Summary
The SARS-CoV-2 immune response in human milk has not yet been examined, although protecting infants and young children from COVID-19 is critical for limiting community transmission and preventing serious illness and death. Here, milk samples from eight COVID-19-recovered and seven COVID-19-suspected donors were tested for antibody (Ab) binding to the SARS-CoV-2 Spike protein. All samples exhibited significant specific IgA reactivity to the full Spike, whereas 80% exhibited significant IgA and secretory (s)Ab binding to the Receptor-Binding Domain (RBD). Additionally, 67% samples exhibited IgG and/or IgM binding to RBD. IgA and sAb titers were highly correlated, indicating most IgA to be sIgA. Overall, these data indicate that a robust sIgA-dominant SARS-CoV-2 Ab response in human milk after infection should be expected in a significant majority of individuals. Further research is highly warranted to determine Ab functionality and the potential for exploiting extracted milk sIgA for therapeutic use.
Subject Areas: Pediatrics, Immunology, Virology
Graphical Abstract
Highlights
-
•
All milk from recovered donors contained significant SARS-CoV-2-specific IgA
-
•
Most IgA could bind the Receptor-Binding Domain (important neutralization epitope)
-
•
Most Receptor-Binding Domain-specific IgA was in secretory (s) form
-
•
sIgA is durable in the mucosa, and thus potentially as a respiratory therapeutic
Pediatrics; Immunology; Virology
Introduction
In the 7 months following the first reported case of coronavirus disease 2019 (COVID-19) in December 2019, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has infected over 21 million people and caused more than 750,000 deaths worldwide (World Health Organization, 2020). Although COVID-19 pathology in children is typically mild compared with adults, approximately 10% of infants younger than 1 year who contract the virus will experience severe COVID-19 illness requiring advanced care (CDC COVID-19 Response Team, 2020; Dong et al., 2020). Given that COVID-19 pathology does not always correlate with transmissibility (Li et al., 2020; Wei et al., 2020), recent studies suggest that infants and young children can transmit SARS-CoV-2 (CDC COVID-19 Response Team, 2020; Lopez et al., 2020). As well, recently it has become evident that a minority of children will experience a “Multisystem Inflammatory Syndrome in Children associated with COVID-19” after SARS-CoV-2 infection, which has been fatal in certain cases (Riphagen et al., 2020; Verdoni et al., 2020). For all these reasons, protecting this population from infection is essential. One potential protective mechanism might be passive immunity via breastfeeding from a previously infected mother or milk donor.
To date, almost nothing is known about the antibody (Ab) response in human milk to SARS-CoV-2 (Lackey et al., 2020). One preprint by Yu et al. (2020) reported that two milk samples produced by a 32-year-old Chinese mother of a 13-month-old boy were positive for SARS-CoV-2 IgG and negative for IgM on days 8 and 24 after hospital admission. Additional research is urgently needed to test human milk for SARS-CoV-2-specific Abs and their functions. Knowing the typology and degree of COVID-19-specific Abs in human milk will help inform smart policy and treatment decisions for the many pregnant and breastfeeding mothers who are or will become infected by SARS-CoV-2.
Certainly, any evidence of SARS-CoV-2-specific Abs in human milk must also be carefully weighed against the risks of potential vertical transmission of SARS-CoV-2 through human milk (for a review, see Centeno-Tablante et al., 2020). At the time of writing, 9 of the 68 milk samples obtained from donors infected with SARS-CoV-2 that have been tested to date were found to contain SARS-CoV-2 RNA, although there is no evidence of SARS-CoV-2 transmission through breastfeeding and no replication-competent virus has been found in any milk samples (Centeno-Tablante et al., 2020; Chambers et al., 2020; Groβ et al., 2020; Wu et al., 2020).
Despite the dearth of research, there are strong reasons to expect some SARS-CoV-2-specific Abs to be present in the milk of previously infected mothers. Given that milk IgG originates predominantly from serum, it follows that specific IgG in milk should appear contemporaneously with the previously reported serum SARS-CoV-2 Ab response, although IgG comprises only ∼2% of milk Ig (Hurley and Theil, 2011). Approximately 90% of human milk Ab is IgA and ∼8% IgM, nearly all in secretory (s) form (sIgA/sIgM; polymeric Abs (Abs) complexed to j-chain and secretory component (SC) proteins (Brandtzaeg, 2010; Demers-Mathieu et al., 2018; Hurley and Theil, 2011). These secretory Abs are marked with SC as part of the mechanism by which they are secreted into the milk, whereby they are actively transported via the polymeric immunoglobulin receptor (pIgR), from which SC is cleaved (Brandtzaeg, 2010). SC is essential for protecting these Abs from relatively harsh mucosal environments such as the infant mouth and gut. The majority of sIgA/sIgM derives from the gut-associated lymphoid tissue (GALT), although there is also homing of B cells from other mucosa (i.e., the respiratory system) to the mammary gland. Therefore, we predicted SARS-CoV-2-specific sIgA/sIgM to be present in the milk of previously infected mothers.
Here, we sought to characterize the types and magnitude of targeted Abs in human milk against SARS-CoV-2. Specifically, this report details the findings regarding SARS-CoV-2-reactive IgA, IgG, IgM, and total sAb in 15 milk samples obtained from donors previously infected with COVID-19, 3–4 weeks after symptoms had abated.
Results
All Human Milk Samples Obtained Form COVID-19-Recovered Donors Contain Significant Levels of SARS-CoV-2-Specific IgA
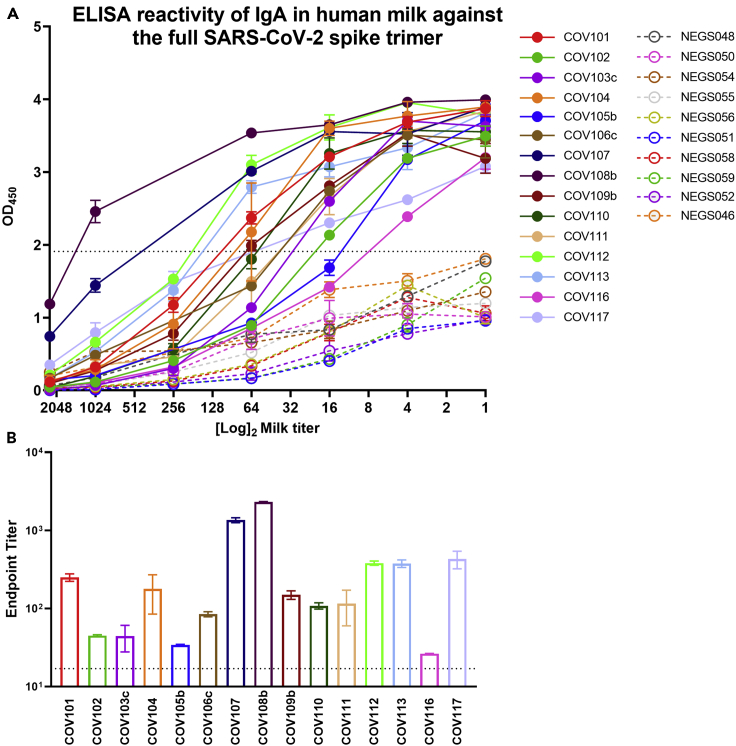
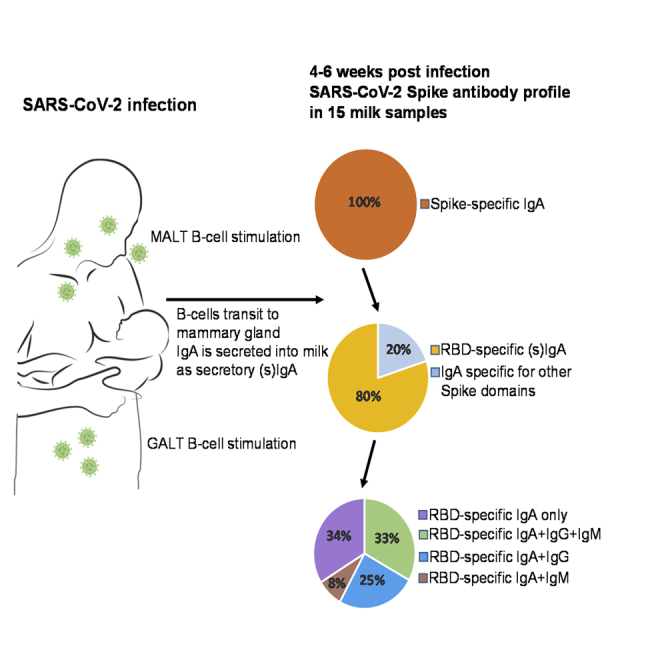
Milk samples were initially evaluated for IgA binding reactivity by human IgA-specific ELISA to the full trimeric SARS-CoV-2 Spike (Figure 1). It was evident that all samples obtained from COVID-19-recovered donors (100%), in undiluted form, exhibited binding activity significantly above that of the pre-pandemic control milk samples, which did exhibit some low-level non-specific or cross-reactive activity (Figure 1A). Milk samples were titrated and endpoint titer values were determined. It was found that all COVID-19-recovered samples exhibited endpoint titers significantly higher than control samples (Figure 1B).
Figure 1.
All Human Milk Samples Collected from COVID-19-Recovered Donors Exhibit IgA Reactivity Against the Full SARS-CoV-2 Spike Trimer
(A) Full titration against Spike. NEG (i.e., negative)/segmented lines: pre-pandemic controls. COV/solid lines: milk from COVID-19-recovered donors.
(B) Endpoint dilution titers. Experiments were performed in duplicate and repeated twice. Mean with SEM is shown. Dotted lines indicate positive cutoff value (mean OD or endpoint titer of negative control milk samples +2 ×SD). Segmented line indicates 10× positive cutoff value.
Most Milk from COVID-19-Recovered Donors Exhibits IgA and Secretory Ab Reactivity against the Receptor-Binding Domain of the SARS-CoV-2 Spike
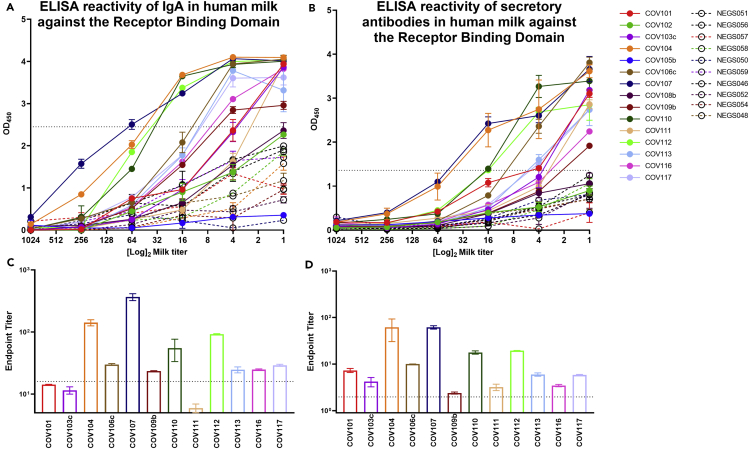
Samples were further tested separately for Ab binding reactivity to the Receptor-Binding Domain (RBD) of the Spike protein. Of 15 samples 12 (80%) obtained from previously COVID-19-infected donors exhibited significant IgA binding activity to RBD, as determined by human IgA-specific ELISA, compared with controls in undiluted form (Figure 2A). Milk samples were titrated, and endpoint titer values were determined. It was found that 9 of the 12 samples with reactive IgA to RBD when undiluted (75%) exhibited significant endpoint titers (Figure 2C). Notably, all 12 of the milk samples with significant IgA reactivity in undiluted form to RBD (100%) were also positive for RBD-specific secretory Ab reactivity as determined by human SC-specific ELISA (Figure 2B). All 12 milk samples also exhibited positive secretory Ab endpoint titers upon dilution compared with controls (Figure 2D).
Figure 2.
Eighty Percent of Human Milk Samples Collected from COVID-19-Recovered Donors Exhibit IgA and Secretory Antibody Reactivity Against the Receptor-Binding Domain (RBD) of the SARS-CoV-2 Spike
(A) Full titration against RBD, measuring IgA binding.
(B) IgA endpoint dilution titers.
(C) Full titration against RBD, measuring secretory antibody binding.
(D) Secretory antibody endpoint dilution titers. NEG (i.e., negative)/segmented lines: pre-pandemic controls. COV/solid lines: milk from COVID-19-recovered donors. Experiments were performed in duplicate and repeated twice. Mean with SEM is shown. Dotted lines indicate positive cutoff value (mean OD or endpoint titer of negative control milk samples +2 × SD).
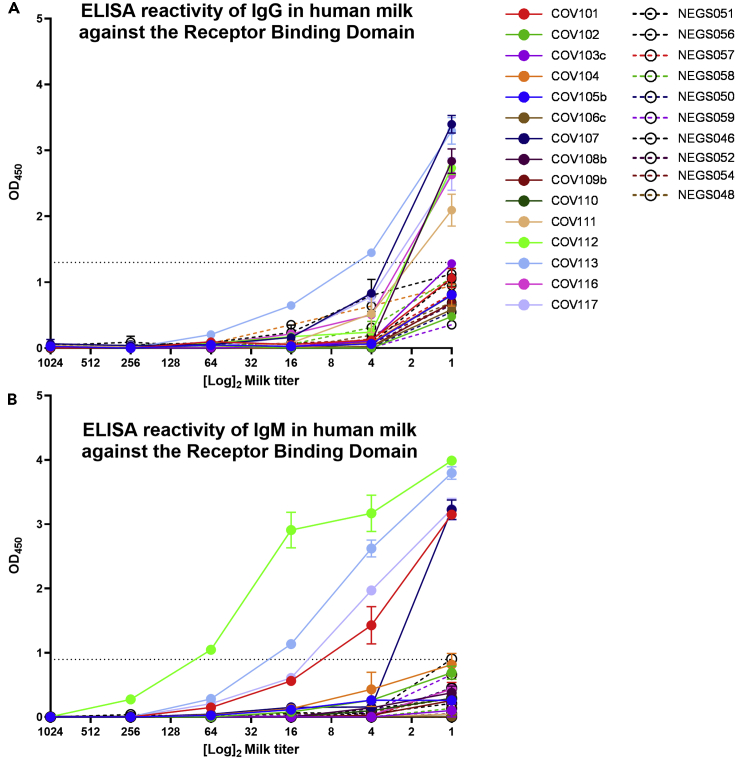
The RBD-Specific IgA Response in Milk Is Dominant and Not Necessarily Concurrent with a Measurable IgG or IgM Response
Individual profiles of Ab subclasses detected in the milk samples from COVID-19-recovered mothers are reported in Table 1. Notably, specific IgG and IgM were only measurable for a subset of samples, as determined by human IgG- and IgM-specific ELISAs, respectively. Of the 12 milk samples shown to be positive for IgA reactivity, 8 were also positive for IgG and/or IgM activity (67%). Four samples exhibited significant IgG and IgM reactivity to RBD (COV107, COV112, COV113, COV117; Figure 3). An additional 3 samples exhibited significant IgG reactivity but not IgM (COV108b, COV111, COV116), and 1 sample exhibited IgM reactivity but not IgG (COV101; Figure 3).
Table 1.
Summary Data
| Sample ID | COVID-19 Confirmed or Suspected (C/S) | Months Post-partum | Infected Ante- or Post-partum (A/P) | Reactivity Against SARS-CoV-2 Spike |
|||
|---|---|---|---|---|---|---|---|
| IgA | SCb | IgM | IgG | ||||
| COV101 | C | 4 | P | + | + | + | – |
| COV102 | C | 1 | Ad | +a | +c | – | – |
| COV103c | C | 4 | P | + | + | – | – |
| COV104 | S | 23 | P | + | + | – | – |
| COV105b | S | 6 | P | +a | +c | – | – |
| COV106c | S | 8 | P | + | + | – | – |
| COV107 | S | 32 | P | + | + | + | + |
| COV108b | C | 4 | P | +a | +c | – | + |
| COV109b | S | 3 | P | + | + | – | – |
| COV110 | S | 14 | P | + | + | – | – |
| COV111 | S | 7 | P | + | + | – | + |
| COV112 | C | 1 | Ad | + | + | + | + |
| COV113 | C | 7 | P | + | + | + | + |
| COV116 | C | 6 | P | + | + | – | + |
| COV117 | C | 4 | P | + | + | + | + |
Sample was positive against Spike but negative against RBD.
Secretory component.
SC reactivity against Spike is presumed based on RBD data, but was not tested.
Participants were infected within the last 6 weeks of pregnancy.
Figure 3.
The RBD-Specific IgA Response in Milk is Dominant and Not Necessarily Concurrent with a Measurable IgG or IgM Response
(A and B) Full titrations against RBD, measuring IgG (A) and IgM (B) binding are shown.
NEG (i.e., negative)/segmented lines: pre-pandemic controls. COV/solid lines: milk from COVID-19-recovered donors. Experiments were performed in duplicate and repeated twice. Mean with SEM is shown. Dotted lines indicate positive cutoff value (mean OD or endpoint titer of negative control milk samples +2 × SD).
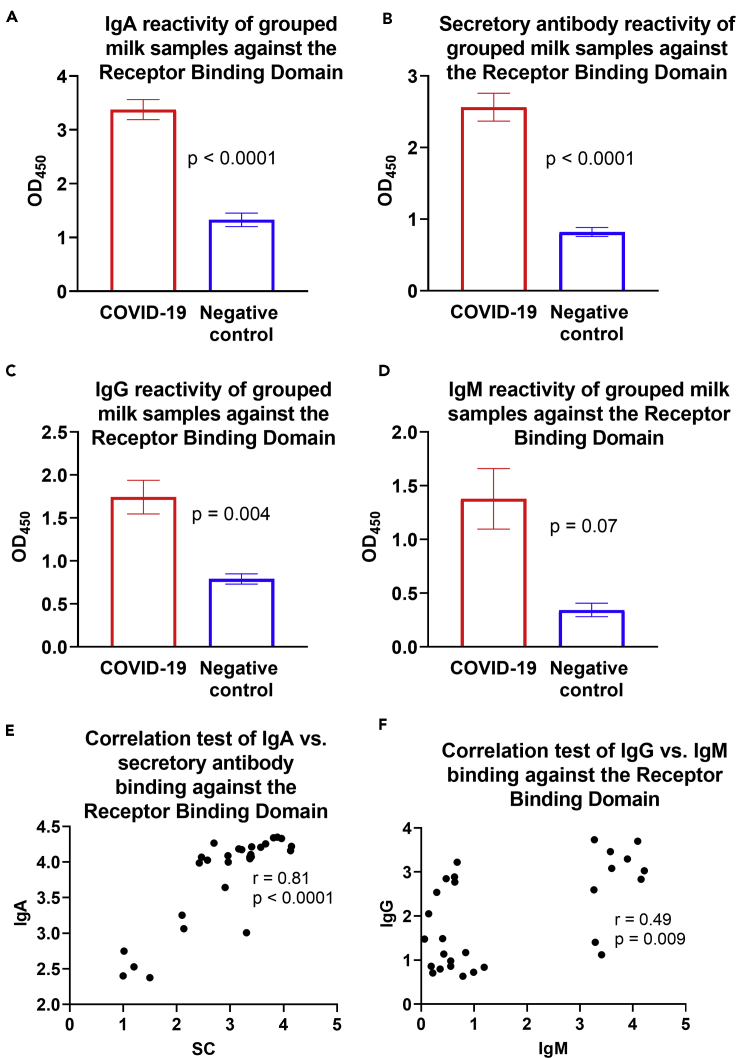
Milk from COVID-19-Recovered Donors Exhibits Significantly Greater IgA, Secretory Ab, and IgG Binding against RBD Compared with Controls
Overall, optical density (OD) values of undiluted milk obtained from COVID-19-recovered donors and pre-pandemic controls for each assay were grouped and compared, and it was found that the COVID-19-recovered group mean values were significantly greater for IgA (p < 0.0001), secretory Abs (p < 0.0001), and IgG (p = 0.004), but not for IgM (Figures 4A–4D). OD values for undiluted milk were compared between each Ab subclass. The IgA and secretory Ab OD values for undiluted milk were found to be highly correlated (r = 0.81, p < 0.0001; Figure 4E). IgM and IgG OD values were found to be modestly positively correlated (r = 0.49, p = 0.009; Figure 4F). No other correlations were found (data not shown).
Figure 4.
Milk from COVID-19-Recovered Donors Exhibits Significantly Greater IgA, Secretory Antibody, and IgG Binding Against RBD Compared with Controls
(A–D) Grouped OD values for undiluted milk are shown for IgA (A), secretory antibody (B), IgG (C), and IgM (D). Experiments were performed in duplicate and repeated twice. Mean with SEM is shown.
(E) Correlated IgA- versus secretory Ab-binding activity.
(F) Correlated IgG- versus IgM binding activity. For correlation tests, OD values for undiluted milk were used in 2-tailed Spearman correlation tests. SC: secretory component.
Discussion
All milk samples obtained from COVID-19-recovered participants were positive for Spike-reactive Ab of at least one subclass, namely, IgA. Eighty percent of these samples were specifically reactive against the RBD, with most (75%) exhibiting RBD binding activity that was quantitatively and/or qualitatively high such that endpoint titers were significantly above the background activity of the pre-pandemic controls. The samples analyzed represent only a snapshot of what is likely a dynamic immune response. A much larger sample size and long-term follow-up study is needed to better understand the time course of SARS-CoV-2 immunity in milk, as well as whether a typical response is truly protective for breastfed babies.
Although it might be expected that the milk Ab response would be reflective of systemic immunity (i.e., milk Ab should generally mirror serum Ab), only a small fraction of milk Ab originates from serum—likely less than 10%, and only ∼2% of milk Ab is IgG (Yu et al., 2020). Human milk Ab is ∼90% IgA and 8% IgM, nearly all sIgA/sIgM. The B cells that ultimately produce sIgA/sIgM originate mainly from the GALT, known as the entero-mammary link, with some proportion originating from other mucosa such as the respiratory system (Ahlstedt et al., 1977; Brandtzaeg, 2010; Kleinman and Walker, 1979). As such, there is much precedent for milk Ab composition and specificity being unique from that found in blood. Although we did not compare the milk donors' blood Ab titers to the milk data, it was evident that most of the samples contained SARS-CoV-2-reactive IgA without necessarily containing measurable IgG and/or IgM, which particularly in the case of IgG would likely be derived in large part from the serum. Notably, IgG and IgM reactivities in undiluted milk exhibited a moderate correlation. It may also be that as total IgG and IgM are so much lower in milk than IgA, this ELISA lacked the sensitivity to pick up very-low-titer responses.
Although it has been determined by previous studies that most IgA in human milk is sIgA, our ELISA could not determine with certainty that the IgA (or IgM) measured was of the secretory type or not (Brandtzaeg, 2010). The assay measuring secretory Ab reactivity employs a secondary Ab specific for the SC, which can be free or bound to Ab. Notably, all samples exhibiting positive IgA reactivity also exhibited positive SC reactivity, and a very strong positive correlation was present when comparing the OD values of undiluted milk for the IgA and SC assays. This suggests that a very high proportion of the SARS-CoV-2-reactive IgA measured herein was sIgA. This is extremely relevant to the possibility of using extracted milk Ab as a COVID-19 therapy—for anyone with severe COVID-19 disease, as sIgA is unique from the IgG-dominant convalescent plasma or purified plasma immunoglobulin being tested currently (Bloch et al., 2020). Extracted milk sIgA used therapeutically would likely survive well upon targeted respiratory administration, with a much lower dose of Ab likely needed for efficacy compared with systemically administered convalescent plasma or purified plasma immunoglobulin. Notably, the purified material would need to be extensively safety-tested, including ensuring it as free of SARS-CoV-2 material. Alternatively, recombinantly produced, monoclonal or polyclonal Spike-specific sIgA could be employed as a similar therapeutic.
As we continue with our comprehensive studies on the human milk immune response to SARS-CoV-2, we aim to ultimately determine the efficacy of “convalescent milk Ab” as a treatment for COVID-19, and the utility of these Abs to prevent or mitigate infant SARS-CoV-2 infection. These data will have implications beyond the pandemic, as they will serve to fill relatively large knowledge gaps regarding human milk immunology.
Limitations of the Study
Not all samples analyzed herein were obtained from participants with PCR-confirmed SARS-CoV-2 infection, and a larger sample set of confirmed infections should be studied to confirm these findings. The samples analyzed represent only a snapshot of what is likely a dynamic immune response. Long-term follow-up is needed to understand the kinetics of this response. Further functional studies, in particular neutralization data, are essential to understand this response.
Resource Availability
Lead Contact
Further information and requests for resources and reagents should be directed to the Lead Contact, Rebecca Powell: Rebecca.Powell@mssm.edu.
Materials Availability
No unique reagents have been generated in this study. The SARS-CoV-2 antigens used in this study are available from the Krammer lab upon reasonable request, and similar antigens are available commercially. All other reagents are commercially available.
Data and Code Availability
The data that support the findings of this study are available from the Lead Contact upon request.
Methods
All methods can be found in the accompanying Transparent Methods supplemental file.
Acknowledgments
As always, we are indebted to the milk donors who make this work possible.
This work was supported by the NIAID Centers of Excellence for Influenza Research and Surveillance (CEIRS) contract HHSN272201400008C and the Department of Medicine, Division of Infectious Diseases, Icahn School of Medicine at Mount Sinai.
Author Contributions
A.F. performed all experiments described in the paper, helped with data analysis, and gave final approval of the final manuscript. J.M. helped with data collection, gave feedback on drafts of the paper, and gave final approval of the final manuscript. F.A. and F.K. developed and provided SARS-CoV-2 antigens and gave final approval of the final manuscript. J.H.-H. helped with data collection, gave feedback on drafts of the paper, and gave final approval of the final manuscript. S.Z.-P. gave feedback on drafts of the paper and gave final approval of the final manuscript. R.L.P. conceived of the research project, oversaw data collection, preformed data analysis and interpretation, drafted and revised manuscript, and gave final approval of the manuscript.
Declaration of Interests
Mount Sinai has licensed serological assays to commercial entities and has filed for patent protection for SARS-CoV-2 serological and milk assays.
Published: November 20, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.isci.2020.101735.
Supplemental Information
References
- Ahlstedt S., Carlsson B., Fällström S.P., Hanson L.A., Holmgren J., Lidin-Janson G., Lindblad B.S., Jodal U., Kaijser B., Solh-Akerlund A. Antibodies in human serum and milk induced by enterobacteria and food proteins. Ciba Found. Symp. 1977;1:115–134. doi: 10.1002/9780470720288.ch6. [DOI] [PubMed] [Google Scholar]
- Bloch E.M., Shoham S., Casadevall A., Sachais B.S., Shaz B., Winters J.L., van Buskirk C., Grossman B.J., Joyner M., Henderson J.P. Deployment of convalescent plasma for the prevention and treatment of COVID-19. J. Clin. Invest. 2020;130:2757–2765. doi: 10.1172/JCI138745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandtzaeg P. The mucosal immune system and its integration with the mammary glands. J. Pediatr. 2010;156:S8–S15. doi: 10.1016/j.jpeds.2009.11.014. [DOI] [PubMed] [Google Scholar]
- CDC COVID-19 Response Team Coronavirus disease 2019 in children - United States, February 12-April 2, 2020. MMWR. Morb. Mortal. Wkly. Rep. 2020;69:422–426. doi: 10.15585/mmwr.mm6914e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centeno-Tablante E., Medina-Rivera M., Finkelstein J.L., Rayco-Solon P., Garcia-Casal M.N., Rogers L., Ghezzi-Kopel K., Ridwan P., Peña-Rosas J.P., Mehta S. Transmission of SARS-CoV-2 through breast milk and breastfeeding: a living systematic review. Ann. N. Y. Acad. Sci. 2020 doi: 10.1111/nyas.14477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers C., Krogstad P., Bertrand K., Contreras D., Tobin N.H., Bode L., Aldrovandi G. Evaluation for SARS-CoV-2 in Breast Milk From 18 Infected Women. JAMA. 2020;324:1347–1348. doi: 10.1001/jama.2020.15580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demers-Mathieu V., Underwood M.A., Beverly R.L., Nielsen S.D., Dallas D.C. Comparison of human milk immunoglobulin survival during gastric digestion between preterm and term infants. Nutrients. 2018;10:631. doi: 10.3390/nu10050631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Y., Mo X., Hu Y., Qi X., Jiang F., Jiang Z., Tong S. Epidemiological characteristics of 2143 pediatric patients with 2019 coronavirus disease in China. Pediatrics. 2020;145:e20200702. [Google Scholar]
- Groß R., Conzelmann C., Müller J.A., Stenger S., Steinhart K., Kirchhoff F., Münch J. Detection of SARS-CoV-2 in human breastmilk. Lancet. 2020;395:1757–1758. doi: 10.1016/S0140-6736(20)31181-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley W.L., Theil P.K. Perspectives on immunoglobulins in colostrum and milk. Nutrients. 2011;3:442–474. doi: 10.3390/nu3040442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinman R.E., Walker W.A. The enteromammary immune system: an important new concept in breast milk host defense. Dig. Dis. Sci. 1979;24:876–882. doi: 10.1007/BF01324906. [DOI] [PubMed] [Google Scholar]
- Lackey K.A., Pace R.M., Williams J.E., Bode L., Donovan S.M., Järvinen K.M., Seppo A., Raiten D.J., Meehan C.L., McGuire M.A. SARS-CoV-2 and human milk: what is the evidence? MedRxiv. 2020 doi: 10.1101/2020.04.07.20056812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R., Pei S., Chen B., Song Y., Zhang T., Yang W., Shaman J. Substantial undocumented infection facilitates the rapid dissemination of novel coronavirus (SARS-CoV-2) Science. 2020;368:489–493. doi: 10.1126/science.abb3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez A., Hill M., Antezano J., Vilven D., Rutner T., Bogdanow L., Claflin C., Kracalik I., Fields V., Dunn A. Transmission dynamics of COVID-19 outbreaks associated with child care Facilities - salt lake city, Utah, April-July 2020. MMWR. Morb. Mortal. Wkly. Rep. 2020;69:1319–1323. doi: 10.15585/mmwr.mm6937e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riphagen S., Gomez X., Gonzalez-Martinez C., Wilkinson N., Theocharis P. Hyperinflammatory shock in children during COVID-19 pandemic. Lancet. 2020;395:1607–1608. doi: 10.1016/S0140-6736(20)31094-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdoni L., Mazza A., Gervasoni A., Martelli L., Ruggeri M., Ciuffreda M., Bonanomi E., D’Antiga L. An outbreak of severe Kawasaki-like disease at the Italian epicentre of the SARS-CoV-2 epidemic: an observational cohort study. Lancet. 2020;395:P1771–P1778. doi: 10.1016/S0140-6736(20)31103-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei W.E., Li Z., Chiew C.J., Yong S.E., Toh M.P., Lee V.J. Presymptomatic transmission of SARS-CoV-2-Singapore, January 23–March 16, 2020. Morb. Mortal. Wkly. Rep. 2020;69:411–415. doi: 10.15585/mmwr.mm6914e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization Coronavirus disease (COVID-2019) situation report - 126. Saf. Risk Pharmacother. 2020;8:3–8. [Google Scholar]
- Wu Y., Liu C., Dong L., Zhang C., Chen Y., Liu J., Zhang C., Duan C., Zhang H., Mol B.W. Viral shedding of COVID-19 in pregnant women. SSRN Electron. J. 2020 doi: 10.2139/ssrn.3562059. [DOI] [Google Scholar]
- Yu Y., Xu J., Li Y., Hu Y., Li B. Breast Milk-Fed Infant of COVID-19 Pneumonia Mother: A Case Report. 2020. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the Lead Contact upon request.