Abstract
Retinal diseases constitute a genetically and phenotypically diverse group of clinical conditions leading to vision impairment or blindness with limited treatment options. Advances in reprogramming of somatic cells to induced pluripotent stem cells and generation of three‐dimensional organoids resembling the native retina offer promising tools to interrogate disease mechanisms and evaluate potential therapies for currently incurable retinal neurodegeneration. Next‐generation sequencing, single‐cell analysis, advanced electrophysiology, and high‐throughput screening approaches are expected to greatly expand the utility of stem cell‐derived retinal cells and organoids for developing personalized treatments. In this review, we discuss the current status and future potential of combining retinal organoids as human models with recent technologies to advance the development of gene, cell, and drug therapies for retinopathies.
Keywords: neural differentiation, retina, retinal photoreceptors, somatic stem cells
Retinal organoids offer a unique human model system to study mechanisms of disease pathogenesis and for developing therapies.
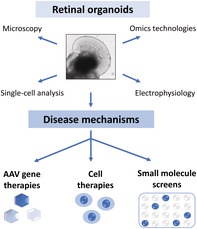
Significance statement.
Most retinal diseases resulting in vision impairment still lack effective treatments. Advances in stem cell technologies now offer methods of differentiating retinal organoids that represent key features of native human retina. Better understanding of human models derived from patient stem cells and use of emerging experimental tools will facilitate elucidation of disease mechanisms and design treatments for retinal neurodegeneration.
1. INTRODUCTION
Loss of sight is one of the most feared disabilities. The retina is often called the window to the brain, and dysfunction of retinal neurons in complex multifactorial diseases such as age‐related macular degeneration (AMD), glaucoma, or diabetic retinopathy is a major cause of incurable blindness worldwide. 1 , 2 Mutations in over 200 genes can lead to inherited retinal and macular diseases (IRDs), and genetic variations at numerous loci have been associated with susceptibility to AMD (RetNet, https://sph.uth.edu/retnet/). 3 , 4 , 5 Identification of causative genes has enabled generation of animal models, resulting in elucidation of physiological functions of underlying gene products and development of gene therapies, in particular using adeno‐associated virus (AAV) vector platform. 6 In addition, fundamental advances in understanding of retinal development has resulted in rapid preclinical translation of potential cell replacement therapies, 7 especially transplantation of photoreceptors 8 , 9 and retinal pigment epithelium (RPE). 10 Although overall morphology, physiology, and molecular profile of the retina are highly conserved, many human clinical phenotypes are not faithfully recapitulated in animal models. Therefore, human stem cell‐based in vitro models that complement the studies in mice and in vivo systems are highly desirable, with renewable sources of human cells needed for developing cell‐based treatments. Breakthroughs in differentiating retinal tissues from patient‐specific induced pluripotent stem cells (iPSCs) offer promising personalized models of human disease as well as provide a resource for evaluating therapeutic strategies. In this article, we will review current status of research on stem cell‐derived retinal organoids, including methods for differentiation, and discuss their utility for identifying potential therapies.
2. DIFFERENTIATION METHODS AND STAGING OF RETINAL ORGANOIDS
Groundbreaking discoveries in reprogramming of somatic cells into iPSCs and their differentiation into retinal lineages have greatly facilitated the study and treatment design of retinopathies. Early differentiation protocols used two‐dimensional (2D) adherent cultures to derive retinal cells from pluripotent stem cells (PSCs). 11 , 12 Sasai laboratory pioneered the methods for 3D optic cup morphogenesis in vitro from mouse embryonic stem cells, 13 followed by adaptation of the protocol to produce human retinal organoids. 14 Subsequently, two major approaches emerged for deriving retinal organoids (Figure 1). In the first approach, PSCs are suspended as single cells, followed by quick aggregation into embryoid bodies in a multiwell format, and then retinal neuroepithelium differentiation is stimulated by addition of basal lamina components present in Matrigel. 14 , 15 As Matrigel composition can vary greatly, low concentration of BMP4 can be used in a chemically defined medium instead. 16 Although this approach is suitable for automation and potential use in high‐throughput screens, laminated retinal morphology appears to be preserved for a limited period. 16 The second set of protocols combine 2‐D and 3‐D approaches with initial formation of embryoid bodies from detached human PSC (hPSC) colonies and neural induction in suspension, then plating and eye field formation in adherent conditions, followed by isolation of retinal domains and suspension culture as retinal organoids. 17 , 18 , 19 The initial step of embryoid body formation can be omitted with growth of hPSCs to confluence and neural induction in adherent cultures. 17 , 20 Another alternative is mixing hPSC clumps with Matrigel to form epithelialized cysts, which can be then plated, later followed by gentle lifting with dispase to form well‐laminated organoids. 21 , 22 Replacement of widely used all‐trans retinoic acid with 9‐cis retinal accelerates differentiation of rod photoreceptors in retinal organoids, 23 whereas signaling by thyroid hormone facilitates the specification of L/M cone subtypes, 24 consistent with S‐cones being the default fate of photoreceptors. 25 , 26 Interestingly, scraping of the adherent culture and selection of subsequently formed optic vesicles improves the yield and efficiency of generating retinal organoids. 27 Typically, the combination of 2‐D and 3‐D approaches achieves higher organization and maturity levels in late‐stage organoids. 15 , 19 , 23 , 24 The use of bioreactors significantly improves the culture of brain organoids with photosensitive cells 28 and has shown promise in culture of retinal organoids. 29 , 30 However, customized and efficient bioreactors are required for their wider acceptance in the field for producing organoids. 31
FIGURE 1.
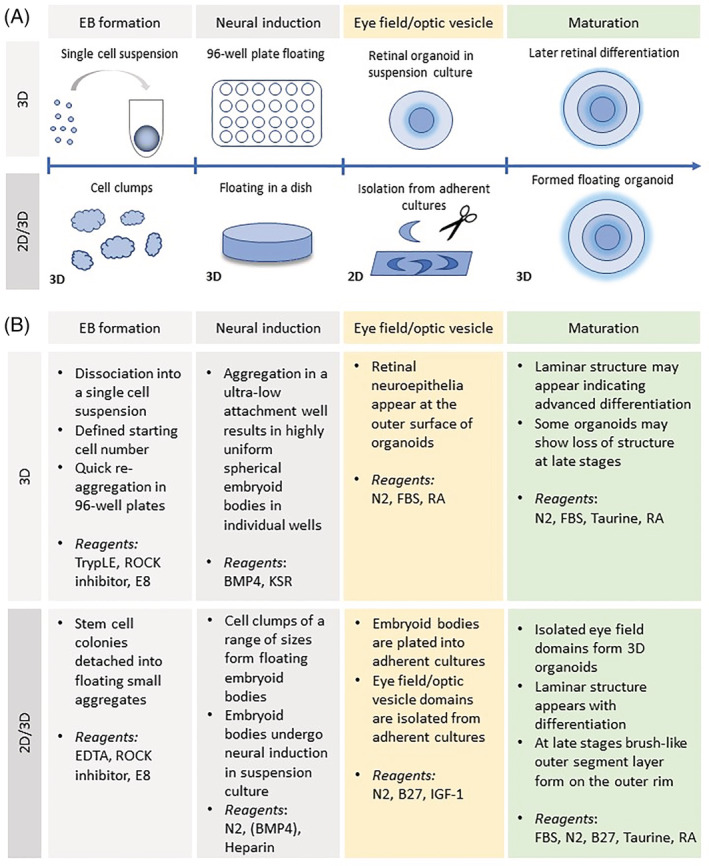
Retinal organoid differentiation methods. A, Two main approaches for generation of retinal organoids: 3D suspension culture throughout, starting from embryoid bodies, which subsequently differentiate into retinal tissues or a mix of 2D/3D culture with isolation of optic regions from adherent cultures. B, Comparison of main steps between the two approaches and key reagents used at each step
The array of methods to derive retinal organoids across independent laboratories has created challenges in comparing distinct studies and assuring faithful recapitulation of native developmental processes. Initially, limited sets of markers of retinal cell types for immunohistochemistry or gene profiling were used to characterize organoid differentiation and correlate it to human eye development. 18 , 20 , 32 , 33 However, different methods often led to variable timing or robustness of marker expression. In order to facilitate cross‐comparison across studies, a light microscopy‐based staging system was developed. 34 In this classification (Figure 2), stage 1 corresponds to phase‐bright early retinal neuroepithelium developing into an opaque intermediate stage 2 and followed by stage 3 characterized by emergence of brush‐like surface protrusions that correspond to developing photoreceptor outer segments. 34 In another study, transcriptomes of developing retinal organoids were compared to developmental epochs in human fetal retina. 35 Expression levels of retinal genes, however, varied considerably among retinal organoids from different iPSCs lines. 23 , 36 Transcriptional profiling alone 21 , 23 , 32 , 36 or in combination with open chromatin analysis 37 revealed similarities to human eye development with stage 1 showing early neurogenesis and retinal progenitor gene signatures, followed by interneuron and synaptogenesis‐related gene expression in stage 2 and, finally, photoreceptor differentiation and maturation prominent in stage 3. These studies highlighted broad similarities of organoids to developing native retina tissue, despite protocol differences, based on light microscopy and/or transcriptome‐based staging.
FIGURE 2.
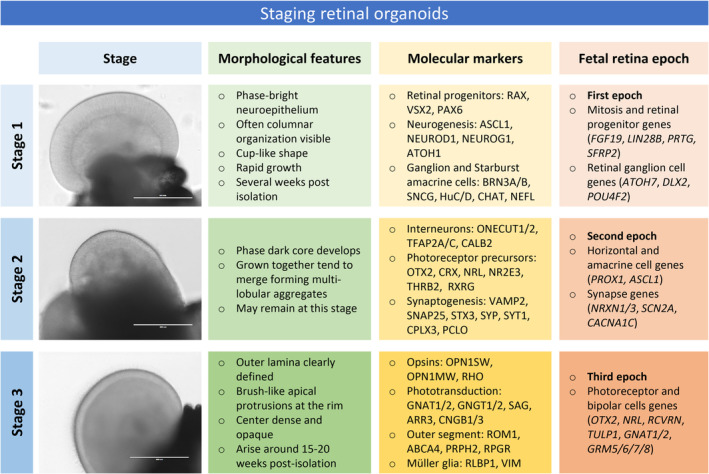
Staging of retinal organoids. Based on histological features and molecular marker expression from organoid transcriptome studies, three major sequential stages of differentiation can be related to molecularly defined epochs in human fetal retina development. Bright‐field images on the left show retinal tissue in organoids at each stage (scale bar 400 μm). Table summarizes key morphological features, molecular markers, and corresponding human fetal development epoch at each organoid differentiation stage
3. RETINAL ORGANOIDS AS DISEASE MODELS
Technologies to convert somatic cells harvested in a patient biopsy into iPSCs have opened new avenues for interrogating disease mechanisms associated with retinopathies. Early studies using iPSC lines from retinitis pigmentosa patients differentiated in adherent conditions identified endoplasmic reticulum stress as a common disease signature. 38 , 39 Subsequently progress in 3‐D cultures allowed examining disease mechanisms in organoids with more native spatial cell arrangement. Defects in early retinogenesis were modeled in cells carrying R200Q mutation in the transcription factor visual system homeobox 2 (VSX2). 40 Consistent with the clinical phenotype of microphthalmia, optic vesicles revealed slower growth and loss of retinal bipolar cells that express VSX2 at later stages. During retinal development, a distinct profile of alternative splicing is established, and retinal organoids have been used for disease modeling of patients with mutations in splicing factor PRPF31 (RP11). 41 Many disease‐causing mutations in IRDs are specifically located in genes related to sensory cilia and can be modeled in organoids. 42 Two of the published studies examined effects of mutations in a centrosome‐cilia protein CEP290. 43 , 44 Cilia formation was shown to be impaired in fibroblasts derived from CEP290 patients with a more severe syndromic disease Joubert syndrome, but not in those diagnosed with Leber congenital amaurosis (LCA), a blinding retinal disease. However, retinal organoids from CEP290‐LCA patients exhibited abnormal ciliogenesis demonstrating tissue‐specific impact of pathogenic mutations and correlating broader defects with increased disease severity. 43 Retinal organoids from patients with an intronic CEP290 mutation common in LCA patients revealed a more frequent cryptic exon inclusion compared to fibroblasts, iPSCs or RPE cells from the same patient. 44 Reduced CEP290 protein levels in CEP290‐LCA organoids led to impaired cilia development. RPGR is another cilia protein commonly mutated in retinitis pigmentosa (RP). 45 , 46 Retinal organoids from patients harboring RPGR mutations displayed impairment of cilia biogenesis and abnormal photoreceptor morphology as well as decreased expression of many phototransduction‐related genes. 47 However, the molecular basis of how a defective ciliary protein leads to photoreceptor cell death still remains elusive. In addition to genes encoding ciliary proteins, mutations in two genes related to structural integrity of the retina have been studied in organoid cultures. Organoids derived from iPSCs of a X‐linked retinoschisis patient with RS1 mutation recapitulated the retinal phenotype. 48 In organoids derived from patient with mutations in the CRB1 gene, the outer limiting membrane was disrupted with ectopic photoreceptor cell bodies detected beyond the apical limit of the neural retina. 49 The aforementioned studies have focused on examining monogenic diseases. With respect to multifactorial traits, such as AMD, glaucoma, or diabetic retinopathy, combining several tissues by incorporation of a vascular network 50 or combining organoids with RPE into a retina‐on‐a‐chip format 51 might be necessary to recreate more physiological interactions. We note that iPSC‐derived RPE from AMD patients with common ARMS2/HTRA1 susceptibility variants has demonstrated upregulation of several disease biomarkers, including complement and pro‐inflammatory factors. 52 These studies clearly demonstrate the utility of patient stem cell‐derived retinal organoids in a dish for modeling endophenotypes of human disease in complement with in vivo animal systems.
4. EVALUATION OF THERAPEUTIC STRATEGIES IN HUMAN RETINAL ORGANOIDS
Most studies to date have used gene editing in iPSCs to rescue disease phenotypes. 41 , 47 , 48 However, such a strategy is not yet applicable for patient treatment. Most translational research so far has focused on gene therapy using AAV vectors and replacement of RPE or photoreceptors 6 , 7 , 10 ; nonetheless, retinal organoid protocols are now being adapted to search for small molecule therapeutic compounds using high‐throughput screening strategies. 53 , 54
4.1. Gene‐based therapies
AAV vector platform has become a gene delivery method of choice for retinal diseases 6 (Figure 3). Transduction of mouse and human retinal organoids as well as RPE tissues has been examined by a panel of commonly used AAV capsid serotypes. 55 Interestingly, ShH10 capsid serotype, which was developed through directed evolution approach of screening AAV mutant libraries, 56 was highly efficient in transducing all stem cell‐derived tissues examined. 55 In mouse retinal organoids, AAV2 vector rescued the CEP290‐associated disease phenotype. 57 For human photoreceptor transduction AAV2 capsid serotype also appears to be highly potent, whereas AAV5 is most effective for RPE cells of both mouse and human origin. Additionally, AAV5 transduced both human photoreceptors as well as Müller glia, albeit at a lower frequency, in a separate study. 49 Interestingly, human cone cells seem to be more effectively transduced by AAV8 vector, 55 suggesting that even cell subtype differences in tropism might exist. Other treatment paradigms are also being explored in organoid models; these include antisense oligonucleotides (ASOs), which are short, chemically modified RNA molecules that can interfere with splicing or translation. ASO treatment is shown to rescue ciliation defects in retinal organoids and RPE cells derived from a patient with a common intronic mutation in the CEP290 gene. 44 , 58
FIGURE 3.
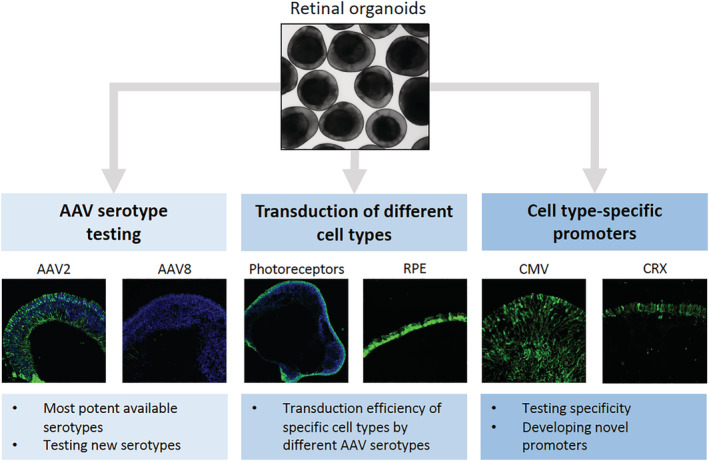
Utility of retinal organoids in gene therapy studies. Whereas most retinal gene therapy studies to date use animal models of inherited retinal and macular diseases, breakthroughs in reprogramming and differentiation of patient cells into retinal organoids now allow testing potential therapeutic approaches in human retinal tissue in vitro. Applications of organoids in retinal gene therapy studies include testing most effective vector serotypes (left panel), determining transduction of target cells in human tissue (middle panel) and establishing promoter specificity and levels of expression for driving therapeutic genes (right panel)
4.2. Cell therapies
Stem cells and organoids have been widely used as a source for evaluating cell replacement therapy approaches. 11 , 20 , 59 , 60 , 61 The degenerative process in retinopathies primarily affects two retinal cell types: the light‐sensitive photoreceptors and RPE. 7 , 10 As a simple monolayer easily distinguishable by pigmentation and cuboidal morphology, RPE has been one of the first cell types to be efficiently differentiated from PSC sources and the first hESC‐derived cell type to be transplanted into humans. 62 Implanting intact RPE sheets alone or grown on a bioscaffold is likely to result in improved outcomes. 59 , 63 , 64
With respect to photoreceptor cells, pioneering studies on neural retina transplantation used primary cells derived from either donor mice 8 or human fetal tissue. 65 Similar to RPE, both suspension as well as whole tissue approaches have been studied; however, unlike RPE, much of the studies so far have focused on animal models. Transplantation of donor‐derived photoreceptor precursor suspensions rescued some level of visual function in a model of IRD with intact but dysfunctional photoreceptors (Gnat1 −/− mice) 8 , 66 ; however, unfortunately rather than true functional replacement, this process involved fusion events and cytoplasmic transfer of reporter and other proteins to the diseased photoreceptors from the graft. 67 , 68 , 69 Notably, transplantation of stem cell‐derived photoreceptors have been performed in both control and models of advanced retinal degeneration where majority of host photoreceptors are lost 20 , 70 and assessment of transplanted cells is not confounded by cell fusion. In such instances, survival of a cell mass in subretinal space, expression of phototransduction‐related proteins, small structures resembling early outer segments (parts of photoreceptors where light is detected and phototransduction initiated) and presence of synapse‐related proteins in the graft have been reported and indicate that the transplanted cells can mature in vivo. 20 , 71 , 72 Recently, rods directly derived from fibroblasts using a combination of five small molecules are shown to exhibit similar characteristics when transplanted into rd1 model of end‐stage retinal degeneration and partially restored some aspects of visual function. 9 These observations suggest that, provided they are capable of connecting to the remaining host visual circuit, these transplanted cells might confer some light sensitivity to severely degenerated retina. Whether such a cell replacement strategy would indeed be applicable for treatment remains to be established. Given that donor‐host species mismatch in human cell transplantation into animal models might affect cell‐to‐cell interactions, an interesting, as yet unexplored, avenue would be to use the organoids as in vitro recipient tissue for dissociated human photoreceptors.
Another approach to treat vision loss in advanced retinal degeneration is transplantation of laminated retinal sheets. 60 , 61 , 65 Here, 3D retinal tissue is dissected out from organoids and transplanted whole into the subretinal space to overlay the degenerated retina. The major setbacks associated with this method include difficulty in maintaining correct morphology and polarity while surgically placing the graft into the recipient eye and presence of interneurons in the graft, which may hinder connectivity with remaining host inner retina. In contrast to dissociated cells, retinal sheets transplanted into rat 60 or nonhuman primate recipients 61 showed clear elaboration of outer segment structures suggesting advanced photoreceptor maturation following grafting. The presence of synaptic connectivity was also detected between the graft and host. However, obtaining a clear evidence for graft‐driven retinal function remains challenging. Experiments performed on explanted retinas from recipient animals using microelectroretinography and microelectrode arrays have shown signals suggestive of light‐evoked responses, 73 but unequivocal evidence for functionality of the graft itself is difficult to obtain in the presence of activity of the host retina. Stronger and more direct evidence are required for both cell suspension as well as retinal sheet transplantation to demonstrate efficacy of photoreceptor replacement therapy.
4.3. Small molecule screening
Retinal neurons and organoids derived from human cells are valuable model systems to discover candidate target drugs for various diseases. Such an approach has been successfully applied to identify inhibitors of Zika virus brain infection 74 and assess efficacy of chemotherapy agents for glioblastoma‐like neoplasms. 75 A 3‐D retinoblastoma organoid model has been used to assess cellular responses to currently used chemotherapy drugs. 76 Challenges of variability and scalability of retinal organoid cultures have so far limited their use in high‐throughput compound screens. Organoids grown in a multiwell format were used to determine impact of seeding density, nutrients added to culture media and targets of ophthalmic toxicity of the antibiotic moxifloxacin. 54 Recent changes in methodology 23 , 27 and use of fluorescent reporter‐based screening platforms 53 , 77 should expedite the use of in vitro‐generated organoids for high content screening of small molecules for treatment of retinal diseases.
5. CURRENT LIMITATIONS OF RETINAL ORGANOID TECHNOLOGY
At present, successful use of retinal organoids in modeling and treatment of retinal diseases is limited by several issues. A hallmark of retinal disease is impairment of visual function, and retinal organoids from even healthy control cell lines typically show poor, if any, response to light. 19 , 54 This is likely related to limited development of outer segment discs, where visual pigments and associated phototransduction proteins are concentrated, because of the absence of direct interaction with RPE. In addition, projection neurons in the retina (the retinal ganglion cells) degenerate in organoids probably due to the lack of connection to brain targets, 19 , 23 , 78 further precluding assessment of functional retinal circuits formation. Other challenges for using the organoid system include a great heterogeneity of cell types, 21 , 28 , 79 their variability in maturation states, 23 and a lack of consistent laminated structure. The diversity and developmental variability pose a challenge to comparative analysis and assessment of therapeutic effects, yet the recent staging methods may alleviate some of these problems. 23 , 34 , 54
6. EMERGING TECHNOLOGIES FOR THE ANALYSIS OF RETINAL ORGANOIDS
Much of the characterization of retinal organoids, at least thus far, has been largely focused on histological and bulk transcriptome examination 23 , 34 , 78 and suggested differentiation of major retinal cell types in organoids similar to the human fetal retina. 35 However, faithfulness of subtype diversification, molecular marker expression within subtypes and relative proportions of various types are still largely missing. Several recent studies took advantage of the advances in single‐cell transcriptional profiling to fill this gap. 21 , 79 , 80 , 81 , 82 First studies with a relatively small number of cells demonstrated robust specification into photoreceptors and Müller glia. 21 , 79 Analysis of cone photoreceptor profiles highlighted their similarity to native human cones. 21 , 24 , 82 Consistent with in vivo mouse retina data, 83 , 84 neurogenic retinal progenitors have been identified in the organoids characterized by ASCL1 expression. More comprehensive single‐cell resolution analyses of developing human retinal organoids performed in comparison with human fetal retina samples at equivalent developmental stages have identified transitional cell populations, similar to the neurogenic retinal progenitors in vivo. 80 The single‐cell transcriptomic studies lay foundation for future use of this new technology in describing pathological changes in stem cell‐derived disease models and more precisely determining the developmental stage of cells for transplantation in cell therapies.
Although transcriptome profiling provides powerful insights into gene expression patterns in retinal organoids, little is known as to how epigenetic regulation of chromatin features contributes to establishment of these transcriptional profiles. In brain organoids derived from human and nonhuman primate stem cells, combining single‐cell profiling with open chromatin analysis using an assay for transposase‐accessible chromatin using sequencing (ATAC‐seq) has identified features of gene expression characteristic of human development, such as expression of cadherin 7 (CDH7) solely in human neurons. 86 One could envisage using similar comparative analysis to decipher regulatory networks for fovea/macula development, 35 a primate‐specific retina structure required for high acuity vision and affected in many retinal diseases.
Important but so far unexplored features of retinal organoids are their proteomic and metabolomic profiles compared to the native retina. Considering limited development of outer segment structures in retinal organoids, it is pertinent to comprehensively assess their levels of phototransduction‐related proteins. Research on kidney organoids has uncovered the extracellular matrix (ECM) formed by stem cell‐derived glomeruli and compared it with native tissue finding an immature ECM profile more consistent with fetal glomeruli. 87 This study demonstrates the potential to characterize the protein composition of cellular compartments in organoids and suggests that an analogous approach could conceivably be used for retinal organoid proteome characterization. Given the small size, isolated nature, and distinct protein composition of the photoreceptor cilium compartment, 88 determining the impact of mutations in ciliary proteins on cilia proteomic profile might shed light on their impact on the rest of the photoreceptor cell, eventually leading to its death. Efficient organoid protocols might allow performing such analysis with IRD patient‐derived samples. 42
The retinal photoreceptors are highly metabolically active 89 , 90 and, because of the nature of light detection machinery, highly enriched in metabolites related to the retinoid visual cycle. RPE, which plays critical roles in the metabolic processes of the visual cycle, is not correctly associated with photoreceptors in current retinal organoid protocols. 15 , 16 , 18 , 19 , 23 Furthermore, vasculature does not form in organoids, potentially limiting oxygenation and impacting on oxidative phosphorylation. A study using human cortical organoids revealed higher cell stress markers in vitro and impaired molecular subtype specification. 91 Cell stress signatures could be alleviated by transplantation into mouse cortex, presumably by improved oxygenation and/or exposure to vasculature‐derived factors. Taking into consideration in vitro culture limitations, it would be interesting to determine how well the metabolic profile of retinal organoids recapitulates the native tissue (for summary of novel analysis approaches, see Figure 4).
FIGURE 4.
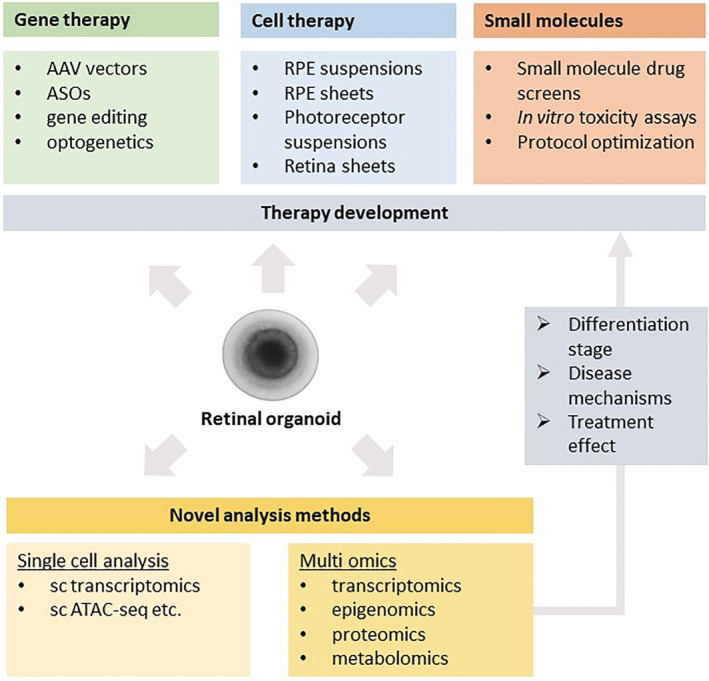
Utility of retinal organoids in developing therapies for retinal diseases. Stem cell‐derived retinal organoids are valuable models of retinal development and disease conditions for studies aiming to identify promising potential gene and cell therapies. Applying emerging technologies such as single‐cell analysis and multi‐omics may further inform about how faithfully organoids recapitulate native tissue and provide insights into molecular mechanisms of disease and therapy development
7. CONCLUSION
Exciting fundamental advances in the differentiation of a plethora of cell lineages and tissues, including the retina, have unlocked new frontiers for deciphering underlying mechanisms of retinopathies and identifying potential therapies. Somatic cells of affected patients can now provide, via in vitro differentiation of iPSCs, retinal organoids, which recapitulate many aspects of gene expression and cell type organization of the native human retina. Broader incorporation of high‐throughput and next‐generation “omics” tools is expected to expand the utility of retinal organoids as disease models, facilitate elucidation of underlying cellular pathways, and assist in discovery of effective treatments.
CONFLICT OF INTEREST
The authors declared no potential conflicts of interest.
AUTHOR CONTRIBUTIONS
A.S.: conception and design, manuscript writing, final approval of manuscript, and financial support and supervision; K.K.: conception and design, manuscript writing.
ACKNOWLEDGMENTS
We thank Holly Y. Chen and Zepeng Qu for helpful comments. Our research is supported by the Intramural Research Program of the National Eye Institute (ZIAEY000450, ZIAEY000474 and ZIAEY000546).
Kruczek K, Swaroop A. Pluripotent stem cell‐derived retinal organoids for disease modeling and development of therapies. Stem Cells. 2020;38:1206–1215. 10.1002/stem.3239
Funding information National Eye Institute, Grant/Award Numbers: ZIAEY000546, ZIAEY000474, ZIAEY000450
DATA AVAILABILITY STATEMENT
Data sharing is not applicable to this article as no new data were created or analyzed in this study.
REFERENCES
- 1. Flaxman SR, Bourne RRA, Resnikoff S, et al. Global causes of blindness and distance vision impairment 1990‐2020: a systematic review and meta‐analysis. Lancet Glob Health. 2017;5(12):e1221‐e1234. [DOI] [PubMed] [Google Scholar]
- 2. Singh M, Tyagi SC. Genes and genetics in eye diseases: a genomic medicine approach for investigating hereditary and inflammatory ocular disorders. Int J Ophthalmol. 2018;11(1):117‐134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Fritsche LG, Igl W, Bailey JN, et al. A large genome‐wide association study of age‐related macular degeneration highlights contributions of rare and common variants. Nat Genet. 2016;48(2):134‐143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Carss KJ, Arno G, Erwood M, et al. Comprehensive rare variant analysis via whole‐genome sequencing to determine the molecular pathology of inherited retinal disease. Am J Hum Genet. 2017;100(1):75‐90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ratnapriya R, Sosina OA, Starostik MR, et al. Retinal transcriptome and eQTL analyses identify genes associated with age‐related macular degeneration. Nat Genet. 2019;51(4):606‐610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Trapani I, Auricchio A. Seeing the light after 25 years of retinal gene therapy. Trends Mol Med. 2018;24:669‐681. [DOI] [PubMed] [Google Scholar]
- 7. Stern JH, Tian Y, Funderburgh J, et al. Regenerating eye tissues to preserve and restore vision. Cell Stem Cell. 2018;22(6):834‐849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. MacLaren RE, Pearson RA, MacNeil A, et al. Retinal repair by transplantation of photoreceptor precursors. Nature. 2006;444(7116):203‐207. [DOI] [PubMed] [Google Scholar]
- 9. Mahato B, Kaya KD, Fan Y, et al. Pharmacologic fibroblast reprogramming into photoreceptors restores vision. Nature. 2020;581(7806):83‐88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Nommiste B, Fynes K, Tovell VE, Ramsden C, Cruz L, Coffey P. Stem cell‐derived retinal pigment epithelium transplantation for treatment of retinal disease. Prog Brain Res. 2017;231:225‐244. [DOI] [PubMed] [Google Scholar]
- 11. Lamba DA, Gust J, Reh TA. Transplantation of human embryonic stem cell‐derived photoreceptors restores some visual function in Crx‐deficient mice. Cell Stem Cell. 2009;4(1):73‐79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Osakada F, Ikeda H, Mandai M, et al. Toward the generation of rod and cone photoreceptors from mouse, monkey and human embryonic stem cells. Nat Biotechnol. 2008;26(2):215‐224. [DOI] [PubMed] [Google Scholar]
- 13. Eiraku M, Takata N, Ishibashi H, et al. Self‐organizing optic‐cup morphogenesis in three‐dimensional culture. Nature. 2011;472(7341):51‐56. [DOI] [PubMed] [Google Scholar]
- 14. Nakano T, Ando S, Takata N, et al. Self‐formation of optic cups and storable stratified neural retina from human ESCs. Cell Stem Cell. 2012;10(6):771‐785. [DOI] [PubMed] [Google Scholar]
- 15. Wahlin KJ, Maruotti JA, Sripathi SR, et al. Photoreceptor outer segment‐like structures in long‐term 3D retinas from human pluripotent stem cells. Sci Rep. 2017;7(1):766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kuwahara A, Ozone C, Nakano T, Saito K, Eiraku M, Sasai Y. Generation of a ciliary margin‐like stem cell niche from self‐organizing human retinal tissue. Nat Commun. 2015;6:6286. [DOI] [PubMed] [Google Scholar]
- 17. Reichman S, Terray A, Slembrouck A, et al. From confluent human iPS cells to self‐forming neural retina and retinal pigmented epithelium. Proc Natl Acad Sci U S A. 2014;111(23):8518‐8523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Meyer JS, Shearer RL, Capowski EE, et al. Modeling early retinal development with human embryonic and induced pluripotent stem cells. Proc Natl Acad Sci U S A. 2009;106(39):16698‐16703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhong X, Gutierrez C, Xue T, et al. Generation of three‐dimensional retinal tissue with functional photoreceptors from human iPSCs. Nat Commun. 2014;5:4047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gonzalez‐Cordero A, Kruczek K, Naeem A, et al. Recapitulation of human retinal development from human pluripotent stem cells generates transplantable populations of cone photoreceptors. Stem Cell Rep. 2017;9(3):820‐837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kim S, Lowe A, Dharmat R, et al. Generation, transcriptome profiling, and functional validation of cone‐rich human retinal organoids. Proc Natl Acad Sci U S A. 2019;116(22):10824‐10833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lowe A, Harris R, Bhansali P, Cvekl A, Liu W. Intercellular adhesion‐dependent cell survival and ROCK‐regulated actomyosin‐driven forces mediate self‐formation of a retinal organoid. Stem Cell Rep. 2016;6(5):743‐756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kaya KD, Chen HY, Brooks MJ, et al. Transcriptome‐based molecular staging of human stem cell‐derived retinal organoids uncovers accelerated photoreceptor differentiation by 9‐cis retinal. Mol Vis. 2019;25:663‐678. [PMC free article] [PubMed] [Google Scholar]
- 24. Eldred KC, Hadyniak SE, Hussey KA, et al. Thyroid hormone signaling specifies cone subtypes in human retinal organoids. Science. 2018;362(6411):eaau6348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Swaroop A, Kim D, Forrest D. Transcriptional regulation of photoreceptor development and homeostasis in the mammalian retina. Nat Rev Neurosci. 2010;11(8):563‐576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ng L, Lu A, Swaroop A, Sharlin DS, Swaroop A, Forrest D. Two transcription factors can direct three photoreceptor outcomes from rod precursor cells in mouse retinal development. J Neurosci. 2011;31(31):11118‐11125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Regent F, Chen HY, Kelley RA, Qu Z, Swaroop A, Li T. A simple and efficient method for generating human retinal organoids. Mol Vis. 2020;26:97‐105. [PMC free article] [PubMed] [Google Scholar]
- 28. Quadrato G, Nguyen T, Macosko EZ, et al. Cell diversity and network dynamics in photosensitive human brain organoids. Nature. 2017;545(7652):48‐53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. DiStefano T, Chen HY, Panebianco C, et al. Accelerated and improved differentiation of retinal organoids from pluripotent stem cells in rotating‐wall vessel bioreactors. Stem Cell Rep. 2018;10(1):300‐313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ovando‐Roche P, West EL, Branch MJ, et al. Use of bioreactors for culturing human retinal organoids improves photoreceptor yields. Stem Cell Res Ther. 2018;9(1):156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Phelan MA, Lelkes PI, Swaroop A. Mini and customized low‐cost bioreactors for optimized high‐throughput generation of tissue organoids. Stem Cell Investig. 2018;5:33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kaewkhaw R, Kaya KD, Brooks M, et al. Transcriptome dynamics of developing photoreceptors in three‐dimensional retina cultures recapitulates temporal sequence of human cone and rod differentiation revealing cell surface markers and gene networks. Stem Cells. 2015;33(12):3504‐3518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Welby E, Lakowski J, Di Foggia V, et al. Isolation and comparative transcriptome analysis of human fetal and iPSC‐derived cone photoreceptor cells. Stem Cell Rep. 2017;9(6):1898‐1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Capowski EE, Samimi K, Mayerl SJ, et al. Reproducibility and staging of 3D human retinal organoids across multiple pluripotent stem cell lines. Development. 2019;146(1):dev171686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hoshino A, Ratnapriya R, Brooks MJ, et al. Molecular anatomy of the developing human retina. Dev Cell. 2017;43:763‐779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mellough CB, Collin J, Queen R, et al. Systematic comparison of retinal organoid differentiation from human pluripotent stem cells reveals stage specific, cell line, and methodological differences. Stem Cells Translational Medicine. 2019;8(7):694‐706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Xie H, Zhang W, Zhang M, et al. Chromatin accessibility analysis reveals regulatory dynamics of developing human retina and hiPSC‐derived retinal organoids. Sci Adv. 2020;6(6):eaay5247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Jin ZB, Okamoto S, Osakada F, et al. Modeling retinal degeneration using patient‐specific induced pluripotent stem cells. PLoS One. 2011;6(2):e17084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Jin ZB, Okamoto S, Xiang P, Takahashi M. Integration‐free induced pluripotent stem cells derived from retinitis pigmentosa patient for disease modeling. Stem Cells Translational Medicine. 2012;1(6):503‐509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Phillips MJ, Perez ET, Martin JM, et al. Modeling human retinal development with patient‐specific induced pluripotent stem cells reveals multiple roles for visual system homeobox 2. Stem Cells. 2014;32(6):1480‐1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Buskin A, Zhu L, Chichagova V, et al. Disrupted alternative splicing for genes implicated in splicing and ciliogenesis causes PRPF31 retinitis pigmentosa. Nat Commun. 2018;9(1):4234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Chen HY, Welby E, Li T, Swaroop A. Retinal disease in ciliopathies: recent advances with a focus on stem cell‐based therapies. Transl Sci Rare Dis. 2019;4(1–2):97‐115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Shimada H, Lu Q, Insinna‐Kettenhofen C, et al. In vitro modeling using ciliopathy‐patient‐derived cells reveals distinct cilia dysfunctions caused by CEP290 mutations. Cell Rep. 2017;20(2):384‐396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Parfitt DA, Lane A, Ramsden CM, et al. Identification and correction of mechanisms underlying inherited blindness in human iPSC‐derived optic cups. Cell Stem Cell. 2016;18(6):769‐781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Branham K, Othman M, Brumm M, et al. Mutations in RPGR and RP2 account for 15% of males with simplex retinal degenerative disease. Invest Ophthalmol Vis Sci. 2012;53(13):8232‐8237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Megaw RD, Soares DC, Wright AF. RPGR: its role in photoreceptor physiology, human disease, and future therapies. Exp Eye Res. 2015;138:32‐41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Deng WL, Gao ML, Lei XL, et al. Gene correction reverses ciliopathy and photoreceptor loss in iPSC‐derived retinal organoids from retinitis pigmentosa patients. Stem Cell Rep. 2018;10(4):1267‐1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Huang KC, Wang ML, Chen SJ, et al. Morphological and molecular defects in human three‐dimensional retinal organoid model of X‐linked juvenile retinoschisis. Stem Cell Rep. 2019;13(5):906‐923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Quinn PM, Buck TM, Mulder AA, et al. Human iPSC‐derived retinas recapitulate the fetal CRB1 CRB2 complex formation and demonstrate that photoreceptors and Muller glia are targets of AAV5. Stem Cell Rep. 2019;12(5):906‐919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wimmer RA, Leopoldi A, Aichinger M, et al. Human blood vessel organoids as a model of diabetic vasculopathy. Nature. 2019;565(7740):505‐510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Achberger K, Probst C, Haderspeck J, et al. Merging organoid and organ‐on‐a‐chip technology to generate complex multi‐layer tissue models in a human retina‐on‐a‐chip platform. Elife. 2019;8:e46188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Saini JS, Corneo B, Miller JD et al. Nicotinamide ameliorates disease phenotypes in a human iPSC model of age‐related macular degeneration. Cell Stem Cell 2017;20(5):635‐647.e637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Vergara MN, Flores‐Bellver M, Aparicio‐Domingo S, et al. Three‐dimensional automated reporter quantification (3D‐ARQ) technology enables quantitative screening in retinal organoids. Development. 2017;144(20):3698‐3705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Hallam D, Hilgen G, Dorgau B, et al. Human‐induced pluripotent stem cells generate light responsive retinal organoids with variable and nutrient‐dependent efficiency. Stem Cells. 2018;36(10):1535‐1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Gonzalez‐Cordero A, Goh D, Kruczek K, et al. Assessment of AAV vector tropisms for mouse and human pluripotent stem cell‐derived RPE and photoreceptor cells. Hum Gene Ther. 2018;29(10):1124‐1139. [DOI] [PubMed] [Google Scholar]
- 56. Klimczak RR, Koerber JT, Dalkara D, Flannery JG, Schaffer DV. A novel adeno‐associated viral variant for efficient and selective intravitreal transduction of rat Muller cells. PLoS One. 2009;4(10):e7467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Mookherjee S, Chen HY, Isgrig K, et al. A CEP290 C‐terminal domain complements the mutant CEP290 of Rd16 mice in trans and rescues retinal degeneration. Cell Rep. 2018;25(3):611‐623.e616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Dulla K, Aguila M, Lane A, et al. Splice‐modulating oligonucleotide QR‐110 restores CEP290 mRNA and function in human c.2991+1655A>G LCA10 models. Mol Ther Nucleic Acids. 2018;12:730‐740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Kamao H, Mandai M, Okamoto S, et al. Characterization of human induced pluripotent stem cell‐derived retinal pigment epithelium cell sheets aiming for clinical application. Stem Cell Rep. 2014;2(2):205‐218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Assawachananont J, Mandai M, Okamoto S, et al. Transplantation of embryonic and induced pluripotent stem cell‐derived 3D retinal sheets into retinal degenerative mice. Stem Cell Rep. 2014;2(5):662‐674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Shirai H, Mandai M, Matsushita K, et al. Transplantation of human embryonic stem cell‐derived retinal tissue in two primate models of retinal degeneration. Proc Natl Acad Sci U S A. 2016;113(1):E81‐E90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Schwartz SD, Hubschman JP, Heilwell G, et al. Embryonic stem cell trials for macular degeneration: a preliminary report. Lancet. 2012;379(9817):713‐720. [DOI] [PubMed] [Google Scholar]
- 63. da Cruz L, Fynes K, Georgiadis O, et al. Phase 1 clinical study of an embryonic stem cell‐derived retinal pigment epithelium patch in age‐related macular degeneration. Nat Biotechnol. 2018;36(4):328‐337. [DOI] [PubMed] [Google Scholar]
- 64. Ben M'Barek K, Habeler W, Plancheron A, et al. Human ESC‐derived retinal epithelial cell sheets potentiate rescue of photoreceptor cell loss in rats with retinal degeneration. Sci Transl Med. 2017;9(421):eaai7471. [DOI] [PubMed] [Google Scholar]
- 65. Radtke ND, Seiler MJ, Aramant RB, Petry HM, Pidwell DJ. Transplantation of intact sheets of fetal neural retina with its retinal pigment epithelium in retinitis pigmentosa patients. Am J Ophthalmol. 2002;133(4):544‐550. [DOI] [PubMed] [Google Scholar]
- 66. Pearson RA, Barber AC, Rizzi M, et al. Restoration of vision after transplantation of photoreceptors. Nature. 2012;485(7396):99‐103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Pearson RA, Gonzalez‐Cordero A, West EL, et al. Donor and host photoreceptors engage in material transfer following transplantation of post‐mitotic photoreceptor precursors. Nat Commun. 2016;7:13029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Santos‐Ferreira T, Llonch S, Borsch O, Postel K, Haas J, Ader M. Retinal transplantation of photoreceptors results in donor‐host cytoplasmic exchange. Nat Commun. 2016;7:13028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Singh MS, Balmer J, Barnard AR, et al. Transplanted photoreceptor precursors transfer proteins to host photoreceptors by a mechanism of cytoplasmic fusion. Nat Commun. 2016;7:13537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Barnea‐Cramer AO, Wang W, Lu SJ, et al. Function of human pluripotent stem cell‐derived photoreceptor progenitors in blind mice. Sci Rep. 2016;6:29784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Hambright D, Park KY, Brooks M, McKay R, Swaroop A, Nasonkin IO. Long‐term survival and differentiation of retinal neurons derived from human embryonic stem cell lines in un‐immunosuppressed mouse retina. Mol Vis. 2012;18:920‐936. [PMC free article] [PubMed] [Google Scholar]
- 72. Kruczek K, Gonzalez‐Cordero A, Goh D, et al. Differentiation and transplantation of embryonic stem cell‐derived cone photoreceptors into a mouse model of end‐stage retinal degeneration. Stem Cell Rep. 2017;8(6):1659‐1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Iraha S, Tu HY, Yamasaki S, et al. Establishment of immunodeficient retinal degeneration model mice and functional maturation of human ESC‐derived retinal sheets after transplantation. Stem Cell Rep. 2018;10(3):1059‐1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Xu M, Lee EM, Wen Z, et al. Identification of small‐molecule inhibitors of Zika virus infection and induced neural cell death via a drug repurposing screen. Nat Med. 2016;22(10):1101‐1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Bian S, Repic M, Guo Z, et al. Genetically engineered cerebral organoids model brain tumor formation. Nat Methods. 2018;15(8):631‐639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Saengwimol D, Rojanaporn D, Chaitankar V, et al. A three‐dimensional organoid model recapitulates tumorigenic aspects and drug responses of advanced human retinoblastoma. Sci Rep. 2018;8(1):15664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Kaewkhaw R, Swaroop M, Homma K, et al. Treatment paradigms for retinal and macular diseases using 3‐D retina cultures derived from human reporter pluripotent stem cell lines. Invest Ophthalmol Vis Sci. 2016;57(5):ORSFl1‐ORSFl11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Langer KB, Ohlemacher SK, Phillips MJ, et al. Retinal ganglion cell diversity and subtype specification from human pluripotent stem cells. Stem Cell Rep. 2018;10(4):1282‐1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Collin J, Queen R, Zerti D, et al. Deconstructing retinal organoids: single cell RNA‐Seq reveals the cellular components of human pluripotent stem cell‐derived retina. Stem Cells. 2019;37(5):593‐598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Sridhar A, Hoshino A, Finkbeiner CR et al. Single‐cell transcriptomic comparison of human fetal retina, hPSC‐derived retinal organoids, and long‐term retinal cultures. Cell Rep 2020;30(5):1644‐1659.e1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Zerti D, Collin J, Queen R, Cockell SJ, Lako M. Understanding the complexity of retina and pluripotent stem cell derived retinal organoids with single cell RNA sequencing: current progress, remaining challenges and future prospective. Curr Eye Res. 2020;45(3):385‐396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Lukowski SW, Lo CY, Sharov AA, et al. A single‐cell transcriptome atlas of the adult human retina. EMBO J. 2019;38(18):e100811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Brooks MJ, Chen HY, Kelley RA, et al. Improved retinal organoid differentiation by modulating signaling pathways revealed by comparative transcriptome analyses with development in vivo. Stem Cell Rep. 2019;13(5):891‐905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Clark BS, Stein‐O'Brien GL, Shiau F, et al. Single‐cell RNA‐Seq analysis of retinal development identifies NFI factors as regulating mitotic exit and late‐born cell specification. Neuron. 2019;102(6):1111‐1126.e1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Mao X, An Q, Xi H, et al. Single‐cell RNA sequencing of hESC‐derived 3D retinal organoids reveals novel genes regulating RPC commitment in early human retinogenesis. Stem Cell Rep. 2019;13(4):747‐760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Kanton S, Boyle MJ, He Z, et al. Organoid single‐cell genomic atlas uncovers human‐specific features of brain development. Nature. 2019;574(7778):418‐422. [DOI] [PubMed] [Google Scholar]
- 87. Hale LJ, Howden SE, Phipson B, et al. 3D organoid‐derived human glomeruli for personalised podocyte disease modelling and drug screening. Nat Commun. 2018;9(1):5167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Liu Q, Tan G, Levenkova N, et al. The proteome of the mouse photoreceptor sensory cilium complex. Mol Cell Proteomics. 2007;6(8):1299‐1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Okawa H, Sampath AP, Laughlin SB, Fain GL. ATP consumption by mammalian rod photoreceptors in darkness and in light. Curr Biol. 2008;18(24):1917‐1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Kooragayala K, Gotoh N, Cogliati T, et al. Quantification of oxygen consumption in retina ex vivo demonstrates limited reserve capacity of photoreceptor mitochondria. Invest Ophthalmol Vis Sci. 2015;56(13):8428‐8436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Bhaduri A, Andrews MG, Mancia Leon W, et al. Cell stress in cortical organoids impairs molecular subtype specification. Nature. 2020;578(7793):142‐148. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing is not applicable to this article as no new data were created or analyzed in this study.


