Abstract
The IL‐7 receptor specific α chain, CD127, can be expressed both as a membrane‐associated (mCD127) and a soluble form (sCD127), however, the mechanisms involved in their regulation remain to be defined. We first demonstrated in primary human CD8+ T cells that IL‐7‐induced downregulation of mCD127 expression is dependent on JAK and PI3K signaling, whereas IL‐7‐induced sCD127 release is also mediated by STAT5. Following stimulation with IL‐7, expression of alternatively spliced variants of the CD127 gene, sCD127 mRNA, is reduced, but to a lesser degree than the full‐length gene. Evaluation of the role of proteases revealed that MMP‐9 was involved in sCD127 release, without affecting the expression of mCD127, suggesting it does not induce direct shedding from the cell surface. Since defects in the IL‐7/CD127 pathway occur in various diseases, including HIV, we evaluated CD8+ T cells derived from HAART‐treated HIV‐infected individuals and found that IL‐7‐induced (1) downregulation of mCD127, (2) release of sCD127, and (3) expression of the sCD127 mRNA were all impaired. Expression of mCD127 and sCD127 is, therefore, regulated by distinct, but overlapping, mechanisms and their impairment in HIV infection contributes to our understanding of the CD8+ T cell dysfunction that persists despite effective antiretroviral therapy.
Keywords: CD127, CD8+ T cells, HAART, HIV, IL‐7
IL‐7 induces sCD127 release through JAK, STAT5, and PI3K signaling, whereas IL‐7‐induced downregulation of mCD127 does not involve STAT5. Moreover, IL‐7 causes an increase in the ratio of sCD127/total CD127 mRNA. In HAART‐treated HIV infection, IL‐7‐induced sCD127 release, mCD127 downregulation, and the increased sCD127/total CD127 mRNA ratio are all impaired.

Introduction
Interleukin (IL)‐7, a pivotal cytokine in cell‐mediated immune responses, regulates homeostasis and development of T cells [1, 2, 3, 4]. This cytokine is recognized by the IL‐7 receptor (IL‐7R), composed of the cytokine‐specific high‐affinity α‐chain (CD127) and the common γ chain (γc; CD132). On binding to its receptor, IL‐7 activates two major pathways: the Janus kinase/signal transducer and activator of transcription 5 (JAK/STAT5) and the Akt/phosphatidylinositol 3‐kinase (Akt‐PI3K) pathways [5, 6]. Since the expression of CD132 is relatively constant, the degree of CD127 expression determines the extent of signaling through the IL‐7R complex [7, 8]. Moreover, IL‐7 is known to regulate CD127 expression and, as such, the IL‐7R complex. In the presence of exogenous IL‐7, human T cells cultured in vitro downregulate the expression of membrane‐bound CD127 (mCD127) and reduce the levels of CD127 mRNA transcripts [9, 10, 11]. In vivo administration of IL‐7 also results in a reduction of the CD127 expression on both human and murine T cells [12, 13].
Both components of the IL‐7 receptor (CD127 and CD132) also exist as soluble entities (i.e., sCD127 and sCD132) in human plasma. When combined, the heterocomplex can bind IL‐7 with an affinity comparable to that of the membrane bound receptor [14]. It has since been shown that the presence of CD132 is not a pre‐requisite for IL‐7‐sCD127 binding [15], however, the importance of IL‐7‐sCD127 binding is unclear and there is a lack of consensus on whether sCD127 increases or inhibits the bioactivity of IL‐7 [16, 17, 18].
Along with its effect on reducing mCD127, IL‐7 can also induce the release of sCD127 from human T cells [11]. Two potential methods for the production of sCD127 have been proposed. First, IL‐7 can induce an alternative mRNA splicing pathway for CD127, leading to the production of sCD127. The splicing event results in the removal of exon 6, which encodes the transmembrane domain, leading to a shift in the reading frame and a premature stop codon. The resulting truncated protein lacks the transmembrane domain and thus exists as a soluble protein [14, 19]. Second, our previous work suggested that the release of sCD127 may occur due to the proteolytic cleavage of membrane‐bound CD127 [11], similar to other soluble cytokine receptors (e.g., TNFR1 and IL‐6R) [20, 21]. Whether one or both of these mechanisms are responsible for IL‐7‐induced expression of sCD127 remains unknown.
A number of nonhuman primate studies have determined that CD8+ T cells are critical in the control of HIV infection and that CD8+ T cell depletion results in uncontrolled viral replication and increase in disease progression [22, 23, 24]. In concordance with these results, subsequent studies in humans have reported the presence of a more robust CD8+ T cell response in HIV nonprogressors [25, 26, 27]. We and others have shown that, in the face of their importance in controlling viral replication, CD8+ T cells, as well as thymocytes and Th17 cells, retain impaired responsivity to IL‐7 in virally suppressed HIV‐infected individuals [28, 29, 30, 31, 32].
In this context, we aimed to study the pathways involved in IL‐7‐mediated regulation of mCD127 and sCD127 expression by CD8+ T cells and to determine whether these pathways are impaired in HAART‐controlled HIV‐infected patients.
Results
IL‐7‐induced downregulation of mCD127 is mediated by JAK and PI3K
To confirm the effect of IL‐7 on the expression of mCD127 on CD8+ T cells, isolated CD8+ T cells were cultured with IL‐7 (1 ng/mL, 10 ng/mL) for up to 72 h. In keeping with previously published data [11, 13], with IL‐7 (10 ng/mL), a significant reduction in the proportion of CD127+ CD8+ T cells was observed and maintained over the 72 h period. The lower concentration of IL‐7 (1 ng/mL) induced a significant, but transient, reduction in this population, that returned to baseline levels by 72 h (Fig. 1A and B).
Figure 1.
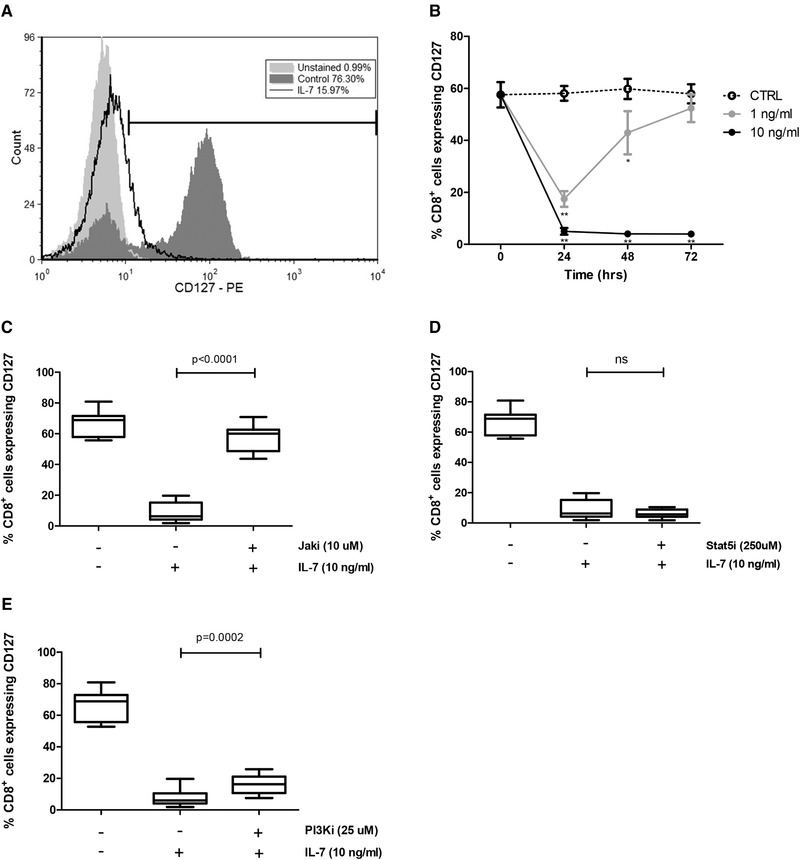
IL‐7‐induced downregulation of mCD127 is dependent on JAK and PI3K but not STAT5. CD8+ T cells were magnetically isolated from PBMCs derived from healthy individuals and cultured with or without IL‐7. For some experiments, inhibitors were used as described. Flow cytometry was performed to evaluate expression of CD127. (A) Representative flow cytometry histogram showing the CD127 median fluorescence intensity (MFI) and the percentage of CD127+ cells among unstained cells (light gray), medium control (dark gray), and the IL‐7‐ (10 ng/mL) treated cells (black unfilled). The gating strategy is shown in Supporting Information Fig. S6. (B) Time course of CD127 expression on CD8+ cells with media control (dotted line), 1 ng/mL IL‐7 (gray line), and 10 ng/mL IL‐7 (black line). Significance was calculated between each IL‐7 concentration in relation to the medium control (*p < 0.05, **p < 0.001, two‐way ANOVA with Bonferroni posttest), n = 8. Data are shown as mean ± SEM of eight samples from four independent experiments. (C) The effect of JAK inhibitor (10 μM) (n = 8), (D) STAT5 inhibitor (250 μM) (n = 8), and € PI3K inhibitor (25 μM) (n = 10) in the IL‐7‐induced reduction in the proportion of CD8+ T cells that express CD127 was evaluated at 48 h. Significance was calculated between IL‐7 alone in relation to IL‐7 plus each inhibitor (paired t test). Graphs show the distribution of samples from five independent experiments.
JAK, STAT5, and PI3K represent the primary signaling pathways that are activated following IL‐7 binding to the IL‐7R complex [5, 6]. Therefore, to investigate the mechanisms by which IL‐7 regulates the surface expression of CD127 on CD8+ T cells, chemical inhibitors were used to block the JAK, STAT5, and PI3K signaling pathways. Inhibition of JAK abrogated IL‐7‐induced mCD127 downregulation on CD8+ T cells (Fig. 1C). STAT5 inhibition, on the other hand, did not prevent the IL‐7‐induced reduction of CD127 surface expression (Fig. 1D), excluding a role for STAT5 in IL‐7‐mediated downregulation of mCD127. PI3K inhibition partially prevented this downregulation (Fig. 1E).
These data indicate that the IL‐7‐induced decrease in CD127 surface expression is mediated by JAK and in part by PI3K, but not STAT5 signaling pathways.
IL‐7‐induced sCD127 release from CD8+ T cells is mediated by JAK, STAT5 and PI3K
IL‐7 is known to induce sCD127 release from CD8+ T cells [11], however, the mechanism by which this occurs has not been established. To address this, CD8+ T cells were first stimulated with IL‐7 (1 ng/mL, 10 ng/mL) for 24, 48, and 72 h. In the presence of a higher concentration of IL‐7 (10 ng/mL), a significant increase in sCD127 was detected at all time points, when compared to untreated cells. When a lower concentration of IL‐7 (1 ng/mL) was used, a significant increase in sCD127 was only detected at the later time points (48 and 72 h) (Fig. 2A). This increase in sCD127 production in response to IL‐7 (1 ng/mL) was significantly less than when cells were treated with 10 ng/mL IL‐7. These data indicate that sCD127 is released in response to IL‐7 in a dose‐dependent manner.
Figure 2.
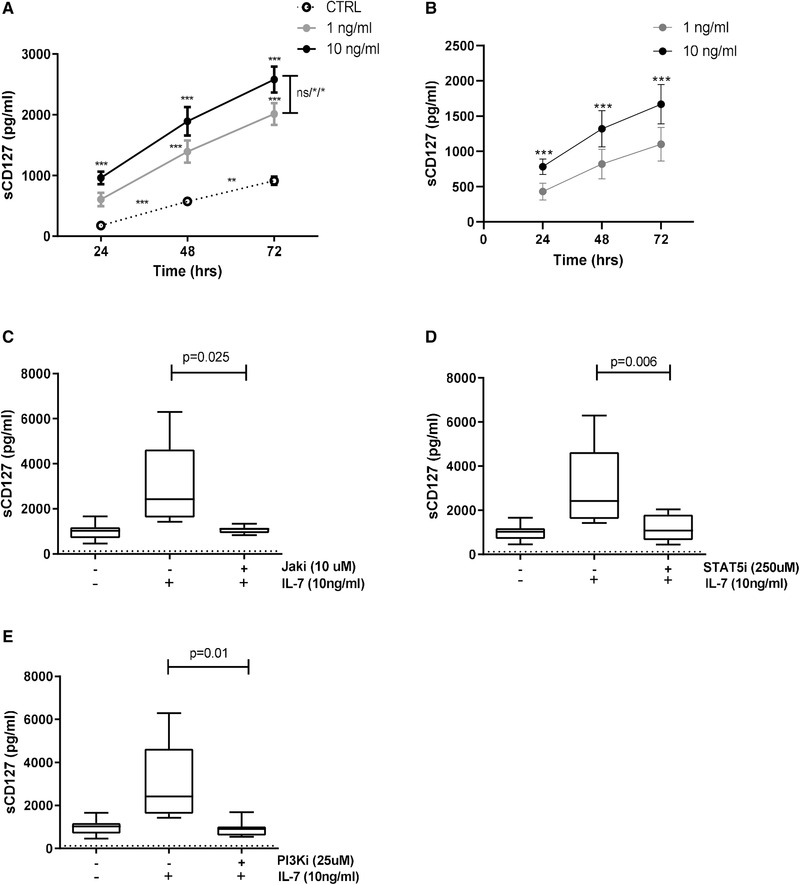
IL‐7‐induced sCD127 release from CD8+ T cells involves JAK, STAT5, and PI3K. CD8+ T cells were magnetically isolated from PBMCs derived from healthy individuals and cultured with or without IL‐7. For some experiments, inhibitors were used as described. ELISA assays were performed to evaluate secretion of sCD127. (A) sCD127 release with media alone (dotted line), 1 ng/mL IL‐7 (gray line), and 10 ng/mL IL‐7 (black line) over time. sCD127 release was compared in the different time points between media alone and 1 ng/mL IL‐7 or media alone and 10 ng/mL IL‐7 (two‐way ANOVA with Bonferroni posttest); between 1 ng/mL IL‐7 and 10 ng/mL IL‐7 on each time point (two‐way ANOVA with Bonferroni posttest); and between the time points in the media alone (one‐way ANOVA with Bonferroni post‐test), n = 4. Data are shown as mean ± SEM of four samples from two independent experiments. (B) sCD127 release was normalized to the control values (values obtained in the media alone cell cultures were subtracted) and compared between cells treated with 1 ng/mL IL‐7 (grey line) or 10 ng/mL IL‐7 (black line) over time (two‐way ANOVA with Bonferroni posttest), n = 4. Data are shown as mean ± SEM of four samples from two independent experiments. (C) The impact of JAK inhibitor (10 μM), (D) STAT5 inhibitor (250 μM), and (E) PI3K (25 μM) inhibitor in the IL‐7‐induced sCD127 secretion in 48 h (paired t test), n = 7. Graphs show the distribution of seven samples from four independent experiments. The dotted line represents the limit of detection for this assay (125 pg/mL) (*p < 0.05, **p < 0.01, ***p < 0.001, ns: not significant).
Further, sCD127 levels were also detected in cultures without addition of IL‐7, and increased significantly over time (Fig. 2A). This increase in the sCD127 over time was also observed when the sCD127 values were normalized to the control (through subtraction from the control values) (Fig. 2B). Together, these data demonstrate the occurrence of spontaneous release of the soluble receptor, consistent with previous observations [15, 33].
In order to determine if the JAK, STAT5, or PI3K signaling proteins play a role in the release of sCD127, CD8+ T cells were incubated for 48 h in the presence of IL‐7 (10 ng/mL) as well as 10 μM JAK inhibitor, 250 μM STAT5 inhibitor, or 25 μM PI3K inhibitor. All three inhibitors completely abrogated the effect of IL‐7 on sCD127 release (Fig. 2C‐E) identifying a role for each of these pathways in IL‐7‐mediated expression of sCD127.
IL‐7 may induce an alternative splicing of CD127 mRNA
It has been shown that sCD127 can be generated from the CD127 gene via alternative splicing [14]. A PCR probe was designed to overlap the exon 5–7 junction to detect the sCD127 alternatively spliced variant. Production of the CD127 mRNA variants (full‐length and the alternatively spliced sCD127 mRNA variant) in response to IL‐7 treatment was then quantified. A significant decrease in levels of total CD127 mRNA (i.e., including RNA encoding both mCD127 and sCD127) was detected at the 4, 8, and 24 h time points following exposure to IL‐7 (1 ng/mL, 10 ng/mL) (Fig. 3A), whereas the sCD127 mRNA variant levels decreased at the 8 and 24 h time points (Fig. 3B). Incubation with the higher concentration of IL‐7 (10 ng/mL) resulted in a continued reduction in total CD127 mRNA to 24 h (Fig. 3B). When the ratio of sCD127 mRNA to total CD127 mRNA was determined, both concentrations of IL‐7 induced an increase in the ratio of sCD127:total CD127 mRNA (Fig. 3C). These data indicate that IL‐7 induces a pathway resulting in the reduction of both total and sCD127 mRNA. While the effect on the sCD127 mRNA variant is somewhat spared, these effects of IL‐7 do not account for the increased levels of sCD127 protein.
Figure 3.

IL‐7 induces a preference for the alternatively spliced CD127 mRNA. CD8+ T cells were magnetically isolated from PBMCs derived from healthy individuals and cultured with or without IL‐7. RNA was extracted, converted to cDNA, and a quantitative probe‐based PCR was performed. (A) Time course of total CD127 mRNA (i.e., including RNA encoding both mCD127 and sCD127 variants) detected following IL‐7 stimulation of CD8+ T cells, n = 8. (B) Time course of alternatively spliced CD127 mRNA (sCD127 mRNA) expression after stimulation with IL‐7, n = 8. (C) The sCD127:total CD127 mRNA ratio over time, n = 8. *p < 0.01, **p < 0.001 by two‐way ANOVA with Bonferroni posttest. Significance was calculated in relation to the medium control. Data are shown as mean ± SEM of eight samples per group from four independent experiments.
Direct shedding from the surface of CD8+ T cells is not the main source of sCD127
Since IL‐7 induced reduction of both total (i.e., including RNA encoding both mCD127 and sCD127) and alternatively spliced sCD127 mRNA levels (Fig. 3a and B), increased gene transcription could not account for the IL‐7‐mediated increase in sCD127 protein (Fig. 2A). We, therefore, decided to investigate whether IL‐7 induces direct shedding of CD127 from the CD8+ T cell membrane, a process that might explain how IL‐7 enhances sCD127 expression.
Cleavage of membrane‐bound cytokine receptors by metalloproteinases, producing their soluble forms, has been demonstrated (e.g., IL‐6R and TNFR1) [20, 21], however, dependence on protease cleavage for the release of sCD127 has yet to be evaluated. Various metalloproteinase inhibitors were added to CD8+ T cell cultures prior to IL‐7 stimulation. While inhibition of MMP‐2 or other metalloproteinases did not impact the release of sCD127 (Fig. 4A and data not shown), inhibition of MMP‐9 activity prevented IL‐7‐induced release of sCD127 (Fig. 4B). The effect of blocking MMP‐9 on mCD127 expression was then assessed to address whether mCD127 was being cleaved by MMP‐9 to produce sCD127. Interestingly, MMP‐9 inhibition did not prevent the IL‐7‐induced reduction in mCD127 expression on CD8+ T cells (Fig. 4C). Together, these results suggest that neither gene transcription nor direct shedding from the cell surface is the main sources of the IL‐7‐induced sCD127 production.
Figure 4.

Role of MMP‐2, MMP‐9, and brefeldin on IL‐7‐induced sCD127 release and impact of MMP‐9 inhibition on IL‐7‐induced reduction of mCD127 expression on CD8+ T cells. CD8+ T cells were magnetically isolated from PBMCs derived from healthy individuals and cultured with or without IL‐7. For some experiments, inhibitors were used as described. Release of sCD127 was evaluated through ELISA. Expression of CD127 was evaluated through flow cytometry. (A) Effect of the addition of MMP‐2 inhibitor (20 μM) (n = 3) and (B) MMP‐9 inhibitor (20 μM) (n = 9) in the IL‐7‐induced production of sCD127 (paired t test). (C) Effect of the addition of MMP‐9 inhibitor in the IL‐7‐induced reduction in the proportion of CD8+ cells expressing CD127 (paired t test), n = 9. (D) Effect of brefeldin A (1 μg/mL) in the IL‐7‐induced sCD127 expression at 24 h (paired t test), n = 6. The dotted line represents the limit of detection for this assay (125 pg/mL). Significance was calculated between IL‐7 alone in relation to IL‐7 plus each inhibitor. Graphs show the distribution of samples from four independent experiments.
One other possible source of sCD127 is preformed protein. If the source of sCD127 is preformed protein, it would need to be transported intracellularly and then secreted. Thus, in order to determine if mobilization of intracellular vesicles was involved with the release of sCD127, the effect of the transport inhibitor, brefeldin A (BFA), was evaluated. BFA acts to inhibit secretion by disassembling the Golgi complex and thus stopping the ER‐to‐Golgi transport. Interestingly, inhibition of ER‐to‐Golgi transport with BFA resulted in the suppression of the IL‐7‐induced sCD127 release by CD8+ T cells (Fig. 4D), indicating that the transport of intracellular vesicles, through the Golgi complex, is required for IL‐7‐induced sCD127 release.
In order to evaluate if suppression of sCD127 transport was the mechanism through which BFA inhibited sCD127 release, CD8+ T cells were cultured with or without BFA (1 μg/mL). Flow cytometry analysis revealed that treatment with BFA had no impact on the surface CD127 expression (Supporting Information Fig. S1A). Interestingly, when cells were permeabilized before being stained with anti‐CD127 antibody, identifying both surface and intracellular expression, a modest increase in CD127 expression by CD8+ T cells was observed (Supporting Information Fig. S1B). Though not quite significant, this observation may suggest that treatment with BFA induces intracellular accumulation of CD127.
Impaired responsiveness to IL‐7 in HAART‐treated HIV infection
CD8+ T cells derived from HIV+ individuals are known to possess impaired responses, including those in the IL‐7 pathway [8, 28, 29], however, whether HIV infection affects the modulation of both mCD127 and sCD127 expression, in response to IL‐7, still remains to be elucidated. We first determined that CD127 was expressed on the surface of the CD8+ T cells isolated from the HIV+ individuals. Indeed, the percentage of CD8+ T cells expressing CD127 was slightly but significantly higher in the HAART‐treated HIV+ individuals studied here, when compared to the healthy controls, indicating that any potential lack in IL‐7 response would not be due to reduced levels of mCD127 (Fig. 5A).
Figure 5.

CD8+ T cells isolated from HIV+ HAART‐treated individuals are less responsive to IL‐7 than those from healthy individuals. CD8+ T cells were magnetically isolated from PBMCs derived from healthy controls and HIV+ HAART‐treated individuals and cultured with or without IL‐7. Expression of CD127 was evaluated through flow cytometry. Release of sCD127 was evaluated through ELISA. RNA was extracted, converted to cDNA, and a quantitative probe‐based PCR was performed to quantify mRNA as described. (A) The percentage of CD127+ cells within the CD8+ cell population in HIV+ individuals compared to healthy individuals (unpaired t test), n = 8 for both cohorts. (B, C) Relative change in the surface CD127 expression over time with 1 ng/mL and 10 ng/mL IL‐7 stimulation (two‐way ANOVA), n = 8. (D, E) IL‐7 induced sCD127 release in HIV+ and healthy individuals. Upon stimulation with IL‐7, sCD127 release was quantified over time in the HIV+ individuals and compared to the healthy individuals (two‐way ANOVA) (1 ng/mL IL‐7 HIV+, n = 6, healthy individuals, n = 7; 10 ng/mL IL‐7 HIV+, n = 9, healthy individuals, n = 7; controls: HIV+, n = 9, healthy individuals, n = 7). (F) Total CD127 mRNA (i.e., includes mCD127 and sCD127 variants) detected following 24 h of IL‐7 stimulation in HIV+ and healthy individuals (HIV+, n = 9, healthy individuals, n = 8) (two‐way ANOVA with Bonferroni posttest). (G) sCD127 mRNA expression following 24 h of 1 ng/mL and 10 ng/mL IL‐7 stimulation in HIV+ and healthy individuals (two‐way ANOVA with Bonferroni posttest) (controls: HIV+, n = 9, healthy, n = 10; IL‐7, 1 ng/mL: HIV+, n = 10, healthy, n = 10; IL‐7, 10 ng/mL: HIV+, n = 10, healthy, n = 4). (H) Ratio of sCD127:mCD127 mRNA induced by 24 h incubation with 1 ng/mL and 10 ng/mL IL‐7 in HIV+ and healthy individuals (two‐way ANOVA with Bonferroni posttests) (controls: HIV+, n = 9, healthy, n = 8; IL‐7, 1 ng/mL and 10 ng/mL: HIV+, n = 10, healthy, n = 8). Data are shown as mean ± SEM from four (A–C) and five (D–H) independent experiments.
Differences in IL‐7 responsiveness between healthy and HIV+ HAART‐treated individuals were then assessed. When CD8+ T cells were stimulated with 1 ng/mL IL‐7, the decrease in mCD127 expression was significantly reduced in CD8+ T cells from the HIV+ cohort (Fig. 5B). However, when CD8+ T cells isolated from healthy and HIV+ individuals were cultured with 10 ng/mL IL‐7, both groups showed a similar pattern in the reduction in mCD127 (Fig. 5C), suggesting a decrease in sensitivity to IL‐7 exists in HIV infection despite effective treatment.
IL‐7‐induced sCD127 release was then assessed in HIV+ and healthy individuals. With the lower IL‐7 concentration (1 ng/mL), significantly less sCD127 was released from CD8+ T cells isolated from HIV+ individuals than from healthy individuals (Fig. 5D). However, with the higher IL‐7 concentration (10 ng/mL), there was no significant difference in sCD127 release between HIV+ individuals and healthy controls (Fig. 5E), suggesting again that CD8+ cells from HIV+ individuals are less sensitive to IL‐7.
The effect of HIV infection on CD127 mRNA expression was then analyzed. Stimulation of CD8+ T cells from healthy and HAART‐treated HIV+ individuals with 1 ng/mL or 10 ng/mL IL‐7 resulted in a significant reduction in both total and sCD127 mRNA expression (Fig. 5F and G). However, while IL‐7 induced an increase in the ratio of sCD127:total CD127 mRNA in CD8+ T cells from healthy individuals (Fig. 5G), this was not observed in HAART‐treated HIV+ individuals. Again, these data suggest that CD8+ T cells isolated from treated HIV+ patients have an impaired ability to respond to IL‐7.
Discussion
CD127 is expressed both as a membrane‐bound surface receptor (mCD127) and in a soluble form (sCD127) [14, 19], however, the mechanisms involved in the regulation of the expression of the two forms are not well understood. This work sought to determine how IL‐7 induces both a reduction in mCD127 and an increase in the release of sCD127. We demonstrate that mCD127 and sCD127 expression in CD8+ T cells are regulated by distinct yet overlapping pathways. Specifically, IL‐7‐induced downregulation of mCD127 is dependent on JAK and PI3K, whereas IL‐7‐induced release of sCD127 involves JAK, PI3K, and STAT5. IL‐7 also induces reduction of both total (i.e., including RNA encoding both mCD127 and sCD127) and alternatively spliced sCD127 mRNA, but with a degree of sparing of the alternatively spliced sCD127 mRNA variant, the gene transcript of which has been implicated in the production of sCD127 protein [14]. The release of sCD127 is dependent on MMP‐9 activity, however, this protease does not impact the expression of mCD127 on the cell surface, suggesting mechanisms of release not related to proteolytic cleavage of the membrane receptor as seen with other cytokine receptors [20, 21]. Lastly, despite long‐term HAART and the high levels of mCD127 expression, CD8+ T cells derived from HIV+ individuals are less sensitive to the effects induced by IL‐7.
In the present work, we demonstrate that IL‐7‐induced sCD127 release by CD8+ T cells is dependent on STAT5 activity, whereas IL‐7‐induced downregulation of mCD127 is found to be STAT5 independent. This finding suggests that at least two different pathways appear to be involved in the release of sCD127 and reduction of mCD127 expression at the cell surface. Further, while IL‐7 induced a pathway favoring the expression of the alternatively spliced version of the CD127 gene, the absolute levels of alternatively spliced sCD127 mRNA did not increase, indicating that IL‐7 does not induce the production of sCD127 through increased gene expression. It would thus appear that IL‐7 induces a pathway resulting in posttranscriptional or translational regulation of the sCD127 gene. Posttranslational regulation of mCD127 has been previously demonstrated, where expression of mCD127 was shown to be more stable on the surface of T cells as a result of interactions with Ephrinb1 and Ephrinb2, also cell surface molecules [34]. Interestingly, it was reported that Ephrin signaling can be modulated by proteases [35, 36]. In this context, although better characterization of the interplay between IL‐7, Ephrins, and proteases is needed, it is likely that following translation of the alternatively spliced sCD127 mRNA variant, IL‐7 induces further pathways that result in an increase in sCD127.
Shedding of a number of cytokine receptors has been shown to be induced by proteases [20, 21]. Here, we demonstrated a role for MMP‐9 in sCD127 release. Inhibition of MMP‐9 had no effect on mCD127, suggesting that the mechanism by which MMP‐9 induces sCD127 expression is not by proteolytic cleavage of mCD127. Therefore, distinct mechanisms appear to be involved in the downregulation of mCD127 and the release of sCD127. Additionally, since MMP‐9 has been shown to be active and able to degrade proteins intracellularly [37], it is possible that MMP‐9 cleaves molecules involved with the intracellular processing of CD127. Beyond its proteolytic activity, MMP‐9 could be contributing to the phenomena (i.e., sCD127 release) observed here through other mechanisms. MMP‐9 has recently been shown to inhibit the type 1 IFN signaling pathway by binding IFNAR1, repressing JAK/STAT pathways [38]. Another study demonstrated that pro‐MMP‐9 binding to α4β7 induces STAT3 and results in an increase in Bcl‐2/Mcl‐2, preventing apoptosis in some cell line models [39]. Since we demonstrated that IL‐7‐induced sCD127 is dependent on both MMP‐9 and STAT5, it is possible that MMP‐9 plays a role in this IL‐7 activity by modulating STAT5.
The sCD127 produced in response to IL‐7, as observed here, may also originate from preformed protein. This is supported by the observation that BFA inhibited sCD127 release, likely as a result of interruption of the transport of sCD127 containing vesicles through the secretory pathway from the Golgi complex, before extracellular secretion (reviewed in [40]). Indeed, we observed that when CD8+ T cells were permeabilized before CD127 staining, making it possible to evaluate both intracellular and surface expression, treatment with BFA appears to induce a modest increase in CD127 (Supporting Information Fig. S1B). Moreover, while previous work from our group did not demonstrate an abundance of intracytoplasmic CD127 [11], the methods used in that work were not developed to identify CD127 within intracellular vesicles. Further studies are needed in order to evaluate the potential importance of preformed sCD127 within CD8+ T cells.
Given its importance in T‐cell homeostasis, IL‐7 has been studied in the context of a number of diseases characterized by impaired cell‐mediated immunity, such as HIV. In the present study, despite the high level of CD127 expression on CD8+ T cells that accompanied long‐term effective HAART, CD8+ T cells derived from HIV+ individuals were less responsive to IL‐7 with respect to downregulation of mCD127 and induction of sCD127. This decreased sensitivity of CD8+ T cells from effectively treated HIV+ individuals is in line with previous work from our group [32]. Since CD132 is constitutively expressed and has even been shown to be upregulated on T cells from HIV+ patients [7], any reduction in IL‐7 response is unlikely due to lack of the IL‐7R complex. Further, following IL‐7 stimulation, CD8+ T cells isolated from HIV+ individuals did not display a preference for the alternatively spliced sCD127 mRNA variant, as observed in cells derived from healthy individuals. This may contribute to the decrease in sCD127 release, in response to IL‐7 stimulation, showing that, even on long term treatment, CD8+ T cells lack the full functions of those from healthy individuals. Other pathogen‐induced impairment of IL‐7 responses has been identified in infections in humans. For example, the immune exhaustion experienced by patients infected with Trypanosoma cruzi has been shown to be mediated, at least in part, by a decrease in IL‐7R signal transduction in CD8+ T cells [41]. In HIV infection, it is well known that CD8+ T cells have alterations in their functionality (reviewed in [42]), and that immune dysfunction can persist despite long term antiretroviral treatment [43, 44, 45, 46]. Very recently, our group demonstrated that Th17 CD4+ T cells isolated from HIV+ patients on long‐term HAART had a reduced proliferative response to IL‐7 [31]. The exact reasons why complete recovery of T‐cell function is not achieved, despite prolonged suppression of viral replication and normalization of other aspects of host immunity, have yet to be elucidated.
Another interesting approach for future studies is to evaluate how the regulation of both sCD127 and mCD127 is affected in different CD8+ T cell subsets. Colle and colleagues demonstrated that distinct CD8+ T cell subsets present different regulation of the expression of mCD127 when treated with IL‐7. The group showed that memory CD8+ T cells from HAART‐treated HIV+ patients regained the ability to downregulate CD127 expression in response to IL‐7 [47]. These results, however, were demonstrated only with 10 ng/mL IL‐7, the higher concentration in the present study. The present work has also some limitations that could be addressed in future studies: small sample size and the difference in age and sex of the groups analyzed.
In summary, we demonstrate here that mCD127 downregulation and sCD127 release by CD8+ T cells, in response to IL‐7, are controlled by different, but overlapping mechanisms. MMP‐9 was shown to be involved with sCD127 release, but not by inducing mCD127 shedding from the cell surface. Moreover, despite the long‐term effective HIV treatment, immune impairment in CD8+ T cell responses to IL‐7 persists and is characterized by decreased sCD127 release, mCD127 downregulation, and expression of the alternatively spliced sCD127 mRNA variant.
Our findings provide further insight into the mechanisms involved in the complex regulation of the membrane bound and the soluble forms of CD127 and how they are impaired in HIV infection. A better understanding of these mechanisms may be important for the development of new therapeutic strategies for restoring CD8+ T cells function in settings in which IL‐7 plays a critical role.
Materials and methods
Sample collection and cell isolation
All research conducted using blood from human subjects was approved by the Ottawa Hospital Research Institute Ethics Board (Ottawa, ON, Canada) and conforms to the provisions of the Declaration of Helsinki. Informed consent was obtained from all participating subjects. Peripheral blood was collected in 100 i.u./mL heparin (LEO Pharma Inc., Thornhill, ON, Canada) from healthy volunteers and HIV+ HAART‐treated individuals (viral load: <40 copies/mL (undetectable) for ≥12 months). Subjects characteristics are shown in Table 1.
Table 1.
Subjects characteristics
| Healthy individuals | HIV+ individuals | |
|---|---|---|
| Age | 38.6 ± 11.1 | 51.0 ± 10.4 |
| Gender | 43% Male (20 female, 15 male) | 55% Male (9 female, 11 male) |
| Duration of infection | 14.3 ± 7.9 | |
| CD4 count | 635.6 ± 155.0 |
Peripheral blood mononuclear cells (PBMC) were isolated by gradient centrifugation using Lymphoprep (Stemcell Technologies Inc., Vancouver, BC, Canada). CD8+ T cells were isolated from fresh PBMCs using the EasySep™ human CD8 Positive Selection Kit (Stemcell Technologies Inc, Vancouver, BC, Canada) according to the manufacturer's instructions and rested overnight in complete medium (RPMI 1640 supplemented with 20% heat inactivated fetal bovine serum [FBS], 100 units/mL penicillin, 100 μg/mL streptomycin, and 0.25 mM l‐glutamine; Life Technologies, Burlington, ON, Canada) before use. Cell purity was assessed by flow cytometry (mouse α‐human α‐CD8 PE, clone HIT8α; BD Biosciences Pharmingen, San Jose, CA, USA) and was 96.5 ± 3.3% for the cells derived from healthy individuals and 96.7 ± 3.7% for the cells derived from HIV+ individuals.
CD8+ T cell stimulation and addition of inhibitors
CD8+ T cells (2 × 106 cells/mL) were stimulated for the indicated time with medium alone, or media with the indicated concentration of IL‐7 (Peprotech, Montreal, QC, Canada).
For the inhibition assays, CD8+ T cells were treated with either 10 μM JAK inhibitor (CAS 457081‐03‐7; Millipore‐Sigma, USA), 250 μM STAT5 inhibitor (CAS 285986‐31‐4; Millipore‐Sigma), 25 μM PI3K inhibitor (LY294002) (CAS 154447‐36‐6; Millipore‐Sigma), 5–50 μM Broad MMP inhibitor GM6001 (364205, Calbiochem; Millipore‐Sigma), 5–50 μM Broad MMP plus ADAM17 inhibitor TAPI‐O (579050, Calbiochem; Millipore‐Sigma), 1–20 μM MMP‐2 inhibitor (444294, Calbiochem; Millipore‐Sigma), 20 μM MMP‐9 inhibitor (444278, Calbiochem; Millipore‐Sigma) or 1 μg/mL BFA (CAS 20350‐15‐6; Millipore‐Sigma) for 2 h before the addition of IL‐7. In each case, the treatment of cells with signaling and protease inhibitors did not affect cell viability as determined by propidium iodide (PI; Biolegend, San Diego, CA, USA) staining (data not shown). The activity of these inhibitors was confirmed by their ability to block IL‐7 activity in a dose‐dependent manner and the concentrations of the inhibitors, for subsequent experiments, were chosen based on the highest concentration with no or little impact on cell viability, as determined by annexin V staining (A13199; Invitrogen, Life Technologies, Carlsbad, CA, USA) (Supporting Information Figs. S2–S4). Inhibitors alone did not affect the expression of mCD127 or sCD127 (Supporting Information Fig. S5).
Expression of surface or intracellular CD127 on CD8+ T‐cells
For the evaluation of surface expression of CD127, 50 μL of isolated CD8+ T cells (2 × 106/mL) were labeled in PBS 1% BSA with 5 μL monoclonal mouse anti‐human CD127‐PE antibody (clone R34.34, Beckman Coulter Inc., Pasadena, CA, USA) for 30 min at room temperature.To evaluate both intracellular and surface expression of CD127, cells were fixed and permeabilized with 200 μL BD fixation‐permeabilization solution for 10 min at 4°C and washed with 1 mL Fix‐Perm wash buffer from BD Fixation/Permeabilization kit (554714; BD Biosciences), and labeled in 100 μL Fix‐Perm wash buffer with Human IL‑7R alpha/CD127 Biotinylated Antibody (250 pg/μL) (BAF306; R&D Systems, Minneapolis, USA). After a 30 min incubation at 4°C, cells were washed with 1 mL BD Fix‐Perm wash buffer and incubated for 10 min at 4°C with 10 μL diluted streptavidin PE‐Cy7 (2 ng/mL) (557598; BD Biosciences) to detect the antibody. After washing the cells, CD127 expression was assessed on a FC500 Beckman Coulter Flow Cytometer. Gating strategy is shown in Supporting Information Fig. S6. All flow cytometry experiments were performed as per the “Guidelines for the use of flow cytometry and cell sorting in immunological studies (second edition)” [48].
Quantification of sCD127
A sandwich ELISA was used to determine the concentration of sCD127 in the culture supernatants of CD8+ T cells cultured with IL‐7 at the indicated time points. The assay was performed as described previously [49]. Briefly, high binding ELISA plates (Sarstedt, Nümbrecht, Germany) were coated overnight with anti‐CD127 antibody (2.5 μg/mL, clone 40131; R&D Systems). Samples and standard (human IL‐7Rα/Fc chimera extracellular domain; R&D Systems) were tested in triplicate. After 2‐h incubation at room temperature, the plate was washed with washing buffer (PBS with 0.05% Tween 20), biotin‐conjugated α‐human CD127 antibody (50 μg/mL) (R&D Systems) was added, and followed by another 2‐h incubation. The reaction was revealed with streptavidin‐HRP (Millipore‐Sigma), using enhanced K‐blue TMB as substrate (Cedarlane, Burlington, Canada). The plates were assayed with a SpectraMAX 190 microplate spectrophotometer and SoftMAX Pro2.4.1 software (Molecular Devices Corp., USA) at 450 nm, and optical density (OD) values were corrected by subtracting OD measurements at 540 nm. The concentrations of sCD127 in sample supernatants were extrapolated from the standard curve by linear regression analysis.
RNA extraction and RT‐PCR
RNA was extracted from 1.5 to 2 × 106 cells with GE Illustra Mini prep kit (GE Healthcare Life Sciences, MA, USA) according to the manufacturer's instructions. RNA was quantified with a NanoDropTM (ThermoFisher Scientific, MA, USA) and converted to cDNA with iScript cDNA synthesis kit (BioRad, CA, USA) according to the manufacturer's instructions.
Quantification of total CD127 mRNA (i.e., including RNA encoding both mCD127 and sCD127) was performed using primers designed with PrimerBlast in order to cover exons 5–7 (accession number NM_002185.4) (CD127ex7Rev1 and CD127ex5For1; Table 2). With these primers, this PCR is able to amplify full‐length CD127 and RNA that skipped exon 6. Glyceraldehyde‐3‐phosphate dehydrogenase (GAPDH) gene was used as the reference gene using PrimePCR™ SYBR® Green Assay (Bio‐Rad). The RT‐PCR reaction for CD127 and GAPDH was performed using SsoAdvanced™ Universal SYBR Green Supermix (BioRad). To evaluate the presence of the alternatively spliced sCD127 mRNA variant, which lacks exon 6 (sCD127 mRNA), a probe‐based assay was used. The probe covered nucleotides 777–815 of the variant (Seq.ID XM_005248299.3, exon 5–7 junction) (Table 2). Following thermal activation of polymerase (95°C for 30 s), 40 cycles of PCR amplification (with each cycle consisting of a 95°C denaturation step for 15 s and a 60°C annealing step for 15 s) were performed using the CFX Connect™ Real‐Time PCR Detection System (BioRad). Analyses were completed using the BioRad CFX Manager Software. The specificity of all primer sets was confirmed by analysis of PCR product amplicon size on agarose gel electrophoresis.
Table 2.
Sequences of primers and probe used for the quantification of total and alternatively spliced CD127 mRNA
| Primers/probe | Sequence (5′‐3′) | |
|---|---|---|
| Primers | CD127ex7Rev1 | TCT TGG TTT CTT ACA AAG ATG TTC C |
| CD127ex5For1 | CCA ACC GGC AGC AAT GTA TG | |
| Probe | CD127 exons 5–7 probe | /56‐FAM/AGA TCA ATA /ZEN/ ATA GCT CAG GAT TAA GCC TAT CGT ATG GCC/3IABkFQ/ |
Statistical analysis
All statistical analyses were conducted using GraphPad Prism 5.0 software (GraphPad, CA, USA). Statistical significance of treatment effects was determined by paired two‐tailed Student's t‐tests. The effect of different concentrations of the inhibitors was evaluated with one‐way ANOVA with Dunnett's multiple comparison. Time and dose‐dependent effects were detected by two‐way ANOVA, with Bonferroni posttest when multiple comparisons were done. Evaluation of sCD127 spontaneous release over time and analysis of CD127 expression in response to treatment with IL‐7 or IL‐7 and BFA was determined by one‐way ANOVA with Bonferroni posttest. Comparison of CD127 expression between the different groups was determined by unpaired two‐tailed Student's t‐tests. p Values ≤ 0.05 were considered statistically significant for all comparisons.
Conflict of interest
The authors declare no commercial or financial conflict of interest.
Abbreviations
- BFA
brefeldin A
- IL‐7R
IL‐7 receptor
- mCD127
membrane‐bound CD127
Supporting information
Supporting information
Acknowledgements
We would like to acknowledge and thank all the participants who generously donated their blood for the study. J.B.A., S.C.C., and A.M.C. conceived and designed the experiments. S.C.C., S.B.S., A.M.D., and M.S.‐V. carried out the experiments. S.C.C. and S.B.S. analyzed the data, S.C.C. and A.M.C. wrote the first drafts of the manuscript, and P.O.B. wrote the final versions of the manuscript. All authors contributed to editing the manuscript. This work was supported by an operating grant from the Canadian Institutes of Health Research (CIHR) (HOP‐98830). The funding agency played no role in study design, collection or analysis of data, writing of the manuscript, or decision to submit for publication. A.M.C. was supported by a CIHR New Investigator Award and an Ontario HIV Treatment Network Junior Investigator Development Award.
The peer review history for this article is available at https://publons.com/publon/10.1002/eji.201948453
References
- 1. Chetoui N., Boisvert M., Gendron S. and Aoudjit F., Interleukin‐7 promotes the survival of human CD4 + effector/memory T cells by up‐regulating Bcl‐2 proteins and activating the JAK/STAT signalling pathway. Immunology 2010. 130: 418–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Li W. Q., Jiang Q., Aleem E., Kaldis P., Khaled A. R. and Durum S. K., IL‐7 promotes T cell proliferation through destabilization of p27Kip1. J. Exp. Med. 2006. 203: 573–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Liu Z.‐H., Wang M.‐H., Ren H.‐J., Qu W., Sun L.‐M., Zhang Q.‐F., Qiu X.‐S. et al., Interleukin 7 signaling prevents apoptosis by regulating bcl‐2 and bax via the p53 pathway in human non‐small cell lung cancer cells. Int. J. Clin. Exp. Pathol. 2014. 7: 870–881. [PMC free article] [PubMed] [Google Scholar]
- 4. Wofford J. A., Wieman H. L., Jacobs S. R., Zhao Y. and Rathmell J. C., IL‐7 promotes Glut1 trafficking and glucose uptake via STAT5‐mediated activation of Akt to support T‐cell survival. Blood 2008. 111: 2101–2111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Pallard C., Stegmann A. P., van Kleffens T., Smart F., Venkitaraman A. and Spits H., Distinct roles of the phosphatidylinositol 3‐kinase and STAT5 pathways in IL‐7‐mediated development of human thymocyte precursors. Immunity 1999. 10: 525–535. [DOI] [PubMed] [Google Scholar]
- 6. Barata J. T., Silva A., Brandao J. G., Nadler L. M., Cardoso A. A. and Boussiotis V. A., Activation of PI3K is indispensable for interleukin 7–mediated viability, proliferation, glucose use, and growth of T cell acute lymphoblastic leukemia cells. J. Exp. Med. 2004. 200: 659–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sasson S. C., Zaunders J. J., Zanetti G., King E. M., Merlin K. M., Smith D. E., Stanley K. K. et al., Increased plasma interleukin‐7 level correlates with decreased CD127 and increased CD132 extracellular expression on T cell subsets in patients with HIV‐1 infection. J. Infect. Dis. 2006. 193: 505–514. [DOI] [PubMed] [Google Scholar]
- 8. Faller E. M., McVey M. J., Kakal J. A. and MacPherson P. A., Interleukin‐7 receptor expression on CD8 T‐cells is downregulated by the HIV Tat protein. J. Acquir. Immune Defic. Syndr. 2006. 43: 257–269. [DOI] [PubMed] [Google Scholar]
- 9. Alves N. L., van Leeuwen E. M. M., Derks I. A. M. and van Lier R. A. W., Differential regulation of human IL‐7 receptor α expression by IL‐7 and TCR signaling. J. Immunol. 2008. 180: 5201–5210. [DOI] [PubMed] [Google Scholar]
- 10. Ghazawi F. M., Faller E. M., Sugden S. M., Kakal J. A. and MacPherson P. A., IL‐7 downregulates IL‐7Rα expression in human CD8 T cells by two independent mechanisms. Immunol. Cell Biol. 2013. 91: 149–158. [DOI] [PubMed] [Google Scholar]
- 11. Vranjkovic A., Crawley A. M., Gee K., Kumar A. and Angel J. B., IL‐7 decreases IL‐7 receptor (CD127) expression and induces the shedding of CD127 by human CD8+ T cells. Int. Immunol. 2007. 19: 1329–1339. [DOI] [PubMed] [Google Scholar]
- 12. Park J.‐H., Yu Q., Erman B., Appelbaum J. S., Montoya‐Durango D., Grimes H. L. and Singer A., Suppression of IL7Rα transcription by IL‐7 and other prosurvival cytokines: a novel mechanism for maximizing IL‐7‐dependent T cell survival. Immunity 2004. 21: 289–302. [DOI] [PubMed] [Google Scholar]
- 13. Sereti I., Dunham R. M., Spritzler J., Aga E., Proschan M. A., Medvik K., Battaglia C. A. et al., IL‐7 administration drives T cell‐cycle entry and expansion in HIV‐1 infection. Blood 2009. 113: 6304–6314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Rose T., Lambotte O., Pallier C., Delfraissy J.‐F. and Colle J.‐H., Identification and biochemical characterization of human plasma soluble IL‐7R: lower concentrations in HIV‐1‐infected patients. J. Immunol. 2009. 182: 7389–7397. [DOI] [PubMed] [Google Scholar]
- 15. Monti P., Brigatti C., Krasmann M., Ziegler A. G. and Bonifacio E., Concentration and activity of the soluble form of the interleukin‐7 receptor α in type 1 diabetes identifies an interplay between hyperglycemia and immune function. Diabetes 2013. 62: 2500–2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Côté S., Matte J., Sad S., Angel J. B. and Crawley A. M., Complexed soluble IL‐7 receptor α and IL‐7 increase IL‐7‐mediated proliferation and viability of CD8+ T‐cells in vitro. Cell. Immunol. 2015. 293: 122–125. [DOI] [PubMed] [Google Scholar]
- 17. Crawley A. M., Faucher S. and Angel J. B., Soluble IL‐7R (sCD127) inhibits IL‐7 activity and is increased in HIV infection. J. Immunol. 2010. 184: 4679–4687. [DOI] [PubMed] [Google Scholar]
- 18. Lundström W., Highfill S., Walsh S. T. R., Beq S., Morse E., Kockum I., Alfredsson L. et al., Soluble IL7Rα potentiates IL‐7 bioactivity and promotes autoimmunity. Proc. Natl. Acad. Sci. USA 2013. 110: E1761–E1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Goodwin R. G., Friend D., Ziegler S. F., Jerzy R., Falk B. A., Gimpel S., Cosman D. et al., Cloning of the human and murine interleukin‐7 receptors: demonstration of a soluble form and homology to a new receptor superfamily. Cell 1990. 60: 941–951. [DOI] [PubMed] [Google Scholar]
- 20. Schumacher N., Meyer D., Mauermann A., von der Heyde J., Wolf J., Schwarz J., Knittler K. et al., Shedding of endogenous interleukin‐6 receptor (IL‐6R) is governed by a disintegrin and metalloproteinase (ADAM) proteases while a full‐length IL‐6R isoform localizes to circulating microvesicles. J. Biol. Chem. 2015. 290: 26059–26071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chanthaphavong R. S., Loughran P. A., Lee T. Y. S., Scott M J. and Billiar T. R., A role for cGMP in inducible nitric‐oxide synthase (iNOS)‐induced tumor necrosis factor (TNF) α‐converting enzyme (TACE/ADAM17) activation, translocation, and TNF receptor 1 (TNFR1) shedding in hepatocytes. J. Biol. Chem. 2012. 287: 35887–35898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Matano T., Shibata R., Siemon C., Connors M, Lane H. C. and Martin M. A., Administration of an anti‐CD8 monoclonal antibody interferes with the clearance of chimeric simian/human immunodeficiency virus during primary infections of rhesus macaques. J. Virol. 1998. 72: 164–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Schmitz J. E., Kuroda M. J., Santra S., Sasseville V. G., Simon M. A., Lifton M. A., Racz P. et al., Control of viremia in simian immunodeficiency virus infection by CD8+ lymphocytes. Science. 1999. 283: 857–860. [DOI] [PubMed] [Google Scholar]
- 24. Veazey R. S., Acierno P. M., McEvers K. J., Baumeister S. H. C., Foster G. J., Rett M. D., Newberg M. H. et al., Increased loss of CCR5+ CD45RA− CD4+ T Cells in CD8+ lymphocyte‐depleted simian immunodeficiency virus‐infected rhesus monkeys. J. Virol. 2008. 82: 5618–5630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Betts M. R, Nason M. C., West S. M., De Rosa S. C., Migueles S. A., Abraham J., Lederman M. M. et al., HIV nonprogressors preferentially maintain highly functional HIV‐specific CD8+ T cells. Blood 2006. 107: 4781–4789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hersperger A. R., Martin J. N., Shin L. Y., Sheth P. M., Kovacs C. M., Cosma G. L., Makedonas G. et al., Increased HIV‐specific CD8+ T‐cell cytotoxic potential in HIV elite controllers is associated with T‐bet expression. Blood 2011. 117: 3799–3808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Migueles S. A., Laborico A. C., Shupert W. L., Sabbaghian M. S., Rabin R., Hallahan C. W., Van Baarle D. et al., HIV‐specific CD8+ T cell proliferation is coupled to perforin expression and is maintained in nonprogressors. Nat. Immunol. 2002. 3: 1061–1068. [DOI] [PubMed] [Google Scholar]
- 28. O'Connor A. M., Crawley A. M. and Angel J. B, Interleukin‐7 enhances memory CD8(+) T‐cell recall responses in health but its activity is impaired in human immunodeficiency virus infection. Immunology 2010. 131: 525–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Vranjkovic A., Crawley A. M., Patey A. and Angel J. B., IL‐7‐dependent STAT‐5 activation and CD8+ T cell proliferation are impaired in HIV infection. J. Leukoc. Biol. 2011. 89: 499–506. [DOI] [PubMed] [Google Scholar]
- 30. Perdomo‐Celis F., Velilla P. A., Taborda N. A. and Rugeles M. T., An altered cytotoxic program of CD8+ T‐cells in HIV‐infected patients despite HAART‐induced viral suppression. PLoS One 2019. 14:e0210540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Côté S. C., Stilla A., Burke Schinkel S. C., Berthoud T. K. and Angel J. B., IL‐7‐induced proliferation of peripheral Th17 cells is impaired in HAART‐controlled HIV infection. AIDS 2019. 33: 985–991. [DOI] [PubMed] [Google Scholar]
- 32. Young C. D. and Angel J. B., HIV infection of thymocytes inhibits IL‐7 activity without altering CD127 expression. Retrovirology 2011. 8: 72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mouillaux J., Allam C., Rimmelé T., Uberti T., Delwarde B., Textoris J., Monneret G. et al., Regulation of soluble CD127 protein release and corresponding transcripts expression in T lymphocytes from septic shock patients. Intensive Care Med. Exp. 2019. 7: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Luo H., Wu Z., Qi S., Jin W., Han B. and Wu J., Ephrinb1 and Ephrinb2 are associated with interleukin‐7 receptor α and retard its internalization from the cell surface. J. Biol. Chem. 2011. 286: 44976–44987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Beauchamp A., Lively M. O., Mintz A., Gibo D., Wykosky J. and Debinski W., EphrinA1 is released in three forms from cancer cells by matrix metalloproteases. Mol. Cell. Biol. 2012. 32: 3253–3264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Georgakopoulos A., Litterst C., Ghersi E., Baki L., Xu C., Serban G. and Robakis N. K., Metalloproteinase/presenilin1 processing of ephrinB regulates EphB‐induced Src phosphorylation and signaling. EMBO J. 2006. 25: 1242–1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zhang Z., Amorosa L. F., Coyle S. M., Macor M. A., Lubitz S. E., Carson J. L., Birnbaum M. J. et al., Proteolytic cleavage of AMPKα and intracellular MMP9 expression are both required for TLR4‐mediated mTORC1 activation and HIF‐1α expression in leukocytes. J. Immunol. 2015. 195: 2452–2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chen J., Xu W., Chen Y., Xie X., Zhang Y., Ma C., Yang Q. et al., Matrix metalloproteinase 9 facilitates hepatitis B virus replication through binding with type I interferon (IFN) receptor 1 to repress IFN/JAK/STAT signaling. J. Virol. 2017. 91: e01824‐16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Redondo‐Muñoz J., Ugarte‐Berzal E., Terol M. J., Van den Steen P. E., Hernández del Cerro M., Roderfeld M., Roeb E. et al., Matrix metalloproteinase‐9 promotes chronic lymphocytic leukemia B cell survival through its hemopexin domain. Cancer Cell 2010. 17: 160–172. [DOI] [PubMed] [Google Scholar]
- 40. Grieve A. G. and Rabouille C., Golgi bypass: skirting around the heart of classical secretion. Cold Spring Harb. Perspect. Biol. 2011. 3: a005298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Albareda M. C., Perez‐Mazliah D., Natale M. A., Castro‐Eiro M., Alvarez M. G., Viotti R., Bertocchi G. et al., Perturbed T cell IL‐7 receptor signaling in chronic Chagas disease. J. Immunol. 2015. 194: 3883–3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kuchroo V. K., Anderson A. C. and Petrovas C., Coinhibitory receptors and CD8 T cell exhaustion in chronic infections. Curr. Opin. HIV AIDS. 2014. 9: 439–445. [DOI] [PubMed] [Google Scholar]
- 43. Emu B., Moretto W. J., Hoh R., Krone M., Martin J. N., Nixon D. F., Deeks S. G. et al., Composition and function of T cell subpopulations are slow to change despite effective antiretroviral treatment of HIV disease. PLoS One 2014. 9: e85613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. French M. A., King M. S., Tschampa J. M., da Silva B. A. and Landay A. L., Serum immune activation markers are persistently increased in patients with HIV infection after 6 years of antiretroviral therapy despite suppression of viral replication and reconstitution of CD4+ T cells. J. Infect. Dis. 2009. 200: 1212–1215. [DOI] [PubMed] [Google Scholar]
- 45. Cobos Jiménez V., Wit F. W. N. M., Joerink M., Maurer I., Harskamp A. M., Schouten J., Prins M. et al., T‐cell activation independently associates with immune senescence in HIV‐infected recipients of long‐term antiretroviral treatment. J. Infect. Dis. 2016. 214: 216–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Rueda C. M., Velilla P. A., Chougnet C. A. and Rugeles M. T., Incomplete normalization of regulatory t‐cell frequency in the gut mucosa of Colombian HIV‐infected patients receiving long‐term antiretroviral treatment. PLoS One 2013. 8: e71062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Colle J.‐H., Moreau J.‐L., Fontanet A., Lambotte O., Joussemet M., Delfraissy J.‐F. and Theze J., CD127 expression and regulation are altered in the memory CD8 T cells of HIV‐infected patients—reversal by highly active anti‐retroviral therapy (HAART). Clin. Exp. Immunol. 2006. 143: 398–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Cossarizza A., Chang H., Radbruch A., Acs A., Adam D., Adam‐Klages S., Agace W. W. et al., Guidelines for the use of flow cytometry and cell sorting in immunological studies (second edition). Eur. J. Immunol. 2019. 49: 1457–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hoe E., McKay F. C., Schibeci S. D., Gandhi K., Heard R. N., Stewart G. J. and Booth D. R., Functionally significant differences in expression of disease‐associated IL‐7 receptor α haplotypes in CD4 T cells and dendritic cells. J. Immunol. 2010. 184: 2512–2517. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting information


