Abstract
Bacterial microcompartments are protein‐based organelles that carry out specialized metabolic functions in diverse bacteria. Their outer shells are built from several thousand protein subunits. Some of the architectural principles of bacterial microcompartments have been articulated, with lateral packing of flat hexameric BMC proteins providing the basic foundation for assembly. Nonetheless, a complete understanding has been elusive, partly owing to polymorphic mechanisms of assembly exhibited by most microcompartment types. An earlier study of one homologous BMC shell protein subfamily, EutS/PduU, revealed a profoundly bent, rather than flat, hexameric structure. The possibility of a specialized architectural role was hypothesized, but artifactual effects of crystallization could not be ruled out. Here we report a series of crystal structures of an orthologous protein, CutR, from a glycyl‐radical type choline‐utilizing microcompartment from the bacterium Streptococcus intermedius. Depending on crystal form, expression construct, and minor mutations, a range of novel quaternary architectures was observed, including two spiral hexagonal assemblies. A new graphical approach helps illuminate the variations in BMC hexameric structure, with results substantiating the idea that the EutS/PduU/CutR subfamily of BMC proteins may endow microcompartment shells with flexible modes of assembly.
Keywords: bacterial microcompartment, bacterial organelle, carboxysome, choline, glycyl radical, polymorphism, shell protein, symmetry breaking
1. INTRODUCTION
Nearly 20% of bacteria species produce giant protein‐based organelles that are used for carrying out sensitive metabolic reactions in a sequestered cellular environment. 1 These extraordinary structures, known as bacterial microcompartments or MCPs (or alternatively BMCs), encapsulate distinct enzyme types in different bacteria. 1 , 2 , 3 , 4 Generally, a small metabolic intermediate (either volatile, toxic, or both) is produced within the MCP and is further metabolized before it can escape into the cytosol or out of the cell entirely. 2 , 5 , 6 , 7 These key metabolic intermediates include CO2 for the case of the carboxysome, the founding member of the MCP family, while various aldehyde intermediates occur in other types of MCPs, including the type under investigation in the present study. Comprised of thousands of protein subunits, MCPs are among the largest and most mechanistically sophisticated protein assemblies to have evolved in nature. 8 , 9
A protein shell, reminiscent of a viral capsid, is the hallmark of MCPs (Figure 1). Different MCP types, even those with divergent metabolic functions, are all assembled from homologous shell proteins. The bulk of the MCP shell is composed of proteins containing a roughly 100‐amino acid BMC protein domain (Pfam PF00936). The recognition that this protein family (first identified by Shively and coworkers 10 ) is encoded across diverse bacterial operons has provided a bioinformatics basis for exploring MCP function and evolution. 1 , 4 , 11 , 12 It is now understood that BMC proteins assemble to form nearly flat faces of the MCP shell, while a second minor and distinct protein (known as a BMV protein, Pfam PF03319) forms vertices. 13 , 14 , 15 , 16 , 17 Multiple paralogs of BMC proteins, typically ranging from two to seven in number, are present in different systems. Different BMC protein paralogs offer modularity in the shell. Some BMC proteins provide pores for substrate and product diffusion across the shell, some are believed to bind and organize interior enzymes, some may serve specialized architectural roles, while others may have yet unrecognized functions.
FIGURE 1.
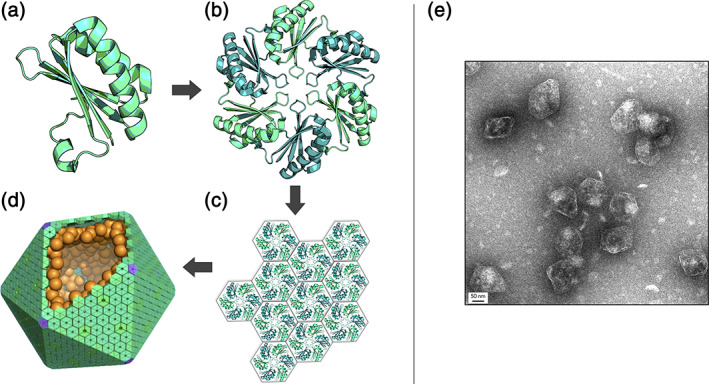
Microcompartment shells are composed of homologous hexameric proteins. For all bacterial microcompartments, the canonical BMC domain (a) oligomerizes to form the traditional flat hexamer (b). These hexameric shell proteins tessellate (c) to form the nearly flat faces of bacterial microcompartments shells. (d) An idealized model of a microcompartment with hexameric BMC proteins that form the face (teal), pentameric BMV proteins that form the vertices (purple), and internal encapsulated enzymes (orange). (e) Negative stain EM of purified Pdu microcompartments (scale bar: 50 nm)
Structural studies on MCP proteins have shed light on architectural and functional mechanisms, with the basic principles emerging from crystallographic studies beginning fifteen years ago. 18 , 19 BMC proteins of the canonical type form flat hexameric units; these are the building blocks for shell assembly. Typically, BMC hexamers possess narrow pores for molecular transport. These hexamers tile side by side to form extended facets of nearly solid protein perforated by small holes. A remarkable feature of BMC shell proteins is the wide range of structural rearrangements that have occurred through evolution, thereby conferring distinct properties on multiple BMC paralogs within an operon. Notable variations include circular permutations of the chain to produce BMC versions with termini in different locations, 20 versions bearing tandem BMC domains, 16 , 21 , 22 , 23 versions bearing iron–sulfur clusters and other small molecules in their central pores, 24 , 25 , 26 , 27 and versions fused to other protein domains. 23 Biochemical, genetic, and engineering studies have provided additional insights into the prospective roles of different BMC shell proteins, 16 , 28 , 29 , 30 , 31 , 32 , 33 but much remains unknown about separate and distinct (or even redundant) functions that might be attributed to different BMC paralogs. Functional distinctions between BMC shell proteins may furthermore vary between different MCP types, based on differences in metabolic purpose, BMC paralog composition, and MCP architecture; for example, some MCP shells, like the carboxysome, appear much more geometrically regular (i.e., icosahedral) (Figure 1d) than those that metabolize various organic metabolites (Figure 1e).
One type of BMC shell protein paralog of special interest is EutS/PduU (from the ethanolamine utilization and propanediol utilization MCPs, respectively). Proteins from this subfamily are a permuted version of the canonical BMC domain. By virtue of their permuted topology, EutS/PduU paralogs present distinct features in the form of a protruding, N‐terminal six‐stranded parallel beta‐barrel that occludes the central pore. Remarkably, an early crystallographic study of EutS from E. coli revealed a unique structure in which the hexamer had undergone an extraordinary twisting deformation of approximately 40°, 23 a stark contrast to other BMC hexamers that are very nearly flat, a general expectation for cyclic homooligomeric assemblies. This peculiar observation invoked the possibility of an important architectural role. However, other structures of this paralog (e.g., PduU and even a point mutant of EutS) revealed a typical flat hexameric assembly, 13 , 20 leaving uncertainty about the meaning and significance of the dramatic departure from the typical BMC structure. The relevance versus artifactual nature of the structural variation observed in EutS has not been revisited since that initial observation a decade ago. In the present work, we undertook a series of crystal structure investigations on an orthologous protein, CutR, from a different type of MCP from Streptococcus intermedius. We present further observations of major polymorphism in this shell protein subfamily and discuss implications for a specialized architectural role in certain bacterial metabolic organelles.
2. RESULTS
2.1. CutR crystallized as five unique structures
In order to investigate the structural polymorphism of a permuted BMC shell protein, we undertook a series of crystallographic studies of CutR, a BMC shell protein from the type 1 choline microcompartment (choline utilization or Cut MCP) from Streptococcus intermedius. When an initial structure revealed a completely unexpected, and likely artificial, dimeric structure, further mutagenesis and structural studies were pursued to dissect the possible effects of individual amino acids and terminal purification tags. We report five crystal structures obtained from four sequence variants. These structural forms include one novel dimer, two traditional flat hexamers, and two novel six‐fold screws of varying pitch. Geometric and computational analyses helped illuminate details of the observed quaternary forms.
The first structure we obtained for CutR revealed a novel dimeric arrangement not previously observed in other BMC proteins (Figure 2). A model was refined to a resolution of 1.8 Å with final Rwork/Rfree values of 0.176/0.210. Interestingly, this unexpected dimeric form of CutR crystallized from gel‐filtration fractions whose elution positions were consistent with that of a BMC hexamer (a molecular weight of approximately 80 kDa based on standard curves). In this unexpected structure, the two monomeric subunits came together, exchanging terminal segments in a fashion characteristic of domain swapping. 34 Moreover, we observed an unexpected intramolecular disulfide bond between C37 and C73 (Figure 2a). The disulfide bond pulls the first N‐terminal beta‐strand (residues G18‐A28) out of its anticipated location and into a position contacting the adjacent monomer (Figure 2a). This domain swap preserves the overall BMC tertiary structure. The N‐terminal beta‐strand from one monomer (residues G18‐A28) packs alongside the remaining residues (A29‐S116) of the second monomer, forming intermolecular interactions that results in a structure resembling the traditional BMC fold [0.50 Å RMSD backbone deviation (Figure 2b)].
FIGURE 2.
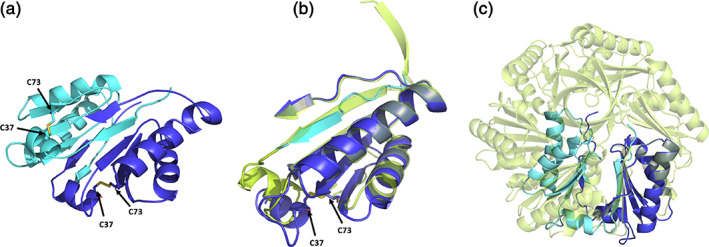
A comparison of a novel CutR dimeric form to the traditional flat hexameric form of the same protein. (a) Cartoon representation of the observed novel dimeric form. A disulfide bridge forms between C37 and C73, pulling the N‐terminus from its traditional position in the BMC domain, causing residues 18–28 to occupy the adjacent monomer by domain swapping. (b) Overlay of a monomer from the hexameric form (lime green) with residues 18–28 of one monomer (cyan) and residues 29–116 from the second monomer (deep blue), with emphasis on regions that differ. (c) Superimposition of one chain from the dimer with one chain from the flat hexamer shows that severe steric clashes would be caused by the presence of the dimeric form in the context of the hexameric assembly
While this dimeric architecture of CutR presented a new and intriguing structure, it could not be reconciled with the quaternary hexameric arrangement understood for BMC shell proteins or their packing in a layer. Aligning one monomer from the dimeric form with one monomer from the hexameric form results in severe steric clashes of their quaternary structures (Figure 2c). Notwithstanding the effects of an apparently adventitious disulfide bond, the unusual quaternary structure that we observed prompted further structural studies to look for potential modes of polymorphic assembly. We thus conducted structural studies on sequence variants of the protein as well as structural studies under reducing conditions. This work led to multiple structures with additional novel quaternary arrangements.
Motivated by the unconventional dimeric form, we pursued mutagenesis work to determine whether the circularly permuted CutR was capable of forming a traditional flat BMC hexamer. We obtained two distinct crystal forms of CutR, with subsequent structural analysis revealing canonical flat hexamers. The first crystal form referred to as Hexamer 1 (Figure 3a) crystallized in space group C2 and provided an atomic structure to a resolution of 2.6 Å with Rwork/Rfree values of 0.201/0.242. We also solved the structure of a second crystal form, referred to as Hexamer 2 (Figure 3b). Hexamer 2 crystallized in space group P42212 and provided a structure to a resolution of 1.5 Å with Rwork/Rfree values of 0.161/0.184. We obtained numerous crystals with varied morphologies including flat hexagonal plates and octahedral bi‐pyramidal forms. Importantly, two separate mutants of CutR each gave rise to traditional flat hexagonal BMC structures. The first mutant retained an N‐terminal His‐6 tag and had a C37A mutation. The second mutant contained a cleavable His‐6 tag and a K66A mutation, instead of the original K66D mutation (Table S1). Hexamer 2 was the most well behaved, presumably owing to the non‐polar residue at position 66 (discussed subsequently) located at the edge and to the lack of obstruction from the N‐terminal His‐6 tag. This version purified with relative ease and formed numerous diffraction quality crystals within 1 month of setting up broad screen crystallization trials. Crystals that formed in 2 M ammonium sulfate, 0.2 M lithium sulfate, and 0.1 M CAPS/sodium hydroxide pH 10.5 had the strongest diffraction pattern and the highest resolution, ultimately giving rise to the Hexamer 2 structure. As with other circularly‐permuted BMC domains, specifically PduU 20 (PDB ID 3CGI) and EutS 23 (PDB ID 3I96), we observed a beta‐barrel on the flat face created by the protruding N‐termini of the six chains (Figure 3a,b). The interior of the beta‐barrel is decorated with the side chains of alternating residues I11, Q13, and S15. As with PduU, 20 the bulky side chains of the interior‐lining residues are unable to occupy symmetry‐equivalent positions. Specifically, the six instances of Q13 adopt two conformations: pointing up toward the N‐terminus and down toward the center of the hexameric disk.
FIGURE 3.
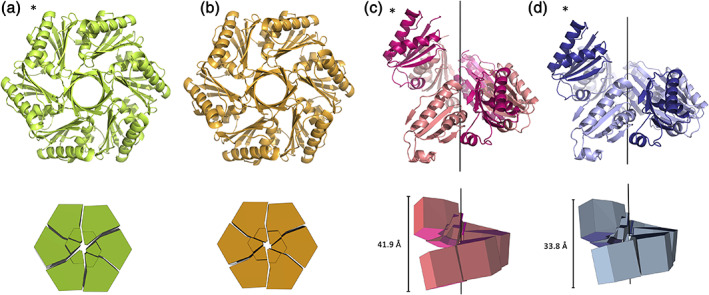
Cartoon and geometric representation of four CutR polymorphs. CutR crystallized as a traditional flat BMC hexamer (a and b) and formed two distinct screws with varying pitch (c and d). The flat hexamers are viewed down the six‐fold axis of symmetry (a and b) and the screws are displayed on their sides to show their pitch (c and d). Geometric representations reveal that in the flat form, Hexamer 1 (a) and Hexamer 2 (b) both deviate slightly from perfect C6 symmetry. Screw 1 (c) has a pitch of 41.9 Å and Screw 2 (d) has a pitch of 33.8 Å. Asterisks indicate constructs that crystalized while retaining an N‐terminal His‐6 tag
We also determined the structure of two novel BMC domain‐based structures in which the quaternary structure formed a six‐fold screw axis. The first hereinafter referred to as Screw 1, came from the same CutR construct that gave rise to the dimer, which still contains a cysteine at position 37, an aspartic acid at position 66 (significance explained below) and an N‐terminal His‐6 tag (Table S1). We solved this structure to 2.8 Å with Rwork/Rfree values of 0.231/0.267. Screw 1 is a right‐handed screw with a pitch of 41.9 Å (Figure 3c). This screw has a 6 sub 1 axis of helical symmetry. Looking down the axis, we observe that the structure forms an apparent hexamer that except for its pitch in the z‐axis, has a similar morphology to Hexamer 2 (Figure S1). Measuring from C‐alpha to C‐alpha of corresponding positions, Screw1 and Hexamer 2 have diameters with maxima of 69.2 and 71.1 Å, respectively (Figure S1). The unexpected nature of the screw form prompted further studies.
Owing to purification challenges from initial work, including aggregation and dynamic oligomerization, and because we observed novel and unexpected structures, we sought to purify an additional mutant version, CutR_D66A, under reducing conditions. CutR_D66A was purified in the presence of TCEP to disrupt the disulfide bridge observed in the initial dimeric structure and has a point mutation at residue 66. In the native form, this residue is normally occupied by a conserved lysine, which participates in intermolecular salt bridges to support the lateral tessellation of BMC hexamers in the shell (Figure 1c); this lysine has been widely mutated in laboratory studies to allow isolation of individual hexamers. 35 , 36 We hypothesized that the aspartic acid in this version played a significant role in our inability to purify well‐behaved, monodisperse species of the CutR construct. This new CutR_D66A mutant retained the His‐6 affinity tag and was purified with relative ease. Surprisingly, this construct gave rise to a second structure having a six‐fold screw axis, hereinafter referred to as Screw 2. Like Screw 1, Screw 2 forms a right‐handed screw, though its pitch of 33.8 Å is considerably shorter than that of Screw 1. We solved the structure to 3.3 Å with Rwork/Rfree values of 0.219/0.265. Like the previously observed screw, Screw 2 also creates an apparent hexamer when looking down the six‐fold axis of symmetry with a similar diameter of 70.1 Å (Figure S1). Both screw structures were obtained with an intact N‐terminal His‐6 tag.
2.2. Analysis of quaternary polymorphisms in CutR
We found value in a new graphical approach for visualizing structural arrangements in the BMC shell protein family. Our prior studies have emphasized the canonical packing of BMC subunits into roughly flat hexameric units. BMC subunits typically fit together like six slightly twisted pie pieces with a central depression commonly found on one side of the full hexamer. Furthermore, as noted in this work, the permuted BMC family bears a small protruding beta‐barrel on the opposite side of the hexameric disk. Based on the well‐established packing of canonical BMC hexamers, we established a coordinate system for mapping a simple pie‐shaped structure onto the conserved secondary structural elements of a BMC protomer. With this mapping established, it was possible to generate diverse packing diagrams to illustrate the wide‐ranging quaternary arrangements observed in the permuted BMC family (Figure 4). This was helpful for interpreting the nature of the nuanced variations and for complementing numerical calculations such as angular rotations and atomic coordinate shifts.
FIGURE 4.
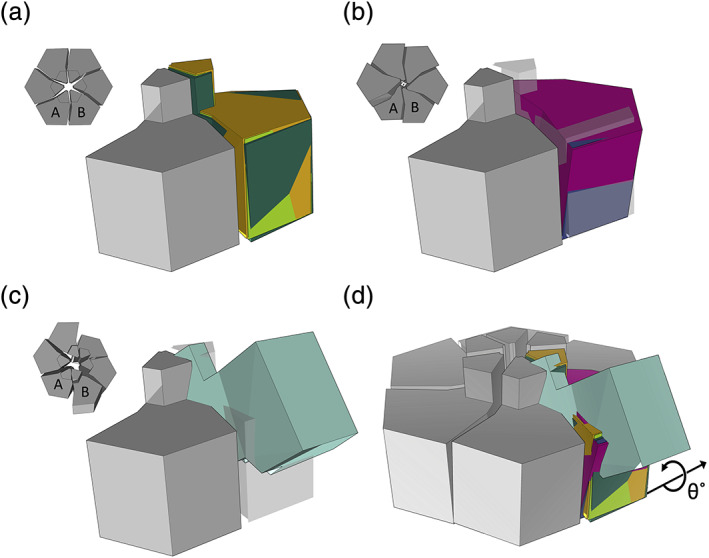
Summary of the quaternary structure deformations observed in the CutR/EutS/PduU subfamily of BMC shell proteins. (a) Geometric representation of adjacent pairs of permuted BMC shell protein subunits from three flat or very nearly flat hexameric structures (CutR Hexamer 1, CutR Hexamer 2, and PduU) after aligning their A subunits (gray). The relative position of the B subunits is shown for CutR Hexamer 1 (lime green), CutR Hexamer 2 (orange), and PduU (forest green). (b) Similar representations for the two screw forms (CutR Screw 1 (magenta) and CutR Screw 2 (purple) relative to the flat CutR form (gray). (c) A similar representation of the bent EutS structure (teal, PDB ID 3I96) relative to the flat CutR form (gray). (d) Overlay of all A monomers relative to the flat hexamer with the B monomers colored as follows: Cut R flat Hexamer 1 (lime green), Cut R flat Hexamer 2 (orange), PduU (forest green), CutR Screw 1 (magenta), CutR Screw 2 (purple), and EutS (teal). The rotation of adjacent subunits in alternate assembly forms, as calculated in Table 1, is diagrammed
Previous studies of the circularly‐permuted BMC proteins PduU (PDB ID 3CGI) and the bent EutS (PDB ID 3I96) have been interesting study cases of the BMC domain. Both maintain the traditional BMC domain fold but comparing adjacent monomer pairs reveals remarkable flexibility. We established a system for evaluating the angular rotations and shifts observed in the EutS structures. We overlapped the A subunit of a given structure with the A subunit of a canonical BMC hexamer, and then evaluated the difference between the adjacent B subunits from the two structures being compared (Figure 4). These comparisons are summarized here and in Table 1. As anticipated, traditional flat hexamers from this permuted BMC family, including PduU and the CutR hexamers from this work, have RMSD values of less than 1 Å, while a comparison of the flat hexamers to EutS (bent) yields RMSD values between 15 and 18 Å. Likewise, the rotation angles required to align corresponding B monomers of flat permuted BMC hexamers were less than 2°, while alignment of the corresponding B monomer from any flat hexamer to the corresponding B monomer from EutS required up to 53° of rotation in order to achieve the optimal overlap (Table 1). Unsurprisingly, a comparison of the two CutR screws shows relatively minor coordinate deviations (0.8 Å RMSD) and angular differences (2.9°), as the two screw forms vary mainly in pitch.
TABLE 1.
Comparison of structural deviations across the permuted BMC family
| – | PduU | Hex1 | Hex2 | Screw1 | Screw2 | EutS |
|---|---|---|---|---|---|---|
| PduU | – | 0.5 Å | 0.7 Å | 3.5 Å | 2.8 Å | 15.8 Å |
| Hex1 | 1.6° | – | 0.2 Å | 3.5 Å | 2.7 Å | 15.6 Å |
| Hex2 | 1.7° | 0.6° | – | 3.4 Å | 2.7 Å | 15.4 Å |
| Screw1 | 14.4° | 16.5° | 16.7° | – | 0.8 Å | 17.1 Å |
| Screw2 | 11.7° | 13.6° | 13.9° | 2.9° | – | 17.1 Å |
| EutS | 42.0° | 40.4° | 40.4° | 55.9° | 52.9° | – |
Note: RMSD values (upper right) shown are based on adjacent AB monomers for four hexameric CutR polymorphs, along with PduU and EutS. The values describe backbone deviations in corresponding B subunits after superimposing A subunits. The rotation angle required to align the B monomers for any given pair of adjacent monomers is reported in the bottom left.
Motivated by the variability observed between the structures of CutR polymorphs, we checked for evidence of dynamic behavior in solution studies. Using another BMC‐domain protein (EutL, a tandem BMC‐domain protein from Clostridium perfringens) as a control, we compared elution profiles of recursive size exclusion chromatography runs of the various CutR constructs (Figure S2). While recursive size exclusion of the EutL control resulted in a single sharp peak, recursive size exclusion of the hexameric peak from CutR constructs resulted in either broad peaks or multiple peaks, supporting a tendency to reequilibrate between multiple conformations or assembly forms in solution.
Finally, the variable packing modes in CutR motivated a study of atomic interactions at the subunit interfaces. A general finding was that for most of the alternate packing variations observed, the bending modes between subunits can occur without substantial perturbations to key atomic interactions. The exceptions to this are the (presumptively artifactual) dimeric form, and the highly bent hexameric form initially observed by Tanaka et al. 23 Structural variation in the latter case, in addition to the bending motion, is accompanied by a twist of roughly 40°, which brings alternative secondary structure packing interactions into play. For the various flat and screw hexamers observed for CutR, major disruptions are not observed. The retention of a basic hexameric (flat or screw) shape despite high flexibility raises a question as to whether a small number of particularly strong interactions in CutR might be important for integrity. A potential role for the beta‐barrel in that regard was noted previously. 20 Our analysis showed that while the extended portions of the beta‐barrel are disrupted in the screw forms, other interactions near the central regions and at the interfaces of the hexamer could be important. Using manual evaluation and the PDBePISA server, 37 we determined that residues S50, E55, D62, S81, Y101, K115, and S116 may play key stabilizing roles. A hydrogen bond network involving these key residues is present in both the flat hexamer and in the screw forms of CutR (Figure 5).
FIGURE 5.
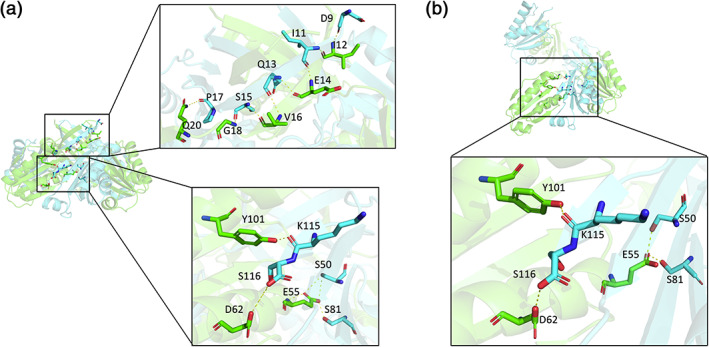
The extensive hydrogen bond network of the beta‐barrel from a flat hexamer compared to the hydrogen bond network of the screw. (a) The flat hexameric form has a beta‐barrel with an extensive hydrogen bond network. There are several key residues essential for maintaining the overall hexameric architecture. (b) In the screw form, the hydrogen bond network of the beta‐barrel is largely lost, but other key intermolecular interactions near the pore are retained
3. DISCUSSION
Since first determining the structures of PduU 20 and EutS, 23 the single‐domain permuted BMC family has been an interesting but under‐studied special class of the BMC family. In this work, we sought to investigate another orthologue from this class, CutR from Streptococcus intermedius. We conclude that the circularly‐permuted BMC protein CutR has an innate and dynamic capacity to sample alternative quaternary forms, with its canonical C6 symmetry breaking down in various ways. Based on these findings, we hypothesize that the permuted BMC family may play a key role in conferring heterogeneity and flexibility in certain types of bacterial microcompartments.
In this work, we elucidated five crystal structures from four CutR constructs, observing a dimer, two flat hexamers, and two six‐fold screws of varying pitch. In addition to variations driven by different crystallization conditions and crystal packing forces, it is likely that variations in the different constructs played important roles in dictating the diverse outcomes. The presence or absence of cysteine (C37), the presence or absence of a charged residue in the traditional edge‐lysine position (K66), and the presence or absence of an N‐terminal His‐6 purification tag (Table S1) are all relevant variations. Certain correlations between constructs and quaternary forms are notable. The formation of a screw was only observed in structures that retained an N‐terminal His‐6 tag (Table S1). However, the presence of an N‐terminal purification tag did not force the formation of a helical arrangement, as evidenced by the formation of flat Hexamer 1. A further intriguing question concerns how (or at what point) helical architectures are formed. We note that the recombinant protein that gave rise to both screw forms, based on size exclusion chromatography, had an estimated molecular weight consistent with a discrete hexamer (roughly 80 kDa). This argues for ordinary (non‐helical) hexameric forms being predominant, yet retaining the flexibility to undergo considerable distortions and interface disruptions under diverse, relatively mild, and solution conditions. Significantly, the screws retained key intermolecular interactions that are shared with the flat hexameric form (Figure 5).
The vast majority of homomeric protein assemblies are arranged in a symmetric fashion. The reasons for this were articulated as early as Crick and Watson, 38 and have been expanded upon and surveyed many times since. 39 , 40 , 41 The evident tendency toward asymmetry, therefore, puts CutR and its orthologues in a unique category. Symmetry breaking in protein assemblies has been described in other areas. In particular, it has been explored in the context of molecular motors where alternating configurations occur during catalytically‐driven motions 42 as well as in viral capsids. 43 In the latter category, as explained by Casper and Klug, alternative conformations that break symmetry are required to accommodate different packing environments. 44 The unusual polymorphism seen in CutR may help explain the heterogeneity and structural irregularity seen in many types of bacterial microcompartments.
Carboxysomes are among the most extensively studied bacterial microcompartments, and they are typically the most regular in shape. EM studies have shown that carboxysome microcompartments exhibit a relatively high degree of homogeneity and geometric regularity. 45 , 46 , 47 , 48 , 49 , 50 , 51 In a few cases, miniaturized versions of microcompartment shells have been produced from synthetic operons. Those have proven to assemble with high levels of order, even obeying icosahedral symmetry, with examples including a beta‐carboxysome shell, a GRM2 microcompartment shell, and a shell from a microcompartment of unknown function, 15 , 30 , 31 but native forms of these MCPs likely diverge in important ways. Unlike their more symmetrical carboxysome counterparts, other native microcompartments have been found to have polymorphic structures 52 , 53 (Figure 1e). These include several MCP types, such as Pdu, Eut, and Cut. Sometimes classified as “metabolosomes”, these MCPs degrade propanediol, ethanolamine, choline, aminoalcohols, and other small metabolites. 1 , 2 , 3 , 4 , 5 , 16 Circularly permuted BMC paralogs (PduU, EutS, CutR) are found in several MCPs of the metabolosome type, but as far as we know these circularly permuted paralogs are not present in carboxysome microcompartments, nor have they been included in the synthetic miniaturized MCP shells that have proven to be geometrically regular. Some insight is available from genetic studies in the Pdu system, which metabolizes 1,2‐propanediol. While the permuted shell protein PduU is not required for the formation of intact microcompartment shells (as it is a relatively low‐abundance component of the shell), PduU deletion mutants showed growth defects, with an increase in lag time while growing to higher cell density compared to their wildtype counterparts. 28 The dynamic nature of permuted BMC proteins, highlighted in the present study, helps explain the impaired growth rates of PduU deletion mutants, considering that dynamic behavior could be important for degradation, recycling, and seeding new microcompartment shells.
The highly twisted EutS structure originally reported by Tanaka 13 and the CutR polymorphs presented here may only be a small population of the potential quaternary structure variations that the permuted BMC family is capable of forming. Beyond a potential role in dynamics, increased shell flexibility could allow for packing a greater variety of internal enzymes in non‐carboxysomal MCPs, or more variable enzyme stoichiometries. The processes that govern MCP assembly and disassembly remain only partially understood, 15 , 54 , 55 , 56 , 57 and flexibility could be important for those processes. The flexibility observed in this protein family invites additional questions about microcompartment structure, evolution, and function. Investigation of additional orthologues could add further insights.
4. MATERIALS AND METHODS
4.1. Negative stain electron microscopy
Intact Pdu microcompartments were purified from S. enterica serovar Typhimurium LT2 and imaged with negative stain electron microscopy as previously described by Havemann et al. and Sinha et al. 58 , 59 Briefly, S. enterica serovar Typhimurium LT2 was grown in 400 ml of growth medium containing 1X NCE, 1 mM MgSO4, 0.5% succinate, and 0.6% 1,2‐PD. Cultures were grown at 37°C overnight, shaking. Cells were harvested by centrifugation (4,000g for 15 min) and washed once with Buffer A, containing 50 mM Tris–HCl (pH 8.0), 500 mM KCl, 12.5 mM MgCl2, and 1.5% 1,2‐PD. The cells were resuspended in a lysis buffer containing a mixture of 40% Buffer A and 60% B‐PER II (Thermo Scientific) supplemented with Pierce Protease Inhibitor tablets (Thermo Scientific), Lysozyme (Sigma‐Aldrich), and DNase I (Sigma‐Aldrich) and incubated at 30°C for 1 hr to lyse. Cell debris was removed by centrifugation at 12,000g for 5 min, and intact Pdu MCPs were pelleted by spinning at 20,000g for 40 min. Pdu MCPs were resuspended in a buffer containing 50 mM Tris–HCl (pH 8.0), 50 mM KCl, 5 mM MgCl2, and 1% 1,2‐PD to a concentration of 0.5 mg/ml. Pdu MCPs were placed on Formvar/Carbon 400 mesh copper grids (Ted Pella), washed twice with 0.1% 1,2‐PD and stained with 5 μl of 2% uranyl acetate. Pdu MCPs were imaged using an FEI Tecnai T12 transmission electron microscope.
4.2. Cloning, protein expression, and purification
We initially purchased a codon‐optimized gBlock Gene Fragment of CutR, a EutS homolog from Streptococcus intermedius, from Integrated DNA Technologies (IDT). This first construct was ordered with a non‐cleavable N‐terminal His‐6 tag and a K66D mutation. Previous work has demonstrated that mutating this edge lysine facilitates recombinant BMC protein expression and purification. We hypothesized that maintaining a charged amino acid in this position would achieve the same facile purification while maintaining structural integrity. We inserted the gBlock into the pET24a expression vector using NdeI and HindIII restriction endonuclease sites. For mutagenesis work, we also purchased primers from IDT. Primers were designed based on the previously published Quikchange protocol. 60 We explored a variety of mutants including C37A, to disrupt the observed disulfide bridge, D66A to further facilitate expression and purification, and insertion of a TEV‐cleavage site (ENLYFQG) just after the His‐6 tag, in order to assess the role of the His‐6 tag in crystal packing.
We used the BL21(DE3) E. coli expression system (New England Biolabs) to express recombinant protein. Briefly, we used 6 ml 1 mg/L ampicillin supplemented LB overnight cultures to inoculate 1 L of autoinduction media. Cells were grown in TB medium supplemented with kanamycin and 5,052 autoinduction sugars. 61 Cultures were grown at 37°C for 6 hr and then 18°C overnight and subsequently harvested by centrifugation for 15 min at 5,000g. For a recombinant protein that resulted in the dimeric form, cells were grown in TB medium supplemented with 1 mg/L kanamycin. At OD600 = 1.0–1.2, we used 1 mM of IPTG to induce expression and proceeded to grow cells at 25°C overnight.
Cell pellets were resuspended in 20 mM Tris pH 8.0, 300 mM NaCl, supplemented with protease inhibitor cocktail tablets (Pierce Protease Inhibitor Tablets, EDTA‐Free, Thermo Scientific), lysozyme (Sigma), DNase (Sigma), and RNase (Sigma). We lysed the resuspended pellets with sonication over ice and clarified the lysate by centrifugation (15,000g for 30 min). The clarified lysate was applied over a pre‐equilibrated Nickel IMAC gravity column (HisPur Ni‐NTA Resin, Thermo Scientific) and His‐tagged samples were eluted using 20 mM Tris pH 8.0, 300 mM NaCl, 300 mM imidazole, and 10% glycerol. Samples that contained a TEV‐cleavage site were subject to TEV‐protease and dialyzed overnight at 4°C into 50 mM Tris pH 8.0 and 150 mM NaCl. All proteins were subject to the second round of purification using gel‐filtration chromatography (Superdex 200 10/300 GL, GE Healthcare) and eluted with a buffer containing 50 mM Tris pH 8.0 and 150 mM NaCl. Fractions from peaks that corresponded to a molecular weight of approximately 80 kDa were pooled together. We verified the presence and purity of protein throughout the purification process using denaturing SDS‐PAGE. We also sought to investigate the role and significance of the observed disulfide bridge by using reducing agents. In one iteration of our purification procedure, we added 10 mM tris(2‐carboxyethyl)phosphine (TCEP) to purification buffers. Following the two‐step purification, proteins were subsequently concentrated to approximately 30 mg/ml using 10KDa MWCO Amicon Ultra concentrators (Millipore) and syringe‐filtered through 0.22 μM filters (Millipore). Following concentration and filtration, this protein was used for crystallization experiments.
4.3. Crystallization
We obtained diffraction quality crystals using a TTP Labtech robotic mosquito and the hanging‐drop vapor diffusion method at the UCLA Crystallization facility. We observed several crystal forms in numerous conditions, the most prominent being flat hexagonal sheets and bi‐pyramidal octahedrons. We collected diffraction data for the CutR dimer that crystallized in 0.1 M Potassium thiocyanate, 30% w/v PEG 2000. Flat Hexamer 1, which contained a C37A mutation, crystallized in 2.0 M ammonium sulfate, 0.1 M BIS‐Tris, pH 5.5. Flat Hexamer 2, with a cleaved His‐6 tag, crystallized in 2 M ammonium sulfate, 0.2 M lithium sulfate, and 0.1 M CAPS/Sodium hydroxide pH 10.5. Screw 1 crystallized in 10% (w/v) PEG 3000, 200 mM sodium chloride, and 100 mM sodium phosphate dibasic/Citric acid pH 4.2. Finally, Screw 2, which was purified in the presence of reducing agents and contained the D66A mutation, crystallized in 0.2 M potassium sodium tartrate tetrahydrate, 0.1 M Bis‐Tris propane, 8.52% w/v PEG 3350.
4.4. Data collection, structure determination, and refinement
X‐ray diffraction datasets were collected at the Advanced Photon Source in Chicago on beamlines 24‐ID‐C and 24‐ID‐E (NE‐CAT) and then indexed, integrated, and scaled using XDS/XSCALE. 62 We used PHASER 63 to first solve the structure of the CutR dimer by molecular replacement using a EutS homolog from Clostridium difficile (PDB ID 4AXI) as a reference model. The model was built using COOT 64 and refined using PHENIX. 65 We also used Refmac 66 and Buster 67 in subsequent rounds of refinement. The CutR dimer was refined to a final model with Rwork/Rfree of 0.176/0.210 at a resolution of 1.8 Å. Subsequent structures were solved by molecular replacement using one monomer of the CutR dimer as a reference using similar model‐building and refinement strategies. We also utilized the PDBePISA server 37 to computationally assess the interfaces created by these various structures.
AUTHOR CONTRIBUTIONS
Jessica Ochoa: Conceptualization; data curation; formal analysis; investigation; methodology; software; visualization; writing‐original draft; writing‐review and editing. Vy Nguyen: Data curation; formal analysis; investigation; validation; visualization; writing‐original draft; writing‐review and editing. Mengxiao Nie: Data curation; formal analysis; investigation. Michael R. Sawaya: Data curation; formal analysis; investigation; methodology; validation; writing‐review and editing. Thomas Bobik: Conceptualization; funding acquisition; investigation; project administration; supervision; writing‐review and editing. Todd Yeates: Conceptualization; data curation; formal analysis; funding acquisition; investigation; methodology; project administration; resources; software; supervision; validation; visualization; writing‐original draft; writing‐review and editing.
CONFLICT OF INTEREST
The authors declare no competing interests.
Data Availability
Coordinates and diffraction data are deposited at the PDB under codes: 6XPH, 6XPI, 6XPJ, 6XPK, and 6XPL.
Supporting information
Appendix S1: Supporting information.
ACKNOWLEDGMENTS
This work was funded by an award from the National Institutes of Health, NIAID R01AI081146 (TAB and TOY). Diffraction studies were supported by research conducted at the Northeastern Collaborative Access Team beamlines (NECAT), which are funded by the National Institute of General Medical Sciences from the National Institutes of Health (P30 GM124165). JMO is a Howard Hughes Medical Institute Gilliam Fellow. The Eiger 16 M detector on 24‐ID‐E is funded by an NIH‐ORIP HEI grant (S10OD021527). This research used resources of the Advanced Photon Source, a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE‐AC02‐06CH11357. We are grateful for the support of the NECAT staff. The authors thank Duilio Cascio and Michael Collazo for crystallographic help and advice.
Ochoa JM, Nguyen VN, Nie M, Sawaya MR, Bobik TA, Yeates TO. Symmetry breaking and structural polymorphism in a bacterial microcompartment shell protein for choline utilization. Protein Science. 2020;29:2201–2212. 10.1002/pro.3941
Funding information National Institute of Allergy and Infectious Diseases, Grant/Award Number: NIAID R01AI081146; National Institute of General Medical Sciences, Grant/Award Number: P30 GM124165; National Institutes of Health, Grant/Award Number: S10OD021527; Office of Science, Grant/Award Number: DE‐AC02‐06CH11357
REFERENCES
- 1. Jorda J, Lopez D, Wheatley NM, Yeates TO. Using comparative genomics to uncover new kinds of protein‐based metabolic organelles in bacteria. Protein Sci. 2013;22:179–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chowdhury C, Sinha S, Chun S, Yeates TO, Bobik TA. Diverse bacterial microcompartment organelles. Microbiol Mol Biol Rev. 2014;78:438–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ravcheev DA, Moussu L, Smajic S, Thiele I. Comparative genomic analysis reveals novel microcompartment‐associated metabolic pathways in the human gut microbiome. Front Genet. 2019;10:636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Axen SD, Erbilgin O, Kerfeld CA. A taxonomy of bacterial microcompartment loci constructed by a novel scoring method. PLoS Comput Biol. 2014;10:e1003898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bobik TA, Lehman BP, Yeates TO. Bacterial microcompartments: Widespread prokaryotic organelles for isolation and optimization of metabolic pathways. Mol Microbiol. 2015;98:193–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kerfeld CA, Heinhorst S, Cannon GC. Bacterial microcompartments. Annu Rev Microbiol. 2010;64:391–408. [DOI] [PubMed] [Google Scholar]
- 7. Kerfeld CA, Aussignargues C, Zarzycki J, Cai F, Sutter M. Bacterial microcompartments. Nat Rev Microbiol. 2018;16:277–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yeates TO, Crowley CS, Tanaka S. Bacterial microcompartment organelles: Protein shell structure and evolution. Annu Rev Biophys. 2010;39:185–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yeates TO, Thompson MC, Bobik TA. The protein shells of bacterial microcompartment organelles. Curr Opin Struct Biol. 2011;21:223–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. English RS, Lorbach SC, Qin X, Shively JM. Isolation and characterization of a carboxysome shell gene from Thiobacillus neapolitanus. Mol Microbiol. 1994;12:647–654. [DOI] [PubMed] [Google Scholar]
- 11. Bobik TA, Xu Y, Jeter RM, Otto KE, Roth JR. Propanediol utilization genes (pdu) of salmonella typhimurium: Three genes for the propanediol dehydratase. J Bacteriol. 1997;179:6633–6639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Beeby M, Bobik TA, Yeates TO. Exploiting genomic patterns to discover new supramolecular protein assemblies. Protein Sci. 2009;18:69–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tanaka S, Kerfeld CA, Sawaya MR, et al. Atomic‐level models of the bacterial carboxysome shell. Science. 2008;319:1083–1086. [DOI] [PubMed] [Google Scholar]
- 14. Wheatley NM, Gidaniyan SD, Liu Y, Cascio D, Yeates TO. Bacterial microcompartment shells of diverse functional types possess pentameric vertex proteins. Protein Sci. 2013;22:660–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sutter M, Greber B, Aussignargues C, Kerfeld CA. Assembly principles and structure of a 6.5‐MDa bacterial microcompartment shell. Science. 2017;356:1293–1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mallette E, Kimber MS. A complete structural inventory of the mycobacterial microcompartment shell proteins constrains models of global architecture and transport. J Biol Chem. 2017;292:1197–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sutter M, Wilson SC, Deutsch S, Kerfeld CA. Two new high‐resolution crystal structures of carboxysome pentamer proteins reveal high structural conservation of CcmL orthologs among distantly related cyanobacterial species. Photosynth Res. 2013;118:9–16. [DOI] [PubMed] [Google Scholar]
- 18. Kerfeld CA, Sawaya MR, Tanaka S, et al. Protein structures forming the shell of primitive bacterial organelles. Science. 2005;309:936–938. [DOI] [PubMed] [Google Scholar]
- 19. Tsai Y, Sawaya MR, Cannon GC, et al. Structural analysis of CsoS1A and the protein shell of the Halothiobacillus neapolitanus carboxysome. PLoS Biol. 2007;5:e144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Crowley CS, Sawaya MR, Bobik TA, Yeates TO. Structure of the PduU shell protein from the Pdu microcompartment of salmonella. Structure. 2008;16:1324–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Klein MG, Zwart P, Bagby SC, et al. Identification and structural analysis of a novel carboxysome shell protein with implications for metabolite transport. J Mol Biol. 2009;392:319–333. [DOI] [PubMed] [Google Scholar]
- 22. Sagermann M, Ohtaki A, Nikolakakis K. Crystal structure of the EutL shell protein of the ethanolamine ammonia lyase microcompartment. Proc Natl Acad Sci U S A. 2009;106:8883–8887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tanaka S, Sawaya MR, Yeates TO. Structure and mechanisms of a protein‐based organelle in Escherichia coli . Science. 2010;327:81–84. [DOI] [PubMed] [Google Scholar]
- 24. Crowley CS, Cascio D, Sawaya MR, Kopstein JS, Bobik TA, Yeates TO. Structural insight into the mechanisms of transport across the Salmonella enterica Pdu microcompartment shell. J Biol Chem. 2010;285:37838–37846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pang A, Warren MJ, Pickersgill RW. Structure of PduT, a trimeric bacterial microcompartment protein with a 4Fe–4S cluster‐binding site. Acta Crystallogr. 2011;D67:91–96. [DOI] [PubMed] [Google Scholar]
- 26. Aussignargues C, Pandelia M‐E, Sutter M, et al. Structure and function of a bacterial microcompartment shell protein engineered to bind a [4Fe‐4S] cluster. J Am Chem Soc. 2016;138:5262–5270. [DOI] [PubMed] [Google Scholar]
- 27. Samborska B, Kimber MS. A dodecameric CcmK2 structure suggests β‐carboxysomal shell facets have a double‐layered organization. Structure. 2012;20:1353–1362. [DOI] [PubMed] [Google Scholar]
- 28. Cheng S, Sinha S, Fan C, Liu Y, Bobik TA. Genetic analysis of the protein shell of the microcompartments involved in coenzyme B12‐dependent 1,2‐propanediol degradation by salmonella. J Bacteriol. 2011;193:1385–1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Parsons JB, Lawrence AD, McLean KJ, Munro AW, Rigby SEJ, Warren MJ. Characterisation of PduS, the pdu metabolosome corrin reductase, and evidence of substructural organisation within the bacterial microcompartment. PLOS ONE. 2010;5:e14009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sutter M, Laughlin TG, Sloan NB, Serwas D, Davies KM, Kerfeld CA. Structure of a synthetic β‐carboxysome shell. Plant Physiol. 2019;181:1050–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kalnins G, Cesle E‐E, Jansons J, Liepins J, Filimonenko A, Tars K. Encapsulation mechanisms and structural studies of GRM2 bacterial microcompartment particles. Nat Commun. 2020;11:388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lee MJ, Mantell J, Hodgson L, et al. Engineered synthetic scaffolds for organizing proteins within the bacterial cytoplasm. Nat Chem Biol. 2018;14:142–147. [DOI] [PubMed] [Google Scholar]
- 33. Kinney JN, Axen SD, Kerfeld CA. Comparative analysis of carboxysome shell proteins. Photosynth Res. 2011;109:21–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bennett MJ, Schlunegger MP, Eisenberg D. 3D domain swapping: A mechanism for oligomer assembly. Protein Sci. 1995;4:2455–2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sinha S, Cheng S, Sung YW, et al. Alanine scanning mutagenesis identifies an asparagine‐arginine‐lysine triad essential to assembly of the shell of the Pdu microcompartment. J Mol Biol. 2014;426:2328–2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Pang A, Frank S, Brown I, Warren MJ, Pickersgill RW. Structural insights into higher order assembly and function of the bacterial microcompartment protein PduA. J Biol Chem. 2014;289:22377–22384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372:774–797. [DOI] [PubMed] [Google Scholar]
- 38. Crick FHC, Watson JD. Structure of small viruses. Nature. 1956;177:473–475. [DOI] [PubMed] [Google Scholar]
- 39. André I, Strauss CEM, Kaplan DB, Bradley P, Baker D. Emergence of symmetry in homooligomeric biological assemblies. Proc Natl Acad Sci U S A. 2008;105:16148–16152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Goodsell DS, Olson AJ. Structural symmetry and protein function. Annu Rev Biophys Biomol Struct. 2000;29:105–153. [DOI] [PubMed] [Google Scholar]
- 41. Cannon KA, Ochoa JM, Yeates TO. High‐symmetry protein assemblies: Patterns and emerging applications. Curr Opin Struct Biol. 2019;55:77–84. [DOI] [PubMed] [Google Scholar]
- 42. Sun SX, Wang H, Oster G. Asymmetry in the F1‐ATPase and its implications for the rotational cycle. Biophys J. 2004;86:1373–1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Harrison SC. The familiar and the unexpected in structures of icosahedral viruses. Curr Opin Struct Biol. 2001;11:195–199. [DOI] [PubMed] [Google Scholar]
- 44. Caspar DLD, Klug A. Physical principles in the construction of regular viruses. Cold Spring Harb Symp Quant Biol. 1962;27:1–24. [DOI] [PubMed] [Google Scholar]
- 45. Shively JM, Ball FL, Kline BW. Electron microscopy of the carboxysomes (polyhedral bodies) of Thiobacillus neapolitanus. J Bacteriol. 1973;116:1405–1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Cannon GC, Bradburne CE, Aldrich HC, Baker SH, Heinhorst S, Shively JM. Microcompartments in prokaryotes: Carboxysomes and related polyhedra. Appl Environ Microbiol. 2001;67:5351–5361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Price GD, Coleman JR, Badger MR. Association of carbonic anhydrase activity with carboxysomes isolated from the cyanobacterium Synechococcus PCC7942. Plant Physiol. 1992;100:784–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Long BM, Badger MR, Whitney SM, Price GD. Analysis of carboxysomes from Synechococcus PCC7942 reveals multiple rubisco complexes with carboxysomal proteins CcmM and CcaA. J Biol Chem. 2007;282:29323–29335. [DOI] [PubMed] [Google Scholar]
- 49. Iancu CV, Morris DM, Dou Z, Heinhorst S, Cannon GC, Jensen GJ. Organization, structure, and assembly of α‐carboxysomes determined by electron cryotomography of intact cells. J Mol Biol. 2010;396:105–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Dai W, Chen M, Myers C, et al. Visualizing individual RuBisCO and its assembly into carboxysomes in marine cyanobacteria by cryo‐electron tomography. J Mol Biol. 2018;430:4156–4167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Dai W, Fu C, Raytcheva D, et al. Visualizing virus assembly intermediates inside marine cyanobacteria. Nature. 2013;502:707–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Bag S, Prentice MB, Liang M, Warren MJ, Roy Choudhury K. Classification of polyhedral shapes from individual anisotropically resolved cryo‐electron tomography reconstructions. BMC Bioinformatics. 2016;17:234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kennedy NW, Hershewe JM, Nichols TM, et al. Apparent size and morphology of bacterial microcompartments varies with technique. PloS One. 2020;15:e0226395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Bonacci W, Teng PK, Afonso B, et al. Modularity of a carbon‐fixing protein organelle. Proc Natl Acad Sci U S A. 2012;109:478–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Hill NC, Tay JW, Altus S, Bortz DM, Cameron JC. Life cycle of a cyanobacterial carboxysome. Sci Adv. 2020;6:eaba1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Cameron JC, Wilson SC, Bernstein SL, Kerfeld CA. Biogenesis of a bacterial organelle: The carboxysome assembly pathway. Cell. 2013;155:1131–1140. [DOI] [PubMed] [Google Scholar]
- 57. Perlmutter JD, Mohajerani F, Hagan MF. Many‐molecule encapsulation by an icosahedral shell. Elife. 2016;5:e14078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Havemann GD, Bobik TA. Protein content of polyhedral organelles involved in coenzyme B12‐dependent degradation of 1,2‐propanediol in Salmonella enterica serovar typhimurium LT2. J Bacteriol. 2003;185:5086–5095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Sinha S, Cheng S, Fan C, Bobik TA. The PduM protein is a structural component of the microcompartments involved in coenzyme B12‐dependent 1,2‐propanediol degradation by salmonella. J Bacteriol. 2012;194:1912–1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Liu H, Naismith JH. An efficient one‐step site‐directed deletion, insertion, single and multiple‐site plasmid mutagenesis protocol. BMC Biotechnol. 2008;8:91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Studier FW. Protein production by auto‐induction in high‐density shaking cultures. Protein Expr Purif. 2005;41:207–234. [DOI] [PubMed] [Google Scholar]
- 62. Kabsch W. XDS. Acta Crystallogr. 2010;D66:125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. McCoy AJ, Grosse‐Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Cryst. 2007;40:658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Emsley P, Cowtan K. Coot: Model‐building tools for molecular graphics. Acta Crystallogr. 2004;D60:2126–2132. [DOI] [PubMed] [Google Scholar]
- 65. Adams PD, Grosse‐Kunstleve RW, Hung LW, et al. PHENIX: Building new software for automated crystallographic structure determination. Acta Crystallogr. 2002;D58:1948–1954. [DOI] [PubMed] [Google Scholar]
- 66. Winn MD, Ballard CC, Cowtan KD, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. 2011;D67:235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Bricogne G, Blanc E, Brandl M, et al. BUSTER version 2.10.3. Cambridge, United Kingdom: Global Phasing Ltd; 2019.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1: Supporting information.
Data Availability Statement
Coordinates and diffraction data are deposited at the PDB under codes: 6XPH, 6XPI, 6XPJ, 6XPK, and 6XPL.


