila genome and are also enriched for CENP‐A binding. It turns out that D. melanogaster centromeres are unexpectedly enriched for non‐LTR retroelements G2/Jockey‐3, instead of satellite sequences183 like humans or yeast.184 In addition, G2/Jockey‐3 elements seem to be only restricted to D. melanogaster subgroup species, indicating the rapid evolution of centromere genomic sequences.
In addition to the new findings regarding the composition of heterochromatin sequences, we also expect more studies characterizing the spatiotemporal changes in heterochromatin in the context of chromatin topologies. Previous epigenomic or chromatin conformation studies in ENCODE or modENCODE consortiums mostly used adult tissues with pooled cell types,29, 31, 183 restricted by the requirement of the large amount of starting materials. However, chromatin domains and TAD structures seem to be dynamically established during embryogenesis, as shown by recently developed single‐cell ChIP and Hi‐C techniques.114, 185 Once established, the TAD structures seem to remain stable during development117 and conserved during evolution.96, 155, 186
For developmental and molecular biologists, the critical questions remaining to be answered include what factors are involved, and how do they interact with each other to recognize the genomic cis elements, probably with a different priority during early embryogenesis to establish the chromatin states and architectures? What are the distinct mechanisms for establishing and maintaining heterochromatin in different genomic regions? For evolutionary biologists, new questions emerge from these new discoveries: What is the resolution for the paradox of fast‐evolving heterochromatin sequences and conserved TAD structures, sometimes even between humans and Drosophila?187 How would the arms race between heterochromatin and its regulatory proteins and RNAs drive other parts of the host genome to change? Do the genomic sequences evolve with their respective regulatory compartments as a module? Answers to these questions will greatly benefit from studying the systems that recently evolved heterochromatin, that is, polymorphic inversions or young sex chromosomes, as well as genetic manipulations that introduce or perturb the heterochromatin, with more insights to come that illuminate the genomic and epigenomic dark matter.
Competing interests
The authors declare no competing interests.
Acknowledgments
We thank Drs. Xiang‐Wei He and Kai Yuan for their helpful comments on the manuscript. Q.Z. is supported by the National Natural Science Foundation of China (Grant nos. 31722050 and 31671319), the Natural Science Foundation of Zhejiang Province (LD19C190001), and the European Research Council Starting Grant (Grant agreement 677696).
[Correction added on February 6, 2020, after online publication: Affiliation of the corresponding author was corrected.]
References
- 1. Heitz, E. 1928. Das Heterochromatin der Moose. Jahrb. Wiss. Bot. 69: 762–818. [Google Scholar]
- 2. Mcclung, C.E. 1902. The accessory chromosome—sex determinant? Biol. Bull. 3: 43–84. [Google Scholar]
- 3. Sutton, W.S. 1902. On the morphology of the chromosome group in Brachystola magna . Biol. Bull. 4: 24–39. [Google Scholar]
- 4. Gatti, M. , Pimpinelli S. & Santini G.. 1976. Characterization of Drosophila heterochromatin. Chromosoma 57: 351–375. [DOI] [PubMed] [Google Scholar]
- 5. Pimpinelli, S. , Gatti M. & De Marco A.. 1975. Evidence for heterogeneity in heterochromatin of Drosophila melanogaster . Nature 256: 335. [DOI] [PubMed] [Google Scholar]
- 6. Pimpinelli, S. , Pignone D., Gatti M., et al 1976. X‐ray induction of chromatid interchanges is somatic cells of Drosophila melanogaster: variations through the cell cycle of the pattern of rejoining. Mutat. Res. 35: 101–110. [DOI] [PubMed] [Google Scholar]
- 7. Brown, S.W. 1966. Heterochromatin. Science 151: 417–425. [DOI] [PubMed] [Google Scholar]
- 8. Heitz, E. 1933. Die somatische Heteropyknose bei Drosophila melanogaster und ihre genetische Bedeutung. Z. Zellforsch. Mikrosk. Anat. 20: 237–287. [Google Scholar]
- 9. Zacharias, H. 1995. Emil Heitz (1892–1965): chloroplasts, heterochromatin, and polytene chromosomes. Genetics 141: 7–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Schultz, J. & Dobzhansky T.. 1934. The relation of a dominant eye color in Drosophila melanogaster to the associated chromosome rearrangement. Genetics 19: 344–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wakimoto, B.T. & Hearn M.G.. 1990. The effects of chromosome rearrangements on the expression of heterochromatic genes in chromosome 2L of Drosophila melanogaster . Genetics 125: 141–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Elgin, S.C. & Reuter G.. 2013. Position‐effect variegation, heterochromatin formation, and gene silencing in Drosophila . Cold Spring Harb. Perspect. Biol. 5: a017780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tschiersch, B. , Hofmann A., Krauss V., et al 1994. The protein encoded by the Drosophila position‐effect variegation suppressor gene Su(var)3‐9 combines domains of antagonistic regulators of homeotic gene complexes. EMBO J. 13: 3822–3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Nakayama, J.I. & Nakayama J.I.. 2001. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science 292: 110–113. [DOI] [PubMed] [Google Scholar]
- 15. Barr, M.L. & Bertram E.G.. 1949. A morphological distinction between neurones of the male and female, and the behaviour of the nucleolar satellite during accelerated nucleoprotein synthesis. Nature 163: 676. [DOI] [PubMed] [Google Scholar]
- 16. Lyon, M.F. 1961. Gene action in the X‐chromosome of the mouse (Mus musculus L.). Nature 190: 372–373. [DOI] [PubMed] [Google Scholar]
- 17. Heard, E. & Disteche C.M.. 2006. Dosage compensation in mammals: fine‐tuning the expression of the X chromosome. Genes Dev. 20: 1848–1867. [DOI] [PubMed] [Google Scholar]
- 18. Lewis, E.B. 1978. A gene complex controlling segmentation in Drosophila . Nature 276: 565–570. [DOI] [PubMed] [Google Scholar]
- 19. Lewis, P. 1947. Melanogaster—new mutants: report of Pamela H. Lewis. Dros. Info. Ser. 21: 69. [Google Scholar]
- 20. Ingham, P. 1983. Differential expression of bithorax complex genes in the absence of the extra sex combs and trithorax genes. Nature 306: 591. [DOI] [PubMed] [Google Scholar]
- 21. Struhl, G. & White R.A.. 1985. Regulation of the Ultrabithorax gene of Drosophila by other bithorax complex genes. Cell 43: 507–519. [DOI] [PubMed] [Google Scholar]
- 22. Kornberg, R.D. & Thonmas J.O.. 1974. Chromatin structure: oligomers of the histones. Science 184: 865–868. [DOI] [PubMed] [Google Scholar]
- 23. Luger, K. , Mäder A.W., Richmond R.K., et al 1997. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 389: 251–260. [DOI] [PubMed] [Google Scholar]
- 24. Jenuwein, T. & Allis C.D.. 2001. Translating the histone code. Science 293: 1074–1080. [DOI] [PubMed] [Google Scholar]
- 25. Allshire, R.C. & Madhani H.D.. 2018. Ten principles of heterochromatin formation and function. Nat. Rev. Mol. Cell Biol. 19: 229–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Clemson, C.M. , Hall L.L., Byron M., et al 2006. The X chromosome is organized into a gene‐rich outer rim and an internal core containing silenced nongenic sequences. Proc. Natl Acad. Sci. USA 103: 7688–7693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Margueron, R. , Justin N., Ohno K., et al 2009. Role of the polycomb protein EED in the propagation of repressive histone marks. Nature 461: 762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mozzetta, C. , Pontis J., Fritsch L., et al 2014. The histone H3 lysine 9 methyltransferases G9a and GLP regulate polycomb repressive complex 2‐mediated gene silencing. Mol. Cell 53: 277–289. [DOI] [PubMed] [Google Scholar]
- 29. ENCODE Project Consortium . 2012. An integrated encyclopedia of DNA elements in the human genome. Nature 489: 57–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Roadmap Epigenomics Consortium ; Kundaje A., Meuleman W., et al 2015. Integrative analysis of 111 reference human epigenomes. Nature 518: 317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ho, J.W.K. , Jung Y.L., Liu T., et al 2014. Comparative analysis of metazoan chromatin organization. Nature 512: 449–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kharchenko, P.V. , Alekseyenko A.A., Schwartz Y.B., et al 2011. Comprehensive analysis of the chromatin landscape in Drosophila melanogaster . Nature 471: 480–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Filion, G.J. , Van Bemmel J.G., Braunschweig U., et al 2010. Systematic protein location mapping reveals five principal chromatin types in Drosophila cells. Cell 143: 212–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Koryakov, D.E. , Walther M., Ebert A., et al 2011. The SUUR protein is involved in binding of SU(VAR)3–9 and methylation of H3K9 and H3K27 in chromosomes of Drosophila melanogaster . Chromosome Res. 19: 235–249. [DOI] [PubMed] [Google Scholar]
- 35. Zhimulev, I.F. , Belyaeva E.S., Makunin I.V., et al 2003. Influence of the SuUR gene on intercalary heterochromatin in Drosophila melanogaster polytene chromosomes. Chromosoma 111: 377–398. [DOI] [PubMed] [Google Scholar]
- 36. Roudier, F. , Ahmed I., Berard C., et al 2011. Integrative epigenomic mapping defines four main chromatin states in Arabidopsis. EMBO J. 30: 1928–1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gorkin, D.U. , Williams B.A., Trout D., et al 2017. Systematic mapping of chromatin state landscapes during mouse development. biorxiv 166652.
- 38. Fanti, L. & Pimpinelli S.. 2008. HP1: a functionally multifaceted protein. Curr. Opin. Genet. Dev. 18: 169–174. [DOI] [PubMed] [Google Scholar]
- 39. Bisht, K.K. , Arora S., Ahmed S., et al 2008. Role of heterochromatin in suppressing subtelomeric recombination in fission yeast. Yeast 25: 537–548. [DOI] [PubMed] [Google Scholar]
- 40. Savitsky, M. , Kravchuk O., Melnikova L., et al 2002. Heterochromatin protein 1 is involved in control of telomere elongation in Drosophila melanogaster . Mol. Cell. Biol. 22: 3204–3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Allshire, R.C. & Ekwall K.. 2015. Epigenetic regulation of chromatin states in Schizosaccharomyces pombe . Cold Spring Harb. Perspect. Biol. 7: a018770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Pidoux, A.L. & Allshire R.C.. 2005. The role of heterochromatin in centromere function. Philos. Trans. R. Soc. Lond. B Biol. Sci. 360: 569–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Allshire, R.C. , Nimmo E.R., Ekwall K., et al 1995. Mutations derepressing silent centromeric domains in fission yeast disrupt chromosome segregation. Genes Dev. 9: 218–233. [DOI] [PubMed] [Google Scholar]
- 44. Ekwall, K. , Javerzat J., Lorentz A., et al 1995. The chromodomain protein Swi6: a key component at fission yeast centromeres. Science 269: 1429–1431. [DOI] [PubMed] [Google Scholar]
- 45. Janssen, A. , Colmenares S.U. & Karpen G.H.. 2018. Heterochromatin: guardian of the genome. Annu. Rev. Cell Dev. Biol. 34: 265–288. [DOI] [PubMed] [Google Scholar]
- 46. Peng, J.C. & Karpen G.H.. 2007. H3K9 methylation and RNA interference regulate nucleolar organization and repeated DNA stability. Nat. Cell Biol. 9: 25–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Peng, J.C. & Karpen G.H.. 2009. Heterochromatic genome stability requires regulators of histone H3 K9 methylation. PLoS Genet. 5: e1000435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Czech, B. , Munafò M., Ciabrelli F., et al 2018. piRNA‐guided genome defense: from biogenesis to silencing. Annu. Rev. Genet. 52: 131–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ozata, D.M. , Gainetdinov I., Zoch A., et al 2019. PIWI‐interacting RNAs: small RNAs with big functions. Nat. Rev. Genet. 20: 89–108. [DOI] [PubMed] [Google Scholar]
- 50. Paro, R. & Hogness D.S.. 1991. The Polycomb protein shares a homologous domain with a heterochromatin‐associated protein of Drosophila. Proc. Natl. Acad. Sci. USA 88: 263–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Aasland, R. , Gibson T.J. & Stewart A.F.. 1995. The PHD finger: implications for chromatin‐mediated transcriptional regulation. Trends Biochem. Sci. 20: 56–59. [DOI] [PubMed] [Google Scholar]
- 52. Baulcombe, D.C. & Dean C.. 2014. Epigenetic regulation in plant responses to the environment. Cold Spring Harb. Perspect. Biol. 6: a019471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Pikaard, C.S. & Mittelsten Scheid O.. 2014. Epigenetic regulation in plants. Cold Spring Harb. Perspect. Biol. 6: a019315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kawasaki, H. & Taira K.. 2004. Induction of DNA methylation and gene silencing by short interfering RNAs in human cells. Nature 431: 211. [DOI] [PubMed] [Google Scholar]
- 55. Sentmanat, M.F. & Elgin S.C.R.. 2012. Ectopic assembly of heterochromatin in Drosophila melanogaster triggered by transposable elements. Proc. Natl. Acad. Sci. USA 109: 14104–14109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Klattenhoff, C. , Xi H., Li C., et al 2009. The Drosophila HP1 homolog Rhino is required for transposon silencing and piRNA production by dual‐strand clusters. Cell 138: 1137–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Mohn, F. , Sienski G., Handler D., et al 2014. The rhino‐deadlock‐cutoff complex licenses noncanonical transcription of dual‐strand piRNA clusters in Drosophila. Cell 157: 1364–1379. [DOI] [PubMed] [Google Scholar]
- 58. Zhang, Z. , Wang J., Schultz N., et al 2014. The HP1 homolog rhino anchors a nuclear complex that suppresses piRNA precursor splicing. Cell 157: 1353–1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Andersen, P.R. , Tirian L., Vunjak M., et al 2017. A heterochromatin‐dependent transcription machinery drives piRNA expression. Nature 549: 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Batki, J. , Schnabl J., Wang J., et al 2019. The nascent RNA binding complex SFiNX licenses piRNA‐guided heterochromatin formation. Nat. Struct. Mol. Biol. 26: 720–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Yu, B. , Cassani M., Wang M., et al 2015. Structural insights into Rhino‐mediated germline piRNA cluster formation. Cell Res. 25: 525–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Yasuhara, J.C. & Wakimoto B.T.. 2006. Oxymoron no more: the expanding world of heterochromatic genes. Trends Genet. 22: 330–338. [DOI] [PubMed] [Google Scholar]
- 63. Weiler, K.S. & Wakimoto B.T.. 1995. Heterochromatin and gene expression in Drosophila. Annu. Rev. Genet. 29: 577–605. [DOI] [PubMed] [Google Scholar]
- 64. Eberl, D.F. , Duyf B.J. & Hilliker A.J.. 1993. The role of heterochromatin in the expression of a heterochromatic gene, the rolled locus of Drosophila melanogaster . Genetics 134: 277–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Dimitri, P. , Corradini N., Rossi F., et al 2005. The paradox of functional heterochromatin. Bioessays 27: 29–41. [DOI] [PubMed] [Google Scholar]
- 66. Vicoso, B. & Bachtrog D.. 2013. Reversal of an ancient sex chromosome to an autosome in Drosophila . Nature 499: 332–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Larsson, J. & Meller V.H.. 2006. Dosage compensation, the origin and the afterlife of sex chromosomes. Chromosome Res. 14: 417–431. [DOI] [PubMed] [Google Scholar]
- 68. Riddle, N.C. & Elgin S.C.R.. 2018. The Drosophila dot chromosome: where genes flourish amidst repeats. Genetics 210: 757–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Hannah, A. 1951. Localization and function of heterochromatin in Drosophila melanogaster . Adv. Genet. 4: 87–125. [DOI] [PubMed] [Google Scholar]
- 70. Davis, R.J. , Belikoff E.J., Scholl E.H., et al 2018. no blokes is essential for male viability and X chromosome gene expression in the Australian Sheep Blowfly. Curr. Biol. 28: 1987–1992.e1983. [DOI] [PubMed] [Google Scholar]
- 71. Cryderman, D.E. , Grade S.K., Li Y., et al 2005. Role of Drosophila HP1 in euchromatic gene expression. Dev. Dyn. 232: 767–774. [DOI] [PubMed] [Google Scholar]
- 72. De Lucia, F. , Ni J.‐Q., Vaillant C., et al 2005. HP1 modulates the transcription of cell‐cycle regulators in Drosophila melanogaster . Nucleic Acids Res. 33: 2852–2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Piacentini, L. , Fanti L., Negri R., et al 2009. Heterochromatin protein 1 (HP1a) positively regulates euchromatic gene expression through RNA transcript association and interaction with hnRNPs in Drosophila . PLoS Genet. 5: e1000670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Johansson, A.‐M. , Stenberg P., Allgardsson A., et al 2012. POF regulates the expression of genes on the fourth chromosome in Drosophila melanogaster by binding to nascent RNA. Mol. Cell. Biol. 32: 2121–2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Riddle, N.C. , Jung Y.L., Gu T., et al 2012. Enrichment of HP1a on Drosophila chromosome 4 genes creates an alternate chromatin structure critical for regulation in this heterochromatic domain. PLoS Genet. 8: e1002954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Wang, S.H. , Nan R., Accardo M.C., et al 2014. A distinct type of heterochromatin at the telomeric region of the Drosophila melanogaster Y chromosome. PLoS One 9: e86451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Dimitri, P. & Pisano C.. 1989. Position effect variegation in Drosophila melanogaster: relationship between suppression effect and the amount of Y chromosome. Genetics 122: 793–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Berloco, M. , Palumbo G., Piacentini L., et al 2014. Position effect variegation and viability are both sensitive to dosage of constitutive heterochromatin in Drosophila . G3 4: 1709–1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Henikoff, S. 1996. Dosage‐dependent modification of position‐effect variegation in Drosophila. Bioessays 18: 401–409. [DOI] [PubMed] [Google Scholar]
- 80. Zuckerkandl, E. 1974. A possible role of “inert” heterochromatin in cell differentiation. Action of and competition for “locking” molecules. Biochimie 56: 937–954. [DOI] [PubMed] [Google Scholar]
- 81. Lemos, B. , Araripe L.O. & Hartl D.L.. 2008. Polymorphic Y chromosomes harbor cryptic variation with manifold functional consequences. Science 319: 91–93. [DOI] [PubMed] [Google Scholar]
- 82. Lemos, B. , Branco A.T. & Hartl D.L.. 2010. Epigenetic effects of polymorphic Y chromosomes modulate chromatin components, immune response, and sexual conflict. Proc. Natl. Acad. Sci. USA 107: 15826–15831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Zhou, J. , Sackton T.B., Martinsen L., et al 2012. Y chromosome mediates ribosomal DNA silencing and modulates the chromatin state in Drosophila. Proc. Natl. Acad. Sci. USA 109: 9941–9946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Sackton, T.B. & Hartl D.L.. 2013. Meta‐analysis reveals that genes regulated by the Y chromosome in Drosophila melanogaster are preferentially localized to repressive chromatin. Genome Biol. Evol. 5: 255–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Francisco, F.O. & Lemos B.. 2014. How do y‐chromosomes modulate genome‐wide epigenetic States: genome folding, chromatin sinks, and gene expression. J. Genomics 2: 94–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Brown, E.J. , Nguyen A.H. & Bachtrog D.. 2020. The Drosophila Y chromosome affects heterochromatin integrity genome‐wide. Mol. Biol. Evol. 10.1093/molbev/msaa082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Reddy, H.M. , Bhattacharya R., Jehan Z., et al 2018. Y chromosomal noncoding RNA regulates autosomal gene expression via piRNAs in mouse testis. bioRxiv 10.1101/285429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Ulianov, S.V. , Doronin S.A., Khrameeva E.E., et al 2019. Nuclear lamina integrity is required for proper spatial organization of chromatin in Drosophila . Nat. Commun. 10: 1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Sexton, T. , Yaffe E., Kenigsberg E., et al 2012. Three‐dimensional folding and functional organization principles of the Drosophila genome. Cell 148: 458–472. [DOI] [PubMed] [Google Scholar]
- 90. Szabo, Q. , Bantignies F. & Cavalli G.. 2019. Principles of genome folding into topologically associating domains. Sci. Adv. 5: eaaw1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Szabo, Q. , Jost D., Chang J.‐M., et al 2018. TADs are 3D structural units of higher‐order chromosome organization in. Sci. Adv. 4: eaar8082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Wang, J. , Lawry S.T., Cohen A.L., et al 2014. Chromosome boundary elements and regulation of heterochromatin spreading. Cell. Mol. Life Sci. 71: 4841–4852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Dong, P. , Tu X., Chu P.‐Y., et al 2017. 3D chromatin architecture of large plant genomes determined by local A/B compartments. Mol. Plant 10: 1497–1509. [DOI] [PubMed] [Google Scholar]
- 94. Lieberman‐Aiden, E. , Van Berkum N.L., Williams L., et al 2009. Comprehensive mapping of long‐range interactions reveals folding principles of the human genome. Science 326: 289–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Simonis, M. , Klous P., Splinter E., et al 2006. Nuclear organization of active and inactive chromatin domains uncovered by chromosome conformation capture–on‐chip (4C). Nat. Genet. 38: 1348–1354. [DOI] [PubMed] [Google Scholar]
- 96. Dixon, J.R. , Selvaraj S., Yue F., et al 2012. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485: 376–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Ramirez, F. , Bhardwaj V., Arrigoni L., et al 2018. High‐resolution TADs reveal DNA sequences underlying genome organization in flies. Nat. Commun. 9: 189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Wang, Q. , Sun Q., Czajkowsky D.M., et al 2018. Sub‐kb Hi‐C in D. melanogaster reveals conserved characteristics of TADs between insect and mammalian cells. Nat. Commun. 9: 188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Rao, S.S.P. , Huntley M.H., Durand N.C., et al 2014. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159: 1665–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. De Wit, E. , Vos E.S., Holwerda S.J., et al 2015. CTCF binding polarity determines chromatin looping. Mol. Cell 60: 676–684. [DOI] [PubMed] [Google Scholar]
- 101. Guo, Y. , Xu Q., Canzio D., et al 2015. CRISPR inversion of CTCF sites alters genome topology and enhancer/promoter function. Cell 162: 900–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Nora, E.P. , Goloborodko A., Valton A.‐L., et al 2017. Targeted degradation of CTCF decouples local insulation of chromosome domains from genomic compartmentalization. Cell 169: 930–944.e922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Wutz, G. , Várnai C., Nagasaka K., et al 2017. Topologically associating domains and chromatin loops depend on cohesin and are regulated by CTCF, WAPL, and PDS5 proteins. EMBO J. 36: 3573–3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Sanborn, A.L. , Rao S.S.P., Huang S.‐C., et al 2015. Chromatin extrusion explains key features of loop and domain formation in wild‐type and engineered genomes. Proc. Natl. Acad. Sci. USA 112: E6456–E6465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Fawcett, D.W. 1966. On the occurrence of a fibrous lamina on the inner aspect of the nuclear envelope in certain cells of vertebrates. Am. J. Anat. 119: 129–145. [DOI] [PubMed] [Google Scholar]
- 106. Van Steensel, B. & Belmont A.S.. 2017. Lamina‐associated domains: links with chromosome architecture, heterochromatin, and gene repression. Cell 169: 780–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Németh, A. , Conesa A., Santoyo‐Lopez J., et al 2010. Initial genomics of the human nucleolus. PLoS Genet. 6: e1000889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Van Koningsbruggen, S. , Gierlinski M., Schofield P., et al 2010. High‐resolution whole‐genome sequencing reveals that specific chromatin domains from most human chromosomes associate with nucleoli. Mol. Biol. Cell 21: 3735–3748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Fraser, J. , Ferrai C., Chiariello A.M., et al 2015. Hierarchical folding and reorganization of chromosomes are linked to transcriptional changes in cellular differentiation. Mol. Syst. Biol. 11: 852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Falk, M. , Feodorova Y., Naumova N., et al 2019. Heterochromatin drives compartmentalization of inverted and conventional nuclei. Nature 570: 395–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Kind, J. , Pagie L., Ortabozkoyun H., et al 2013. Single‐cell dynamics of genome–nuclear lamina interactions. Cell 153: 178–192. [DOI] [PubMed] [Google Scholar]
- 112. Strom, A.R. , Emelyanov A.V., Mir M., et al 2017. Phase separation drives heterochromatin domain formation. Nature 547: 241–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Larson, A.G. , Elnatan D., Keenen M.M., et al 2017. Liquid droplet formation by HP1α suggests a role for phase separation in heterochromatin. Nature 547: 236–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Hug, C.B. & Vaquerizas J.M.. 2018. The birth of the 3D genome during early embryonic development. Trends Genet. 34: 903–914. [DOI] [PubMed] [Google Scholar]
- 115. Schier, A.F. 2007. The maternal–zygotic transition: death and birth of RNAs. Science 316: 406–407. [DOI] [PubMed] [Google Scholar]
- 116. Li, X.Y. , Harrison M.M., Villalta J.E., et al 2014. Establishment of regions of genomic activity during the Drosophila maternal to zygotic transition. elife 3 10.7554/eLife.03737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Hug, C.B. , Grimaldi A.G., Kruse K., et al 2017. Chromatin architecture emerges during zygotic genome activation independent of transcription. Cell 169: 216–228.e219. [DOI] [PubMed] [Google Scholar]
- 118. Rudolph, T. , Yonezawa M., Lein S., et al 2007. Heterochromatin formation in Drosophila is initiated through active removal of H3K4 methylation by the LSD1 homolog SU(VAR)3‐3. Mol. Cell 26: 103–115. [DOI] [PubMed] [Google Scholar]
- 119. Sanulli, S. , Trnka M., Dharmarajan V., et al 2019. HP1 reshapes nucleosome core to promote heterochromatin phase separation. Nature 575: 390–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Seller, C.A. , Cho C.‐Y. & O'Farrell P.H.. 2019. Rapid embryonic cell cycles defer the establishment of heterochromatin by Eggless/SetDB1 in Drosophila . Genes Dev. 33: 403–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Yuan, K. & O'Farrell P.H.. 2016. TALE‐light imaging reveals maternally guided, H3K9me2/3‐independent emergence of functional heterochromatin in Drosophila embryos. Genes Dev. 30: 579–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Gu, T. & Elgin S.C.R.. 2013. Maternal depletion of Piwi, a component of the RNAi system, impacts heterochromatin formation in Drosophila . PLoS Genet. 9: e1003780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Riddle, N.C. , Minoda A., Kharchenko P.V., et al 2011. Plasticity in patterns of histone modifications and chromosomal proteins in Drosophila heterochromatin. Genome Res. 21: 147–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Makova, K.D. & Hardison R.C.. 2015. The effects of chromatin organization on variation in mutation rates in the genome. Nat. Rev. Genet. 16: 213–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Schuster‐Bockler, B. & Lehner B.. 2012. Chromatin organization is a major influence on regional mutation rates in human cancer cells. Nature 488: 504–507. [DOI] [PubMed] [Google Scholar]
- 126. Chuong, E.B. , Elde N.C. & Feschotte C.. 2017. Regulatory activities of transposable elements: from conflicts to benefits. Nat. Rev. Genet. 18: 71–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Bourque, G. , Burns K.H., Gehring M., et al 2018. Ten things you should know about transposable elements. Genome Biol. 19: 199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Sharma, S. & Raina S.N.. 2005. Organization and evolution of highly repeated satellite DNA sequences in plant chromosomes. Cytogenet. Genome Res. 109: 15–26. [DOI] [PubMed] [Google Scholar]
- 129. Wei, K.H. , Lower S.E., Caldas I.V., et al 2018. Variable rates of simple satellite gains across the Drosophila phylogeny. Mol. Biol. Evol. 35: 925–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Jagannathan, M. , Warsinger‐Pepe N., Watase G.J., et al 2017. Comparative analysis of satellite DNA in the species complex. G3 7: 693–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Kapusta, A. & Suh A.. 2017. Evolution of bird genomes—a transposon's‐eye view. Ann. N.Y. Acad. Sci. 1389: 164–185. [DOI] [PubMed] [Google Scholar]
- 132. Cechova, M. , Harris R.S., Tomaszkiewicz M., et al 2019. High satellite repeat turnover in great apes studied with short‐ and long‐read technologies. Mol. Biol. Evol. 36: 2415–2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Naville, M. , Henriet S., Warren I., et al 2019. Massive changes of genome size driven by expansions of non‐autonomous transposable elements. Curr. Biol. 29: 1161–1168.e1166. [DOI] [PubMed] [Google Scholar]
- 134. Leung, W. , Shaffer C.D., Chen E.J., et al 2017. Retrotransposons are the major contributors to the expansion of the Muller F element. G3 7: 2439–2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Powell, J.R. , Dion K., Papaceit M., et al 2011. Nonrecombining genes in a recombination environment: the Drosophila “dot” chromosome. Mol. Biol. Evol. 28: 825–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Larracuente, A.M. , Noor M. & Clark A.G.. 2010. Translocation of Y‐linked genes to the dot chromosome in Drosophila pseudoobscura . Mol. Biol. Evol. 27: 1612–1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Larracuente, A.M. & Clark A.G.. 2014. Recent selection on the Y‐to‐dot translocation in Drosophila pseudoobscura . Mol. Biol. Evol. 31: 846–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Chang, C.‐H. & Larracuente A.M.. 2017. Genomic changes following the reversal of a Y chromosome to an autosome in Drosophila pseudoobscura . Evolution 71: 1285–1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Bellott, D.W. , Hughes J.F., Skaletsky H., et al 2014. Mammalian Y chromosomes retain widely expressed dosage‐sensitive regulators. Nature 508: 494–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Skaletsky, H. , Kuroda‐Kawaguchi T., Minx P.J., et al 2003. The male‐specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature 423: 825–837. [DOI] [PubMed] [Google Scholar]
- 141. Gläser, B. , Grützner F., Willmann U., et al 1998. Simian Y chromosomes: species‐specific rearrangements of DAZ, RBM, and TSPY versus contiguity of PAR and SRY. Mamm. Genome 9: 226–231. [DOI] [PubMed] [Google Scholar]
- 142. Hughes, J.F. , Skaletsky H., Pyntikova T., et al 2010. Chimpanzee and human Y chromosomes are remarkably divergent in structure and gene content. Nature 463: 536–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Soh, Y.Q. , Alfoldi J., Pyntikova T., et al 2014. Sequencing the mouse Y chromosome reveals convergent gene acquisition and amplification on both sex chromosomes. Cell 159: 800–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Yasuhara, J.C. , Decrease C.H. & Wakimoto B.T.. 2005. Evolution of heterochromatic genes of Drosophila . Proc. Natl. Acad. Sci. USA 102: 10958–10963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Rebollo, R. , Karimi M.M., Bilenky M., et al 2011. Retrotransposon‐induced heterochromatin spreading in the mouse revealed by insertional polymorphisms. PLoS Genet. 7: e1002301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Lee, Y.C.G. & Karpen G.H.. 2017. Pervasive epigenetic effects of Drosophila euchromatic transposable elements impact their evolution. eLife 6: e2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Baimaj, V. 1977. Chromosomal polymorphisms of constitutive heterochromatin and inversions in Drosophila. Genetics 85: 85–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Charlesworth, B. , Langley C.H. & Stephan W.. 1986. The evolution of restricted recombination and the accumulation of repeated DNA sequences. Genetics 112: 947–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Ebert, A. 2004. Su(var) genes regulate the balance between euchromatin and heterochromatin in Drosophila . Genes Dev. 18: 2973–2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Renauld, H. , Aparicio O.M., Zierath P.D., et al 1993. Silent domains are assembled continuously from the telomere and are defined by promoter distance and strength, and by SIR3 dosage. Genes Dev. 7: 1133–1145. [DOI] [PubMed] [Google Scholar]
- 151. Hecht, A. , Strahl‐Bolsinger S. & Grunstein M.. 1996. Spreading of transcriptional repressor SIR3 from telomeric heterochromatin. Nature 383: 92–96. [DOI] [PubMed] [Google Scholar]
- 152. Helleu, Q. & Levine M.T.. 2018. Recurrent amplification of the heterochromatin protein 1 (HP1) gene family across Diptera. Mol. Biol. Evol. 35: 2375–2389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Levine, M.T. , Mccoy C., Vermaak D., et al 2012. Phylogenomic analysis reveals dynamic evolutionary history of the Drosophila heterochromatin protein 1 (HP1) gene family. PLoS Genet. 8: e1002729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Wang, J. , Reddy B.D. & Jia S.. 2015. Rapid epigenetic adaptation to uncontrolled heterochromatin spreading. elife 4: e06179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Vietri Rudan, M. , Barrington C., Henderson S., et al 2015. Comparative Hi‐C reveals that CTCF underlies evolution of chromosomal domain architecture. Cell Rep. 10: 1297–1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Ferree, P.M. & Barbash D.A.. 2009. Species‐specific heterochromatin prevents mitotic chromosome segregation to cause hybrid lethality in Drosophila . PLoS Biol. 7: e1000234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Bayes, J.J. & Malik H.S.. 2009. Altered heterochromatin binding by a hybrid sterility protein in Drosophila sibling species. Science 326: 1538–1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Lee, Y.C. , Leek C. & Levine M.T.. 2017. Recurrent innovation at genes required for telomere integrity in Drosophila. Mol. Biol. Evol. 34: 467–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Lewis, S.H. , Salmela H. & Obbard D.J.. 2016. Duplication and diversification of Dipteran Argonaute genes, and the evolutionary divergence of Piwi and Aubergine. Genome Biol. Evol. 8: 507–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Parhad, S.S. & Theurkauf W.E.. 2019. Rapid evolution and conserved function of the piRNA pathway. Open Biol. 9: 180181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Vermaak, D. , Henikoff S. & Malik H.S.. 2005. Positive selection drives the evolution of rhino, a member of the heterochromatin protein 1 family in Drosophila . PLoS Genet. 1: 96–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Simkin, A. , Wong A., Poh Y.P., et al 2013. Recurrent and recent selective sweeps in the piRNA pathway. Evolution 67: 1081–1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Bachtrog, D. 2013. Y‐chromosome evolution: emerging insights into processes of Y‐chromosome degeneration. Nat. Rev. Genet. 14: 113–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Chang, C.H. & Larracuente A.M.. 2019. Heterochromatin‐enriched assemblies reveal the sequence and organization of the Drosophila melanogaster Y chromosome. Genetics 211: 333–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Adams, M.D. 2000. The genome sequence of Drosophila melanogaster . Science 287: 2185–2195. [DOI] [PubMed] [Google Scholar]
- 166. Ming, R. , Bendahmane A. & Renner S.S.. 2011. Sex chromosomes in land plants. Annu. Rev. Plant Biol. 62: 485–514. [DOI] [PubMed] [Google Scholar]
- 167. Pennell, M.W. , Mank J.E. & Peichel C.L.. 2018. Transitions in sex determination and sex chromosomes across vertebrate species. Mol. Ecol. 27: 3950–3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Jeffries, D.L. , Lavanchy G., Sermier R., et al 2018. A rapid rate of sex‐chromosome turnover and non‐random transitions in true frogs. Nat. Commun. 9: 4088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Pala, I. , Naurin S., Stervander M., et al 2012. Evidence of a neo‐sex chromosome in birds. Heredity 108: 264–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170. Zhou, Q. , Wang J., Huang L., et al 2008. Neo‐sex chromosomes in the black muntjac recapitulate incipient evolution of mammalian sex chromosomes. Genome Biol. 9: R98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Zhang, W. , Wang X., Yu Q., et al 2008. DNA methylation and heterochromatinization in the male‐specific region of the primitive Y chromosome of papaya. Genome Res. 18: 1938–1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Bachtrog, D. & Charlesworth B.. 2002. Reduced adaptation of a non‐recombining neo‐Y chromosome. Nature 416: 323–326. [DOI] [PubMed] [Google Scholar]
- 173. Steinemann, M. & Steinemann S.. 1992. Degenerating Y chromosome of Drosophila miranda: a trap for retrotransposons. Proc. Natl. Acad. Sci. USA 89: 7591–7595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Bachtrog, D. 2003. Accumulation of Spock and Worf, two novel non‐LTR retrotransposons, on the neo‐Y chromosome of Drosophila miranda . Mol. Biol. Evol. 20: 173–181. [DOI] [PubMed] [Google Scholar]
- 175. Zhou, Q. , Ellison C.E., Kaiser V.B., et al 2013. The epigenome of evolving Drosophila neo‐sex chromosomes: dosage compensation and heterochromatin formation. PLoS Biol. 11: e1001711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176. Zhou, Q. & Bachtrog D.. 2015. Ancestral chromatin configuration constrains chromatin evolution on differentiating sex chromosomes in Drosophila . PLoS Genet. 11: e1005331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Hoskins, R.A. , Carlson J.W., Wan K.H., et al 2015. The Release 6 reference sequence of the Drosophila melanogaster genome. Genome Res. 25: 445–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Hughes, J.F. & Rozen S.. 2012. Genomics and genetics of human and primate y chromosomes. Annu. Rev. Genomics Hum. Genet. 13: 83–108. [DOI] [PubMed] [Google Scholar]
- 179. Kuderna, L.F.K. , Lizano E., Julià E., et al 2019. Selective single molecule sequencing and assembly of a human Y chromosome of African origin. Nat. Commun. 10: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180. Gordon, D. , Huddleston J., Chaisson M.J.P., et al 2016. Long‐read sequence assembly of the gorilla genome. Science 352: aae0344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181. Tomaszkiewicz, M. , Rangavittal S., Cechova M., et al 2016. A time‐ and cost‐effective strategy to sequence mammalian Y chromosomes: an application to the de novo assembly of gorilla Y. Genome Res. 26: 530–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182. Mckinley, K.L. & Cheeseman I.M.. 2016. The molecular basis for centromere identity and function. Nat. Rev. Mol. Cell Biol. 17: 16–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183. Talbert, P.B. , Kasinathan S. & Henikoff S.. 2018. Simple and complex centromeric satellites in Drosophila sibling species. Genetics 208: 977–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184. Chang, C.H. , Chavan A., Palladino J., et al 2019. Islands of retroelements are major components of Drosophila centromeres. PLoS Biol. 17: e3000241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185. Xu, Q. & Xie W.. 2018. Epigenome in early mammalian development: inheritance, reprogramming and establishment. Trends Cell Biol. 28: 237–253. [DOI] [PubMed] [Google Scholar]
- 186. Yakushiji‐Kaminatsui, N. , Lopez‐Delisle L., Bolt C.C., et al 2018. Similarities and differences in the regulation of HoxD genes during chick and mouse limb development. PLoS Biol. 16: e3000004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187. Harmston, N. , Ing‐Simmons E., Tan G., et al 2017. Topologically associating domains are ancient features that coincide with Metazoan clusters of extreme noncoding conservation. Nat. Commun. 8: 441. [DOI] [PMC free article] [PubMed] [Google Scholar]


