Abstract
l‐Lysine oxidase (LysOX) is a FAD‐dependent homodimeric enzyme that catalyzes the oxidative deamination of l‐lysine to produce α‐keto‐ε‐aminocaproate with ammonia and hydrogen peroxide. LysOX shows strict substrate specificity for l‐lysine, whereas most l‐amino acid oxidases (LAAOs) exhibit broad substrate specificity for l‐amino acids. Previous studies of LysOX showed that overall structural similarity to the well‐studied snake venom LAAOs. However, the molecular mechanism of strict specificity for l‐lysine was still unclear. We here determined the structure of LysOX in complex with l‐lysine at 1.7 Å resolution. The structure revealed that the hydrogen bonding network formed by D212, D315, and A440 with two water molecules is responsible for the recognition of the side chain amino group. In addition, a narrow hole formed by five hydrophobic residues in the active site contributes to strict substrate specificity. Mutation studies demonstrated that D212 and D315 are essential for l‐lysine recognition, and the D212A/D315A double mutant LysOX showed different substrate specificity from LysOX. Moreover, the structural basis of the substrate specificity change has also been revealed by the structural analysis of the mutant variant and its substrate complexes. These results clearly explain the molecular mechanism of the strict specificity of LysOX and suggest that LysOX is a potential candidate for a template to design LAAOs specific to other l‐amino acids.
Keywords: crystal structure, L‐lysine α‐oxidase, substrate recognition
Short abstract
Abbreviations
- LAAO
l ‐amino acid oxidase
- LysOX
l ‐lysine α‐oxidase
1. INTRODUCTION
l‐Amino acid oxidase (LAAO) is a flavoenzyme that catalyzes the oxidative deamination of an l‐amino acid to produce a 2‐oxo acid with byproducts of ammonia and hydrogen peroxide. 1 LAAO is widely distributed in various organisms, such as venomous snakes, mammals, insects, fishes, mollusks, fungi, and bacteria. 2 l‐Lysine oxidase (LysOX) is an LAAO family protein that has strict substrate specificity for l‐lysine. LysOX is a unique LAAO because most of the known LAAOs show broad substrate specificity for l‐amino acids whereas almost no activity for l‐lysine. 1 , 2
The first LysOX was isolated from Trichoderma viride during the search for an antitumor agent and showed broad antitumor activity. 3 T. viride secrets LysOX to the outside of the cell, but its physiological function is still unknown. LysOX was also found in marine organisms, such as fish and aplysia. Chub mackerel (Scomber japonicus) infected with larval nematode produces LysOX, which induces apoptosis of the fish cell to impede nematode infection and therefore is termed apoptosis inducing protein (AIP). 4 The skin mucosal of rockfish Sebastes schlegeli contains LysOX as an antibacterial protein. 5 Aplysia californica has LysOX, called escapin, in the ink and uses it as a chemical defense weapon to escape from predators. 6 The enzyme properties of AIP have been characterized, and two aspartic acid residues are predicted to be involved in substrate binding. 7 However, molecular mechanism of strict substrate specificity of LysOX is not known.
LysOX from T. viride is a 540 amino acid protein and forms a homo‐dimer. It is synthesized as a precursor protein composed of 617 amino acids, and its N‐terminal 77 residues are removed for maturation. 8 LysOX is stable against heat and pH change. 3 LysOX retains more than 90% of the original activity after incubation at 60°C for 30 min in 0.1 M potassium phosphate buffer (KBP; pH 7.4). The activity of the enzyme does not significantly change in the pH range from 7 to 10. Thus, LysOX from T. viride is a potential candidate for an enzyme‐based amino acid sensor. 9 The crystal structure of LysOX without substrate revealed that LysOX is composed of three domains, the FAD‐binding domain, the substrate‐binding domain, and the helical domain, like snake venom LAAOs. 8 Comparison of the substrate‐binding site of LysOX with those of other known LAAOs 10 , 11 , 12 , 13 , 14 , 15 revealed that the residues involved in recognition of the l‐amino acid backbone are structurally conserved although the amino acid sequence identity to the snake LAAOs is only around 20%, suggesting that oxidative deamination reaction of LysOX proceeds in the same manner as other LAAOs. In contrast, the residues involved in the substrate side chain recognition of other LAAOs are not conserved in LysOX. Moreover, LysOX has a negative charge on the surface of the entrance to the active site, which lies deep inside the molecule. Substrate docking simulation suggests that D212 may interact with the ε‐amino group of l‐lysine and contribute strict substrate specificity of LysOX. 8 However, the molecular mechanism of the strict substrate specificity of LysOX is still obscure due to a lack of information on the substrate‐binding structure of LysOX.
Here, we show the structure of LysOX in complex with l‐lysine at 1.7 Å resolution. The structure and following mutation analyses based on the complex structure have revealed that the triangular arrangement of D212 and the two water molecules, which form hydrogen bonds to D315 and to the carbonyl oxygen of A440, around the ε‐amino group of l‐lysine is a key structure for strict recognition of l‐lysine. Interestingly, double mutation of these residues changes the substrate specificity of LysOX to l‐amino acids with aromatic rings. The structures of the double mutant protein and its substrate complexes revealed the structural basis of the substrate specificity change. These results suggest that LysOX can be used as a template to design LAAOs specific to other l‐amino acids.
2. RESULTS
2.1. Preparation of the LysOX substrate complex crystals
LysOX expressed in Streptomyces lividans TK24 was purified as previously described 8 and used for crystallization. We initially tried the conventional soaking method to prepare the substrate complex crystals. However, the crystals were immediately cracked or dissolved after soaking in the reservoir solution containing l‐lysine. Thus, we grew the crystals in a high‐concentration agarose gel to protect the crystals from the soaking damage. 16 The crystals were grown in a condition similar to that applied for the previous LysOX crystals 8 but exhibited a different space group of orthorhombic C2221. After soaked in l‐lysine solution for 2.5 hr, the color of the crystals grown in the gel was changed from yellow to colorless, suggesting that FAD is converted from the oxidized form to the reduced form, as reported for the preparation of the phenylalanine complex crystals of LAAO from Calloselasma rhodostoma venom (CrLAAO‐Phe) 12 and the alanine complex crystals of LAAO from Rhodococcus opacus (RoLAAO‐Ala). 17 We used the colorless crystals for X‐ray diffraction data collection.
2.2. Overall structure of recombinant LysOX in complex with l‐lysine
The structure of the l‐lysine complex of recombinant LysOX (LysOX‐Lys) has been determined at 1.7 Å resolution (Figure 1, Table 1). The N‐terminal three, the C‐terminal 28, and a short loop segment of Residues 388–391 are not modeled due to poor electron density. The crystal contains a single dimer of LysOX‐Lys in an asymmetric unit, and the dimer subunits have an almost identical structure with the root mean square deviation (RMSD) of 0.08 Å for Cα atoms. The dimer forms a tetramer with the neighboring dimer related by crystallographic twofold symmetry, similar to the tetramer of ligand‐free LysOX. 8 LysOX‐Lys is composed of three domains: the FAD‐binding domain (G3‐G47, G230‐L312, and F442‐L512), the substrate‐binding domain (R48‐A109, P222‐G229, and H313‐L441) and the helical domain (K110‐K221). No significant difference was observed in the backbone structures of LysOX‐Lys and the ligand‐free LysOX. 8 The LysOX‐Lys structure was superimposed onto the ligand‐free LysOX structure with the RMSD of 0.14 Å for corresponding Cα atoms.
FIGURE 1.
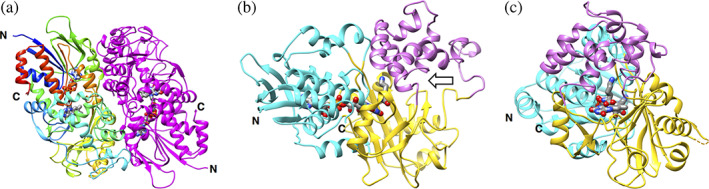
Structure of l‐lysine oxidase (LysOX) in complex with l‐lysine (LysOX‐Lys). (a) Ribbon representation of the dimer of LysOX‐Lys in the crystallographic asymmetric unit. One subunit is colored in rainbow from the N‐terminus (blue) to the C‐terminus (red), and the other in magenta. (b) A ribbon diagram of a single subunit of LysOX‐Lys. The domain structure is depicted in different color:cyan, the FAD‐binding domain; yellow, the substrate‐binding domain; magenta, the helical domain. FAD and the substrate lysine are shown in stick model colored by element: red, oxygen; blue, nitrogen; gray, carbon. The entrance of the funnel is shown by an arrow. (c) The view from the funnel (from the right of (b)). The invisible loop segment of 388–391 are shown by yellow dotted line
TABLE 1.
Data collection and refinement statistics
| LysOX‐Lys | D212A/D315A | D212A/D315A‐Phe | D212A/D315A‐Tyr | |
|---|---|---|---|---|
| Data collection | ||||
| Space group | C2221 | C2221 | C2221 | C2221 |
| Cell dimensions | ||||
| a, b, c (Å) | 116.1, 170.2, 119.5 | 116.5, 170.3, 119.6 | 114.9, 169.7, 118.5 | 117.1, 170.5, 120.4 |
| α, β, γ (°) | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 | 90, 90, 90 |
| Resolution (Å) | 24.0–1.7 (1.79–1.70) a | 85.2–2.3 (2.37–2.3) | 95.2–2.2 (2.26–2.20) a | 75.3–1.8 (1.83–1.80) |
| R merge | 0.097 (0.404) | 0.096 (0.273 | 0.110 (0.334) | 0.095 (0.260) |
| I/σI | 10.2 (3.8) | 8.3 (4.2) | 7.0 (3.3) | 9.9 (5.0) |
| Completeness (%) | 99.7 (99.9) | 99.9 (100) | 99.1 (98.7) | 99.8 (99.8) |
| Redundancy | 6.0 (6.2) | 3.6 (3.8) | 4.4 (4.3) | 5.8 (6.2) |
| Refinement | ||||
| Resolution (Å) | 24.0–1.7 (1.72–1.70) | 74.9–2.3 (2.34–2.30) | 69.0–2.2 (2.24–2.20) | 69.6–1.8 (1.82–1.8) |
| No. reflections | 128,462 (4,260) | 52,468 (2,767) | 58,264 (2,769) | 110,620 (3.635) |
| R work/R free | 17.5/20.9 (20.7/25.3) | 17.9/22.5 (20.8/25.5) | 18.6/23.6 (23.3/29.3) | 15.9/19.0 (20.4/24.1) |
| No. atoms | ||||
| Protein | 8,134 | 8,020 | 8,087 | 8,068 |
| Ligand/ion | 180 | 158 | 164 | 166 |
| Water | 918 | 710 | 647 | 1,347 |
| B‐factors | ||||
| Protein | 14.1 | 20.4 | 22.8 | 17.7 |
| Ligand/ion | 14.3 | 21.9 | 20.5 | 18.7 |
| Water | 21.9 | 26.0 | 26.7 | 19.7 |
| RMSD | ||||
| Bond lengths (Å) | 0.019 | 0.005 | 0.003 | 0.008 |
| Bond angles (°) | 1.438 | 0.706 | 0.635 | 1.137 |
| Ramachandran plot (%) | ||||
| Preferred regions | 97.0 | 97.1 | 96.6 | 96.9 |
| Allowed regions | 2.9 | 2.9 | 3.3 | 3.1 |
| Outliers | 0.1 | 0 | 0.1 | 0 |
Abbreviations: LysOX, l‐lysine oxidase; RMSD, root mean square deviation.
Values in parentheses are for highest‐resolution shell.
2.3. Structural basis of l‐lysine recognition
l‐Lysine is bound in a space at the bottom of the funnel formed between the substrate‐binding domain and the helical domain and lies on the isoalloxazine ring of FAD (Figure 1b,c). The amino acid backbone of l‐lysine interacts with LysOX in the same manner as other LAAOs (Figure 2a,b). The α‐carboxy group of l‐lysine forms hydrogen bonds with R68 and Y369. The α‐amino group hydrogen bonds with the carbonyl oxygen of A475 and forms cation‐pi interaction with the indole ring of W476. These interactions are conserved in other LAAOs (Figure 2a,b), such as CrLAAO 12 and RoLAAO. 17
FIGURE 2.
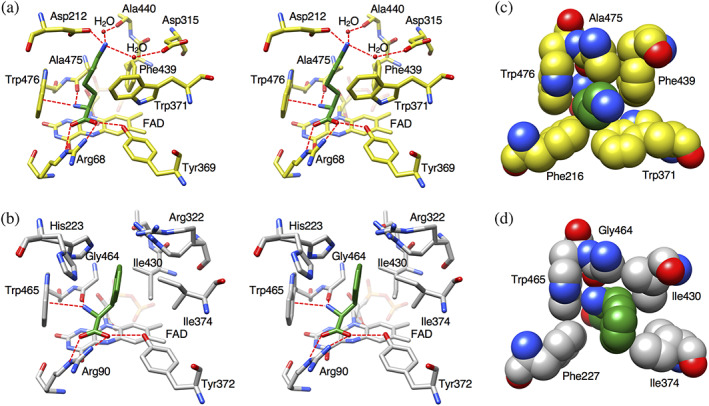
Comparison of the substrate‐binding site structure of l‐lysine oxidase (LysOX) with that of CrLAAO. Stick representation of the substrate‐binding structure (stereo view). LysOX‐Lys (a) and CrLAAO‐Phe (b). Close up view of the substrate molecules in the hydrophobic hole. (c) LysOX‐Lys and (d) CrLAAO‐Phe. The structures are represented by balls. The substrate molecules are colored in green. The protein residues are shown in yellow for LysOX and gray for CrLAAO. The nitrogen and oxygen atoms are colored in blue and red, respectively. The bound water molecules are shown as red balls. Possible hydrogen bonds are indicated by red broken lines
The side chain of l‐lysine adopts an extended conformation in a narrow hole formed by the hydrophobic side chains of F216, W371, F439, A475, and W476 (Figure 2c). Compared with the structure of CrLAAO, LysOX has more bulky residues around the substrate side chain (Figure 2c,d). The residues corresponding to W371, F439, and A475 of LysOX are I374, I430, and G464 in CrLAAO, respectively. Therefore, the hole for the substrate side chain is long and narrow in LysOX. The side chain amino (ε‐amino) group of l‐lysine interacts with the side chain carboxy group of D212 and two water molecules: one is bound to the carbonyl oxygen of A440, and the other to the side chain carboxy group of D315 (Figure 2a). One of the side chain carboxy oxygen of D212 and the two water molecules are triangularly arranged around the ε‐amino group of l‐lysine. This arrangement is reasonable because the ε‐amino group is protonated in the experimental condition (pH 7). These structural features provide a high affinity to l‐lysine.
The substrate binding induces conformational changes of the side chains of W371, Y369, and D212 (Figure 3a). The side chains of D212 and Y369 turn to make hydrophilic interactions with the ε‐amino group and the α‐carboxy group of l‐lysine, respectively. The indole ring of W371 moves toward l‐lysine to directly interact with the aliphatic part of the lysine side chain. This movement blocks the path from the funnel to the active site and seals the active site from the outside (Figure 3b). No other molecule is accessible to the active site through the long funnel after binding of the substrate in the proper position. Therefore, W371 acts as a gate to the active site.
FIGURE 3.
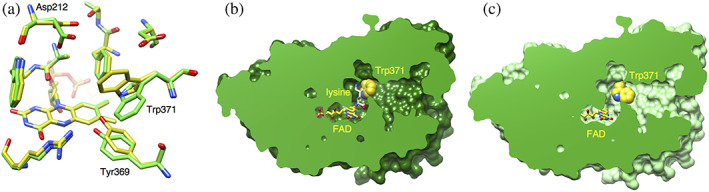
Structural change induced by the l‐lysine binding. (a) Superimposition of the substrate‐binding site structure of l‐lysine oxidase (LysOX)‐Lys on that of ligand‐free LysOX. The carbon atoms of LysOX‐Lys and the ligand‐free LysOX are colored in yellow and green, respectively. The nitrogen and oxygen atoms are colored in blue and red, respectively. (b) Vertical section of the LysOX‐Lys surface model. Trp371 of LysOX‐Lys blocks the path to the active site. (c) Vertical section of the ligand‐free LysOX surface model. FAD and the substrate lysine are represented by stick model. Trp371 is shown by ball model
2.4. Preparation of recombinant LysOX from Escherichia coli
We previously succeeded in recombinant expression of LysOX in S. lividans TK24. 8 However, we have to cultivate the cells more than 20 days to obtain enough amount of LysOX for biochemical assay and crystallization. Therefore, we tried to express LysOX in E. coli. LysOX was successfully expressed with prosequence but not expressed without prosequence, like in S. lividans. LysOX expressed in S. lividans was secreted in culture media as a mature form. 8 However, LysOX was expressed as a precursor enzyme with prosequence in E. coli and showed low activity. Therefore, we removed the N‐terminal prosequence using a metalloprotease from Streptomyces griseus for maturation and then purified the mature LysOX proteins. The properties of the purified LysOX, such as specific activity, kinetic parameters, and substrate specificity, are almost the same as those of the native LysOX and recombinant LysOX from S. lividans transformant (Tables 2 and 3).
TABLE 2.
Kinetic parameters of LysOX and LysOX mutant variants for l‐lysine
| Specific activity (U/mg) | K m (mM) | k cat (s−1) | k cat/K m (s/mM) | |
|---|---|---|---|---|
| Recombinant LysOX | ||||
| From Streptomyces lividans | 62.5 | 1.3 × 10−2 | 59.1 | 4.5 × 103 |
| From Escherichia coli | 66.1 | 1.3 × 10−2 | 62.8 | 4.8 × 103 |
| LysOX D212A | 18.5 | 0.56 | 26.4 | 47 |
| LysOX D315A | 22.2 | 0.28 | 24.3 | 87 |
| LysOX D212A/D315A | 6.4 | 7.6 | 17.0 | 2.2 |
| LysOX M67A | 0.7 | ND | ND | ND |
Abbreviations: LysOX, l‐lysine oxidase; ND, not determined.
TABLE 3.
Specific activity (U/mg) of LysOX and LysOX mutant variants for various substrates
| l‐Arg | l‐Phe | l‐Tyr | l‐Trp | |
|---|---|---|---|---|
| LysOX | 4.8 | 1.3 | 0.9 | 0.0 |
| LysOX D212A | 9.4 | 8.6 | 8.5 | 2.3 |
| LysOX D315A | 7.2 | 8.5 | 1.9 | 0.8 |
| LysOX D212A/D315A | 14.3 | 20.5 | 13.4 | 10.0 |
Abbreviation: LysOX, l‐lysine oxidase.
2.5. Mutation analysis
To confirm the contribution of D212 and D315 to the l‐lysine recognition, we expressed D212A, D315A, and D212A/D315A mutant variants of LysOX in E. coli and determined their kinetic parameters (Table 1). The K m values of D212A and D315A mutant enzymes for l‐lysine were increased 43‐fold and 22‐fold, respectively, to the wild‐type enzyme, whereas the k cat values were slightly decreased, indicating that these mutations reduce the binding affinity for l‐lysine. The D212A mutation showed more effect on K m than the D315A mutation. This is probably because that D212 directly interacts with the ε‐amino group whereas D315 indirectly. The K m value of the double mutant variant increased 585‐fold and k cat/K m decreased more than 2,000‐fold to the wild‐type enzyme. These results indicate that D212 and D315 are important for recognition of l‐lysine. However, single mutation to D212 or D315 does not much affect substrate preference (Figure 4). Interestingly, the D212A/D315A double mutation increased relative activity for aromatic l‐amino acids (l‐phenylalanine, l‐tyrosine, and l‐tryptophan) and l‐arginine while reduced for l‐lysine (Figure 4, Tables 3 and 4). Thus, the D212A/D315A double mutation changed the substrate specificity of LysOX.
FIGURE 4.
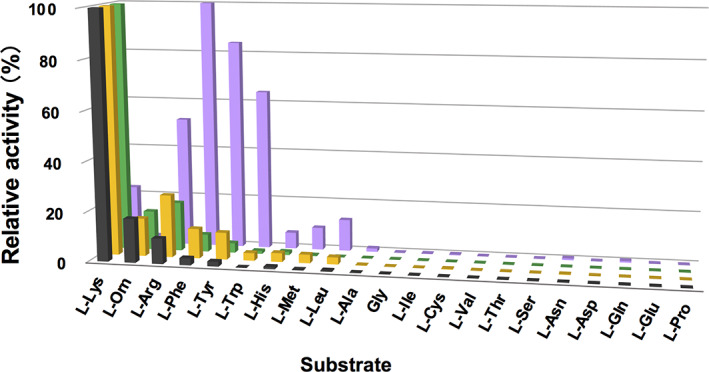
Comparison of substrate specificity profiles of l‐lysine oxidase (LysOX) and its variants. Relative oxidative activities of wild‐type, D212A, D315A, and D212A/D315A LysOX are indicated in black, yellow, green, and purple bars, respectively. The substrate that showed the highest activity was set as 100% relative activity. Enzyme activities were measured with 1 mM substrates at 40°C and pH 7.4
TABLE 4.
Kinetic parameters of LysOX D212A/D315A for various substrates
| Substrate | Specific activity (U/mg) | K m (mM) | k cat (s−1) | k cat/K m (s/mM) |
|---|---|---|---|---|
| l‐Arg | 14.3 | 5.6 | 30.1 | 5.4 |
| l‐Phe | 20.5 | 3.8 | 36.7 | 9.6 |
| l‐Tyr | 13.4 | 1.9 | 18.5 | 9.7 |
| l‐Trp | 10.0 | 1.0 | 15.3 | 15.3 |
Abbreviation: LysOX, l‐lysine oxidase.
The Lys‐H2O‐flavin N5 motif is conserved in various amine and amino acid oxidases and is proposed to be important for the hydrolytic attack on the imino‐intermediate 18 (Figure 5). The lysine residue is thought to increases the nucleophilicity of the water molecule, although no experimental evidence has been reported yet. 10 , 18 The Lys‐H2O‐flavin N5 motif is conserved in the ligand free LysOX structure, but the water molecule is lost in the LysOX‐Lys structure. M67 adopts an alternative conformation in the crystal and one of the side chains occupies the position of the water molecule. M67 is well conserved in various LAAOs, thus M67 may be involved in the regulation of the reaction (Figure 5). We prepared the M67A mutant variant of LysOX (LysOX M67A) and examined its enzyme activity. The activity of LysOX M67A was greatly reduced (Table 2) and was too low to determine the kinetic parameters for l‐lysine, suggesting that M67 is involved in the reaction of LAAOs. However, further experiments are needed to clarify the role of M67 in the enzymatic reaction.
FIGURE 5.
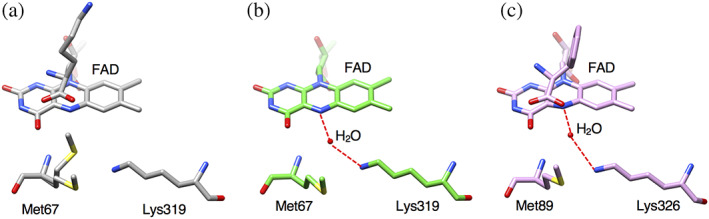
Structural comparison around the Lys‐H2O‐flavin N5 motif in LAAO. (a) The structure of l‐lysine oxidase (LysOX)‐Lys. The nucleophilic water molecule is lost in LysOX‐Lys. Instead, one of the side chains of M67 alternative conformation occupies the corresponding position. (b) The structure of ligand‐free LysOX. LysOX conserves the Lys‐H2O‐flavin N5 motif in the ligand‐free form. (c) The structure of Cr‐LAAO in complex with l‐phenylalanine shows conserved Lys‐H2O‐flavin N5 motif. Possible hydrogen bonds are indicated by red broken lines
2.6. Structure of the LysOX D212A/D315A variant and its substrate complex
To elucidate the structural basis of the effect of the D212A/D315A mutation on the substrate specificity of LysOX, we determined the crystal structures of the D212A/D315A mutant LysOX (LysOX D212A/D315A) and its l‐phenylalanine and l‐tyrosine complexes. The substrate complex crystals of the mutant LysOX were prepared by the soaking method. LysOX D212A/D315A crystals belong to the same space group as the LysOX‐Lys crystal with similar cell dimensions and molecular packing (Table 1). The LysOX D212A/D315A structure is almost the same as that of wild‐type LysOX with the RMSD value for corresponding Cα atoms of 0.18 Å. The mutation does not affect the conformation of the residues in the substrate‐binding pocket except for W371, which interacts with the hydrophobic side chain arm of l‐lysine in LysOX. The side chain of W371 adopts an alternative conformation in the substrate‐binding site of LysOX D212A/D315A (Figure 6a,b). One conformation resembles W371 in ligand‐free LysOX, and therefore the gate to the active site is open (Figure 6a). The other conformation shows a T‐stacking interaction with F439, and the gate is fully closed (Figure 6b). Thus, the gate to the active site sometimes closes in the ligand‐free state of LysOX D212A/D315A.
FIGURE 6.
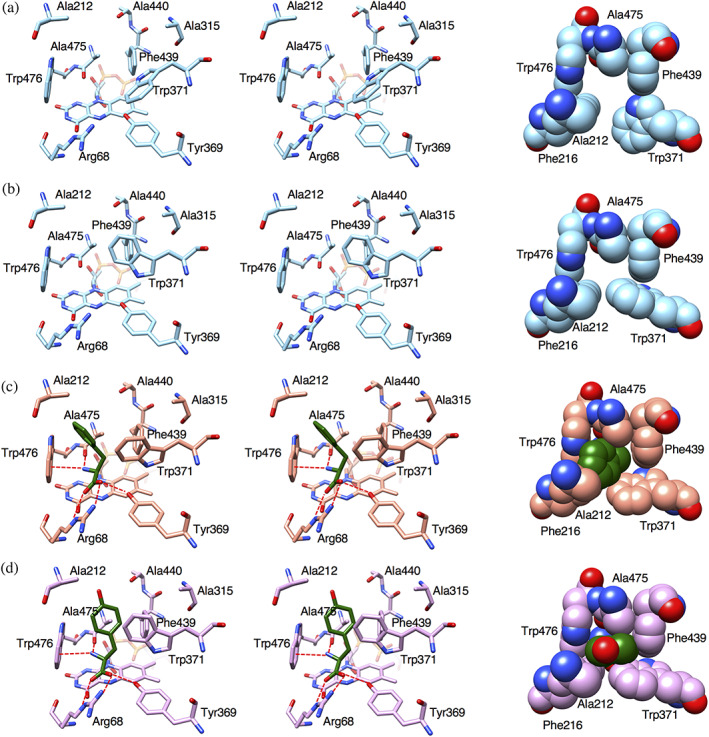
Structures of the substrate‐binding site of l‐lysine oxidase (LysOX) D212A/D315A variant and its substrate complexes. (a,b) W371 adopts two conformations; the gate‐open form (a) and the gate‐closed form (b). (c) Substrate‐binding site of LysOX D212A/D315A in complex with l‐phenylalanine. (d) Substrate‐binding site of LysOX D212A/D315A in complex with l‐tyrosine. Stereo views of the substrate‐binding sites are shown in stick representation in left and middle panels. The hydrophobic holes are shown in ball representation in right panel. The substrate molecules are colored in green. The nitrogen and oxygen atoms are colored in blue and red, respectively. Possible hydrogen bonds are indicated by red broken lines
The substrate complex structures of LysOX D212A/D315A revealed that the aromatic side chain rings of l‐phenylalanine and l‐tyrosine are accommodated in the hydrophobic hole formed by F216 W371 F439, A475, and W476, like the hydrophobic arm of the l‐lysine side chain in the LysOX‐Lys structure. However, the ring conformations of both substrates are totally different (Figure 6c,d). The aromatic ring of l‐phenylalanine directly contacts with Cβ of A212 whereas it has no direct contact with F439. The aromatic ring plane is nearly parallel to the W476 ring and perpendicular to the W371 ring (Figure 6c). In contrast, the aromatic ring plane of l‐tyrosine turns 90° from that of l‐phenylalanine. Thus, the ring plane is nearly perpendicular to the W476 ring and parallel to the W371 ring. The ring does not contact with Cβ of A212 but closely contacts with F439 (Figure 6d). l‐Tyrosine is unable to adopt the same conformation as l‐phenylalanine due to steric hindrance between the hydroxy group of l‐tyrosine and the methyl group of A212. Thus, l‐tyrosine and l‐phenylalanine show different conformation in the active site.
l‐Tyrosine is more closely packed in the hydrophobic hole than l‐phenylalanine. This is consistent that K m for l‐tyrosine is twice of that for l‐phenylalanine (Table 4). However, the backbone atoms of l‐phenylalanine are almost in the same positions as those of l‐lysine bound in LysOX whereas those of l‐tyrosine are in slightly different positions. The LysOX‐Lys structure showed that the distance between the carbonyl oxygen (O) of A475 and the backbone nitrogen (N) of the substrate is 2.69 Å, the angle defined by O, N, and Cα of the substrate is 118.1°, and the torsion angle defined by O, N, Cα and the carboxyl carbon atom of the substrate backbone is −174.9°. The corresponding distance and angles of the l‐phenylalanine complex are 2.66 Å, 121.7°, and −173.1°, and those of the l‐tyrosine complex are 2.73 Å, 113.3°, and −151.9°, indicating that the amino acid backbone of l‐phenylalanine adopts a very similar orientation to l‐lysine in the active site whereas that of l‐tyrosine is slightly distorted. Therefore, l‐phenylalanine has a more favorable orientation for enzymatic reaction than l‐tyrosine. This is consistent that k cat for l‐phenylalanine is twice of that for l‐tyrosine (Table 4).
3. DISCUSSION
Unlike other LAAOs, LysOX displays strict substrate specificity for l‐lysine. 3 The K m and k cat/K m values of LysOX for l‐lysine are 0.013 mM and 4,700 s/mM, respectively, whereas those of Cr‐LAAO are 10 mM and 0.165 s/mM, respectively. 19 Our substrate complex structure has revealed how LysOX exerts high specificity for l‐lysine. A narrow hydrophobic hole formed by F216 W371 F439, A475, and W476 and the acidic surface produced by D212 and D315 are responsible for the strict side chain recognition (Figure 2). The narrow hydrophobic hole prefers binding of l‐amino acids with long linear aliphatic side chains, such as l‐lysine, l‐arginine, l‐ornithine, and l‐methionine. The acidic surface above the hydrophobic hole accepts basic amino acid side chains but refuses others. Therefore, l‐lysine, l‐arginine, and l‐ornithine can be substrates for LysOX, but hydrophobic l‐methionine cannot (Figure 4). The triangular arrangement of oxygen atoms of D212 and the two water molecules best fits the protonated ε‐amino group of l‐lysine, whereas the guanidino group of l‐arginine may not form tight interactions with these atoms. The side chain amino group of l‐ornithine is a little too far to make tight interaction with D212 because its side chain arm is shorter than l‐lysine. Thus, the structure of the substrate‐binding site of LysOX is optimized for l‐lysine binding. LysOX does not react with amino acids with small side chains. This is probably because they are not firmly fixed in the ligand‐binding site with the proper orientation for the reaction.
Recently, the ligand complex structure of laao/monooxygenase (MOG) from Pseudomonas sp. AIU 813 has been determined. 20 L‐AAO/MOG shows strict substrate specificity for l‐lysine, similar to LysOX but rather high relative activities for other positively charged l‐amino acids, such as l‐arginine and l‐ornithine, compared with LysOX. L‐AAO/MOG has an aspartic acid (D238) that directly interacts with ε‐amino group of l‐lysine in the substrate‐binding pocket. D238 lies above the ε‐amino group of l‐lysine, whereas D212 of LysOX locates on the side and surrounds the ε‐amino group with two water molecules. Although some water molecules are present near the ε‐amino group in the L‐AAO/MOG structure, they are not triangularly arranged. Therefore, LysOX shows strict specificity for l‐lysine more than L‐AAO/MOG.
The D212A/D315A double mutation increased activity for amino acids with aromatic rings. The structures of the variant revealed that the mutation does not largely change the conformation of the residues in the substrate‐binding pocket. A212 directly interacts with the aromatic ring of l‐phenylalanine but has no interaction with l‐tyrosine. Both substrates have no interaction with A315. These facts suggest that direct hydrophobic interaction is not the primary factor for the increased activity for aromatic amino acids. The hydrophobic hole of LysOX can accommodate the aromatic side chain by changing the W371 conformation caused by rotation of the Chi2 angle (Figure 7), although it may not be well adjusted to the aromatic amino acids. However, the negatively charged surface around the substrate‐binding site produced by D212 and D315 may not allow the binding of aromatic amino acids in the wild‐type LysOX. The double mutation eliminated the negative charge and enabled the binding of the aromatic amino acids.
FIGURE 7.
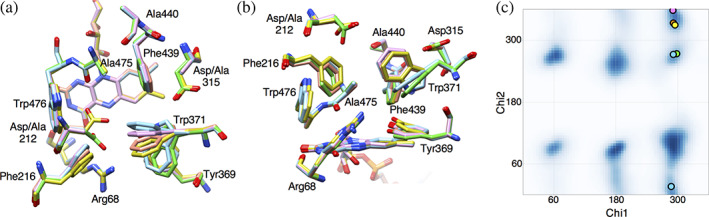
Conformational change of W371. (a) Superimposition of the substrate‐binding site of l‐lysine oxidase (LysOX), LysOX D212A/D315A and their substrate complexes. The nitrogen and oxygen atoms are colored in blue and red, respectively. The carbon atoms are color coded: ligand‐free LysOX, green; LysOX‐Lys, yellow; ligand‐free LysOX D212A/D315A, cyan; the l‐phenylalanine complex of LysOX D212A/D315A, orange; and the l‐tyrosine complex of LysOX D212A/D315A, magenta. (b) View from the bottom of (a). (c) The Chi1–Chi2 plot of W371. The favorable angle regions for tryptophan are shaded in blue. The Chi1 and Chi2 angles of W371 are plotted with circles with the same color code as (a)
The LAAOs of snake venom show broad substrate specificity for l‐amino acids with aromatic or hydrophobic side chains. 21 The snake LAAOs, such as CrLAAO, LAAO from Agkistrodon halys pallas venom, LAAO from Bothrops jararacussu venom, and LAAO from Vipera ammodytes ammodytes venomic, also have a hydrophobic hole in the ligand‐binding site but its size is wider than that of LysOX because some residues that form the hydrophobic hole are replaced by small residues. 11 , 12 , 13 , 14 Although the residues corresponding to F216 and W476 of LysOX are conserved in these LAAOs, W371 and F439 are replaced by isoleucine, and A475 is replaced by glycine. As a result, the relatively wide hydrophobic hole of the snake LAAOs prefers amino acids with bulky side chains. 11 , 12 , 13 , 14
The color of the l‐lysine complex crystals was colorless, indicating that the FAD is reduced form and retains the reduced state with the substrate. A similar phenomenon was observed in the CrLAAO‐Phe and RoLAAO‐Ala crystals. 12 , 17 The concentration of l‐lysine was 50 mM, which is much higher than the concentration of oxygen dissolved in the solution (0.3 mM). This condition is very similar to that of the CrLAAO‐Phe crystal. Therefore, as discussed by Moustafa et al., the substrate may be bound to LysOX before reoxidation of FAD because of high substrate concentration. Structural comparison of the CrLAAO with its substrate complex revealed that the reduced form of the isoalloxazine ring of FAD is more bent than the oxidized form. 12 Although the bending angle was much smaller than that of CrLAAO, LysOX also showed more bending in the ligand‐binding form than that in the ligand free form, supporting that FAD is in the reduced form in the LysOX‐Lys crystal. The N, Cβ, and C atoms of the bound ligand in the CrLAAO‐Phe structure are not in the same plane with the Cα atom but arrange in a near‐tetrahedral geometry around the Cα atom, indicating that the ligand is not in imino form but in amino form in the ligand complex structure. 12 The corresponding atoms of the ligand in the LysOX‐Lys crystal adopt tetrahedral geometry. Thus, the ligand is in amino form in the LysOX‐Lys crystal.
4. MATERIALS AND METHODS
4.1. Expression and purification of recombinant LysOX
Recombinant LysOX is expressed in S. lividans TK24 and purified as the mature form as previously described. 8
4.2. Preparation of LysOX crystals
The crystals were grown in a high‐concentration agarose gel according to the method previously described. 15 Agarose powder (SeaPlaque agarose, Lonza, Switzerland) was added to the water with a final concentration of 4% (wt/vol), stirred at room temperature and completely dissolved at 373 K. The agarose solution was then cooled down to gelate and stored at 277 K until use it. The 4% (wt/vol) agarose gel was melted at 316 K and mixed with LysOX solution (7.1 mg/ml, 20 mM HEPES pH 7.1) in the ratio of 1:1, and the mixture was immediately sucked into a glass capillary. The capillary was placed in a vessel with a reservoir solution containing 2 M ammonium sulfate, 4% (vol/vol) PEG400, and 0.1 M HEPES at pH 7.0, and the vessel was sealed and stored at 293 K for 2 months.
4.3. Preparation, data collection, and structure determination of the LysOX‐Lys crystal
The gel containing LysOX crystals was pulled out from the capillary and cut. The crystals in the gel were soaked in a solution prepared by mixing the reservoir solution containing 50 mM l‐lysine with 10% glycerol for 2.5 hr. The crystals were then transferred into liquid nitrogen for freezing. The diffraction data were collected at Spring‐8 beamline BL41XU with the approval of the Japan Synchrotron Radiation Research Institute (Proposal Nos. 2014A1388 and 2017A2585) under 90 K nitrogen gas flow. The data were processed and scaled using MOSFLM 22 and SCALA 23 from the CCP4 program suite, respectively. The structure was determined by molecular replacement using the structure of LysOX (PDB ID: 3X0V) as a search model with Phaser. 24 The molecular model was built with Coot 25 and refined to 1.7 Å resolution with PHENIX. 26 The statistics of the data collection and the structure refinement are summarized in Table 1.
4.4. Enzyme assay
The enzyme activity of LysOX was measured by detecting hydrogen peroxide using the color development method with 4‐aminoantipyrine, phenol, and horseradish peroxidase. 27 One unit of enzyme activity was defined as the amount of enzyme catalyzing the formation of 1 μmol of hydrogen peroxide per minute. The assay mixture contained 100 mM KPB at pH 7.4, 10 mM l‐lysine, 300 U·ml−1 horseradish peroxidase, 15 mM 4‐aminoantipyrine, 50 mM phenol and an appropriate amount of enzyme in a total volume of 1.0 ml. The reaction was monitored by measuring absorbance at 505 nm using a spectrophotometer (Shimadzu UV‐1240, Shimadzu Corp., Japan) at 40°C for 3 min. Protein concentration was determined with Bio‐Rad Protein Assay (Bio‐Rad) using BSA as a standard. The apparent kinetic parameters, k cat and K m, were calculated from reaction rates determined at various concentrations of substrate l‐amino acids.
4.5. Construction of the plasmid for expression in E. coli
The codon usage of the lysox gene encoding LysOX of T. viride was optimized for expression in E. coli (Figure S1). The codon‐optimized gene, lysox3, was synthesized by GeneScript Inc. (Tokyo). lysox3 was cloned into the Nde I/EcoR I‐digested pCold IV, yielding an expression plasmid, pCold IV‐lysox3. The lysox3 gene was under the control of the promoter of the cold shock gene (cspA) and the lac operator inserted downstream of the cspA promoter.
4.6. Site‐directed mutagenesis
Site‐directed mutagenesis was conducted by inverse PCR using a pair of oligonucleotide primers containing a point mutation and pCold IV‐lysox3. The PCR program was as follows: 25 cycles of 10 s at 98°C, 30 s at 61°C, and 3.5 min at 68°C. The PCR products were treated with DpnIat 37°C for 1.5 hr to digest the template. Then the PCR products were transfected into competent cells of E. coli Top10. The transformants were cultivated at 37°C with shaking at 230 rpm in Luria–Bertani medium containing 50 μg/ml ampicillin, and the plasmids were extracted from the cells. After confirmation of the sequence, the extracted plasmids were transfected into E. coli SoluBL21 as a host strain for heterologous expression in E. coli.
4.7. Expression and purification of recombinant LysOX and its variants in E. coli
E. coli SoluBL21 cells harboring expression plasmid encoding wild‐type or each mutant LysOX protein were cultured in 20 ml of 2× yeast extract‐tryptone (YT) medium supplemented with 50 μg/ml ampicillin at 37°C with shaking at 230 rpm for 16 hr. Then, 10 ml culture was inoculated into 1.2 L of 2 × YT medium supplemented with 50 μg/ml ampicillin. Cells were cultivated at 37°C with shaking at 230 rpm until the culture density reached OD600 of around 0.4. The expression of LysOX was induced by incubation at 15°C for 30 min followed by the addition of 1 mM IPTG. The culture was continued for 24 hr at 15°C with shaking at 180 rpm. Cells were harvested by centrifugation (10,000g for 20 min at 4°C), suspended in 20 mM KPB at pH 7.4, and disrupted by sonication on ice. After removal of the cell debris by centrifugation (10,000g for 20 min at 4°C), metalloprotease from Streptomyces griseus (Sigma‐Aldrich, USA) (50 mg/ml) was added to the supernatant in 1/50 amount of the protein to cleave the prosequence. The reactant was stored at 37°C for 16 hr and then heated at 60°C for 15 min to inactivate the protease, because the enzyme was stable at 60°C for, at least, 15 min. After centrifugation at 10,000g for 20 min at 4°C, ammonium sulfate was added at 65% saturation to the supernatant, and the solution was stored at 4°C for 30 min. After centrifugation at 10,000g for 20 min at 4°C, the precipitate was dissolved in 20 mM KPB (pH 7.4) and dialyzed against 20 mM KPB (pH 7.4) containing ammonium sulfate at 20% saturation. The protein solution was loaded on a Butyl‐Toyopearl 650M column (Tosoh Bioscience, Japan) equilibrated with 20 mM KPB (pH 7.4) containing 20% saturated ammonium sulfate. The bound protein was eluted with 20 mM KPB at pH 7.4 containing 10% saturated ammonium sulfate. The enzymatically active fractions were concentrated using Amicon Ultra centrifugal filters (30 kDa cut‐off) (Merck Millipore), and dialyzed against 20 mM KPB (pH 7.4). The dialyzed solution was applied to a DEAE‐Toyopearl 650M column (Tosoh Bioscience, Japan) equilibrated with 20 mM KPB (pH 7.4). The bound protein was eluted with 20 mM KPB (pH 7.4) containing 200 mM NaCl. The enzymatically active fractions were concentrated using Amicon Ultra centrifugal filters. The purity of the protein was examined by sodium dodecyl sulfate polyacrylamide gel electrophoresis.
4.8. Crystallization, data collection, and structure determination of LysOX (D212A/D315A) and its substrate complexes
Crystallization was carried out using the sitting‐drop vapor‐diffusion method. Crystallization drops were prepared by mixing the protein solution (3.1 or 7.5 mg/ml) with an equal volume of reservoir solution. Crystals were grown at 20°C from drops prepared by mixing 1 μl protein solution (3.1 or 7.5 mg/ml) containing 20 mM potassium phosphate at pH 7.4 with the equivalent volume of reservoir solution containing 2–2.1 M ammonium sulfate, 1% (vol/vol) PEG400 and 0.1 M Tris–HCl (pH 7.5) or HEPES (pH 7.4). The crystals belong to the space group C2221 with unit cell dimensions of a = 116.5, b = 170.3, and c = 119.6 Å.
The substrate complex crystals were prepared by the soaking method. The double mutant variant crystals were transferred into a reservoir solution containing 10% (vol/vol) glycerol and 15 mM l‐phenylalanine or 2.4 mM l‐tyrosine and were stored at 20°C until the yellow color disappeared (typically 5 min).
X‐ray diffraction data were collected at Spring‐8 (Harima, Japan) beamline BL41XU (Proposal No. 2017A2585). Crystals were frozen in liquid nitrogen and mounted in nitrogen gas flow at 100 K. The diffraction data were processed with MOSFLM 22 and were scaled with AIMLESS. 28 The statistics of the diffraction data are summarized in Table 1. The structures were determined by the molecular replacement method using the structure of wild‐type LysOX (PDB ID: 3X0V) as a search model with Phaser. 24 The atomic models were built with Coot 25 and refined to 2.3, 2.2, and 1.8 Å resolution for LysOX (D212A/D315A), its l‐phenylalanine complex and l‐tyrosine complex, respectively, with PHENIX. 26 The refinement statistics are summarized in Table 1.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
AUTHOR CONTRIBUTIONS
Katsumi Imada and Kenji Inagaki designed research; Masaki Kitagawa, Hiroki Kondo, Yuya Matsumoto, Masaya Saito, Marie Amano, Takashi Tamura, Hitoshi Kusakabe, and Shigeru Sugiyama performed experiments, Masaki Kitagawa, Hiroki Kondo, and Katsumi Imada analyzed the crystal structure; and Katsumi Imada and Kenji Inagaki wrote the paper.
Supporting information
Figure S1 The nucleotide sequence of the codon‐optimized gene of LysOX, lysox3. The substituted nucleotides are shown in red.
ACKNOWLEDGMENTS
The authors thank Spring‐8 beamline staffs for technical help in use of beamlines. This work was supported in part by JSPS KAKENHI grant numbers 24560962 (to K. Inagaki) and 25286051 (to S. S.).
Kondo H, Kitagawa M, Matsumoto Y, et al. Structural basis of strict substrate recognition of l‐lysine α‐oxidase from Trichoderma viride . Protein Science. 2020;29:2213–2225. 10.1002/pro.3946
Hiroki Kondo, Masaki Kitagawa, and Yuya Matsumoto contributed equally to this work.
Enzymes: l‐Lysine α‐oxidase [EC 1.4.3.14].
Databases: The atomic coordinates have been deposited in Protein Data Bank, www.pdb.org.
Funding information Japan Society for the Promotion of Science KAKENHI, Grant/Award Numbers: 24560962, 25286051
REFERENCES
- 1. Pollegioni L, Motta P, Molla G. l‐Amino acid oxidase as biocatalyst: A dream too far? Appl Microbiol Biotechnol. 2013;97:9323–9341. [DOI] [PubMed] [Google Scholar]
- 2. Yu Z, Qiao H. Advances in non‐snake venom l‐amino acid oxidase. Appl Biochem Biotechnol. 2012;167:1–13. [DOI] [PubMed] [Google Scholar]
- 3. Kusakabe H, Kodama K, Kuninaka A, Yoshino H, Misono H, Soda K. A new antitumor enzyme, l‐lysine oxidase from Trichoderma viride. Purification and enzymological properties. J Biol Chem. 1980;255:976–981. [PubMed] [Google Scholar]
- 4. Jung SK, Mai A, Iwamoto M, et al. Purification and cloning of an apoptosis‐inducing protein derived from fish infected with Anisakis simplex, a causative nematode of human Anisakiasis. J Immunol. 2000;165:1491–1497. [DOI] [PubMed] [Google Scholar]
- 5. Nagashima Y, Kikuchi N, Shimakura K, Shiomi K. Purification and characterization of an antibacterial protein in the skin secretion of rockfish Sebastes schlegeli . Comp Biochem Physiol C Toxicol Pharmacol. 2003;36:63–71. [DOI] [PubMed] [Google Scholar]
- 6. Yang H, Johnson M, Ko C, et al. Cloning, characterization and expression of escaping, a broadly antimicrobial FAD‐containing l‐amino acid oxidase from ink of the sea hare Aplysia californica . J Exp Biol. 2005;208:3609–3622. [DOI] [PubMed] [Google Scholar]
- 7. Tani Y, Omatsu K, Saito S, et al. Heterologous expression of L‐lysine a‐oxidase from Scomber japonicus in Pichia pastoris and functional characterization of the recombinant enzyme. J Biochem. 2015;157:201–210. [DOI] [PubMed] [Google Scholar]
- 8. Amano M, Mizuguchi H, Sano T, et al. Recombinant expression, molecular characterization and crystal structure of antitumor enzyme, l‐lysine α‐oxidase from Trichoderma viride . J Biochem. 2015;157:549–559. [DOI] [PubMed] [Google Scholar]
- 9. Pohlmann A, Stamn W, Kusakabe H, Kula M‐R. Enzymatic determination of l‐lysine by flow‐injection techniques. Analyt Chim Acta. 1990;235:329–335. [Google Scholar]
- 10. Pawelek PD, Cheah J, Coulombe R, Macheroux P, Ghisla S, Vrielink A. The structure of l‐amino acid oxidase reveals the substrate trajectory into an enantiomerically conserved active site. EMBO J. 2000;19:4204–4215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhang H, Teng M, Niu L, et al. Purification, partial characterization, crystallization and structural determination of AHP‐LAAO, a novel l‐amino‐acid oxidase with cell apoptosis‐inducing activity from Agkistrodon halys pallas venom. Acta Crystallogr. 2004;D60:974–977. [DOI] [PubMed] [Google Scholar]
- 12. Moustafa IM, Foster S, Lyubimov AY, Vrielink A. Crystal structure of LAAO from Calloselasma rhodostoma with an l‐phenylalanine substrate: Insights into structure and mechanism. J Mol Biol. 2006;364:991–1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ullah A, Souza TA, Abrego JR, Betzel C, Murakami MT, Arni RK. Structural insights into selectivity and cofactor binding in snake venom l‐amino acid oxidases. Biochem Biophys Res Commun. 2012;421:124–128. [DOI] [PubMed] [Google Scholar]
- 14. Georgieva D, Murakami M, Perband M, Arni R, Betzel C. The structure of a native l‐amino acid oxidase, the major component of the Vipera ammodytes ammodytes venomic, reveals dynamic active site and quaternary structure stabilization by divalent ions. Mol Biosyst. 2011;7:379–384. [DOI] [PubMed] [Google Scholar]
- 15. Arima J, Sasaki C, Sakaguchi C, et al. Structural characterization of l‐glutamate oxidase from Streptomyces sp. X‐119‐6. FEBS J. 2009;276:3894–3903. [DOI] [PubMed] [Google Scholar]
- 16. Sugiyama S, Maruyama M, Sazaki G, et al. Growth of protein crystals in hydrogels prevents osmotic shock. J Am Chem Soc. 2012;134:5786–5789. [DOI] [PubMed] [Google Scholar]
- 17. Faust A, Niefind K, Hummel W, Schomburg D. The structure of a bacterial l‐amino acid oxidase from Rhodococcus opacus gives new evidence for the hydride mechanism for dehydrogenation. J Mol Biol. 2007;367:234–248. [DOI] [PubMed] [Google Scholar]
- 18. Binda C, Mattevi A, Edmondson DE. Structure‐function relationships in flavoenzyme‐dependent amine oxidations: A comparison of polyamine oxidase and monoamine oxidase. J Biol Chem. 2002;277:23973–23976. [DOI] [PubMed] [Google Scholar]
- 19. Ponnudurai G, Chung MC, Tan NH. Purification and properties of the l‐amino acid oxidase from Malayan pit viper (Calloselasma rhodostoma) venom. Arch Biochem Biophys. 1994;313:373–378. [DOI] [PubMed] [Google Scholar]
- 20. Im D, Matsui D, Arakawa T, Isobe K, Asano Y, Fushinobu S. Ligand complex structures of l‐amino acid oxidase/monooxygenase from Pseudomonas sp. AIU 813 and its conformational change. FEBS Open Bio. 2018;8:314–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Du XY, Clemetson KJ. Snake venom l‐amino acid oxidases. Toxicon. 2002;40:659–665. [DOI] [PubMed] [Google Scholar]
- 22. Battye TGG, Kontogiannis L, Johnson O, Powell HR, Leslie AGW. iMOSFLM: A new graphical interface for diffraction image processing with MOSFLM. Acta Cryst. 2011;D67:271–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Evans P. Scaling and assessment of data quality. Acta Crystallogr. 2006;D62:72–82. [DOI] [PubMed] [Google Scholar]
- 24. McCoy AJ, Grosse‐Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Cryst. 2007;40:658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of coot. Acta Crystallogr. 2010;D66:486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Adams PD, Afonine PV, Bunkóczi G, et al. PHENIX: A comprehensive Python‐based system for macromolecular structure solution. Acta Crystallogr. 2010;D58:1948–1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Job V, Marcone GL, Pilone MS, Pollegioni L. Glycine oxidase from Bacillus subtilis . J Biol Chem. 2002;277:6985–6993. [DOI] [PubMed] [Google Scholar]
- 28. Evans PR, Murshudov GN. How good are my data and what is the resolution? Acta Cryst. 2013;D69:1204–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 The nucleotide sequence of the codon‐optimized gene of LysOX, lysox3. The substituted nucleotides are shown in red.


