Summary
Plasma membrane (PM) intrinsic proteins (PIPs) are aquaporins facilitating the diffusion of water and small solutes. The functional importance of the PM organisation of PIPs in the interaction with other cellular structures is not completely understood.
We performed a pull‐down assay using maize (Zea mays) suspension cells expressing YFP‐ZmPIP2;5 and validated the protein interactions by yeast split‐ubiquitin and bimolecular fluorescence complementation assays. We expressed interacting proteins tagged with fluorescent proteins in Nicotiana benthamiana leaves and performed water transport assays in oocytes. Finally, a phylogenetic analysis was conducted.
The PM‐located ZmPIP2;5 physically interacts with the endoplasmic reticulum (ER) resident ZmVAP27‐1. This interaction requires the ZmVAP27‐1 cytoplasmic major sperm domain. ZmPIP2;5 and ZmVAP27‐1 localise in close vicinity in ER–PM contact sites (EPCSs) and endocytic structures upon exposure to salt stress conditions. This interaction enhances PM water permeability in oocytes. Similarly, the Arabidopsis ZmVAP27‐1 paralogue, AtVAP27‐1, interacts with the AtPIP2;7 aquaporin.
Together, these data indicate that the PIP2–VAP27 interaction in EPCSs is evolutionarily conserved, and suggest that VAP27 might stabilise the aquaporins and guide their endocytosis in response to salt stress.
Keywords: aquaporin, endocytosis, endoplasmic reticulum (ER), endoplasmic reticulum–plasma membrane (ER–PM) contact sites (EPCSs), plant vesicle‐associated membrane protein (VAMP)‐associated protein (VAP27), plasma membrane intrinsic protein (PIP)
Introduction
Plasma membrane (PM) intrinsic proteins (PIPs) are aquaporins that facilitate the diffusion of water and small solutes through lipid bilayers (Maurel et al., 2015; Fox et al., 2017). PIP main function is exerted at the PM, where different lateral channel diffusion patterns exist (Li et al., 2011). Some channels move freely, whereas others localise in restricted lateral diffusion regions such as microdomains (Li et al., 2011). Interestingly, other cellular structures in addition to the PM, such as the cell wall (Martiniere et al., 2012) and actin filaments (Hosy et al., 2015), are involved in PIP lateral diffusion restriction, which reflects PIP close interaction with other cellular structures. The functional importance of PIP PM organisation in interactions with proteins/lipids from other cellular structures is not completely understood, but the role of PIP microdomains in the regulation of exo/endocytosis was proposed (Takano et al., 2017).
Interorganelle contacts through membrane contact sites are involved in the maintenance of phospholipid homeostasis and trafficking events (Pérez‐Sancho et al., 2016). These contact types among organelles are not randomly established but follow patterns that change when cellular nutrient status is modified or the cytoskeleton disassembled (Valm et al., 2017). Recently, it was shown that ER–PM contact sites (EPCSs) were enhanced by ionic stress (Lee et al., 2019), which is especially relevant for plant cells. EPCS proteomes are not yet clearly established, but evidence points to the existence of different EPCS populations (Siao et al., 2016; Stefano et al., 2018). In recent years, plant vesicle‐associated membrane protein (VAMP)‐associated proteins (VAP27s) and synaptotagmins (SYTs) have been characterised as evolutionarily conserved ER tethers that maintain the ER membrane in close proximity to the PM without fusing them (Wang et al., 2014, 2016; Pérez‐Sancho et al., 2015). Arabidopsis STY1 interacts with the SNARE SYP121 and is involved in endocytosis and endosome recycling to the PM (Lewis & Lazarowitz, 2010; Kim et al., 2016). Also, plant VAP interaction with a specific phosphoinositide (PI) subset in clathrin‐mediated endocytosis has been reported (Stefano et al., 2018). Consistently, in ER–endosome membrane contact sites from mammal cells, VAPs are necessary to downregulate specific PI endosome levels and to ensure correct actin filament deposition (Dong et al., 2016). Markedly, PIP endocytosis and autophagic degradation occurs in a PI‐dependent manner (Ueda et al., 2016; Jurkiewicz et al., 2020), and different PIP isoforms are highly expressed in cell elongation zones (Heinen et al., 2009), regions where the EPCSs are abundant (McFarlane et al., 2017). However, evidence for PIP presence at EPCSs has not been shown yet.
Complex posttranslational regulation of eukaryotic aquaporins is mediated by other proteins (Roche & Törnroth‐Horsefield, 2017). For instance, hundreds of putative AtPIP interactors were found (Bellati et al., 2016). PIP cytosolic domains (i.e. the N‐ and C‐termini, loops B and D) are involved in the regulation of channel trafficking, degradation, and gating upon interaction with specific proteins (Lee et al., 2009; Zelazny et al., 2009; Wu et al., 2013; Hachez et al., 2014a; Grondin et al., 2015; Li et al., 2015; Afzal et al., 2016; Bellati et al., 2016). In the present work, we explored which are the main partners of maize (Zea mays) ZmPIP2;5, the most highly expressed PIP isoform in roots involved in radial water movement (Hachez et al., 2006; Ding et al., 2020). We found that ZmPIP2;5 interacts with VAP27s inserted in the ER membrane, and that this interaction positively influences PM water permeability. We propose that VAP27s stabilise PIP2s at the PM and might guide their endocytosis in response to salt stress. This interaction seems to be conserved through land plant evolution.
Materials and Methods
Genetic constructs
Total RNA was extracted from 6‐d‐old maize Black Mexican Sweet (BMS) cells using the RNeasy Plant Mini Kit (Qiagen, Hilden, Germany) following the manufacturer’s instructions. Total cDNA was synthesised from RNA (1 µg) in a 25‐μl reaction volume using M‐MLV Reverse Transcriptase (Promega, Madison, WI, USA) and the oligo(dT)15 primer. For the SUS assay, ZmVAP27‐1 and ZmVAP27‐2 cDNAs were PCR amplified from BMS cell total cDNA, and ZmPIP2;5 cDNA, optimised for expression in yeast, was PCR amplified from pYeDP60u‐ZmPIP2;5OPT (Bienert et al., 2014) using the corresponding att1B and att2B primers (Supporting Information Table S1). The PCR products were cloned with the Gateway® system (Invitrogen, Waltham, MA, USA) into the pDONR221P1P2 entry vector before their integration into the SUS destination vectors. pMetYC‐DEST and pNX35‐DEST (Grefen et al., 2009) were used to produce the Met‐repressible bait construct ZmPIP2;5‐Cub‐PLV and the prey constructs NubG‐ZmVAP27‐1 or NubG‐ZmVAP27‐2, respectively. The NubWT control fragment was obtained from the pNubWT‐Xgate vector (Grefen et al., 2009).
To carry out bimolecular fluorescence complementation (BiFC) and localisation assays the cDNAs were cloned with the Gateway® system (Invitrogen). Briefly, ZmPIP2;5 and ZmVAP27‐1 were PCR amplified using the corresponding attB1 and attB4 primers and cloned into the pDONR221P1P4 entry vector. ZmSYP121, ZmVAP27‐1, ZmVAP27‐2, AtVAP27‐1, NpPMA2 and the deleted ZmVAP27‐1 versions (ΔMSD and ΔTMD) were PCR amplified using the corresponding attB3 and attB2 primers. For the ΔCCD mutant genetic construct, two PCR fragments obtained with the attB3AtVAP27‐1NFw/RvVAP11deltaCCD and attB2AtVAP27‐1NRv/FwVAP11deltaCCD primer pairs were assembled in a second PCR with the attB3 and attB2 primers. All these PCR fragments were cloned into the pDONR221P3P2 entry vector. Details of the plasmid pDONR221P1P4‐AtPIP2;7 were previously published by our laboratory (Hachez et al., 2014a). The vector Gateway® cassettes were transferred to the pBIFCt‐2in1‐NN destination vector (Grefen & Blatt, 2012) and/or the pFRETtv‐2in1‐NN (Hecker et al., 2015).
For the oocyte swelling assays, the pT7Ts‐derived vector containing the T7 RNA polymerase promoter and carrying the 5′ and 3′ translated regions of the Xenopus laevis β‐globin gene (Gorgoni et al., 1995) was modified to add new cloning sites. Briefly, overlapping oligos with the recognition sites for NotI, XhoI, and SacII (Table S1) were annealed and cloned into the vector previously linearised using the BglII and SpeI enzymes. The ZmPIP2;5 and ZmVAP27‐1 cDNAs were PCR amplified from BiFC vectors using specific primers to add restriction enzyme sites (Table S1). ZmPIP2;5 was cloned into the BglII and SpeI sites, and ZmVAP27‐1 was cloned into the SacII and SpeI sites. All cloned products were checked by sequencing.
Transfection of maize BMS cells
BMS cells were cultivated in the dark with constant shaking at 90 rpm and 25°C, in BMS medium (50 ml) (4.4 g l−1 Murashige and Skoog basal salts with minimal organics; M6899, Sigma Aldrich, St. Louis, MO, USA), 30 g l−1 sucrose, 3 mg l−1 2,4‐dichlorophenoxyacetic acid (D8407, Sigma Aldrich), and 0.2 g l−1 of L‐Asn (A4284, Sigma Aldrich), pH 5.8). The cells were subcultured every 14 d at a 1 : 10 ratio with fresh BMS medium at room temperature.
Stable transfected BMS cells were obtained using the PDS‐1000/He Biolistic particle bombardment delivery system (Bio‐Rad, Hercules, CA, USA). Briefly, pCAMBIA35Su:YFP‐PIP2;5 (Chevalier et al., 2014) and pCAMBIA35Su:YFP (Ding et al., 2020) DNAs were precipitated onto 0.6 µm gold particles. These DNA‐coated particles were used to bombard 9‐d‐old BMS cells spread over Whatman filters on top of BMS medium‐agar plates. After 3 d the filters were transferred to selective BMS medium plates (kanamycin 100 µg ml−1 for pCAMBIA35Su:YFP‐PIP2;5 and Bialaphos 3 µg ml−1 for pCAMBIA35Su:YFP). Every 15 d the filters were transferred to fresh plates. Fluorescent calli were selected after 4–6 wk.
Protoplast swelling assay
BMS protoplast isolation was performed as described previously (Moshelion et al., 2004). Briefly, protoplasts were isolated from 2 ml of 8‐d‐old BMS cell suspensions. The cells were left to sediment and the BMS medium was replaced with cell wall digestion solution (1 ml) (1.5% w/v cellulase Y‐C; Kyowa Chemical Products, Japan), 0.3% w/v Macerozyme R‐10 (Duchefa Biochemie, Haarlem, the Netherlands) in isotonic solution (to be described later). The cells were placed on a rotary shaker (90 rpm) for 2 h 30 min at 26°C. Then, the cells were passed through a nylon filter (20 μm pore size) and washed two times with isotonic solution. The protoplast swelling experiments were performed as described previously (Moshelion et al., 2004). The isotonic and hypotonic solutions (10 mM KCl, 1 mM CaCl2, 8 mM MES, pH 5.75) were adjusted with sorbitol to 300–330 and 150–160 mOsm, respectively. The Advanced Instruments 3300 Micro‐Osmometer (Advanced Instruments, MA, USA) served for all osmolarity measurements.
Plant microsomal fraction preparation, immunoprecipitation and LC‐MS assays
BMS cells were grown in 200 ml cultures and harvested after 7 d. The cells were washed with ice‐cold homogenisation buffer (250 mM sorbitol, 50 mM Tris‐HCl pH 8, 2 mM 2‐[2‐[bis(carboxymethyl)amino]ethyl‐(carboxymethyl)amino]acetic acid (EDTA), 10 mM dithiothreitol (DTT), 1 mM phenylmethylsulphonylfluoride (PMSF), and 2 µg ml−1 each of leupeptin, pepstatin, aprotinin, antipain and chymostatin) and ground with glass beads. Cell debris was removed by centrifugation for 5 min at 3500 g. The supernatant was then centrifuged for 30 min at 100 000 g. Finally, the pellet corresponding to the microsomal fraction was suspended in resuspension buffer (250 mM sucrose, 20 mM HEPES pH7.4, 10 mM KCl, 1.5 mM MgCl2, 1 mM EDTA, 1 mM 2‐[2‐[2‐[2‐[bis(carboxymethyl)amino]ethoxy]ethoxy]ethyl‐(carboxymethyl)amino]acetic acid (EGTA), 10 mM DTT, 1 mM PMSF, and 2 µg ml−1 each of leupeptin, pepstatin, aprotinin, antipain and chymostatin). Proteins were quantified by the Bradford assay (Bradford, 1976) and adjusted to 3 mg ml−1 and 1% w/v octyl‐β‐d‐glucopyranoside. Protein extracts (15 mg) were mixed with 30 μl agarose beads (Chromotek, Planegg, Germany) and incubated in a rotor wheel for 1 h at 4°C. Then, the beads were pelleted by centrifugation at 2000 g for 10 s and the supernatant was incubated with 30 μl Chromotek GFP‐Trap® agarose beads in a rotor wheel at 4°C overnight (ON). The beads were washed five times with 700 μl resuspension buffer supplemented with 300 mM NaCl. Bound proteins were eluted from the beads by adding 20 μl Laemmli buffer and incubating for 15 min at 65°C. This step was repeated twice. ZmPIP2;5 immunoprecipitation was confirmed in aliquots (2 µl) by immunoblot. The remaining samples were electrophoresed by SDS‐PAGE just to stack the proteins that were in‐gel digested with trypsin, and peptides recovered for the nano‐ultra‐high performance liquid chromatography coupled with electrospray ionisation‐quadrupole‐time of flight‐mass spectrometry (nano‐ULPC‐ESI‐QTOF‐MS) analysis at the MASSPROT facility (UCLouvain, detailed protocol in Methods S1). For each sample, three technical replicates were run in the nano‐ULPC‐ESI‐QTOF‐MS. Progenesis QI (v.2.0, Nonlinear Dynamics) was used to analyse the spectrometry data. The runs were normalised to YFP levels. For peptide identification the maize UP000007305 reference proteome was used (UniProt). Detailed information of the ions to peptides and peptides to proteins association is listed in Table S2. Progenesis QI software is not prepared to handle technical and biological replicates in statistical analysis. Therefore, to score the differential abundance of proteins between YFP‐ZmPIP2;5 and YFP samples including technical replicates, the online freely available RepExplore software (Glaab & Schneider, 2015) was used. In this software the variance across technical replicates was included in the analysis using a Bayesian approach named probability of positive log ratio (PPLR) statistic (Glaab & Schneider, 2015).
Split‐ubiquitin assays (SUS)
The haploid yeast strain THY.AP4 was cotransformed by electroporation with the Nub and Cub constructs of interest. Yeast colonies coexpressing the bait and prey constructs were recovered 48 h after transfer to selective medium (CSM, ‐Leu−, Trp−) (Grefen et al., 2009). Growth assays were performed as detailed in Hachez et al. (2014a). Yeast coexpressing the Met‐repressible bait construct ZmPIP2;5‐Cub‐PLV and the prey constructs NubG‐ZmVAP27‐1, NubG‐ZmVAP27‐2, NubG (negative control), or NubWT (positive control) were grown on selective medium. The next day, a dilution series (OD600 nm 0.5, 0.05, and 0.005) of the cultures was dropped onto interaction‐selective (CSM, ‐Leu−, Trp−, Ade−, His−, Met−) medium containing 100 µM methionine to repress bait expression. Control plates without methionine serve to verify that an equal yeast amount had been dropped. Yeast growth was recorded after incubation for 48 h at 30°C. Protein expression was verified via immunoblotting using a rat monoclonal antibody against the haemagglutinin (HA) tag (Roche, Mannheim, Germany) and a rabbit polyclonal antibody against VP16 (Abcam, Cambridge, UK), as previously described (Grefen et al., 2009). Protein loading on a PVDF membrane was revealed by Coomassie R250 staining (Goldman et al., 2016).
Confocal microscopy
A Zeiss LSM710 confocal microscope equipped with a spectral detector module (Carl Zeiss, Oberkochen, Germany) was used for confocal image acquisition. Imaging of BMS cells transformed with YFP‐ZmPIP2;5 was achieved with a C‐Apochromat ×40/1.20 water‐immersion objective. The mYFP molecule was excited with the 514‐nm laser lines. Emitted light was collected through a dichroic mirror on detector 520–560 nm. The cell PM was stained with FM4‐64 dye (16 µM, Invitrogen). This dye was excited at 514 nm and detected from 600 to 760 nm.
The BiFC and localisation assays were performed in tobacco (N. benthamiana) epidermal cells transformed by Agrobacterium tumefaciens infiltration (Batoko et al., 2000). Samples were analysed 3 d after infiltration. For fluorophore excitation and emission, the following laser settings were used: for mRFP1 excitation 561 nm and emission 560 to 615 nm, for mYFP excitation at 514 nm and emission 522 to 553 nm, for mTRQ2 excitation 445 nm and emission 463 to 520 nm, for mVenus excitation 514 nm and emission 520 to 574 nm. The BiFC analysis was performed using a ×40/0.75 water‐immersion objective. Images were taken with standardised excitation intensities and photomultiplier gains. Relative fluorescence was analysed using the ZEN 2 Profile module (blue edn, Carl Zeiss). Five lines of five to six µm length were drawn through the PM of one cell. The fluorescence intensity of each line for each fluorophore was obtained. Then, the mYFP to mRFP1 fluorescence ratio of each line was calculated and averaged for that cell. The neighbour cells were not used to measure the fluorescence intensity to avoid including twice the values of a single cell. When specified, the Airyscan module (Carl Zeiss) and Plan‐Apochromat ×63/1.40 oil immersion objective were used.
In vitro RNA synthesis and oocyte water transport assays
The capped cRNA encoding for ZmPIP2;5 was synthesised in vitro using the mMESSAGE mMACHINET7 High Yield Capped RNA Transcription Kit (Ambion, Austin, TX, USA), and the capped ZmVAP27‐1 cRNA was synthesised with the mMESSAGE mMACHINE T7 High Yield Capped RNA ULTRA Transcription Kit (Ambion), as described previously (Jozefkowicz et al., 2013). The synthesised products were resuspended in RNase‐free water. cRNA integrity was checked on agarose gel and quantified in a NanoDropTM 2000 spectrophotometer (Thermo Fisher Scientific).
Defolliculated Xenopus laevis oocytes (stages V–VI) were microinjected with cRNAs and incubated in ND96 buffer (96 mM NaCl, 2 mM KCl, 1 mM MgCl2, 1.8 mM CaCl2, and 5 mM HEPES pH 7.5; c. 200 mOsmol kg−1 H2O) for 3 d at 18°C. The osmotic water permeability coefficient (P f) of oocytes injected or noninjected (NI) with cRNA was determined by measuring the oocyte swelling rate in response to ND96 buffer diluted five‐fold with distilled water, as explained previously (Fortuna et al., 2019). For pH inhibition experiments, the oocyte internal proton concentration was modified as explained previously (Bellati et al., 2010), and the swelling response was induced by transferring the oocytes to an incubation solution diluted five‐fold with distilled water. Briefly, oocytes were preincubated in solutions of different pH (MES for the 5.8 to 6.7 pH interval and HEPES for the 7.0–7.5 pH interval) containing 50 mM sodium acetate, 20 mM MES or HEPES, and supplemented with 1 M mannitol to adjust the osmolarity to c. 200 mOsmol kg−1 H2O. To calculate the final internal pH in oocytes treated with acetate buffers, we used the calibration curve performed by Bellati et al. (2010). Noninjected oocytes, preincubated in a pH 7.5 solution for pH experiments, were used as negative controls. All osmolarities were measured in a vapour pressure osmometer (5600C; Wescor Inc., Logan, UT, USA).
Bioinformatics analysis of ZmVAP proteins
Protein domains were identified with the Simple Modular Architecture Research Tool, Smart (Schultz et al., 1998). Maize proteins containing a major sperm domain (MSD) domain were found in the B73 reference genome (RefGen_v4) via a Blast search with ZmVAP27‐1 MSD, and also were found in the Smart database. ZmVAP27 homologous gene families were identified in the Monocots Plaza 4.0 platform (Van Bel et al., 2018). For the phylogenetic tree, to obtain a harmonic representation of the different plant species, the protein sequences were retrieved from the family HOM04000421 (Monocots database), Dicots Plaza 4.0, and Gymno Plaza 1.0. Multiple sequence alignments (MSA) were assembled in the Guidance2 server (Sela et al., 2015) using the Mafft (FFT‐NS‐100) algorithm. Unreliable sequences below a 0.60 confidence score were removed and the alignment redone. The sequences were trimmed with TrimAl (Capella‐Gutierrez et al., 2009) removing unreliable columns below a 0.80 confidence score conserving 35% of the sequence. The best‐fit model was found using ModelFinder (Kalyaanamoorthy et al., 2017) implemented in the IQ‐Tree (v.1.10) Phylogenomic software. Branch support was calculated with the ultrafast bootstrap (Hoang et al., 2018) and the SH‐aLRT branch test (Guindon et al., 2010). The best‐fit model was JTT+F+R7. Trees were edited using the Interactive Tree of Life tool (Letunic & Bork, 2016). ZmVAP transcriptomic data were obtained and analysed using the Genevestigator ® v.4 platform (https://genevestigator.com) (Zimmermann et al., 2004).
Results
Identification of ZmPIP2;5 interactors associated with the cytoskeleton
We previously reported that the ZmPIP2;5 physical interaction with the syntaxin SYP121 regulates its subcellular trafficking and water transport activity (Besserer et al., 2012). To identify other ZmPIP2;5 partners we designed a pull‐down assay using BMS suspension cells. We generated transgenic lines that overexpressed YFP‐ZmPIP2;5 under the control of the 35S promoter and selected lines with high and stable protein expression levels. Confocal microscopy analysis showed that YFP‐ZmPIP2;5 was localised in the PM (Fig. 1a). We then determined the cell membrane P f by protoplast swelling assays and observed that the P f was greater in the ZmPIP2;5 overexpression cells compared with wild‐type cells (Fig. 1b), demonstrating that YFP‐ZmPIP2;5 was a functional water channel in these cells. We used this YFP‐ZmPIP2;5 overexpression cell line and a cell line overexpressing YFP (negative control) to perform a pull‐down assay. With a P‐like value < 0.05 and fold change (FC)> 2, 138 proteins were defined as putative ZmPIP2;5 interactors (Table S3). Then, we created a shorter interest list, which also included some proteins with a FC > 1.5 (Table 1).
Fig. 1.

Localisation and activity of YFP‐ZmPIP2;5 expressed in Black Mexican Sweet (BMS) cells. (a) Confocal images of maize BMS cells stably transformed with the 35S:YFP‐ZmPIP2;5 construct. A clear colocalisation of the YFP (green) and the plasma membrane dye FM4‐64 (magenta) was observed. Bars, 10 µm. (b) Water permeability coefficient (P f) values of nontransformed BMS cells (wild‐type (WT)) and cells expressing YFP‐ZmPIP2;5 (ZmPIP2;5OE). For each scatter plot the lines indicate the mean ± SE. An unpaired t‐test was used to test the statistical difference between lines (***, P < 0.001).
Table 1.
Potential ZmPIP2;5 interacting proteins.
| Identifier | Description | P‐like value | eBayes adj. P‐value | Peptide count | Unique peptides | FC |
|---|---|---|---|---|---|---|
| Plasma membrane proteins | ||||||
| Zm00001d017526 | Aquaporin PIP1‐2 | 1.6E‐06 | 1.01E‐04 | 9 | 2 | 8.3 |
| Zm00001d017526 | Aquaporin PIP1‐3/PIP1‐4 | 0.0E + 00 | 2.61E‐05 | 10 | 3 | 17.2 |
| Zm00001d019565 | Aquaporin PIP2‐6 | 4.2E‐10 | 1.48E‐04 | 7 | 3 | 7.4 |
| Zm00001d008178 | ABC transporter B family member 21 | 5.2E‐08 | 2.01E‐03 | 29 | 7 | 2.1 |
| Zm00001d013254 | ABC transporter B family member 27 | 8.7E‐09 | 2.36E‐04 | 9 | 3 | 3.3 |
| Zm00001d043766 | ABC transporter B family member 9 | 1.8E‐08 | 1.80E‐03 | 5 | 2 | 2.2 |
| Zm00001d048823 | Monosaccharide‐sensing protein 2 | 6.9E‐03 | 8.27E‐05 | 2 | 2 | 26.2 |
| Zm00001d029762 | Hexose transporter | 1.5E‐03 | 1.74E‐02 | 4 | 3 | 1.6 |
| Zm00001d005451 | Cellulose synthase A5 | 3.3E‐02 | 2.00E‐02 | 10 | 3 | 1.7 |
| Proteins involved in trafficking | ||||||
| Zm00001d047468 | Aberrant pollen transmission1 | 0.0E + 00 | 1.18E‐05 | 23 | 7 | 21.1 |
| Zm00001d037842 | B‐cell receptor‐associated 31‐like | 1.4E‐04 | 4.70E‐03 | 7 | 6 | 2.4 |
| Zm00001d025817 | CLIP‐associated protein (CLASP) | 8.6E‐05 | 2.65E‐03 | 12 | 4 | 2.1 |
| Zm00001d014526 | EH domain‐containing protein 1 | 5.0E‐07 | 1.95E‐03 | 26 | 2 | 2.4 |
| Zm00001d015102 | Golgi SNAP receptor complex member 1 | 4.3E‐03 | 2.27E‐02 | 5 | 2 | 5.2 |
| Zm00001d035041 | Phospholipase SGR2 | 5.1E‐03 | 4.82E‐03 | 4 | 2 | 3.0 |
| Zm00001d014159 | Protein Networked 1A | 2.0E‐03 | 2.45E‐03 | 5 | 2 | 2.3 |
| Zm00001d014616 | Transducin family protein/ WD‐40 repeat family protein | 0.0E + 00 | 2.86E‐06 | 2 | 2 | 46.2 |
| Zm00001d042180 | Vesicle‐associated protein 27‐2 | 1.4E‐10 | 8.21E‐03 | 4 | 2 | 6.5 |
| Zm00001d025939 | Vesicle‐associated protein 27‐1 | 2.6E‐02 | 2.65E‐03 | 3 | 3 | 2.3 |
| Zm00001d038808 | Vesicle‐fusing ATPase | 8.7E‐09 | 2.80E‐03 | 35 | 3 | 2.2 |
| Zm00001d041443 | Protein Networked 1A | 8.3E‐04 | 6.60E‐03 | 10 | 2 | 1.8 |
| Zm00001d021551 | Ras‐related protein RABH1b | 2.3E‐04 | 1.68E‐02 | 16 | 7 | 1.7 |
Proteins in bold are involved in plasma membrane (PM)–cytoskeleton organisation (Pietra et al., 2013; Paredez et al., 2006; Deeks et al., 2012; Wang et al., 2014; Kirik et al., 2007).
P‐like value, transformation of the probability of positive log ratio (PPLR) into a P‐like significance score; eBayes, empirical Bayes moderated t‐statistic; FC, fold change.
A cellulose synthase subunit (CESA) was pulled‐down with ZmPIP2;5, similarly to that reported in a previous pull‐down assay with AtPIP2;1 (Bellati et al., 2016). Interestingly, the CESA complex is one of the few identified mediators of the interaction between the PM and cortical microtubules (Krtková et al., 2016), the latter guiding cellulose deposition (Paredez et al., 2006). Then, we hypothesised that PIPs may also interact with the cytoskeleton, and that this interaction may underlie specific events regulating PIP function in cell water and/or solute homeostasis. In support of this hypothesis: (1) we observed the pull‐down of proteins involved in PM–cytoskeleton association together with ZmPIP2;5 (Table 1); (2) in a previous pull‐down assay using ZmPIP2;6 as bait, we identified kinesin, dynamin, actin, and tubulin as putative interactors (Hachez et al., 2014a; Table S4); and (3) the reported AtPIP2;1 pull‐down assay also identified several dynamin and tubulin isoforms as interactors (Bellati et al., 2016; Table S4). Markedly, we also identified two members of the plant‐specific Networked (NET) actin‐binding proteins superfamily (Deeks et al., 2012), and two VAPs, ZmVAP27‐1 and ZmVAP27‐2. VAPs and NETs, together with the cytoskeleton, are involved in EPCS organisation (Wang et al., 2014). This evidence points to an association of the PM‐localised ZmPIP2;5 with the cortical cytoskeleton and ER, possibly through an interaction with ZmVAP27‐1 and ZmVAP27‐2. Therefore, we decided to investigate the interaction between PIPs and VAP27s and its functional implications.
ZmPIP2;5 interacts with ZmVAP27 proteins
To confirm the ZmPIP2;5 interaction with ZmVAP27‐1 and ZmVAP27‐2, we performed a yeast SUS. The cDNAs of ZmVAP27‐1 and ZmVAP27‐2 were cloned using BMS cell total RNA as a template. ZmVAP27s were N‐terminal tagged with NubG (prey) and ZmPIP2;5 was C‐terminal tagged with Cub–PLV (bait). NubG and NubWT fragments served as negative and positive controls of the Nub–Cub interaction, respectively (Grefen et al., 2009). The specificity in the interaction was observed by reducing bait protein expression with methionine, which inhibited the met25 promoter. Yeast growth was observed when ZmPIP2;5‐Cub‐PVL was expressed with both VAP27 proteins or with the positive NubWT control, whereas it was not observed for the negative control (Fig. 2a), indicating that ZmPIP2;5 is able to physically interact with ZmVAP27‐1 or ZmVAP27‐2. Bait and prey protein expression in the transformed yeast was confirmed by immunoblots (Fig. 2b).
Fig. 2.
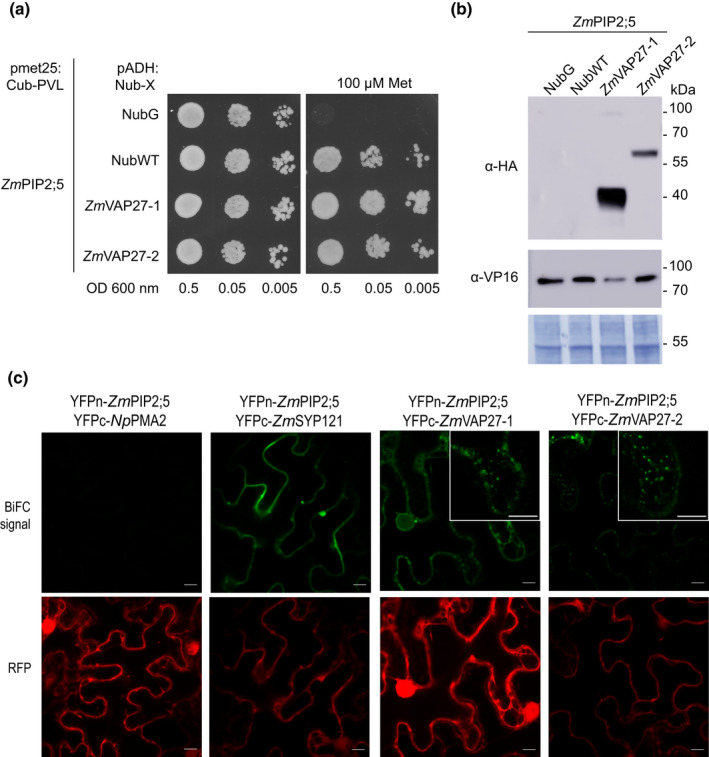
ZmPIP2;5 interacts with ZmVAP27‐1 and ZmVAP27‐2. (a) Spilt‐ubiquitin assay (SUS). Yeast coexpressing the ZmPIP2;5‐Cub‐PLV Met‐repressible bait construct and the prey constructs NubG‐ZmVAP27‐1, NubG‐ZmVAP27‐2, NubG, or NubWT were dropped in a dilution series (OD 0.5, 0.05 and 0.005) onto synthetic medium with or without 100 µM methionine to repress bait expression. Yeast growth was recorded after incubation for 48 h. This experiment was repeated with three independent transformed yeast lines for each construct pair. (b) Immunoblot to verify bait and prey fusion protein expression in yeast used for the SUS. The preys were revealed using anti‐haemaglutinin (anti‐HA) antibody and the bait (ZmPIP2;5) was revealed using an anti‐VP16 antibody. NubG and NubWT do not contain an HA‐tag. The expected molecular weights of the proteins were: NubG‐ZmVAP27‐1, 40 kDa; NubG‐ZmVAP27‐2, 60 kDa, and ZmPIP2;5‐Cub‐PLV, 77 kDa. Polyvinylidene difluoride (PVDF) membrane Coomassie R250 staining (bottom) was used to control the protein loading. (c) Bimolecular fluorescence complementation (BiFC) signals for the pairs YFPn‐ZmPIP2;5/YFPc‐NpPMA2 (negative control), YFPn‐ZmPIP2;5/YFPc‐ZmSYP121 (positive control), YFPn‐ZmPIP2;5/YFPc‐ZmVAP27‐1, YFPn‐ZmPIP2;5/YFPc‐ZmVAP27‐2. BiFC signal (YFP) is shown in green, and the RFP signal in red serves as a transfection control. Bars, 10 µm. YFP signal was detected with a bright dot pattern for the YFPn‐ZmPIP2;5/YFPc‐ZmVAP27‐1 and YFPn‐ZmPIP2;5/YFPc‐ZmVAP27‐2 pairs (insets).
Additionally, the physical interaction between ZmPIP2;5 and ZmVAP27s was validated by a BiFC assay using the pBiFCt‐2in1 vector (Grefen & Blatt, 2012). This vector carries each protein of interest in frame with half of the YFP protein (YFPn and YFPc) and a soluble monomeric red fluorescent protein (mRFP1) as an internal transformation and expression control. ZmSYP121 and NpPMA2 were used as the positive and negative ZmPIP2;5 interaction controls, respectively (Besserer et al., 2012). The constructs were agro‐infiltrated in N. benthamiana leaves and the fluorescence was observed by confocal microscopy after 3 d. For both the YFPn‐ZmPIP2;5/YFPc‐ZmVAP27‐1 and YFPn‐ZmPIP2;5/YFPc‐ZmVAP27‐2 pairs, a YFP fluorescent signal was detected with a particular bright dot pattern (insets Fig. 2c). As expected, a YFP fluorescent signal was also observed when YFPn‐ZmPIP2;5 was co‐expressed with YFPc‐ZmSYP121, whereas no signal was observed when YFPn‐ZmPIP2;5 was co‐expressed with YFPc‐NpPMA2 (Fig. 2c). Additionally, we tested the specificity of the interaction between ZmVAP27s and ZmPIP2;5 by analysing the YFPn‐ZmVAP27‐1/YFPc‐NpPMA2 pair, and did not detect any YFP signal, indicating that ZmVAP27‐1 is not broadly interacting with all PM proteins (Fig. S1). Altogether, the assays in the yeast heterologous system and in plant cells confirmed the pull‐down data showing that ZmVAP27‐1 and ZmVAP27‐2 interacted with ZmPIP2;5.
The MSD of VAP27s is required for PIP2–VAP27 interaction
Both ZmVAP27‐1 and ZmVAP27‐2 present the three characteristic VAP protein domains (Wang et al., 2016): the cytoplasmic MSD, the coiled‐coil domain (CCD), and the transmembrane domain (TMD) that anchors the protein to the ER (Fig. 3a). To determine which domain is involved in the interaction with ZmPIP2;5, we generated ZmVAP27‐1 mutants and tested their physical interaction with ZmPIP2;5 by means of BiFC assays. Ratiometric fluorescence quantification resulted in a significant increase in the YFP/RFP ratio when the MSD was present in ZmVAP27‐1 (i.e. for the pairs YFPn‐ZmPIP2;5/YFPc‐ZmVAP27‐1, YFPn‐ZmPIP2;5/YFPc‐ZmVAP27‐1ΔTMD, and YFPn‐ZmPIP2;5/YFPc‐ZmVAP27‐1ΔCCD), whereas the YFP/RFP ratio of the cells expressing YFPn‐ZmPIP2;5/YFPc‐ZmVAP27‐1ΔMSD was not significantly different from the negative control (Fig. 3b; representative confocal images in Fig. S2a). The expression of ZmVAP27‐1ΔMSD was confirmed by confocal microscopy in tobacco leaves transiently expressing mTRQ2 tagged protein (Fig. S2b). Altogether, these data suggest that the VAP27 MSD domain is required for their interaction with ZmPIP2;5.
Fig. 3.
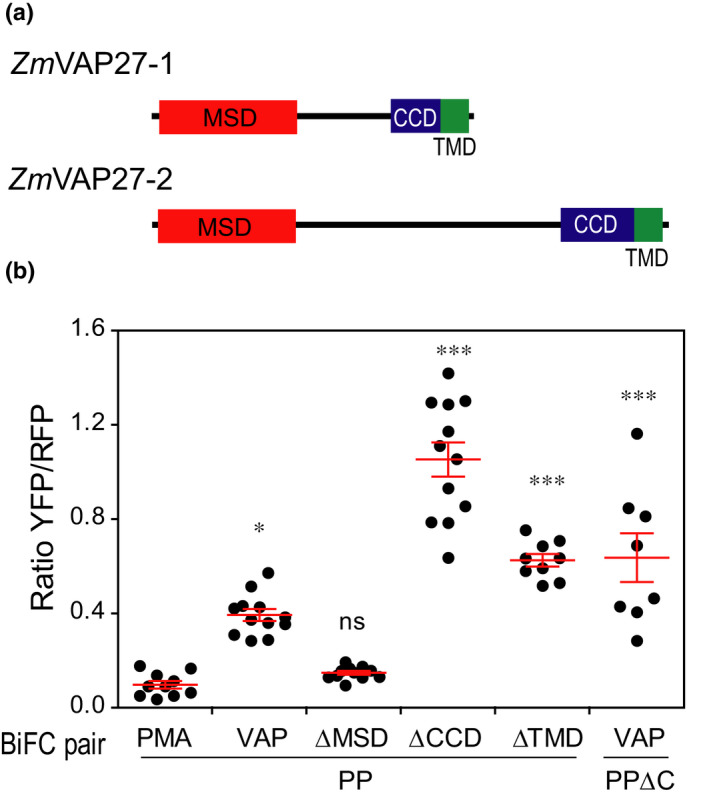
ZmVAP27‐1 major sperm domain (MSD) is required for the interaction with ZmPIP2;5. (a) Domain composition of the VAP27 proteins that interact with ZmPIP2;5. CCD, coiled‐coil domain; TMD, transmembrane domain. (b) Fluorescence ratiometric quantification from bimolecular fluorescence complementation (BiFC) experiments with the pairs PIP/PMA (YFPn‐ZmPIP2;5/YFPc‐NpPMA2), PIP/VAP (YFPn‐ZmPIP2;5/YFPc‐ZmVAP27‐1), PIP/ΔMSD (YFPn‐ZmPIP2;5/YFPc‐ZmVAP27‐1ΔMSD), PIP/ΔTMD (YFPn‐ZmPIP2;5/YFPc‐ZmVAP27‐1ΔTMD), PIP/ΔCCD (YFPn‐ZmPIP2;5/YFPc‐ZmVAP27‐1ΔCCD), and PIPΔC/VAP (YFPn‐ZmPIP2;5ΔC/YFPc‐ZmVAP27‐1). For each scatter plot, the lines indicate the mean ± SE. The Kruskal–Wallis and Dunn’s multiple comparison test were used to calculate the statistical difference between the negative control and the remaining constructs (***, P < 0.001; *, P < 0.05; ns, not significant). This experiment was repeated twice.
PIP2 C‐terminal domain is not required for PIP2–VAP27 interaction
Most of the reported VAP interactors are cytoplasmic proteins with a binding motif of two phenylalanines in an acidic tract (FFAT) or a FFAT‐like motif (Murphy & Levine, 2016). Using a mammal and yeast VAP interactome, an algorithm to predict VAP binding motifs was proposed (Murphy & Levine, 2016). We tested ZmPIP2;5 cytoplasmic domains with that algorithm and found no putative interaction motifs within these sequences. However, this algorithm also predicted a weak interaction of the mammalian potassium channel Kv2.1, which is known to interact with VAP proteins through a noncanonical C‐terminal domain binding motif that is rich in serine residues (Johnson et al., 2018). As the ZmPIP2;5 C‐terminus is also serine rich, we tested whether this domain was required for the interaction with ZmVAP27‐1. We generated the ZmPIP2;5ΔC deletion mutant and tested its physical interaction with ZmVAP27‐1 by means of BiFC assays. The ZmPIP2;5ΔC mutant still interacted with ZmVAP27‐1 (Figs 3b, S2a), suggesting that another ZmPIP2;5 cytosolic domain should be involved in the interaction with ZmVAP27‐1.
ZmPIP2;5 and ZmVAP27s are in close vicinity in EPCS and endocytic structures
To study VAP27 protein intracellular localisation relative to the localisation of ZmPIP2;5, ZmVAP27‐1 or ZmVAP27‐2 and ZmPIP2;5 were tagged with mTRQ2 and mVenus, respectively, and transiently expressed in N. benthamiana leaves. mTRQ2‐ZmVAP signals were found close to the PM and around the nucleus in a reticular network, characteristic of the ER (Fig. 4a,b). This was better observed in high magnification images, in which the mTRQ2‐ZmVAP27‐1 fluorescent signals were in the cortical ER structure near to the PM and in punctuated structures (Fig. 4c), as previously described for AtVAP27s present in EPCS (Wang et al., 2014, 2016). The expression of both VAP27s did not modify ZmPIP2;5 PM localisation (Fig. 4a,b).
Fig. 4.
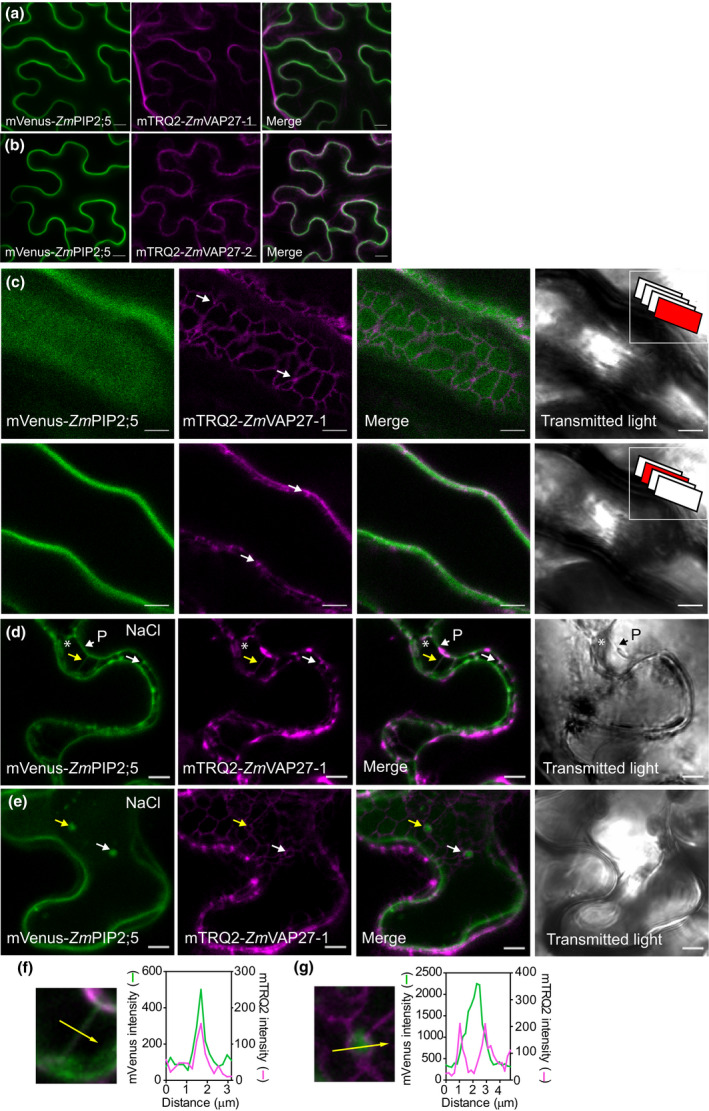
ZmVAP27‐1 and ZmVAP27‐2 are in the endoplasmic reticulum (ER) in close proximity to the plasma membrane (PM)‐localised ZmPIP2;5. (a, b) Confocal images (Z‐stack projection) of tobacco leaf cells transiently overexpressing mVenus‐ZmPIP2;5 and mTRQ2‐ZmVAP27‐1 (a) or mVenus‐ZmPIP2;5 and mTRQ2‐ZmVAP27‐2 (b). Bars, 10 µm. (c) Inset in two confocal planes of cells overexpressing mVenus‐ZmPIP2;5 and mTRQ2‐ZmVAP27‐1. Insets in the transmitted light panels represent in red the scanned plane (1.02 µm thin) position in the Z‐stack. Arrows point to VAP27 dots. (d, e) Confocal images of cells overexpressing mVenus‐ZmPIP2;5 and mTRQ2‐ZmVAP27‐1 after treatment with NaCl (4%) for 15 min. (d) Arrows point to Hetchian strands, asterisks indicate the periplasmic space, and P is the receding protoplast. Arrows in (e) point to mVenus‐ZmPIP2;5 vesicles surrounded by mTRQ2 signal. (f, g) Insets and fluorescence intensity profiles (arbitrary units) for mVenus and mTRQ2 in the structures pointed out with the yellow arrows in (d, e): an Hetchian strand (f), and endocytic vesicle (g). Bars, 5 µm.
PIPs are internalised in response to salt stress (Boursiac et al., 2005; Prak et al., 2008; Pou et al., 2016; Ueda et al., 2016), and VAP27 proteins are involved in endocytic traffic (Stefano et al., 2018). To obtain insights into the nature of the contact between ZmVAP27‐1 with ZmPIP2;5, we challenged the cells with a short but strong hyperosmotic salt stress, aiming to plasmolyse the cells but also to observe endocytosis. After 15 min in 4% NaCl, protoplast detachment from the cell wall occurred and both proteins were detected in Hechtian strands (arrows, Figs 4d,f, S3), which connect the cell wall to the protoplast (Lang‐Pauluzzi & Gunning, 2000). Interestingly, characteristic endocytosed intracellular vesicles containing the aquaporin in response to NaCl were surrounded by the mTRQ2‐ZmVAP27‐1 signal (arrows, Fig. 4e,g).
To further investigate the interaction between ZmPIP2;5 and ZmVAP27‐1 we used the Airyscan confocal microscope super‐resolution module (Zeiss LSM710). This approach revealed a patched arrangement of mVenus‐ZmPIP2;5 in the PM (Fig. 5a), suggesting that ZmPIP2;5 organises in PM domains as previously shown for AtPIP2;1 (Li et al., 2011). This irregular pattern colocalised partially with the nested arrangement of ZmVAP27‐1 in the ER (Fig. 5a). Regarding the mVenus‐ZmPIP2;5‐labelled intracellular vesicles that were near to the PM, we were not able to observe mTRQ2 and mVenus signal colocalisation (Fig. 5b). Cell membrane reorganisation upon NaCl treatment highlighted mVenus‐ZmPIP2;5 and mTRQ2‐ZmVAP27‐1 colocalisation in Hechtian strands (Fig. 5c,f; zoom out of this panel in Fig. S3b) and ZmPIP2;5‐labelled vesicles (Fig. 5d,g), as already shown. Also, we detected colocalisation of both proteins in globular structures (Fig. 5e,h), which resemble the PM invaginations labelled with Venus‐AtPIP2;7 induced in response to salt stress conditions (Pou et al., 2016). Additional images showing mVenus‐ZmPIP2;5 and mTRQ2‐ZmVAP27‐1 colocalisation upon NaCl stress are shown in Fig. S4. Altogether, these data suggested that ZmVAP27‐1‐labelled ER is organised close to the different structures involved in ZmPIP2;5 internalisation.
Fig. 5.
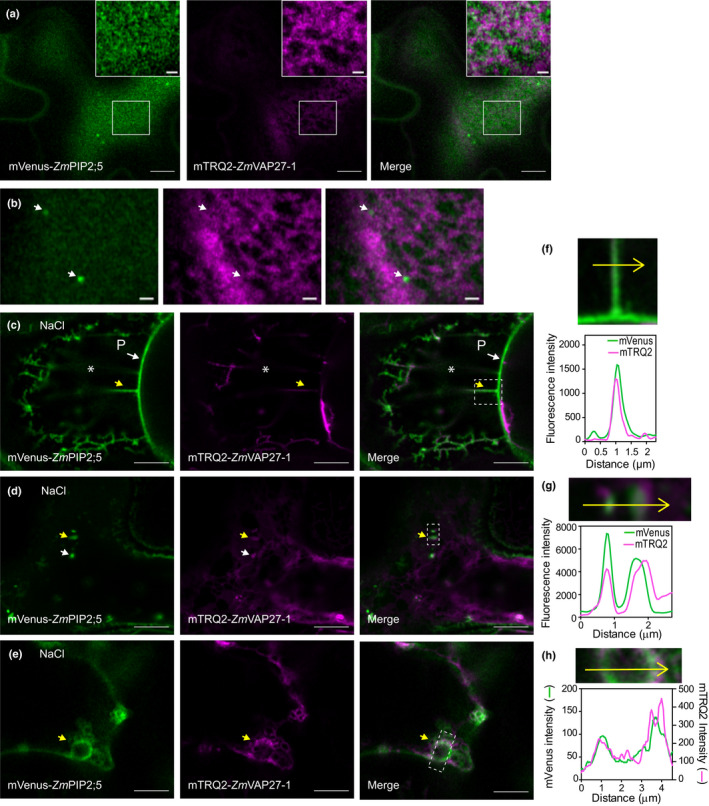
ZmVAP27‐1 colocalises with ZmPIP2;5 in NaCl stress‐related structures. (a) Airyscan confocal images of cells overexpressing mVenus‐ZmPIP2;5 and mTRQ2‐ZmVAP27‐1. Bars, 5 µm. Inset bars, 1 µm. (b) Airyscan confocal images of the same cell as in (a), but a different Z‐position. Bars, 1 µm. (c–e) Airyscan confocal images of cells overexpressing mVenus‐ZmPIP2;5 and mTRQ2‐ZmVAP27‐1 after treatment with NaCl (4%) for 15 min. (c) P, the receding protoplast, asterisks indicate to the periplasmic space. Arrows point to NaCl‐induced: Hetchian strands (c), endocytic vesicles (d), and globular structure (e). (f–h) Insets and fluorescence intensity profiles (arbitrary units) for mVenus and mTRQ2 in the structures pointed with the yellow arrows in (c–e): an Hetchian strand (f), endocytic vesicles (g), and globular structure (h). Bars, 5 µm.
ZmVAP27‐1 positively modifies the osmotic membrane water permeability of ZmPIP2;5 expressing oocytes
To determine whether ZmVAP27‐1 expression modified ZmPIP2;5 water transport activity, we performed Xenopus oocyte swelling assays. Both ZmPIP2;5 and ZmVAP27‐1 coding cRNAs were injected in oocytes and, after 3 d, the latter was subjected to hypo‐osmotic shock. The P f values of oocytes injected with ZmVAP27‐1 cRNA alone were not different from the P f of NI oocytes, whereas a significant P f increase was observed when the oocytes were injected with ZmPIP2;5 cRNA. ZmPIP2;5 and ZmVAP27‐1 cRNA co‐injection in a 1 : 1 ratio did not modify the mean P f value in comparison with ZmPIP2;5 expression alone. However, the co‐injection of ZmPIP2;5 and ZmVAP27‐1 cRNA in a 1 : 10 ratio caused a mean P f value increase over 35% (Figs 6a, S5a,b), indicating a positive synergistic effect on the cell water permeability.
Fig. 6.
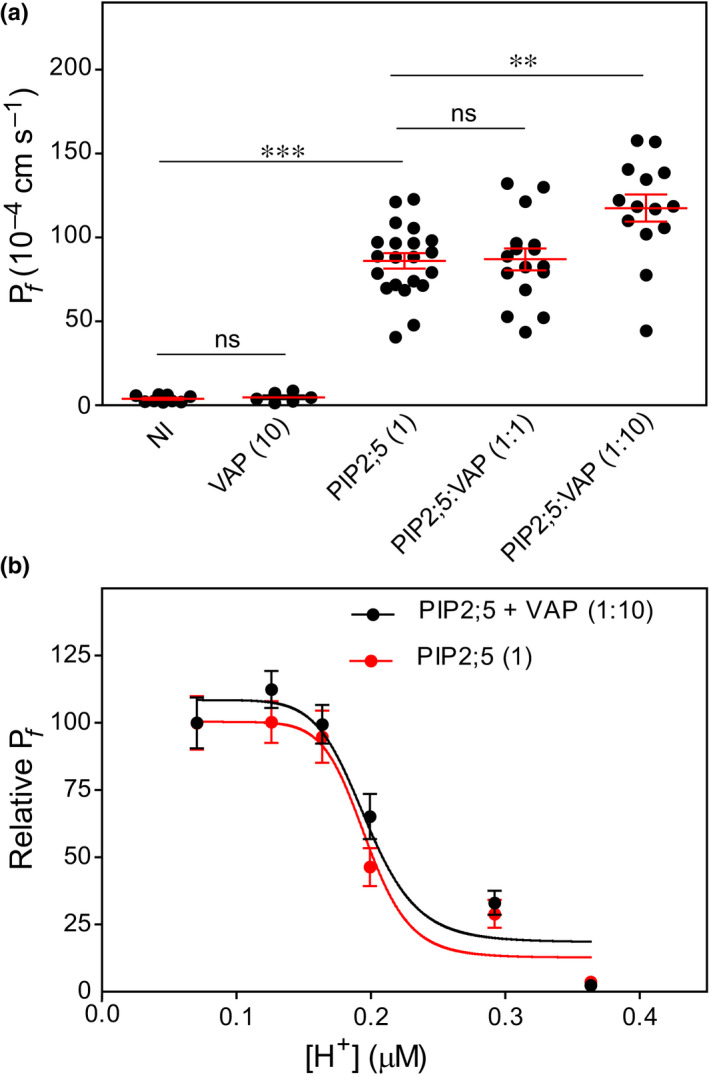
ZmVAP27‐1 increases Xenopus oocyte P f when co‐expressed with ZmPIP2;5. (a) Water permeability coefficient (P f) of noninjected oocytes (NI, control) or injected with ZmVAP27:1 cRNA alone (7.5 ng), ZmPIP2;5 cRNA alone (0.75 ng), or coinjected with both cRNAs in different mass ratios (1 : 1 or 1 : 10). For each scatter plot the lines indicate the mean ± SE. ANOVA and Bonferroni’s multiple comparison test were used to calculate the statistical difference between treatments (***, P < 0.001; **, P < 0.01; ns, not significant). The differences reported here were observed with three independent oocyte batches. (b) Relative P f after cytosolic acidification tested in oocytes injected with ZmPIP2;5 cRNA alone (0.75 ng), or with ZmPIP2;5 (0.75 ng) and ZmVAP27:1 (7.5 ng) cRNAs (1 : 10 ratio). Data points are representative values obtained from the same oocyte batch (mean relative P f ± SE). This is one representative experiment of two independent experiments.
The water permeation through PIP pores is regulated by the cytosolic proton concentration ([H+]i) (Tournaire‐Roux et al., 2003). Under acid conditions, residues located in cytosolic loop D are protonated, and concomitant loop reorganisation leads to PIP pore closure (Törnroth‐Horsefield et al., 2006; Frick et al., 2013). A P f vs [H+]i sigmoidal curve characterises the cooperative behaviour in proton sensing of the channels that integrate the tetramer (Bellati et al., 2010; Yaneff et al., 2014; Jozefkowicz et al., 2016). To evaluate if the PIP–VAP27 interaction affected proton sensing, as it may involve PIP cytosolic domains, we compared the relative‐ P f vs [H+]i curves for ZmPIP2;5 expressed alone and co‐expressed with ZmVAP27‐1 (1 : 10) (Fig. 6b; absolute P f values reported in Fig. S5c,d). The [H+]0.5 remained constant among the assays (0.21 ± 0.02 vs 0.24 ± 0.01, mean ± standard error (SE), n = 2), suggesting that the interaction between ZmPIP2;5 and ZmVAP27‐1 did not alter the aquaporin structural elements involved in pH sensing.
The PIP2–VAP27 interaction is conserved among different angiosperms
Currently our understanding about plant VAPs mainly arises from research in the plant model Arabidopsis (Wang et al., 2014, 2016; Pérez‐Sancho et al., 2015), whereas maize VAP proteins have not been characterised yet. Intending to better understand the PIP‐VAP27 interaction we identified maize VAPs and studied VAP27 phylogeny in green plants (Viridiplantae). We identified 19 proteins containing a MSD in the B73 maize genome (Fig. S6a). The MSD is a conserved feature among VAPs, but proteins from other families may also contain this domain. Indeed, 17 of the 19 proteins were homologues of the previously characterised AtVAP27s, whereas the remaining two proteins were nonrelated to VAP27s. The ZmPIP2;5 interactors (ZmVAP27‐1 and ZmVAP27‐2) are part of the same homology group with an MSD amino acid sequence identity over 50% (Fig. S6b). Publicly available transcriptome data analysis showed that six of 14 group members are highly expressed in most plant tissues (Fig. S6c). Both ZmVAP27‐1 and ZmVAP27‐2 are among these highly expressed genes, and exhibit the greatest expression levels in roots (Fig. S6c), similar to ZmPIP2;5 (Hachez et al., 2006, 2008).
To obtain insights into the family evolution of the ZmVAP27‐1 and ZmVAP27‐2 homology group, we performed a phylogenetic analysis. A balanced selection of species and genes allowed an alignment of 254 proteins from 41 green plant species (see Materials and methods section). The reconstructed phylogenetic tree showed that this VAP27 homology group can be further subdivided into two paralogue groups (Fig. 7a, the full phylogeny is shown in Fig. S7), which correspond with the previously defined AtVAP27s clades I and III (Wang et al., 2016). The presence of these clades can be traced back to Gymnosperms, suggesting that both clades were present in the Spermatophyta ancestor. Within each of these two main clades, two subgroups can be distinguished for Angiosperms, showing family‐specific expansion events. ZmVAP27‐1, together with other seven ZmVAP27s, clearly group in the AtVAP27 clade I, the closer Arabidopsis ZmVAP27‐1 orthologues being the isoforms AtVAP27‐1/3/5/7. Conversely, ZmVAP27‐2, and five other ZmVAP27s, group with the AtVAP27 clade III, the closer Arabidopsis ZmVAP27‐2 orthologue being the isoform AtVAP27‐2 (Fig. 7a).
Fig. 7.
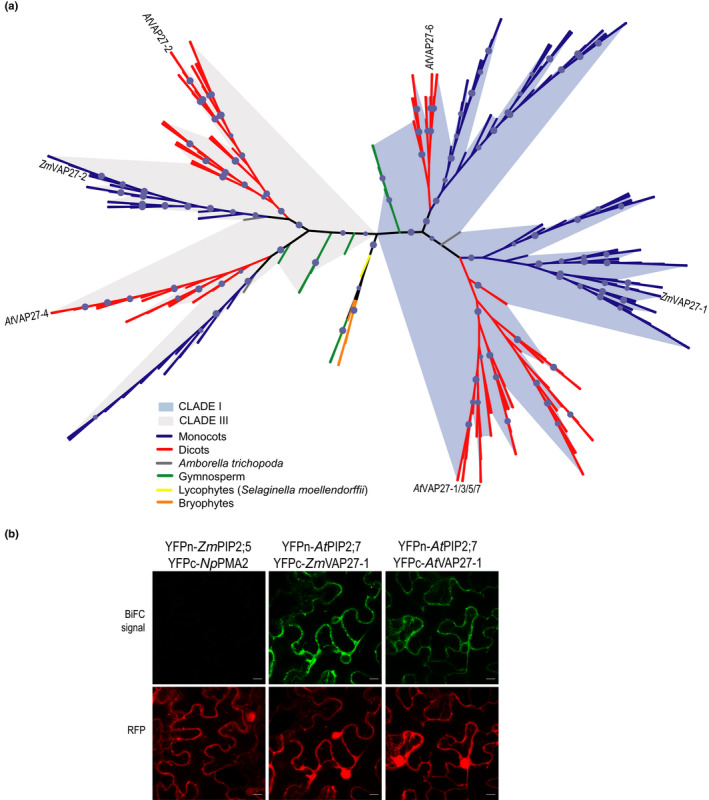
VAP27 protein phylogenetic analysis and conservation of the PIP–VAP27 interaction in Arabidopsis. (a) ZmVAP27‐1 and ZmVAP27‐2 homology group phylogenetic tree reconstructed by maximum likelihood. Branch support was assessed by the ultrafast bootstrap approximation with 1000 replicates (values ≥ 95 are represented by a blue circle). Maize VAP27‐1, VAP27‐2 and Arabidopsis VAP27s are shown in the tree. Branches are coloured according to the taxonomy in the accompanying legends. The shadows emphasise the expansion of two different clades. (b) AtPIP2;7 interacts with the Arabidopsis and maize VAP27‐1 proteins. Bimolecular fluorescence complementation (BiFC) signals for the pairs YFPn‐ZmPIP2;5/YFPc‐NpPMA2 (negative control), YFPn‐AtPIP2;7/YFPc‐ZmVAP27‐1, and YFPn‐AtPIP2;7/YFPc‐AtVAP27‐1. BiFC signal (YFP) is in green, and the RFP signal is in red and serves as a transfection control. Bars, 10 µm.
Regarding PIP2 channel evolution, their origin can be traced back to the Embryophyta ancestor (Soto et al., 2012; Abascal et al., 2014). We hypothesised that the PIP2–VAP27 interaction is conserved through evolution. We looked for evidence to support this hypothesis in the published Arabidopsis data. We localised in the Arabidopsis Interactome Map (Consortium, 2011) AtVAP27‐1 interaction with AtPIP2;3, AtPIP2;7, and AtPIP2;8 and the interaction of AtVAP27‐4 with AtPIP2;7. Also, the AtVAP27‐3 isoform was pulled‐down with AtPIP2;1 and not with AtPIP1;2 (Bellati et al., 2016). We validated the interaction between AtPIP2;7 and AtVAP27‐1 by BiFC (Fig. 7b), and we also tested whether AtPIP2;7 was able to interact with ZmVAP27‐1. As observed in Fig. 7(b), co‐expression of both proteins led to a fluorescent signal, demonstrating their interaction. Altogether, these data showed that the PIP2–VAP27 interaction is an ancient land plant feature, being at least present in the common Angiosperm ancestor. Future work will elucidate whether this interaction is also present in bryophytes, or if it is a distinctive vascular plant feature.
Discussion
To unravel the different aquaporin roles and regulation mechanisms in water and/or solute homeostasis in plant cells, it is crucial to identify their intracellular partners. The pull‐down assays presented here, together with previous assays using tagged PIPs (Hachez et al., 2014a; Bellati et al., 2016), allowed protein identification from the cytoskeleton and ER as PIP partners (Table S4), suggesting a close interaction between PIPs and those cellular structures. Moreover, we showed that PIP2s and the ER resident VAP27s interact, and presented novel evidence of plant aquaporin activity regulation by EPC residents.
To our knowledge, ZmPIP2;5 is the first plant PM protein identified as a partner of ER‐located VAP27s. We confirmed these interactions by SUS and BiFC experiments (Fig. 2). Interestingly, ER‐located VAP27s are anchored to the cell wall by unknown PM proteins (Wang et al., 2016). We speculate that ZmPIP2;5 might be one of the proteins anchoring ER‐located ZmVAP27s to the cell wall. Actually, the cell wall restricts AtPIP2;1 mobility (Martiniere et al., 2012) and, using fluorescence recovery after photobleaching assays (Methods S2), we also showed that ZmPIP2;5 mobility in the PM is restricted in comparison with other proteins that expose few amino acids to the apoplast, like ZmLTi6A (Fig. S8). Also, we demonstrated that the ZmVAP27‐1 MSD is essential for the PIP–VAP27 interaction, whereas the CCD and TMD are not (Fig. 3b). It is important to point out that ZmPIP2;5 and ZmSYP121 are both PM residents and their BiFC signals reconstitute at the PM, whereas the BiFC signal of the ZmPIP2;5‐ZmVAP27‐1/2 pairs reconstitutes in dotted structures (Fig. 2c), reminiscent of the AtVAP27‐1 bright dotted pattern (Wang et al., 2016). Conversely, mTRQ2‐ZmVAP27‐1/2 expression in the ER did not prevent mVenus‐ZmPIP2;5 PM localisation (Figs 4, 5). These results, together with the central role of the MSD in the interaction, demonstrate that the interaction between VAP27‐1/2 and ZmPIP2;5 occurs at EPCSs. We also showed that AtVAP27‐1 interacted with AtPIP2;7 (Fig. 7), and AtVAP27‐3 was also detected in an AtPIP2;1 interactomic study performed by Bellati et al. (2016). All these observations, together with the fact that ZmVAP27‐1 was not interacting in BiFC assays with the H+‐ATPase PMA2 (Fig. S1), another protein that is enriched in PM microdomains just like PIP channels (Laloi et al., 2007), might indicate that PIP aquaporins play a role in anchoring the EPCSs to the cell wall.
Endocytosis is a rapid way to adjust PIP protein abundance in the PM in response to an osmotic or salt stress (Chaumont & Tyerman, 2014; Pou et al., 2016). Salt stress‐induced AtPIP2;1 internalisation is a process that depends on two kinases: phosphatidylinositol 3‐kinase (PI3K) and phosphatidylinositol 4‐kinase (PI4K) (Ueda et al., 2016). Interestingly, AtVAP27‐1/3 bind with high affinity to different phosphorylated PI (Stefano et al., 2018), among them the PI3K and PI4K products. In addition, endocytosis is partially impaired in the plant double mutant vap27‐1/vap27‐3 (Stefano et al., 2018), as it is impaired in mammal cells lacking VAPs (Dong et al., 2016). Here, we showed that salt stress‐induced vesicles carrying ZmPIP2;5 are wrapped with ZmVAP27‐1‐labelled ER (Figs 4, 5, 5d,e). Therefore, PIP interaction with VAP27s can facilitate PIP loading into endocytic structures enriched in specific PIs.
Upon osmotic stress, AtPIP2;7 interaction with the tryptophan‐rich sensory protein/translocator (TSPO) leads to aquaporin level reduction in the PM, through degradation by the autophagic pathway (Hachez et al., 2014b; Jurkiewicz et al., 2020). How the internalisation of this complex occurs is still unknown. Interestingly, TSPO and PIP interaction occurs only if TSPO binds PI(4,5)P2 (Jurkiewicz et al., 2020) and VAP27s also bind PI(4,5)P2 (Stefano et al., 2018). Moreover, AtVAP27‐1 interacts with essential proteins involved in endocytic component autophagy (Wang & Hussey, 2019). As AtPIP2;7 interacts with AtVAP27‐1 (Fig. 7b), we propose that PIP autophagy may be initiated at EPCSs through its interaction with TSPO and VAP27s. This hypothesis will need to be addressed.
The PIP–VAP27 interaction increases oocyte membrane water permeability compared with oocytes expressing PIP alone (Fig. 6a). We found no evidence of a modification in PIP proton sensing by VAP27s (Fig. 6b), suggesting that the structural elements involved in pH sensing are not affected by the interaction. Still, we cannot discard a conformational pore modification. Therefore, the P f increase could be explained either by a greater intrinsic permeability of PIP channels due to a conducting pore conformational change, or by a greater active channel number in the PM. Under normal conditions, when no stress signals are perceived by cells to induce PIP internalisation, VAP27s could recruit and stabilise the channels in EPCSs similar to the VAPs clustering of the mammal potassium channel Kv2.1 (Kirmiz et al., 2018). Interestingly, cells with genetically elevated PI(4,5)P2 levels also have greater P f than control cells (Ma et al., 2015). The authors were able to link the PI effect on P f to aquaporins, but they were not able to discern between an increase in channel abundance in the PM or an increase in their water channel activity. Further research will help to understand if VAP27s stabilise PIPs in the PM and guide the channels to rapid endocytosis in response to a stimulus.
We recently reported how ZmPIP2;5 expression levels affect water relationships and plant growth (Ding et al., 2020). ZmPIP2;5 is the most highly expressed PIP2 isoform in roots. Interestingly, the two different ZmVAP27s that interact with ZmPIP2;5 are also highly expressed in roots (Fig. S6c), and belong to different clades that we traced back to the Spermatophyta ancestor (c. 319 million years ago (Ma); Jiao et al., 2011) (Fig. 7a). The challenge of future research will be to understand: (1) the role of the interaction in plant water movement, especially in response to osmotic and salt stress; (2) whether these interactions are linked to the acquisition of new functionalities in planta; and (3) whether the ZmPIP2;5/ZmVAP27‐1 and ZmPIP2;5/ZmVAP27‐2 interactions form part of different EPCSs types or not.
Author contributions
ARF and FC designed the experiments. ARF, TL, FS, KF and HD performed the experiments. ARF, TL, FS, HD, PM, KA and FC analysed the data. ARF and FC wrote the manuscript. All the authors contributed to the discussion and revision of the manuscript.
Supporting information
Fig. S1 ZmVAP27‐1 does not interact with NpPMA2, a H+‐ATPase PM resident.
Fig. S2 MSD of VAPs is required for the interaction with ZmPIP2;5.
Fig. S3 ZmVAP27‐1 colocalises with ZmPIP2;5 in Hetchian strands.
Fig. S4 ZmVAP27‐1 colocalises with ZmPIP2;5 in NaCl stress‐related structures.
Fig. S5 ZmVAP27‐1 increases Xenopus oocyte P f when co‐expressed with ZmPIP2;5.
Fig. S6 VAP27s in the maize genome.
Fig. S7 VAP27 protein family phylogenetic tree reconstructed by maximum likelihood.
Fig. S8 ZmPIP2;5 mobility in the PM is restricted in comparison with ZmLTi6A.
Methods S1 Mass spectrometry analysis.
Methods S2 Fluorescence recovery after photobleaching assay.
Table S1 Primers used in this work.
Table S2 Mass spectrometry peptides information.
Table S3 Potential ZmPIP2;5 interacting proteins.
Table S4 Proteins associated with the cytoskeleton that were pulled‐down by different PIP2s.
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Acknowledgements
We thank MASSPROT, IMABIOL, Marie‐Christine Eloy for confocal expertise and Joseph Nader for his helpful advice in benchwork. This work was supported by the Belgian National Fund for Scientific Research (FNRS, FRFC 2.4.501.06F), the ‘Communauté française de Belgique‐Actions de Recherches Concertées’ (grant ARC16/21‐075), the Pierre and Colette Bauchau Award, the Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET PIP 2014‐0206), and the Agencia Nacional de Promoción Científica y Tecnológica (PICT 2017‐0244). ARF was supported by an Incoming Post‐doc Move‐in Louvain Fellowship co‐funded by the Marie Curie Actions. FS was supported by a research fellow at the Universidad de Buenos Aires. TL was supported by a research fellow at the Fonds de Formation à la Recherche dans l’Industrie et l’Agriculture.
References
- Abascal F, Irisarri I, Zardoya R. 2014. Diversity and evolution of membrane intrinsic proteins. Biochimica et Biophysica Acta 1840: 1468–1481. [DOI] [PubMed] [Google Scholar]
- Afzal Z, Howton T, Sun Y, Mukhtar M. 2016. The roles of aquaporins in plant stress responses. Journal of Developmental Biology 4: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batoko H, Zheng H‐Q, Hawes C, Moore I. 2000. A Rab1 GTPase is required for transport between the endoplasmic reticulum and golgi apparatus and for normal golgi movement in plants. Plant Cell 12: 2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellati J, Alleva K, Soto G, Vitali V, Jozefkowicz C, Amodeo G. 2010. Intracellular pH sensing is altered by plasma membrane PIP aquaporin co‐expression. Plant Molecular Biology 74: 105–118. [DOI] [PubMed] [Google Scholar]
- Bellati J, Champeyroux C, Hem S, Rofidal V, Krouk G, Maurel C, Santoni V. 2016. Novel aquaporin regulatory mechanisms revealed by interactomics. Molecular & Cellular Proteomics 15: 3473–3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besserer A, Burnotte E, Bienert GP, Chevalier AS, Errachid A, Grefen C, Blatt MR, Chaumont F. 2012. Selective regulation of maize plasma membrane aquaporin trafficking and activity by the SNARE SYP121. Plant Cell 24: 3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bienert GP, Heinen RB, Berny MC, Chaumont F. 2014. Maize plasma membrane aquaporin ZmPIP2;5, but not ZmPIP1;2, facilitates transmembrane diffusion of hydrogen peroxide. Biochimica et Biophysica Acta – Biomembranes 1838: 216–222. [DOI] [PubMed] [Google Scholar]
- Boursiac Y, Chen S, Luu D‐T, Sorieul M, van den Dries N, Maurel C. 2005. Early effects of salinity on water transport in Arabidopsis roots. Molecular and cellular features of aquaporin expression. Plant Physiology 139: 790–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein‐dye binding. Analytical Biochemistry 72: 248–254. [DOI] [PubMed] [Google Scholar]
- Capella‐Gutierrez S, Silla‐Martinez JM, Gabaldon T. 2009. trimAl: a tool for automated alignment trimming in large‐scale phylogenetic analyses. Bioinformatics 25: 1972–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaumont F, Tyerman SD. 2014. Aquaporins: highly regulated channels controlling plant water relations. Plant Physiology 164: 1600–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevalier AS, Bienert GP, Chaumont F. 2014. A new LxxxA motif in the transmembrane Helix3 of maize aquaporins belonging to the plasma membrane intrinsic protein PIP2 group is required for their trafficking to the plasma membrane. Plant Physiology 166: 125–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium AIM . 2011. Evidence for network evolution in an Arabidopsis interactome map. Science 333: 601–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deeks MJ, Calcutt JR, Ingle EKS, Hawkins TJ, Chapman S, Richardson AC, Mentlak DA, Dixon MR, Cartwright F, Smertenko AP et al 2012. A superfamily of actin‐binding proteins at the actin‐membrane nexus of higher plants. Current Biology 22: 1595–1600. [DOI] [PubMed] [Google Scholar]
- Ding L, Milhiet T, Couvreur V, Nelissen H, Meziane A, Parent B, Aesaert S, Lijsebettens MV, Inzé D, Tardieu F et al 2020. Modification of the expression of the aquaporin ZmPIP2;5 affects water relations and plant growth. Plant Physiology 182: 2154–2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong R, Saheki Y, Swarup S, Lucast L, Harper JW, De Camilli P. 2016. Endosome‐ER contacts control actin nucleation and retromer function through VAP‐dependent regulation of PI4P. Cell 166: 408–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortuna AC, Palma GZD, Car LA, Armentia L, Vitali V, Zeida A, Estrin DA, Alleva K. 2019. Gating in plant plasma membrane aquaporins: the involvement of leucine in the formation of a pore constriction in the closed state. FEBS Journal 286: 3473–3487. [DOI] [PubMed] [Google Scholar]
- Fox AR, Maistriaux LC, Chaumont F. 2017. Toward understanding of the high number of plant aquaporin isoforms and multiple regulation mechanisms. Plant Science 264: 179–187. [DOI] [PubMed] [Google Scholar]
- Frick A, Järvå M, Törnroth‐Horsefield S. 2013. Structural basis for pH gating of plant aquaporins. FEBS Letters 587: 989–993. [DOI] [PubMed] [Google Scholar]
- Glaab E, Schneider R. 2015. RepExplore: addressing technical replicate variance in proteomics and metabolomics data analysis. Bioinformatics 31: 2235–2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman A, Harper S, Speicher DW. 2016. Detection of proteins on blot membranes. Current Protocols in Protein Science 86: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorgoni B, Fiorentino L, Marchioni M, Carnevali F. 1995. Cloning, expression and functional role of XRPFI α and β Subunits in Xenopus laevis oocyte. Biochemical and Biophysical Research Communications 215: 1088–1095. [DOI] [PubMed] [Google Scholar]
- Grefen C, Blatt MR. 2012. A 2in1 cloning system enables ratiometric bimolecular fluorescence complementation (rBiFC). BioTechniques 53: 311–314. [DOI] [PubMed] [Google Scholar]
- Grefen C, Obrdlik P, Harter K. 2009. The determination of protein‐protein interactions by the mating‐based split‐ubiquitin system (mbSUS). Methods in Molecular . Biology (Clifton, N.J.) 479: 217–233. [DOI] [PubMed] [Google Scholar]
- Grondin A, Rodrigues O, Verdoucq L, Merlot S, Leonhardt N, Maurel C. 2015. Aquaporins contribute to ABA‐triggered stomatal closure through OST1‐mediated phosphorylation. Plant Cell 27: 1945–1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Dufayard J‐F, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum‐likelihood phylogenies: assessing the performance of PhyML 3.0. Systematic Biology 59: 307–321. [DOI] [PubMed] [Google Scholar]
- Hachez C, Heinen RB, Draye X, Chaumont F. 2008. The expression pattern of plasma membrane aquaporins in maize leaf highlights their role in hydraulic regulation. Plant Molecular Biology 68: 337. [DOI] [PubMed] [Google Scholar]
- Hachez C, Laloux T, Reinhardt H, Cavez D, Degand H, Grefen C, Rycke RD, Inzé D, Blatt MR, Russinova E et al 2014a. Arabidopsis SNAREs SYP61 and SYP121 coordinate the trafficking of plasma membrane aquaporin PIP2;7 to modulate the cell membrane water permeability. Plant Cell 26: 3132–3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hachez C, Moshelion M, Zelazny E, Cavez D, Chaumont F. 2006. Localization and quantification of plasma membrane aquaporin expression in maize primary root: A clue to understanding their role as cellular plumbers. Plant Molecular Biology 62: 305–323. [DOI] [PubMed] [Google Scholar]
- Hachez C, Veljanovski V, Reinhardt H, Guillaumot D, Vanhee C, Chaumont F, Batoko H. 2014b. The Arabidopsis abiotic stress‐induced TSPO‐related protein reduces cell‐surface expression of the aquaporin PIP2;7 through protein‐protein interactions and autophagic degradation. Plant Cell 26: 4974–4990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecker A, Wallmeroth N, Peter S, Blatt MR, Harter K, Grefen C. 2015. Binary 2in1 vectors improve in planta (co)localization and dynamic protein interaction studies. Plant Physiology 168: 776–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinen RB, Ye Q, Chaumont F. 2009. Role of aquaporins in leaf physiology. Journal of Experimental Botany 60: 2971–2985. [DOI] [PubMed] [Google Scholar]
- Hoang DT, Chernomor O, von Haeseler A, Minh BQ, Vinh LS. 2018. UFBoot2: improving the ultrafast bootstrap approximation. Molecular Biology and Evolution 35: 518–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosy E, Martinière A, Choquet D, Maurel C, Luu D‐T. 2015. Super‐resolved and dynamic imaging of membrane proteins in plant cells reveal contrasting kinetic profiles and multiple confinement mechanisms. Molecular Plant 8: 339–342. [DOI] [PubMed] [Google Scholar]
- Jiao Y, Wickett NJ, Ayyampalayam S, Chanderbali AS, Landherr L, Ralph PE, Tomsho LP, Hu Y, Liang H, Soltis PS et al 2011. Ancestral polyploidy in seed plants and angiosperms. Nature 473: 97–100. [DOI] [PubMed] [Google Scholar]
- Johnson B, Leek AN, Solé L, Maverick EE, Levine TP, Tamkun MM. 2018. Kv2 potassium channels form endoplasmic reticulum/plasma membrane junctions via interaction with VAPA and VAPB. Proceedings of the National Academy of Sciences, USA 115: E7331–E7340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jozefkowicz C, Rosi P, Sigaut L, Soto G, Pietrasanta LI, Amodeo G, Alleva K. 2013. Loop A is critical for the functional interaction of two Beta vulgaris PIP aquaporins. PLoS ONE 8: e57993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jozefkowicz C, Sigaut L, Scochera F, Soto G, Ayub N, Pietrasanta LI, Amodeo G, González Flecha FL, Alleva K. 2016. PIP water transport and its pH dependence are regulated by tetramer stoichiometry. Biophysical Journal 110: 1312–1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurkiewicz P, Senicourt L, Ayeb H, Lequin O, Lacapere J‐J, Batoko H. 2020. A plant‐specific N‐terminal extension reveals evolutionary functional divergence within translocator proteins. iScience 23: 100889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nature Methods 14: 587–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Kwon H, Kim S, Kim MK, Botella MA, Yun HS, Kwon C. 2016. Synaptotagmin 1 negatively controls the two distinct immune secretory pathways to powdery mildew fungi in Arabidopsis. Plant and Cell Physiology 57: 1133–1141. [DOI] [PubMed] [Google Scholar]
- Kirik V, Herrmann U, Parupalli C, Sedbrook JC, Ehrhardt DW, Hülskamp M. 2007. CLASP localizes in two discrete patterns on cortical microtubules and is required for cell morphogenesis and cell division in Arabidopsis. Journal of Cell Science 120: 4416–4425. [DOI] [PubMed] [Google Scholar]
- Kirmiz M, Vierra NC, Palacio S, Trimmer JS. 2018. Identification of VAPA and VAPB as Kv2 channel‐interacting proteins defining endoplasmic reticulum‐plasma membrane junctions in mammalian brain neurons. Journal of Neuroscience 38: 7562–7584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krtková J, Benáková M, Schwarzerová K. 2016. Multifunctional microtubule‐associated proteins in plants. Frontiers in Plant Science 7: 474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laloi M, Perret A‐M, Chatre L, Melser S, Cantrel C, Vaultier M‐N, Zachowski A, Bathany K, Schmitter J‐M, Vallet M et al 2007. Insights into the role of specific lipids in the formation and delivery of lipid microdomains to the plasma membrane of plant cells. Plant Physiology 143: 461–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang‐Pauluzzi I, Gunning BES. 2000. A plasmolytic cycle: The fate of cytoskeletal elements. Protoplasma 212: 174–185. [Google Scholar]
- Lee E, Vanneste S, Pérez‐Sancho J, Benitez‐Fuente F, Strelau M, Macho AP, Botella MA, Friml J, Rosado A. 2019. Ionic stress enhances ER–PM connectivity via phosphoinositide‐associated SYT1 contact site expansion in Arabidopsis. Proceedings of the National Academy of Sciences, USA 116: 1420–1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HK, Cho SK, Son O, Xu Z, Hwang I, Kim WT. 2009. Drought stress‐induced Rma1H1, a RING membrane‐anchor E3 ubiquitin ligase homolog, regulates aquaporin levels via ubiquitination in transgenic Arabidopsis plants. Plant Cell 21: 622–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I, Bork P. 2016. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Research 44: W242–W245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis JD, Lazarowitz SG. 2010. Arabidopsis synaptotagmin SYTA regulates endocytosis and virus movement protein cell‐to‐cell transport. Proceedings of the National Academy of Sciences, USA 107: 2491–2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Wang H, Gago J, Cui H, Qian Z, Kodama N, Ji H, Tian S, Shen D, Chen Y et al 2015. Harpin Hpa1 interacts with aquaporin PIP1;4 to promote the substrate transport and photosynthesis in Arabidopsis. Scientific Reports 5: 17207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Wang X, Yang Y, Li R, He Q, Fang X, Luu D‐T, Maurel C, Lin J. 2011. Single‐molecule analysis of PIP2;1 dynamics and partitioning reveals multiple modes of Arabidopsis plasma membrane aquaporin regulation. Plant Cell 23: 3780–3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martiniere A, Lavagi I, Nageswaran G, Rolfe DJ, Maneta‐Peyret L, Luu D‐T, Botchway SW, Webb SED, Mongrand S, Maurel C et al 2012. Cell wall constrains lateral diffusion of plant plasma‐membrane proteins. Proceedings of the National Academy of Sciences, USA 109: 12805–12810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurel C, Boursiac Y, Luu D‐T, Santoni V, Shahzad Z, Verdoucq L. 2015. Aquaporins in plants. Physiological Reviews 95: 1321–1358. [DOI] [PubMed] [Google Scholar]
- Ma X, Shatil‐Cohen A, Ben‐Dor S, Wigoda N, Perera IY, Im YJ, Diminshtein S, Yu L, Boss WF, Moshelion M et al 2015. Do phosphoinositides regulate membrane water permeability of tobacco protoplasts by enhancing the aquaporin pathway? Planta 241: 741–755. [DOI] [PubMed] [Google Scholar]
- McFarlane HE, Lee EK, van Bezouwen LS, Ross B, Rosado A, Samuels AL. 2017. Multiscale structural analysis of plant ER–PM contact sites. Plant and Cell Physiology 58: 478–484. [DOI] [PubMed] [Google Scholar]
- Moshelion M, Moran N, Chaumont F. 2004. Dynamic changes in the osmotic water permeability of protoplast plasma membrane. Plant Physiology 135: 2301–2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy SE, Levine TP. 2016. VAP, a versatile access point for the endoplasmic reticulum: review and analysis of FFAT‐like motifs in the VAPome. Biochimica et Biophysica Acta – Molecular and Cell Biology of Lipids 1861: 952–961. [DOI] [PubMed] [Google Scholar]
- Paredez AR, Somerville CR, Ehrhardt DW. 2006. Visualization of cellulose synthase demonstrates functional association with microtubules. Science 312: 1491–1495. [DOI] [PubMed] [Google Scholar]
- Pérez‐Sancho J, Tilsner J, Samuels AL, Botella MA, Bayer EM, Rosado A. 2016. Stitching organelles: organization and function of specialized membrane contact sites in plants. Trends in Cell Biology 26: 705–717. [DOI] [PubMed] [Google Scholar]
- Pérez‐Sancho J, Vanneste S, Lee E, McFarlane HE, del Valle AE, Valpuesta V, Friml J, Botella MA, Rosado A. 2015. The Arabidopsis synaptotagmin1 is enriched in endoplasmic reticulum‐plasma membrane contact sites and confers cellular resistance to mechanical stresses. Plant Physiology 168: 132–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietra S, Gustavsson A, Kiefer C, Kalmbach L, Hörstedt P, Ikeda Y, Stepanova AN, Alonso JM, Grebe M. 2013. ArabidopsisSABRE and CLASP interact to stabilize cell division plane orientation and planar polarity. Nature Communications 4: 2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pou A, Jeanguenin L, Milhiet T, Batoko H, Chaumont F, Hachez C. 2016. Salinity‐mediated transcriptional and post‐translational regulation of the Arabidopsis aquaporin PIP2;7. Plant Molecular Biology 92: 731–744. [DOI] [PubMed] [Google Scholar]
- Prak S, Hem S, Boudet J, Viennois G, Sommerer N, Rossignol M, Maurel C, Santoni V. 2008. Multiple phosphorylations in the C‐terminal tail of plant plasma membrane aquaporins: Role in subcellular trafficking of At PIP2;1 in response to salt stress. Molecular & Cellular Proteomics 7: 1019–1030. [DOI] [PubMed] [Google Scholar]
- Roche JV, Törnroth‐Horsefield S. 2017. Aquaporin protein–protein interactions. International Journal of Molecular Sciences 18: 2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz J, Milpetz F, Bork P, Ponting CP. 1998. SMART, a simple modular architecture research tool: identification of signaling domains. Proceedings of the National Academy of Sciences, USA 95: 5857–5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sela I, Ashkenazy H, Katoh K, Pupko T. 2015. GUIDANCE2: accurate detection of unreliable alignment regions accounting for the uncertainty of multiple parameters. Nucleic Acids Research 43: W7–W14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siao W, Wang P, Voigt B, Hussey PJ, Baluska F. 2016. Arabidopsis SYT1 maintains stability of cortical endoplasmic reticulum networks and VAP27‐1‐enriched endoplasmic reticulum–plasma membrane contact sites. Journal of Experimental Botany 67: 6161–6171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soto G, Alleva K, Amodeo G, Muschietti J, Ayub ND. 2012. New insight into the evolution of aquaporins from flowering plants and vertebrates: orthologous identification and functional transfer is possible. Gene 503: 165–176. [DOI] [PubMed] [Google Scholar]
- Stefano G, Renna L, Wormsbaecher C, Gamble J, Zienkiewicz K, Brandizzi F. 2018. Plant endocytosis requires the ER membrane‐anchored proteins VAP27‐1 and VAP27‐3. Cell Reports 23: 2299–2307. [DOI] [PubMed] [Google Scholar]
- Takano J, Yoshinari A, Luu D‐T. 2017. Plant aquaporin trafficking In: Chaumont F, Tyerman SD, eds. Plant aquaporins. Cham, Switzerland: Springer International Publishing, 47–81. [Google Scholar]
- Törnroth‐Horsefield S, Wang Y, Hedfalk K, Johanson U, Karlsson M, Tajkhorshid E, Neutze R, Kjellbom P. 2006. Structural mechanism of plant aquaporin gating. Nature 439: 688–694. [DOI] [PubMed] [Google Scholar]
- Tournaire‐Roux C, Sutka M, Javot H, Gout E, Gerbeau P, Luu D‐T, Bligny R, Maurel C. 2003. Cytosolic pH regulates root water transport during anoxic stress through gating of aquaporins. Nature 425: 393–397. [DOI] [PubMed] [Google Scholar]
- Ueda M, Tsutsumi N, Fujimoto M. 2016. Salt stress induces internalization of plasma membrane aquaporin into the vacuole in Arabidopsis thaliana . Biochemical and Biophysical Research Communications 474: 742–746. [DOI] [PubMed] [Google Scholar]
- Valm AM, Cohen S, Legant WR, Melunis J, Hershberg U, Wait E, Cohen AR, Davidson MW, Betzig E, Lippincott‐Schwartz J. 2017. Applying systems‐level spectral imaging and analysis to reveal the organelle interactome. Nature 546: 162–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Bel M, Diels T, Vancaester E, Kreft L, Botzki A, Van de Peer Y, Coppens F, Vandepoele K. 2018. PLAZA 4.0: an integrative resource for functional, evolutionary and comparative plant genomics. Nucleic Acids Research 46: D1190–D1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Hawkins TJ, Richardson C, Cummins I, Deeks MJ, Sparkes I, Hawes C, Hussey PJ. 2014. The plant cytoskeleton, NET3C, and VAP27 mediate the link between the plasma membrane and endoplasmic reticulum. Current Biology 24: 1397–1405. [DOI] [PubMed] [Google Scholar]
- Wang P, Hussey PJ. 2019. Plant ER‐PM contact sites in endocytosis and autophagy: does the local composition of membrane phospholipid play a role? Frontiers in Plant Science. doi: 10.3389/fpls.2019.00023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Richardson C, Hawkins TJ, Sparkes I, Hawes C, Hussey PJ. 2016. Plant VAP27 proteins: domain characterization, intracellular localization and role in plant development. New Phytologist 210: 1311–1326. [DOI] [PubMed] [Google Scholar]
- Wu XN, Rodriguez CS, Pertl‐Obermeyer H, Obermeyer G, Schulze WX. 2013. Sucrose‐induced receptor kinase SIRK1 regulates a plasma membrane aquaporin in Arabidopsis. Molecular & Cellular Proteomics 12: 2856–2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaneff A, Sigaut L, Marquez M, Alleva K, Pietrasanta LI, Amodeo G. 2014. Heteromerization of PIP aquaporins affects their intrinsic permeability. Proceedings of the National Academy of Sciences, USA 111: 231–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelazny E, Miecielica U, Borst JW, Hemminga MA, Chaumont F. 2009. An N‐terminal diacidic motif is required for the trafficking of maize aquaporins ZmPIP2;4 and ZmPIP2;5 to the plasma membrane. The Plant Journal 57: 346–355. [DOI] [PubMed] [Google Scholar]
- Zimmermann P, Hirsch‐Hoffmann M, Hennig L, Gruissem W. 2004. GENEVESTIGATOR. Arabidopsis microarray database and analysis toolbox. Plant Physiology 136: 2621–2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 ZmVAP27‐1 does not interact with NpPMA2, a H+‐ATPase PM resident.
Fig. S2 MSD of VAPs is required for the interaction with ZmPIP2;5.
Fig. S3 ZmVAP27‐1 colocalises with ZmPIP2;5 in Hetchian strands.
Fig. S4 ZmVAP27‐1 colocalises with ZmPIP2;5 in NaCl stress‐related structures.
Fig. S5 ZmVAP27‐1 increases Xenopus oocyte P f when co‐expressed with ZmPIP2;5.
Fig. S6 VAP27s in the maize genome.
Fig. S7 VAP27 protein family phylogenetic tree reconstructed by maximum likelihood.
Fig. S8 ZmPIP2;5 mobility in the PM is restricted in comparison with ZmLTi6A.
Methods S1 Mass spectrometry analysis.
Methods S2 Fluorescence recovery after photobleaching assay.
Table S1 Primers used in this work.
Table S2 Mass spectrometry peptides information.
Table S3 Potential ZmPIP2;5 interacting proteins.
Table S4 Proteins associated with the cytoskeleton that were pulled‐down by different PIP2s.
Please note: Wiley Blackwell are not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.


