Abstract
Since swine influenza virus was first isolated in 1930, it has become endemic in pigs worldwide. Although large amount of swine influenza vaccines have been used in swine industry, swine influenza still cannot be efficiently controlled and has been an important economic disease for swine industry. The high diversity and varied distribution of different subtypes and genotypes of swine influenza viruses circulating in pigs globally is a major challenge to produce broadly effective vaccines and control disease. Importantly, swine influenza virus is able to cross species barrier to infect humans and even caused influenza pandemic in 2009. Herein, current status and challenge of swine influenza viruses is reviewed and discussed.
Keywords: swine influenza virus, current status, challenge
Introduction
Swine influenza is an important respiratory disease in pigs and epidemic in most of areas worldwide, which is caused by influenza A virus (IAV) belonging to the family of Orthomyxoviridae. Swine influenza normally causes a high morbidity (up to 100%) and low mortality in infected pigs; however, it can lead to 10–15% mortality in naïve pigs. Infected pigs show clinical signs that are similar to those found in humans, this is why swine have been considered to be a good animal model for influenza research. The clinical signs are normally displayed as acute respiratory disease characterized by fever, lethargy, sneezing, coughing, difficulty breathing and decreased appetite (Alexander and Brown, 2000; McQueen et al., 1968). Swine influenza causes significant economic losses for pig producers primarily due to weight loss and reduced weight gain of infected pigs; however some cases can be much more severe with co-infection with other pathogens. Swine influenza virus together with other swine pathogens such as porcine reproductive and respiratory syndrome virus, porcine circovirus 2 and Mycoplasma hyopneumoniae, Bordetella bronchiseptica, and Actinobacillus pleuropneumoniae can cause porcine respiratory disease complex, resulting in significantly enhanced mortalities and economic losses annually for swine industry (Schultz-Cherry et al., 2012). Although swine influenza has been considered to be a seasonal disease like human season influenza, which majorly happens in winter and spring, it is likely not any more due to highly concentrated raising model in swine industry.
IAVs are single-strand negative-sense RNA viruses that infect a broad range of avian and mammalian species. The genome of IAVs is approximately 13.6 kb in total with 8 gene segments that encode at least 12 viral proteins. Based upon the major antigenic differences within the surface hemagglutinin (HA) and neuraminidase (NA) proteins, IAVs are divided into 18 HA and 11 NA subtypes. The H1-H16 and N1-N9 subtypes of viruses have been identified in aquatic birds which are normally considered to be the natural reservoir of IAVs (Fouchier et al., 2005; Rohm et al., 1996; Webster et al., 1992); while H17-H18 and N10-N11 subtype sequences were detected in bats (Tong et al., 2012; Tong et al., 2013). There are only a limited number of subtypes that have been established in mammals, such as H1 and H3 subtypes of viruses which have been established and circulating in both humans and swine (Ma et al., 2009). Two major mechanisms called antigenic drift and antigenic shift are involved in fast evolution of IAVs. Antigenic drift is due to the gradual accumulation of mutations in the surface HA and NA proteins resulting in antigenic changes of IAV. Antigenic drift is responsible for human seasonal influenza viruses. Antigenic shift also named reassortment is because of the segmented nature of the influenza virus genome; when two or more different IAVs infect a cell or host, viral gene segments can be reshuffled and recombined to generate different reassortant viruses. Antigenic shift is responsible to generate three (1957 Asian Flu H2N2, 1968 Hong Kong flu H3N2 and 2009 pandemic H1N1) of four pandemic influenza viruses in human history. Swine are the nature host of IAVs and have been hypothesized as the mixing vessel of human, avian and swine IAVs with potential to generate a pandemic virus (Kida et al., 1994; Ma et al., 2009; Scholtissek, 1990). The 2009 influenza pandemic has further demonstrated this hypothesis, since it was caused by a reassortant H1N1 virus from pigs that contains NA and M genes from the Eurasian swine virus and remaining six genes from the North American triple reassortant virus (Garten et al., 2009; Smith et al., 2009b). In addition, swine can also be an adaptive host of human and avian IAVs. For example, both the human 1918 Spanish-flu H1N1 and the whole avian H1N1 viruses have been two major lineages of viruses circulating in pigs after adaptation, which are currently named classic H1N1 (cH1N1) and Eurasian avian-like H1N1 swine viruses.
Current status of swine influenza viruses
Currently, three major subtypes of IAVs including H1N1, H3N2 and H1N2 have established and are circulating in swine herds globally. After 1918 human pandemic, the 1918-like cH1N1 virus was detected and became endemic in pigs worldwide (Schultz-Cherry et al., 2012). The cH1N1 swine virus has been maintained and circulating in swine populations globally for several decades, and has disappeared and been replaced by newly emerged swine influenza viruses in some areas such as Europe (Brown, 2000). This review focuses on current status of swine influenza viruses in North America, Europe and Asia since influenza surveillance in pigs have been performed in these areas for many years and more data are available.
Swine influenza viruses in North America
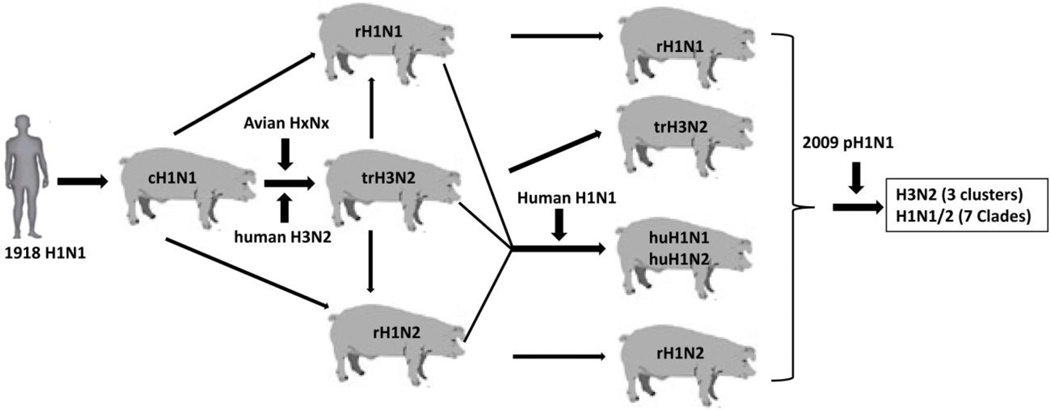
Prior to 1990s, the cH1N1 swine influenza virus that emerged around the 1918 pandemic was the only major subtype of virus detected in North American swine herds for more than 70 years. This virus likely did not cause major problems to swine industry. With introduction of human seasonal and avian influenza viruses into swine herds, multiple genotype and clades of H3 and H1 viruses were generated through reassortment with the endemic cH1N1 virus (Fig. 1), and started to circulate in swine populations (Vincent et al., 2008b). A novel triple-reassortant H3N2 virus emerged in the late 1990’s in North America and became dominant in swine populations, which had human seasonal virus HA, NA and PB1 genes, avian virus PA and PB2 genes and 3 internal genes from the cH1N1 swine virus (Olsen, 2002; Zhou et al., 1999). Subsequently, multiple clusters of antigenically distinct H3N2, reassortant H1N1 (rH1N1) and reassortant H1N2 (rH1N2) strains were detected in swine (Choi et al., 2002; Karasin et al., 2002; Webby et al., 2004; Webby et al., 2000). The triple-reassortant internal gene (TRIG) constellation, which contains gene segments from classical swine-, human- and avian-origin IAVs, was found in all detected triple reassortant viruses; this TRIG cassette is very stable and has capability to support different HA and NA combinations of the viruses circulating in pigs in the U.S. (Anderson et al., 2013; Lorusso et al., 2013; Vincent et al., 2009), resulting in antigenically different lineages of H1 and H3 swine viruses (Vincent et al., 2009). With introductions of human seasonal H1N1 virus into swine herds, reassortant human-like H1N1 (huH1N1) and H1N2 (huH1N2) viruses (Fig. 1), which have the HA and/or NA genes from human seasonal viruses and remaining six internal genes from contemporary triple reassortant swine viruses, were generated by reassorting with endemic triple reassortant viruses (Vincent et al., 2008b). These viruses rapidly spread across the U.S. in swine herds and co-circulated with contemporary swine influenza viruses as a distinct genetic clades from the cH1 swine virus (Vincent et al., 2009). After 2009 pandemic H1N1 (pH1N1) was introduced into swine populations, reassortant viruses between pH1N1 and endemic circulating swine viruses were detected in pigs (Fig. 1) (Ducatez et al., 2011; Kitikoon et al., 2013; Liu et al., 2011; Ma et al., 2014; Nelson et al., 2015). Noticeably, the M gene of TRIG constellation was replaced by the Eurasian swine M gene from the 2009 pH1N1 virus in the majority of viruses detected in the U.S. (Ma et al., 2019; Nelson et al., 2015). So far, there are at least 7 genetically and antigenically distinct clades of H1 viruses (Gao et al., 2017; Rajao et al., 2018) and 4 antigenically different clusters of H3 viruses (Anderson et al., 2013; Richt et al., 2003; Webby et al., 2004; Webby et al., 2000) which have presented and co-circulate in the U.S. swine (Fig. 2A and B). The cluster IV H3 virus currently contains several clades based on genetic and antigenic analysis (Fig. 2B) and is one of major dominant and endemic viruses in swine in the U.S. (Bolton et al., 2018; Kitikoon et al., 2013). In contrast, the cluster II H3 virus has died out from the pig populations. There are no or very limited cross protection between each clades (clusters) of H1 or H3 viruses. The current status of swine influenza viruses circulating in North American swine comprises a highly genetic and antigenic diverse of viruses, making more challenge for industry to produce on-time and effective vaccines to control swine influenza.
Figure 1: Major subtypes and genotype of swine influenza viruses in North America.
The 1918 Spanish flu H1N1 virus was transmitted to pigs and evolved into the cH1N1 swine virus. Introductions of avian and human seasonal influenza viruses into the swine herds resulted in generation triple reassortant H1 and H3 viruses that are maintained in pigs. After 2009, reassortment between 2009 pH1N1 and endemic triple reassortant H1 and H3 viruses generated different genotypes of viruses that are circulating in swine herds in North America.
Figure 2: Phylogenetic trees of H1 and H3 genes of swine influenza viruses in North America.
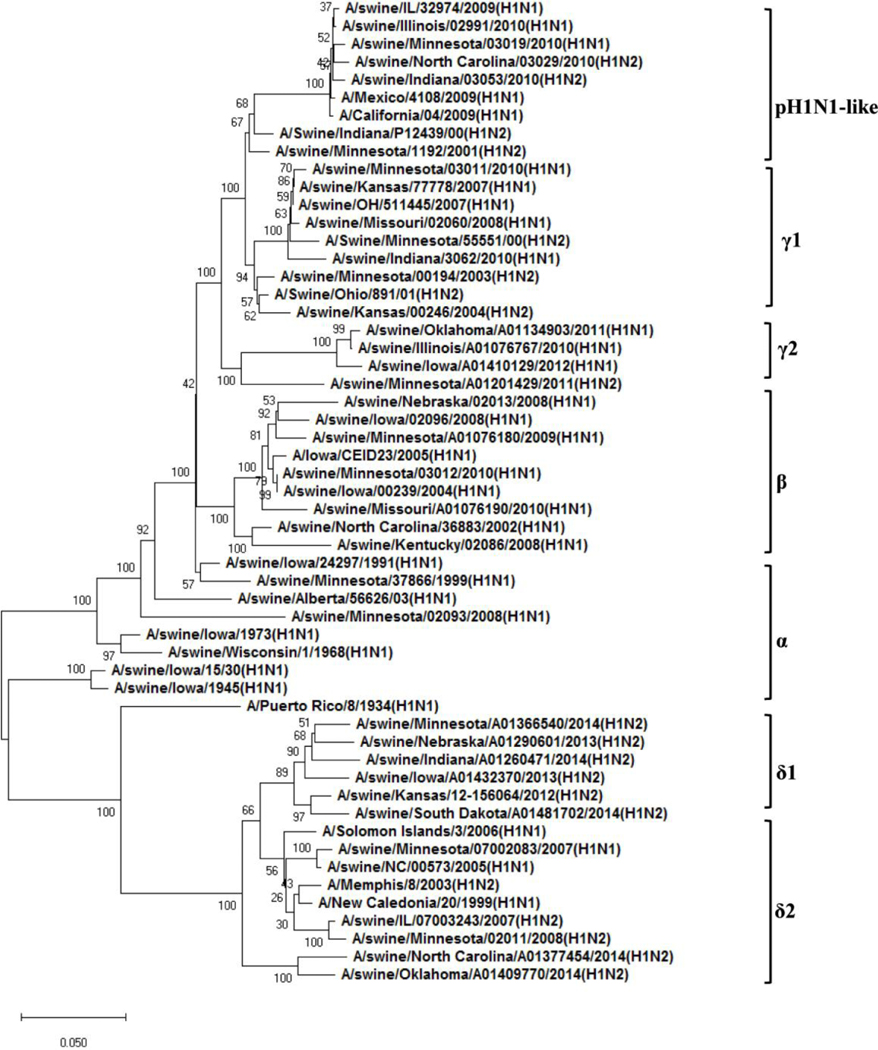
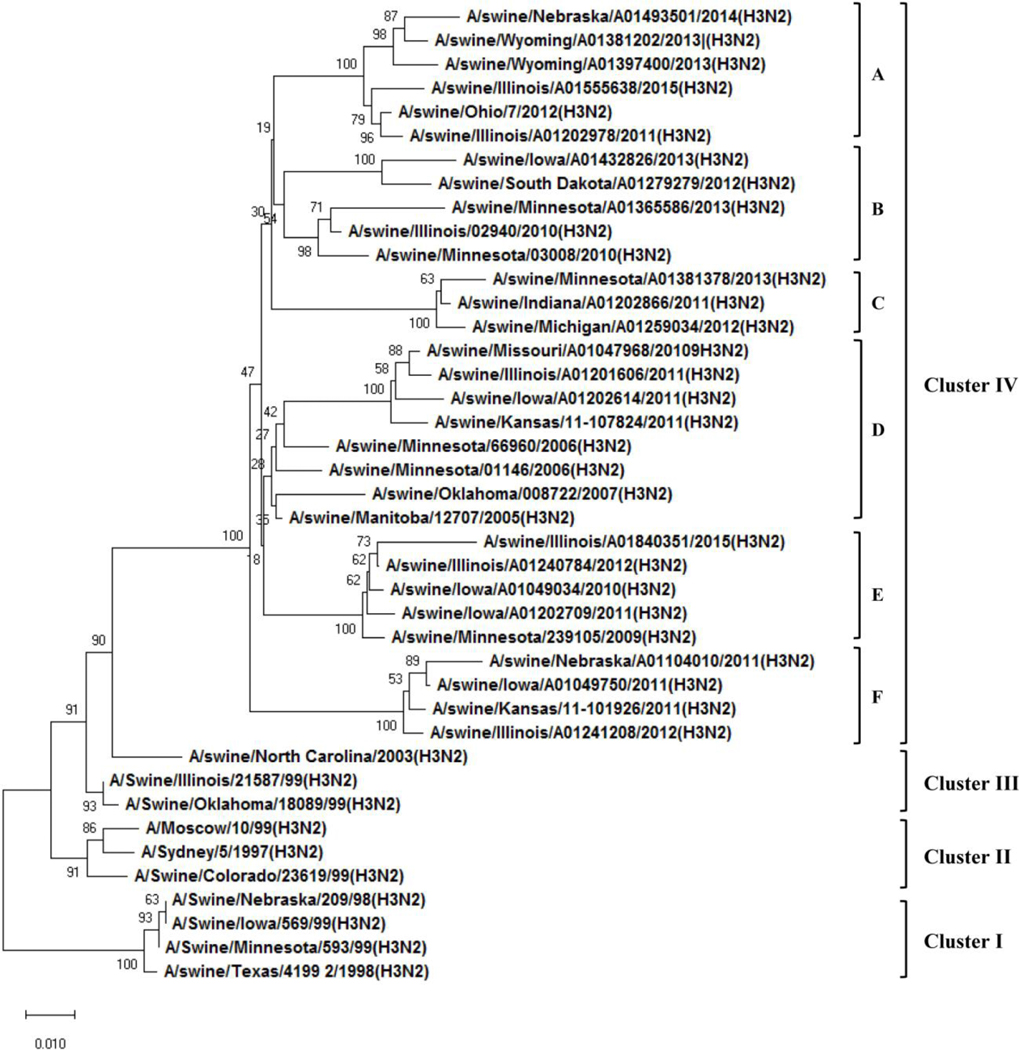
A) Phylogenetic tree of HA of H1 viruses; B) Phylogenetic tree of HA of H3 viruses. The trees were generated by software MEGA 4.1 with the distance-based neighbor-joining method. The reliability of the tree was assessed by bootstrap analysis with 1,000 replications. The H1 viruses have at least 7 genetically and antigenically distinct clades and the H3 viruses have 4 antigenically distinct clusters. The cluster IV H3 viruses have several genetically different (a-f) clades.
Swine influenza viruses in Europe
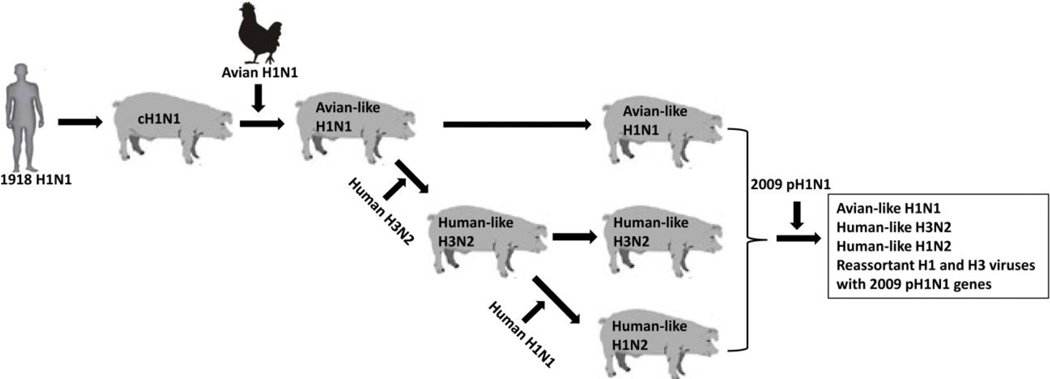
In Europe, H1N1 and H3N2 subtypes of IAVs, which include swine cH1N1, avian-like H1N1 and human-like H3N2 viruses (Fig. 3), have been widely reported and detected in European pigs that displayed clinical symptoms (Brown, 2012). Similar as the North American swine herds, the cH1N1 swine virus also circulated in European pigs for many years, but was replaced by an avian-like H1N1 virus that emerged in European pig populations in 1979 (Pensaert et al., 1981). The avian-like H1N1 (later called Eurasian H1N1) viruses have been highly adapted and transmissible among pigs and spread to all major European pig-producing countries, resulting in significant economic losses for pig producers (Zell et al., 2012). Later, a human-origin H3N2 virus, which has the HA and NA derived from the 1968 H3N2-like pandemic virus and six internal genes from the endemic avian-like H1N1 virus (Castrucci et al., 1994; Haesebrouck et al., 1985), became to circulate in European pigs (de Jong et al., 1999; Simon et al., 2014). Human-like swine H1N2 that became prevalent and endemic in Europe later (Marozin et al., 2002) was first detected in 1994, which contains the H1 that evolved from a 1980 human seasonal H1N1 virus and a human-origin N2 that is distinct from the previously introduced H3N2 human-like virus and the internal gene constellation of the 1979 avian-like virus (Brown et al., 1998) (Fig. 3). All these three subtypes of IAVs have established and become prevalent in the European pig populations (Fig. 3) and they successively replaced the circulating cH1N1 swine virus (Simon et al., 2014; Zell et al., 2012). After the 2009 pandemic occurred, the 2009 pH1N1 virus has been transmitted into swine in Europe (Chastagner et al., 2019; Lange et al., 2013; Welsh et al., 2010; Williamson et al., 2012) and was firstly detected in pigs in Northern Ireland and become endemic (Welsh et al., 2010). Subsequently, reassortant viruses incorporating the H1 and N2 from the endemic H1N2 swine virus, or the N2 from the endemic swine H1N2 virus, or the N1 from the avian-like H1N1 virus and the remaining genes from 2009 pH1N1 virus were detected in 2000 in the United Kingdom (Howard et al., 2011) and in Italy (Moreno et al., 2011) and in Germany (Lange et al., 2013; Starick et al., 2011). Later, a triple-reassortant H3N2 virus with a human-origin HA from a 2004–2005 seasonal virus, N2 from the endemic swine viruses, and the internal genes from 2009 pH1N1 has spread in Denmark swine herds (Krog et al., 2017). Reassortment between 2009 pH1N1 virus with contemporary swine viruses in European pig populations has increased the genotypes of circulating swine influenza viruses (Fig. 3), which represents a significant challenge for disease control and pig production (Beato et al., 2016; Pippig et al., 2016; Simon et al., 2014; Watson et al., 2015).
Figure 3: Major subtypes and genotype of swine influenza viruses in Europe.
A complete avian-like H1N1 virus emerged and became to be dominant virus in pigs after the cH1N1 has been circulated in pigs for several decades. Transmission of human seasonal H3N2 and H1N1 viruses to pigs resulted in human-like H3N2 and H1N2 viruses. Multiple genotype viruses have been produced by reassortment between 2009 pH1N1 and endemic swine viruses with introduction of 2009 pH1N1 into European swine herds.
Swine influenza viruses in Asia
In Asian countries, the cH1N1 swine virus has been considered to be present in pigs in China during 1918–1919 human pandemic (Fig. 4), but it was first isolated in Asia in 1974 (Zhu et al., 2011a). Influenza surveillance in the early 1980’s reveals that the cH1N1 virus is widely distributed in pigs in many Asian regions and countries (Arikawa et al., 1979; Nerome et al., 1982; Shortridge and Webster, 1979). Subsequently, this virus was found to be endemic in many Asian countries including China, Thailand, India, and Korea (Chatterjee et al., 1995; Guan et al., 1996; Kupradinun et al., 1991; Lee et al., 2008; Song et al., 2007). In addition, phylogenetic and antigenic analysis revealed that some cH1N1 isolates in Hong Kong and Japan were introduced from North America through importation of pigs (Nerome et al., 1982; Vijaykrishna et al., 2011). A reassortant H1N2 virus, which has the N2 segment from the early human-like H3N2 and seven remaining segments from the cH1N1 viruses (Sugimura et al., 1980), caused a big outbreak in Japan from winter 1989 to spring 1990 in pigs with a typical influenza illness (Ouchi et al., 1996). Similar reassortant viruses were also detected in Hong Kong from pigs imported from China during 1999 to 2004 (Vijaykrishna et al. 2011), indicating that this kind of virus has spread in Asian countries (Fig. 4). The H3N2 human-like Influenza viruses were first isolated in Asia from pigs in Taiwan soon after the Hong Kong pandemic (Kundin, 1970). The human-like H3N2 variants were also detected in pigs in several Asian countries including mainland China, Korea and Japan (Arikawa et al., 1982; Nerome et al., 1981; Shortridge and Webster, 1979; Shortridge et al., 1977; Song et al., 2003); however, these viruses reassorted with endemic viruses including cH1N1 and later imported European avian-like H1N1 viruses (Yu et al., 2008), and failed to be maintained and established in swine herds (Zhu et al., 2011a).
Figure 4: Major subtypes and genotype of swine influenza viruses in Asia.

With introduction of the human H3N2 virus into pigs, reassortant H1N2 and human-like H3N2 virus were produced by reassorting with the endemic cH1N1 virus, the latter one died out from the swine herds. Later, the Eurasian avian-like H1N1 and North American triple reassortant H1 and H3 swine viruses were introduced into the pigs in Asian countries by importing breeding pigs and became prevalent and co-circulated with the endemic viruses in pigs. Endemic H1 and H3 viruses as well as multiple reassortant viruses produced by 2009 pH1N1 reassorting with endemic swine viruses were detected and circulated in pigs after 2009.
European avian-like H1N1 viruses were detected in China in 2001 (Smith et al., 2009a) and in Thailand in 2008 (Takemae et al., 2008), most likely the virus was introduced via importing breeding pigs from European countries (Zhu et al., 2011a). This kind of virus started to co-circulate with swine cH1N1 and human-like H3N2 viruses in these areas and gradually replaced the cH1N1 swine virus to be an endemic and predominant virus in around 2006 (Zhu et al., 2011a). Later with importing breeding pigs from the USA, North American triple reassortant H1N2 virus was first identified in pigs in China in 2002 (Vijaykrishna et al., 2011), then both H1N2 and H3N2 triple reassortant viruses were isolated from pigs in Korea since 2004 (Jung and Chae, 2004; Pascua et al., 2008). These viruses circulate and reassort with locally endemic swine influenza viruses (Fig. 4).
During 2009 pandemic, the 2009 pH1N1 virus was repeatedly detected in pigs in Asian countries (Chen et al., 2012; Hiromoto et al., 2012; Kirisawa et al., 2014; Mine et al., 2018; Nagarajan et al., 2010; Song et al., 2010; Sun et al., 2016; Trevennec et al., 2012; Vijaykrishna et al., 2010; Zhao et al., 2011), resulting in reasssortment with locally endemic viruses to generate different variant viruses (Fig. 4). The 2009 pH1N1 virus and its reassortant virus as well as endemic swine influenza viruses co-circulate in Asian swine populations (Baudon et al., 2014; Kim et al., 2013; Kirisawa et al., 2014; Mine et al., 2018; Song et al., 2010; Sun et al., 2016; Zhu et al., 2011b). In China, the dominant circulating swine influenza virus was a typical Eurasian avian-like H1N1 viruses before 2015 although other subtypes and genotypes of swine influenza viruses were also identified in pig populations (Yang et al., 2015). To date, in addition to the parental Eurasian avian-like H1N1 virus, at least 5 genotypes of reassortant Eurasian H1N1-like viruses, which have at least surface HA and NA from Eurasian avian-like H1N1 viruses and internal genes from other viruses, have been detected in swine herds in China (Cao et al., 2019; Sun et al., 2020; Sun et al., 2016; Yang et al., 2015). The genotype Eurasian H1N1 viruses with 2009 pH1N1 and triple-reassortant derived internal genes have been predominant in swine populations since 2016 (Cao et al., 2019; Sun et al., 2020) and have capability to bind to human-type receptors, and display efficient infectivity and aerosol transmission in ferrets (Sun et al., 2020). Several human infection cases with this genotype of virus have been documented in China (Li et al., 2019; Xie et al., 2018; Zhu et al., 2016). Furthermore, more than 10% of swine workers sero-positive for this genotype of H1N1 virus have been reported, indicating that the predominant genotype Eurasian H1N1 virus has acquired increased human infectivity (Sun et al., 2020). Although avian influenza viruses including H9N2 and H5N1 viruses had been reported to infect humans (Butt et al., 2005; Dong et al., 2020; Guo et al., 1999; Lin et al., 2000; Webster et al., 2006; WHO, 2020) and been frequently detected in pigs in Asian countries (Cao et al., 2013; Cong et al., 2008; He et al., 2013; Lee et al., 2009; Nidom et al., 2010; Yu et al., 2010; Zhou et al., 2014), these viruses did not cause severe consequences in infected animals and establish in pigs. With increasing introductions of avian influenza viruses into swine, it is possible to generate a novel virus that can be fully adapted to pigs, similar to the Eurasian avian-like H1N1 virus (Sun et al., 2020; Yang et al., 2015; Zhu et al., 2011a); or the avian viruses reassort with endemic viruses resulting in new reassortant viruses with efficient transmissibility and potential to threat public health (Ma et al., 2007; Sun et al., 2020).
Two-way transmission of IAVs between humans and swine
IAVs have been recorded in a two-way transmission between humans and swine. In 1919, Koen, an inspector with the U.S. Bureau of Animal Industry, pointed out that influenza outbreaks began with either pigs or people, but were then rapidly transferred to each other (Koen, 1919). Subsequently, infections of humans with either swine influenza H1N1 or H3N2 virus have been reported worldwide due to directly exposure to swine in most cases and also resulted in fatality in several cases (Myers et al., 2007). Noticeably, these swine influenza viruses have almost no or very limited human-to-human transmission capability although they caused disease in infected humans (Myers et al., 2007). Human seasonal IAVs have been repeatedly transmitted to swine herds (Nelson et al., 2015; Nelson et al., 2014; Vincent et al., 2008b). Although so far likely no complete human seasonal virus has established in pigs, it has resulted in the establishment of multiple genetically and antigenically distinct human-origin virus lineages through reassortment with endemic swine influenza viruses globally (Anderson et al., 2020; Nelson et al., 2015). Studies on antigenic characterization of swine influenza viruses have further demonstrated that the antigenic diversity of H1 and H3 viruses circulating in swine herds worldwide was largely due to the frequent introductions of human-origin IAVs into pigs (Lewis et al., 2016). The 2009 pH1N1 virus has further exemplified two way transmission of IAVs between humans and swine (Chastagner et al., 2019). This virus originated from swine and transmitted to humans, resulting in 2009 influenza pandemic (Mena et al., 2016). 2009 pH1N1 virus has been shown to infect and cause disease in pigs without prior adaptation (Ma et al., 2011) and was detected in swine herds worldwide (Nelson and Vincent, 2015), even in countries which were previously considered to be swine influenza free such as Australia and Norway (Hofshagen et al., 2009; Holyoake et al., 2011). It has been documented throughout the world in swine with frequent reassortment with locally endemic viruses (Nelson et al., 2015; Pereda et al., 2011; Zhu et al., 2011b). The most well-known cases were H3N2 and H1N2 variant swine viruses which were reassortants with 7 gene from contemporary North American triple reassortant viruses and the M gene from 2009 pH1N1 virus, leading to more than 300 human infections and several deaths in the U.S. (Bowman et al., 2014; Jhung et al., 2013; Nelson et al., 2016). Importantly, the similar H3N2 variant swine influenza virus had been detected in pigs in Korea (Kim et al., 2013) although no human infected cases were found in Korea, most likely this virus was transmitted to Korea through importation of pigs from the United States. IAVs two-way transmission has expedited IAV evolution in both species and could also enhance severity of influenza virus infections as most of people do not have preexisting immunity against the swine virus.
Challenge to control swine influenza
Multiple highly diverse swine influenza viruses exist and circulate in swine populations worldwide (Anderson et al., 2020; Lewis et al., 2016) and novel genotypes and subtypes of viruses could emerge at any time in pigs due to infections with avian and/or human influenza viruses. For example, the unique H2N3 virus was detected in the US swine in 2006, which was infectious and highly transmissible in swine and ferrets and had undergone some adaptation to the mammalian host (Ma et al., 2007). In addition, live pigs are globally moving with increasing international trades, which has accelerated virus evolution and complicated swine influenza situations worldwide. This has resulted in introductions of novel swine influenza viruses into naïve swine herds such as North American triple reassortant and Eurasian avian-like H1N1 swine influenza viruses which have been introduced into China from North American and European countries (Zhu et al., 2011a). This is a huge challenge to produce effective vaccines and control swine influenza for swine industry.
Vaccination is the most efficient means to prevent and control influenza in humans and animals. Commercially available whole inactivated virus (WIV) vaccines have been shown effective to protect pigs against homologous or genetically similar viruses’ infections and are widely used in swine industry in many countries (Van Reeth and Ma, 2013). However, swine influenza has not been controlled partially due to lacking heterovariant and heterosubtypic protection of WIV vaccines (Ma and Richt, 2010). The efficacy of WIV vaccines can also be affected by maternally derived antibodies (Ma and Richt, 2010; Van Reeth and Ma, 2012). In addition, vaccine associated enhanced respiratory disease has been observed in WIV vaccine immunized pigs when vaccine strains mismatch with experimentally infecting viruses (Vincent et al., 2008a). Due to limited protection of commercial WIV vaccines against contemporary swine influenza viruses, a live-attenuated influenza virus (LAIV) vaccine based on NS1 deletion has been licensed and available to use in pigs in the U.S. (Genzow et al., 2017). The advantage of live-virus vaccines is that it is able to provide good heterovariant and partial heterosubtypic protection, and does not enhance disease (Richt et al., 2006; Vincent et al., 2007). The disadvantage of LAIV vaccines is that they have the potential to reassort with circulating endemic viruses (Vincent et al., 2016). Novel variant viruses have been detected in swine farms in which the LAIV vaccine was used due to reassortment with endemic viruses (Sharma et al., 2020). Different from IAVs in humans, evolution of IAVs in pigs shows a unique trend which is subject to the geographic locations, depending on the country, region, and even at the farm level (Rajao and Perez, 2018). Therefore, the manufacturers for producing swine influenza vaccines cannot make a “universal” vaccine for all farms in different areas like the human influenza seasonal vaccine and have to make independent decisions regarding the strains used in their products. A non-replicating alphavirus RNA vectored influenza vaccine is used to produce autogenous vaccines that match the circulating virus to improve protection and have been widely used in U.S. swine farms (Vander Veen et al., 2012). Although different swine influenza vaccines are widely used in pigs like seasonal influenza vaccine used in humans yearly, swine influenza is still one of major challenges and causes significant economic losses to swine industry.
Swine influenza cannot be efficiently controlled despite of large amount of vaccines used, similar as human seasonal influenza. Very important is that a novel virus could emerge in pigs, which might has capacity to infect humans and potential to adapt to humans to induce epidemic and pandemic in future like the 2009 pH1N1 virus. The 2009 pH1N1 virus originated from North America, likely from Mexico (Mena et al., 2016), not from Southeast Asian “wet market” that has been considered to be epicenter of the pandemic influenza virus (Shortridge, 1992). This indicates that the potential pandemic virus can be generated in anywhere and underscores importance of surveillance in pigs in other areas. In addition, other existing swine viruses such as North American triple reassortant H3N2 virus in the US and Eurasian avian-like H1N1 virus in China have acquired increased human infectivity through reassortment with other viruses, resulting in H3N2 variant virus and reassortant Eurasian H1N1-like virus in respective areas which caused human infections (Nelson et al., 2016; Sun et al., 2020). Therefore, strengthened surveillance of IAVs in pigs worldwide is needed to better understand the ecology of IAVs, and more effective swine vaccines need to be produced in order to protect animal and public health.
Highlights.
The manuscript has reviewed and discussed current status and challenge of swine influenza virus.
Different swine influenza viruses circulating in pigs globally is a major challenge to produce broadly effective vaccines and control disease.
Enhanced surveillance of IAVs in pigs worldwide is needed to better understand the ecology of IAVs in order to protect animal and public health.
Acknowledgement
The author thanks Dr. Tavis K. Anderson for critical discussions and Dr. Wenyu Yang for generation of the Figure 2. This work was partially supported by NIAID funded Centers of Excellence for Influenza Research and Surveillance under contract numbers HHSN 272201400006C and NIAID R21AI121906 and R01AI134768.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alexander DJ and Brown IH (2000) Recent zoonoses caused by influenza A viruses. Rev Sci Tech 19(1), 197–225. [DOI] [PubMed] [Google Scholar]
- Anderson TK, Chang J, Arendsee ZW, Venkatesh D, Souza CK, Kimble JB, Lewis NS, Davis CT and Vincent AL (2020) Swine Influenza A Viruses and the Tangled Relationship with Humans. Cold Spring Harb Perspect Med. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson TK, Nelson MI, Kitikoon P, Swenson SL, Korslund JA and Vincent AL (2013) Population dynamics of cocirculating swine influenza A viruses in the United States from 2009 to 2012. Influenza Other Respir Viruses 7 Suppl 4, 42–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arikawa J, Yamane N, Odagiri T. and Ishida N. (1979) Serological evidence of H1 influenza virus infection among Japanese hogs. Acta Virol 23(6), 508–11. [PubMed] [Google Scholar]
- Arikawa J, Yamane N, Totsukawa K. and Ishida N. (1982) The follow-up study of swine and Hong Kong influenza virus infection among Japanese hogs. Tohoku J Exp Med 136(4), 353–8. [DOI] [PubMed] [Google Scholar]
- Baudon E, Poon LL, Dao TD, Pham NT, Cowling BJ, Peyre M, Nguyen KV and Peiris M. (2014) Detection of Novel Reassortant Influenza A (H3N2) and H1N1 2009 Pandemic Viruses in Swine in Hanoi, Vietnam. Zoonoses Public Health 62(6), 429–34. [DOI] [PubMed] [Google Scholar]
- Beato MS, Tassoni L, Milani A, Salviato A, Di Martino G, Mion M, Bonfanti L, Monne I, Watson SJ and Fusaro A. (2016) Circulation of multiple genotypes of H1N2 viruses in a swine farm in Italy over a two-month period. Vet Microbiol 195, 25–29. [DOI] [PubMed] [Google Scholar]
- Bolton MJ, Abente EJ, Venkatesh D, Stratton JA, Zeller M, Anderson TK, Lewis NS and Vincent AL (2018) Antigenic evolution of H3N2 influenza A viruses in swine in the United States from 2012 to 2016. Influenza Other Respir Viruses 13(1), 83–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman AS, Nelson SW, Page SL, Nolting JM, Killian ML, Sreevatsan S. and Slemons RD (2014) Swine-to-human transmission of influenza A(H3N2) virus at agricultural fairs, Ohio, USA, 2012. Emerg Infect Dis 20(9), 1472–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown IH (2000) The epidemiology and evolution of influenza viruses in pigs. Vet Microbiol 74(1–2), 29–46. [DOI] [PubMed] [Google Scholar]
- Brown IH (2012) History and epidemiology of Swine influenza in Europe. Curr Top Microbiol Immunol 370, 133–46. [DOI] [PubMed] [Google Scholar]
- Brown IH, Harris PA, McCauley JW and Alexander DJ (1998) Multiple genetic reassortment of avian and human influenza A viruses in European pigs, resulting in the emergence of an H1N2 virus of novel genotype. J Gen Virol 79 ( Pt 12), 2947–55. [DOI] [PubMed] [Google Scholar]
- Butt KM, Smith GJ, Chen H, Zhang LJ, Leung YH, Xu KM, Lim W, Webster RG, Yuen KY, Peiris JS and Guan Y. (2005) Human infection with an avian H9N2 influenza A virus in Hong Kong in 2003. J Clin Microbiol 43(11), 5760–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao N, Zhu W, Chen Y, Tan L, Zhou P, Cao Z, Ke C, Li Y, Wu J, Qi W, Jiao P. and Zhang G. (2013) Avian influenza A (H5N1) virus antibodies in pigs and residents of swine farms, southern China. J Clin Virol 58(4), 647–51. [DOI] [PubMed] [Google Scholar]
- Cao Z, Zeng W, Hao X, Huang J, Cai M, Zhou P. and Zhang G. (2019) Continuous evolution of influenza A viruses of swine from 2013 to 2015 in Guangdong, China. PLoS One 14(7), e0217607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castrucci MR, Campitelli L, Ruggieri A, Barigazzi G, Sidoli L, Daniels R, Oxford JS and Donatelli I. (1994) Antigenic and sequence analysis of H3 influenza virus haemagglutinins from pigs in Italy. J Gen Virol 75 ( Pt 2), 371–9. [DOI] [PubMed] [Google Scholar]
- Chastagner A, Enouf V, Peroz D, Herve S, Lucas P, Queguiner S, Gorin S, Beven V, Behillil S, Leneveu P, Garin E, Blanchard Y, van der Werf S. and Simon G. (2019) Bidirectional Human-Swine Transmission of Seasonal Influenza A(H1N1)pdm09 Virus in Pig Herd, France, 2018. Emerg Infect Dis 25(10), 1940–1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee S, Mukherjee KK, Mondal MC, Chakravarti SK and Chakraborty MS (1995) A serological survey of influenza a antibody in human and pig sera in Calcutta. Folia Microbiol (Praha) 40(3), 345–8. [DOI] [PubMed] [Google Scholar]
- Chen Y, Zhang J, Qiao C, Yang H, Zhang Y, Xin X. and Chen H. (2012) Co-circulation of pandemic 2009 H1N1, classical swine H1N1 and avian-like swine H1N1 influenza viruses in pigs in China. Infect Genet Evol 13, 331–8. [DOI] [PubMed] [Google Scholar]
- Choi YK, Goyal SM and Joo HS (2002) Prevalence of swine influenza virus subtypes on swine farms in the United States. Arch Virol 147(6), 1209–20. [DOI] [PubMed] [Google Scholar]
- Cong YL, Wang CF, Yan CM, Peng JS, Jiang ZL and Liu JH (2008) Swine infection with H9N2 influenza viruses in China in 2004. Virus Genes 36(3), 461–9. [DOI] [PubMed] [Google Scholar]
- de Jong JC, van Nieuwstadt AP, Kimman TG, Loeffen WL, Bestebroer TM, Bijlsma K, Verweij C, Osterhaus AD and Class EC (1999) Antigenic drift in swine influenza H3 haemagglutinins with implications for vaccination policy. Vaccine 17(11–12), 1321–8. [DOI] [PubMed] [Google Scholar]
- Dong X, Xiong J, Huang C, Xiang J, Wu W, Chen N, Wen D, Tu C, Qiao X, Kang L, Yao Z, Zhang D. and Chen Q. (2020) Human H9N2 Avian Influenza Infection: Epidemiological and Clinical Characterization of 16 Cases in China. Virol Sin. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducatez MF, Hause B, Stigger-Rosser E, Darnell D, Corzo C, Juleen K, Simonson R, Brockwell-Staats C, Rubrum A, Wang D, Webb A, Crumpton JC, Lowe J, Gramer M. and Webby RJ (2011) Multiple reassortment between pandemic (H1N1) 2009 and endemic influenza viruses in pigs, United States. Emerg Infect Dis 17(9), 1624–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouchier RA, Munster V, Wallensten A, Bestebroer TM, Herfst S, Smith D, Rimmelzwaan GF, Olsen B. and Osterhaus AD (2005) Characterization of a novel influenza A virus hemagglutinin subtype (H16) obtained from black-headed gulls. J Virol 79(5), 2814–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao S, Anderson TK, Walia RR, Dorman KS, Janas-Martindale A. and Vincent AL (2017) The genomic evolution of H1 influenza A viruses from swine detected in the United States between 2009 and 2016. J Gen Virol 98(8), 2001–2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garten RJ, Davis CT, Russell CA, Shu B, Lindstrom S, Balish A, Sessions WM, Xu X, Skepner E, Deyde V, Okomo-Adhiambo M, Gubareva L, Barnes J, Smith CB, Emery SL, Hillman MJ, Rivailler P, Smagala J, de Graaf M, Burke DF, Fouchier RA, Pappas C, Alpuche-Aranda CM, Lopez-Gatell H, Olivera H, Lopez I, Myers CA, Faix D, Blair PJ, Yu C, Keene KM, Dotson PD Jr., Boxrud D, Sambol AR, Abid SH, St George K, Bannerman T, Moore AL, Stringer DJ, Blevins P, Demmler-Harrison GJ, Ginsberg M, Kriner P, Waterman S, Smole S, Guevara HF, Belongia EA, Clark PA, Beatrice ST, Donis R, Katz J, Finelli L, Bridges CB, Shaw M, Jernigan DB, Uyeki TM, Smith DJ, Klimov AI and Cox NJ (2009) Antigenic and genetic characteristics of swine-origin 2009 A(H1N1) influenza viruses circulating in humans. Science 325(5937), 197–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genzow M, Goodell C, Kaiser TJ, Johnson W. and Eichmeyer M. (2017) Live attenuated influenza virus vaccine reduces virus shedding of newborn piglets in the presence of maternal antibody. Influenza Other Respir Viruses 12(3), 353–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Y, Shortridge KF, Krauss S, Li PH, Kawaoka Y. and Webster RG (1996) Emergence of avian H1N1 influenza viruses in pigs in China. J Virol 70(11), 8041–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Li J. and Cheng X. (1999) [Discovery of men infected by avian influenza A (H9N2) virus]. Zhonghua Shi Yan He Lin Chuang Bing Du Xue Za Zhi 13(2), 105–8. [PubMed] [Google Scholar]
- Haesebrouck F, Biront P, Pensaert MB and Leunen J. (1985) Epizootics of respiratory tract disease in swine in Belgium due to H3N2 influenza virus and experimental reproduction of disease. Am J Vet Res 46(9), 1926–8. [PubMed] [Google Scholar]
- He L, Zhao G, Zhong L, Liu Q, Duan Z, Gu M, Wang X. and Liu X. (2013) Isolation and characterization of two H5N1 influenza viruses from swine in Jiangsu Province of China. Arch Virol 158(12), 2531–41. [DOI] [PubMed] [Google Scholar]
- Hiromoto Y, Parchariyanon S, Ketusing N, Netrabukkana P, Hayashi T, Kobayashi T, Takemae N. and Saito T. (2012) Isolation of the pandemic (H1N1) 2009 virus and its reassortant with an H3N2 swine influenza virus from healthy weaning pigs in Thailand in 2011. Virus Res 169(1), 175–81. [DOI] [PubMed] [Google Scholar]
- Hofshagen M, Gjerset B, Er C, Tarpai A, Brun E, Dannevig B, Bruheim T, Fostad IG, Iversen B, Hungnes O. and Lium B. (2009) Pandemic influenza A(H1N1)v: human to pig transmission in Norway? Euro Surveill 14(45). [DOI] [PubMed] [Google Scholar]
- Holyoake PK, Kirkland PD, Davis RJ, Arzey KE, Watson J, Lunt RA, Wang J, Wong F, Moloney BJ and Dunn SE (2011) The first identified case of pandemic H1N1 influenza in pigs in Australia. Aust Vet J 89(11), 427–31. [DOI] [PubMed] [Google Scholar]
- Howard WA, Essen SC, Strugnell BW, Russell C, Barass L, Reid SM and Brown IH (2011) Reassortant Pandemic (H1N1) 2009 virus in pigs, United Kingdom. Emerg Infect Dis 17(6), 1049–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jhung MA, Epperson S, Biggerstaff M, Allen D, Balish A, Barnes N, Beaudoin A, Berman L, Bidol S, Blanton L, Blythe D, Brammer L, D’Mello T, Danila R, Davis W, de Fijter S, Diorio M, Durand LO, Emery S, Fowler B, Garten R, Grant Y, Greenbaum A, Gubareva L, Havers F, Haupt T, House J, Ibrahim S, Jiang V, Jain S, Jernigan D, Kazmierczak J, Klimov A, Lindstrom S, Longenberger A, Lucas P, Lynfield R, McMorrow M, Moll M, Morin C, Ostroff S, Page SL, Park SY, Peters S, Quinn C, Reed C, Richards S, Scheftel J, Simwale O, Shu B, Soyemi K, Stauffer J, Steffens C, Su S, Torso L, Uyeki TM, Vetter S, Villanueva J, Wong KK, Shaw M, Bresee JS, Cox N. and Finelli L. (2013) Outbreak of variant influenza A(H3N2) virus in the United States. Clin Infect Dis 57(12), 1703–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung K. and Chae C. (2004) Phylogenetic analysis of an H1N2 influenza A virus isolated from a pig in Korea. Brief Report. Arch Virol 149(7), 1415–22. [DOI] [PubMed] [Google Scholar]
- Karasin AI, Landgraf J, Swenson S, Erickson G, Goyal S, Woodruff M, Scherba G, Anderson G. and Olsen CW (2002) Genetic characterization of H1N2 influenza A viruses isolated from pigs throughout the United States. J Clin Microbiol 40(3), 1073–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kida H, Ito T, Yasuda J, Shimizu Y, Itakura C, Shortridge KF, Kawaoka Y. and Webster RG (1994) Potential for transmission of avian influenza viruses to pigs. J Gen Virol 75 ( Pt 9), 2183–8. [DOI] [PubMed] [Google Scholar]
- Kim SH, Kim HJ, Jin YH, Yeoul JJ, Lee KK, Oem JK, Lee MH and Park CK (2013) Isolation of influenza A(H3N2)v virus from pigs and characterization of its biological properties in pigs and mice. Arch Virol 158(11), 2351–7.23674250 [Google Scholar]
- Kirisawa R, Ogasawara Y, Yoshitake H, Koda A. and Furuya T. (2014) Genomic reassortants of pandemic A (H1N1) 2009 virus and endemic porcine H1 and H3 viruses in swine in Japan. J Vet Med Sci 76(11), 1457–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitikoon P, Nelson MI, Killian ML, Anderson TK, Koster L, Culhane MR and Vincent AL (2013) Genotype patterns of contemporary reassorted H3N2 virus in US swine. J Gen Virol 94(Pt 6), 1236–41. [DOI] [PubMed] [Google Scholar]
- Koen JS (1919) A practical method for field diagnosis of swine disease. Amer J Vet Sci 14, 468. [Google Scholar]
- Krog JS, Hjulsager CK, Larsen MA and Larsen LE (2017) Triple-reassortant influenza A virus with H3 of human seasonal origin, NA of swine origin, and internal A(H1N1) pandemic 2009 genes is established in Danish pigs. Influenza Other Respir Viruses 11(3), 298–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kundin WD (1970) Hong Kong A-2 influenza virus infection among swine during a human epidemic in Taiwan. Nature 228(5274), 857. [DOI] [PubMed] [Google Scholar]
- Kupradinun S, Peanpijit P, Bhodhikosoom C, Yoshioka Y, Endo A. and Nerome K. (1991) The first isolation of swine H1N1 influenza viruses from pigs in Thailand. Arch Virol 118(3–4), 289–97. [DOI] [PubMed] [Google Scholar]
- Lange J, Groth M, Schlegel M, Krumbholz A, Wieczorek K, Ulrich R, Koppen S, Schulz K, Appl D, Selbitz HJ, Sauerbrei A, Platzer M, Zell R. and Durrwald R. (2013) Reassortants of the pandemic (H1N1) 2009 virus and establishment of a novel porcine H1N2 influenza virus, lineage in Germany. Vet Microbiol 167(3–4), 345–56. [DOI] [PubMed] [Google Scholar]
- Lee CS, Kang BK, Kim HK, Park SJ, Park BK, Jung K. and Song DS (2008) Phylogenetic analysis of swine influenza viruses recently isolated in Korea. Virus Genes 37(2), 168–76. [DOI] [PubMed] [Google Scholar]
- Lee JH, Pascua PN, Song MS, Baek YH, Kim CJ, Choi HW, Sung MH, Webby RJ, Webster RG, Poo H. and Choi YK (2009) Isolation and genetic characterization of H5N2 influenza viruses from pigs in Korea. J Virol 83(9), 4205–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis NS, Russell CA, Langat P, Anderson TK, Berger K, Bielejec F, Burke DF, Dudas G, Fonville JM, Fouchier RA, Kellam P, Koel BF, Lemey P, Nguyen T, Nuansrichy B, Peiris JM, Saito T, Simon G, Skepner E, Takemae N, Webby RJ, Van Reeth K, Brookes SM, Larsen L, Watson SJ, Brown IH and Vincent AL (2016) The global antigenic diversity of swine influenza A viruses. Elife 5, e12217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Guo L, Liu C, Cheng Y, Kong M, Yang L, Zhuang Z, Liu J, Zou M, Dong X, Su X. and Gu Q. (2019) Human infection with a novel reassortant Eurasian-avian lineage swine H1N1 virus in northern China. Emerg Microbes Infect 8(1), 1535–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YP, Shaw M, Gregory V, Cameron K, Lim W, Klimov A, Subbarao K, Guan Y, Krauss S, Shortridge K, Webster R, Cox N. and Hay A. (2000) Avian-to-human transmission of H9N2 subtype influenza A viruses: relationship between H9N2 and H5N1 human isolates. Proc Natl Acad Sci U S A 97(17), 9654–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Ma J, Liu H, Qi W, Anderson J, Henry SC, Hesse RA, Richt JA and Ma W. (2011) Emergence of novel reassortant H3N2 swine influenza viruses with the 2009 pandemic H1N1 genes in the United States. Arch Virol 157(3), 555–62. [DOI] [PubMed] [Google Scholar]
- Lorusso A, Vincent AL, Gramer ME, Lager KM and Ciacci-Zanella JR (2013) Contemporary epidemiology of North American lineage triple reassortant influenza A viruses in pigs. Curr Top Microbiol Immunol 370, 113–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J, Shen H, Liu Q, Bawa B, Qi W, Duff M, Lang Y, Lee J, Yu H, Bai J, Tong G, Hesse RA, Richt JA and Ma W. (2014) Pathogenicity and transmissibility of novel reassortant H3N2 influenza viruses with 2009 pandemic H1N1 genes in pigs. J Virol 89(5), 2831–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J, Shen H, McDowell C, Liu Q, Duff M, Lee J, Lang Y, Hesse D, Richt JA and Ma W. (2019) Virus survival and fitness when multiple genotypes and subtypes of influenza A viruses exist and circulate in swine. Virology 532, 30–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma W, Belisle SE, Mosier D, Li X, Stigger-Rosser E, Liu Q, Qiao C, Elder J, Webby R, Katze MG and Richt JA (2011) 2009 pandemic H1N1 influenza virus causes disease and upregulation of genes related to inflammatory and immune responses, cell death, and lipid metabolism in pigs. J Virol 85(22), 11626–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma W, Kahn RE and Richt JA (2009) The pig as a mixing vessel for influenza viruses: Human and veterinary implications. J Mol Genet Med 3(1), 158–66. [PMC free article] [PubMed] [Google Scholar]
- Ma W. and Richt JA (2010) Swine influenza vaccines: current status and future perspectives. Anim Health Res Rev 11(1), 81–96. [DOI] [PubMed] [Google Scholar]
- Ma W, Vincent AL, Gramer MR, Brockwell CB, Lager KM, Janke BH, Gauger PC, Patnayak DP, Webby RJ and Richt JA (2007) Identification of H2N3 influenza A viruses from swine in the United States. Proc Natl Acad Sci U S A 104(52), 20949–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marozin S, Gregory V, Cameron K, Bennett M, Valette M, Aymard M, Foni E, Barigazzi G, Lin Y. and Hay A. (2002) Antigenic and genetic diversity among swine influenza A H1N1 and H1N2 viruses in Europe. J Gen Virol 83(Pt 4), 735–45. [DOI] [PubMed] [Google Scholar]
- McQueen JL, Steele JH and Robinson RQ (1968) Influenza in animals. Adv Vet Sci 12, 285–336. [PubMed] [Google Scholar]
- Mena I, Nelson MI, Quezada-Monroy F, Dutta J, Cortes-Fernandez R, Lara-Puente JH, Castro-Peralta F, Cunha LF, Trovao NS, Lozano-Dubernard B, Rambaut A, van Bakel H. and Garcia-Sastre A. (2016) Origins of the 2009 H1N1 influenza pandemic in swine in Mexico. Elife 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mine J, Abe H, Parchariyanon S, Boonpornprasert P, Ubonyaem N, Nuansrichay B, Takemae N, Tanikawa T, Tsunekuni R, Uchida Y. and Saito T. (2018) Genetic and antigenic dynamics of influenza A viruses of swine on pig farms in Thailand. Arch Virol 164(2), 457–472. [DOI] [PubMed] [Google Scholar]
- Moreno A, Di Trani L, Faccini S, Vaccari G, Nigrelli D, Boniotti MB, Falcone E, Boni A, Chiapponi C, Sozzi E. and Cordioli P. (2011) Novel H1N2 swine influenza reassortant strain in pigs derived from the pandemic H1N1/2009 virus. Vet Microbiol 149(3–4), 472–7. [DOI] [PubMed] [Google Scholar]
- Myers KP, Olsen CW and Gray GC (2007) Cases of swine influenza in humans: a review of the literature. Clin Infect Dis 44(8), 1084–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagarajan K, Saikumar G, Arya RS, Gupta A, Somvanshi R. and Pattnaik B. (2010) Influenza A H1N1 virus in Indian pigs & its genetic relatedness with pandemic human influenza A 2009 H1N1. Indian J Med Res 132, 160–7. [PubMed] [Google Scholar]
- Nelson MI, Stratton J, Killian ML, Janas-Martindale A. and Vincent AL (2015) Continual Reintroduction of Human Pandemic H1N1 Influenza A Viruses into Swine in the United States, 2009 to 2014. J Virol 89(12), 6218–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson MI, Stucker KM, Schobel SA, Trovao NS, Das SR, Dugan VG, Nelson SW, Sreevatsan S, Killian ML, Nolting JM, Wentworth DE and Bowman AS (2016) Introduction, Evolution, and Dissemination of Influenza A Viruses in Exhibition Swine in the United States during 2009 to 2013. J Virol 90(23), 10963–10971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson MI and Vincent AL (2015) Reverse zoonosis of influenza to swine: new perspectives on the human-animal interface. Trends Microbiol 23(3), 142–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson MI, Wentworth DE, Culhane MR, Vincent AL, Viboud C, LaPointe MP, Lin X, Holmes EC and Detmer SE (2014) Introductions and evolution of human-origin seasonal influenza a viruses in multinational swine populations. J Virol 88(17), 10110–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nerome K, Ishida M, Nakayama M, Oya A, Kanai C. and Suwicha K. (1981) Antigenic and genetic analysis of A/Hong Kong (H3N2) influenza viruses isolated from swine and man. J Gen Virol 56(Pt 2), 441–5. [DOI] [PubMed] [Google Scholar]
- Nerome K, Ishida M, Oya A. and Oda K. (1982) The possible origin H1N1 (Hsw1N1) virus in the swine population of Japan and antigenic analysis of the isolates. J Gen Virol 62 (Pt 1), 171–5. [DOI] [PubMed] [Google Scholar]
- Nidom CA, Takano R, Yamada S, Sakai-Tagawa Y, Daulay S, Aswadi D, Suzuki T, Suzuki Y, Shinya K, Iwatsuki-Horimoto K, Muramoto Y. and Kawaoka Y. (2010) Influenza A (H5N1) viruses from pigs, Indonesia. Emerg Infect Dis 16(10), 1515–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen CW (2002) The emergence of novel swine influenza viruses in North America. Virus Res 85(2), 199–210. [DOI] [PubMed] [Google Scholar]
- Ouchi A, Nerome K, Kanegae Y, Ishida M, Nerome R, Hayashi K, Hashimoto T, Kaji M, Kaji Y. and Inaba Y. (1996) Large outbreak of swine influenza in southern Japan caused by reassortant (H1N2) influenza viruses: its epizootic background and characterization of the causative viruses. J Gen Virol 77 ( Pt 8), 1751–9. [DOI] [PubMed] [Google Scholar]
- Pascua PN, Song MS, Lee JH, Choi HW, Han JH, Kim JH, Yoo GJ, Kim CJ and Choi YK (2008) Seroprevalence and genetic evolutions of swine influenza viruses under vaccination pressure in Korean swine herds. Virus Res 138(1–2), 43–9. [DOI] [PubMed] [Google Scholar]
- Pensaert M, Ottis K, Vandeputte J, Kaplan MM and Bachmann PA (1981) Evidence for the natural transmission of influenza A virus from wild ducts to swine and its potential importance for man. Bull World Health Organ 59(1), 75–8. [PMC free article] [PubMed] [Google Scholar]
- Pereda A, Rimondi A, Cappuccio J, Sanguinetti R, Angel M, Ye J, Sutton T, Dibarbora M, Olivera V, Craig MI, Quiroga M, Machuca M, Ferrero A, Perfumo C. and Perez DR (2011) Evidence of reassortment of pandemic H1N1 influenza virus in swine in Argentina: are we facing the expansion of potential epicenters of influenza emergence? Influenza Other Respir Viruses 5(6), 409–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pippig J, Ritzmann M, Buttner M. and Neubauer-Juric A. (2016) Influenza A Viruses Detected in Swine in Southern Germany after the H1N1 Pandemic in 2009. Zoonoses Public Health 63(7), 555–568. [DOI] [PubMed] [Google Scholar]
- Rajao DS, Anderson TK, Kitikoon P, Stratton J, Lewis NS and Vincent AL (2018) Antigenic and genetic evolution of contemporary swine H1 influenza viruses in the United States. Virology 518, 45–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajao DS and Perez DR (2018) Universal Vaccines and Vaccine Platforms to Protect against Influenza Viruses in Humans and Agriculture. Front Microbiol 9, 123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richt JA, Lager KM, Janke BH, Woods RD, Webster RG and Webby RJ (2003) Pathogenic and antigenic properties of phylogenetically distinct reassortant H3N2 swine influenza viruses cocirculating in the United States. J Clin Microbiol 41(7), 3198–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richt JA, Lekcharoensuk P, Lager KM, Vincent AL, Loiacono CM, Janke BH, Wu WH, Yoon KJ, Webby RJ, Solorzano A. and Garcia-Sastre A. (2006) Vaccination of pigs against swine influenza viruses by using an NS1-truncated modified live-virus vaccine. J Virol 80(22), 11009–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohm C, Zhou N, Suss J, Mackenzie J. and Webster RG (1996) Characterization of a novel influenza hemagglutinin, H15: criteria for determination of influenza A subtypes. Virology 217(2), 508–16. [DOI] [PubMed] [Google Scholar]
- Scholtissek C. (1990) Pigs as ‘Mixing Vessels’ for the Creation of New Pandemic Influenza A Viruses. Med Principles Pract 2, 65–71. [Google Scholar]
- Schultz-Cherry S, Olsen CW and Easterday BC (2012) History of Swine influenza. Curr Top Microbiol Immunol 370, 21–8. [DOI] [PubMed] [Google Scholar]
- Sharma A, Zeller MA, Li G, Harmon KM, Zhang J, Hoang H, Anderson TK, Vincent AL and Gauger PC (2020) Detection of live attenuated influenza vaccine virus and evidence of reassortment in the U.S. swine population. J Vet Diagn Invest, 1040638720907918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shortridge KF (1992) Pandemic influenza: a zoonosis? Semin Respir Infect 7(1), 11–25. [PubMed] [Google Scholar]
- Shortridge KF and Webster RG (1979) Geographical distribution of swine (Hsw1N1) and Hong Kong (H3N2) influenza virus variants in pigs in Southeast Asia. Intervirology 11(1), 9–15. [DOI] [PubMed] [Google Scholar]
- Shortridge KF, Webster RG, Butterfield WK and Campbell CH (1977) Persistence of Hong Kong influenza virus variants in pigs. Science 196(4297), 1454–5. [DOI] [PubMed] [Google Scholar]
- Simon G, Larsen LE, Durrwald R, Foni E, Harder T, Van Reeth K, Markowska-Daniel I, Reid SM, Dan A, Maldonado J, Huovilainen A, Billinis C, Davidson I, Aguero M, Vila T, Herve S, Breum SO, Chiapponi C, Urbaniak K, Kyriakis CS, Brown IH and Loeffen W. (2014) European surveillance network for influenza in pigs: surveillance programs, diagnostic tools and Swine influenza virus subtypes identified in 14 European countries from 2010 to 2013. PLoS One 9(12), e115815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith GJ, Bahl J, Vijaykrishna D, Zhang J, Poon LL, Chen H, Webster RG, Peiris JS and Guan Y. (2009a) Dating the emergence of pandemic influenza viruses. Proc Natl Acad Sci U S A 106(28), 11709–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith GJ, Vijaykrishna D, Bahl J, Lycett SJ, Worobey M, Pybus OG, Ma SK, Cheung CL, Raghwani J, Bhatt S, Peiris JS, Guan Y. and Rambaut A. (2009b) Origins and evolutionary genomics of the 2009 swine-origin H1N1 influenza A epidemic. Nature 459(7250), 1122–5. [DOI] [PubMed] [Google Scholar]
- Song DS, Lee CS, Jung K, Kang BK, Oh JS, Yoon YD, Lee JH and Park BK (2007) Isolation and phylogenetic analysis of H1N1 swine influenza virus isolated in Korea. Virus Res 125(1), 98–103. [DOI] [PubMed] [Google Scholar]
- Song DS, Lee JY, Oh JS, Lyoo KS, Yoon KJ, Park YH and Park BK (2003) Isolation of H3N2 swine influenza virus in South Korea. J Vet Diagn Invest 15(1), 30–4. [DOI] [PubMed] [Google Scholar]
- Song MS, Lee JH, Pascua PN, Baek YH, Kwon HI, Park KJ, Choi HW, Shin YK, Song JY, Kim CJ and Choi YK (2010) Evidence of human-to-swine transmission of the pandemic (H1N1) 2009 influenza virus in South Korea. J Clin Microbiol 48(9), 3204–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starick E, Lange E, Fereidouni S, Bunzenthal C, Hoveler R, Kuczka A, grosse Beilage E, Hamann HP, Klingelhofer I, Steinhauer D, Vahlenkamp T, Beer M. and Harder T. (2011) Reassorted pandemic (H1N1) 2009 influenza A virus discovered from pigs in Germany. J Gen Virol 92(Pt 5), 1184–8. [DOI] [PubMed] [Google Scholar]
- Sugimura T, Yonemochi H, Ogawa T, Tanaka Y. and Kumagai T. (1980) Isolation of a recombinant influenza virus (Hsw 1 N2) from swine in Japan. Arch Virol 66(3), 271–4. [DOI] [PubMed] [Google Scholar]
- Sun H, Xiao Y, Liu J, Wang D, Li F, Wang C, Li C, Zhu J, Song J, Jiang Z, Liu L, Zhang X, Wei K, Hou D, Pu J, Sun Y, Tong Q, Bi Y, Chang KC, Liu S. and Gao GF (2020) Prevalent Eurasian avian-like H1N1 swine influenza virus with 2009 pandemic viral genes facilitating human infection. Proc Natl Acad Sci U S A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun YF, Wang XH, Li XL, Zhang L, Li HH, Lu C, Yang CL, Feng J, Han W, Ren WK, Tian XX, Tong GZ, Wen F, Li ZJ, Gong XQ, Liu XM, Ruan BY, Yan MH and Yu H. (2016) Novel triple-reassortant H1N1 swine influenza viruses in pigs in Tianjin, Northern China. Vet Microbiol 183, 85–91. [DOI] [PubMed] [Google Scholar]
- Takemae N, Parchariyanon S, Damrongwatanapokin S, Uchida Y, Ruttanapumma R, Watanabe C, Yamaguchi S. and Saito T. (2008) Genetic diversity of swine influenza viruses isolated from pigs during 2000 to 2005 in Thailand. Influenza Other Respir Viruses 2(5), 181–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong S, Li Y, Rivailler P, Conrardy C, Castillo DA, Chen LM, Recuenco S, Ellison JA, Davis CT, York IA, Turmelle AS, Moran D, Rogers S, Shi M, Tao Y, Weil MR, Tang K, Rowe LA, Sammons S, Xu X, Frace M, Lindblade KA, Cox NJ, Anderson LJ, Rupprecht CE and Donis RO (2012) A distinct lineage of influenza A virus from bats. Proc Natl Acad Sci U S A 109(11), 4269–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong S, Zhu X, Li Y, Shi M, Zhang J, Bourgeois M, Yang H, Chen X, Recuenco S, Gomez J, Chen LM, Johnson A, Tao Y, Dreyfus C, Yu W, McBride R, Carney PJ, Gilbert AT, Chang J, Guo Z, Davis CT, Paulson JC, Stevens J, Rupprecht CE, Holmes EC, Wilson IA and Donis RO (2013) New world bats harbor diverse influenza A viruses. PLoS Pathog 9(10), e1003657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trevennec K, Leger L, Lyazrhi F, Baudon E, Cheung CY, Roger F, Peiris M. and Garcia JM (2012) Transmission of pandemic influenza H1N1 (2009) in Vietnamese swine in 2009–2010. Influenza Other Respir Viruses 6(5), 348–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Reeth K. and Ma W. (2012) Swine influenza virus vaccines: to change or not to change-that’s the question. Curr Top Microbiol Immunol 370, 173–200. [DOI] [PubMed] [Google Scholar]
- Van Reeth K. and Ma W. (2013) Swine influenza virus vaccines: to change or not to change-that’s the question. Curr Top Microbiol Immunol 370, 173–200. [DOI] [PubMed] [Google Scholar]
- Vander Veen RL, Loynachan AT, Mogler MA, Russell BJ, Harris DL and Kamrud KI (2012) Safety, immunogenicity, and efficacy of an alphavirus replicon-based swine influenza virus hemagglutinin vaccine. Vaccine 30(11), 1944–50. [DOI] [PubMed] [Google Scholar]
- Vijaykrishna D, Poon LL, Zhu HC, Ma SK, Li OT, Cheung CL, Smith GJ, Peiris JS and Guan Y. (2010) Reassortment of pandemic H1N1/2009 influenza A virus in swine. Science 328(5985), 1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijaykrishna D, Smith GJ, Pybus OG, Zhu H, Bhatt S, Poon LL, Riley S, Bahl J, Ma SK, Cheung CL, Perera RA, Chen H, Shortridge KF, Webby RJ, Webster RG, Guan Y. and Peiris JS (2011) Long-term evolution and transmission dynamics of swine influenza A virus. Nature 473(7348), 519–22. [DOI] [PubMed] [Google Scholar]
- Vincent AL, Lager KM, Janke BH, Gramer MR and Richt JA (2008a) Failure of protection and enhanced pneumonia with a US H1N2 swine influenza virus in pigs vaccinated with an inactivated classical swine H1N1 vaccine. Vet Microbiol 126(4), 310–23. [DOI] [PubMed] [Google Scholar]
- Vincent AL, Ma W, Lager KM, Gramer MR, Richt JA and Janke BH (2009) Characterization of a newly emerged genetic cluster of H1N1 and H1N2 swine influenza virus in the United States. Virus Genes 39, 176–185. [DOI] [PubMed] [Google Scholar]
- Vincent AL, Ma W, Lager KM, Janke BH and Richt JA (2008b) Swine influenza viruses a North American perspective. Adv Virus Res 72, 127–54. [DOI] [PubMed] [Google Scholar]
- Vincent AL, Ma W, Lager KM, Janke BH, Webby RJ, Garcia-Sastre A. and Richt JA (2007) Efficacy of intranasal administration of a truncated NS1 modified live influenza virus vaccine in swine. Vaccine 25(47), 7999–8009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent AL, Perez DR, Rajao D, Anderson TK, Abente EJ, Walia RR and Lewis NS (2016) Influenza A virus vaccines for swine. Vet Microbiol 206, 35–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson SJ, Langat P, Reid SM, Lam TT, Cotten M, Kelly M, Van Reeth K, Qiu Y, Simon G, Bonin E, Foni E, Chiapponi C, Larsen L, Hjulsager C, Markowska-Daniel I, Urbaniak K, Durrwald R, Schlegel M, Huovilainen A, Davidson I, Dan A, Loeffen W, Edwards S, Bublot M, Vila T, Maldonado J, Valls L, Brown IH, Pybus OG and Kellam P. (2015) Molecular Epidemiology and Evolution of Influenza Viruses Circulating within European Swine between 2009 and 2013. J Virol 89(19), 9920–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webby RJ, Rossow K, Erickson G, Sims Y. and Webster R. (2004) Multiple lineages of antigenically and genetically diverse influenza A virus co-circulate in the United States swine population. Virus Res 103(1–2), 67–73. [DOI] [PubMed] [Google Scholar]
- Webby RJ, Swenson SL, Krauss SL, Gerrish PJ, Goyal SM and Webster RG (2000) Evolution of swine H3N2 influenza viruses in the United States. J Virol 74(18), 8243–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster RG, Bean WJ, Gorman OT, Chambers TM and Kawaoka Y. (1992) Evolution and ecology of influenza A viruses. Microbiol Rev 56(1), 152–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster RG, Peiris M, Chen H. and Guan Y. (2006) H5N1 outbreaks and enzootic influenza. Emerg Infect Dis 12(1), 3–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsh MD, Baird PM, Guelbenzu-Gonzalo MP, Hanna A, Reid SM, Essen S, Russell C, Thomas S, Barrass L, McNeilly F, McKillen J, Todd D, Harkin V, McDowell S, Choudhury B, Irvine RM, Borobia J, Grant J. and Brown IH (2010) Initial incursion of pandemic (H1N1) 2009 influenza A virus into European pigs. Vet Rec 166(21), 642–5. [DOI] [PubMed] [Google Scholar]
- WHO. (2020) Cumulative number of confirmed human cases of avian influenza A(H5N1) reported to WHO, Cumulative number of confirmed human cases for avian influenza A(H5N1) reported to WHO, 2003–2020. Vol. 2020 https://www.who.int/influenza/human_animal_interface/2020_MAY_tableH5N1.pdf. [Google Scholar]
- Williamson SM, Tucker AW, McCrone IS, Bidewell CA, Brons N, Habernoll H, Essen SC, Brown IH and Wood JL (2012) Descriptive clinical and epidemiological characteristics of influenza A H1N1 2009 virus infections in pigs in England. Vet Rec 171(11), 271. [DOI] [PubMed] [Google Scholar]
- Xie JF, Zhang YH, Zhao L, Xiu WQ, Chen HB, Lin Q, Weng YW and Zheng KC (2018) Emergence of Eurasian Avian-Like Swine Influenza A (H1N1) Virus from an Adult Case in Fujian Province, China. Virol Sin 33(3), 282–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Chen Y, Qiao C, He X, Zhou H, Sun Y, Yin H, Meng S, Liu L, Zhang Q, Kong H, Gu C, Li C, Bu Z, Kawaoka Y. and Chen H. (2015) Prevalence, genetics, and transmissibility in ferrets of Eurasian avian-like H1N1 swine influenza viruses. Proc Natl Acad Sci U S A 113(2), 392–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Hua RH, Zhang Q, Liu TQ, Liu HL, Li GX and Tong GZ (2008) Genetic evolution of swine influenza A (H3N2) viruses in China from 1970 to 2006. J Clin Microbiol 46(3), 1067–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Zhou YJ, Li GX, Ma JH, Yan LP, Wang B, Yang FR, Huang M. and Tong GZ (2010) Genetic diversity of H9N2 influenza viruses from pigs in China: a potential threat to human health? Vet Microbiol 149(1–2), 254–61. [DOI] [PubMed] [Google Scholar]
- Zell R, Scholtissek C. and Ludwig S. (2012) Genetics, evolution, and the zoonotic capacity of European Swine influenza viruses. Curr Top Microbiol Immunol 370, 29–55. [DOI] [PubMed] [Google Scholar]
- Zhao G, Fan Q, Zhong L, Li Y, Liu W, Liu X, Gao S. and Peng D. (2011) Isolation and phylogenetic analysis of pandemic H1N1/09 influenza virus from swine in Jiangsu province of China. Res Vet Sci 93(1), 125–32. [DOI] [PubMed] [Google Scholar]
- Zhou NN, Senne DA, Landgraf JS, Swenson SL, Erickson G, Rossow K, Liu L, Yoon K, Krauss S. and Webster RG (1999) Genetic reassortment of avian, swine, and human influenza A viruses in American pigs. J Virol 73(10), 8851–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P, Hong M, Merrill MM, He H, Sun L. and Zhang G. (2014) Serological report of influenza A (H7N9) infections among pigs in Southern China. BMC Vet Res 10, 203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Webby R, Lam TT, Smith DK, Peiris JS and Guan Y. (2011a) History of Swine influenza viruses in Asia. Curr Top Microbiol Immunol 370, 57–68. [DOI] [PubMed] [Google Scholar]
- Zhu H, Zhou B, Fan X, Lam TT, Wang J, Chen A, Chen X, Chen H, Webster RG, Webby R, Peiris JS, Smith DK and Guan Y. (2011b) Novel reassortment of Eurasian avian-like and pandemic/2009 influenza viruses in swine: infectious potential for humans. J Virol 85(20), 10432–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W, Zhang H, Xiang X, Zhong L, Yang L, Guo J, Xie Y, Li F, Deng Z, Feng H, Huang Y, Hu S, Xu X, Zou X, Li X, Bai T, Chen Y, Li Z, Li J. and Shu Y. (2016) Reassortant Eurasian Avian-Like Influenza A(H1N1) Virus from a Severely Ill Child, Hunan Province, China, 2015. Emerg Infect Dis 22(11), 1930–1936. [DOI] [PMC free article] [PubMed] [Google Scholar]