Abstract
We performed swine influenza virus (SIV) surveillance in Midwest USA and isolated 100 SIVs including endemic and reassortant H1 and H3 viruses with 2009 pandemic H1N1 genes. To determine virus evolution when different genotypes and subtypes of influenza A viruses circulating in the same swine herd, a virus survival experiment was conducted in pigs mimicking field situations. Five different SIVs were used to infect five pigs individually, then two groups of sentinel pigs were introduced to investigate virus transmission. Results showed that each virus replicated efficiently in lungs of each infected pig, but only reassortant H3N2 and H1N2v viruses transmitted to the primary contact pigs. Interestingly, the parental H1N2v was the majority of virus detected in the second group of sentinel pigs. These data indicate that the H1N2v seems to be more viable in swine herds than other SIV genotypes, and reassortment can enhance viral fitness and transmission.
Keywords: swine influenza virus, surveillance, virus competition and survival, pigs, H1N2v
1. Introduction
Swine influenza is a zoonotic disease caused by influenza A viruses that belong to the Orthomyxoviridae family. The segmented feature of influenza virus genome allows reassortment during replication when two or more different influenza virus strains infect the same cell or host at the same time, resulting in generation of progeny viruses containing a novel combination of genes (Essere et al., 2013; Marshall et al., 2013). Pigs have been considered as a “mixing vessel” for influenza A viruses, because they have receptors for both avian and human influenza viruses (Ito et al., 1998). The pigs’ role in ecology of influenza A viruses has been emphasized with the emerging of the swine-origin A(H1N1)pdm09 virus (Mena et al., 2016; Nelson et al., 2016) as well as the H3N2 and H1 variant viruses that caused human infections (Biggerstaff et al., 2013; Jhung et al., 2013; Pulit-Penaloza et al., 2018), which induced great concerns of public health (Vincent et al., 2014).
The H1N1, H1N2, and H3N2 are major subtypes frequently found in the North American pig farms (Corzo et al., 2013). North American triple reassortant SIVs have become the predominant viruses circulating in the US swine since 1990s (Vincent et al., 2008). These viruses contain PA and PB2 genes from the avian influenza virus, PB1 from the human influenza virus, and NP, M and NS from the classical SIV; this unique 6 internal gene constellation in North American triple reassortant SIVs was named as the ‘TRIG’ cassette that can accept different HA and NA combinations such as H1N1, H1N2, H3N1 and H3N2 (Vincent et al., 2008). Triple reassortant SIVs have been endemic in the swine herds until 2009 (Kong et al., 2015; Pasma and Joseph, 2010), when the swine-origin pandemic virus A(H1N1)pdm09 emerged and spread globally in 2009 causing human infections and deaths. Different from other sporadic SIV infections in humans (Myers et al., 2007; Shinde et al., 2009), the A(H1N1)pdm09 not only infected humans, but also crossed the species barrier to infect other species including pigs (Nelson et al., 2012; Song et al., 2010). With introduction of A(H1N1)pdm09 into swine herds, reassortant viruses between endemic SIVs and A(H1N1)pdm09 have been found in pigs in many countries (Charoenvisal et al., 2013; Ducatez et al., 2011; He et al., 2018; Howard et al., 2011; Kitikoon et al., 2012; Moreno et al., 2011; Schaefer et al., 2015). The reassortant H1N1 subtype viruses with A(H1N1)pdm09 genes were first found in Hong Kong, Germany, and Thailand (Kitikoon et al., 2011; Starick et al., 2011; Vijaykrishna et al., 2010); The reassortant H1N2 viruses containing genes from A(H1N1)pdm09 have also been found in UK, Italy, and US (Ducatez et al., 2011; Howard et al., 2011; Moreno et al., 2011); reassortant H3N2 subtype viruses with different genes from A(H1N1)pdm09 have been reported in the US (Kitikoon et al., 2012; Liu et al., 2012), which are becoming endemic in the US swine. Noticeably, SIV isolates in the US with the M gene from the A(H1N1)pdm09 are increasing (Nelson et al., 2016), and both H3N2 and H1N2 variant SIVs that carry the A(H1N1)pdm09 M gene have caused human infections in the US (Bowman et al., 2014; Lindstrom et al., 2012). This fact suggests that the TRIG cassette with the Eurasian M gene might be more feasible for evolution of SIVs in pigs and pose more threats to public health. Multiple genotypes and subtypes of SIVs exist and are circulating in the swine populations, posing a major challenge for the swine industry to predict which subtypes will be more predominant in order to produce vaccine to protect animal health.
In this study, we performed SIV surveillance in the Midwest US pig herds during 2010–2015. Endemic and reassortant H1N1, H1N2, and H3N2 viruses with the single or multiple A(H1N1)pdm09 genes were isolated and further analyzed. In addition, we selected five isolates with different genetic constellations to infect five pigs, then introduced 2 groups of contact animals to determine which virus will be predominant virus and be maintained in swine herds mimicking field situations. Further analysis of transmitted viruses detected in the sentinel animals indicates the H1N2 variant is the most transmissible and viable virus and able to outcompete other viruses, and viral reassortment enhances virus fitness and transmission.
2. Materials and methods
2.1. Ethics statements
The pig study was conducted at the Large Animal Research Center (Biosafety level 2+ facility) at Kansas State University in accordance with the Guide for the Care and Use of Agricultural Animals in Research and Teaching of the U.S. Department of Agriculture. The protocol was approved by the Intuitional Animal Care and Use Committee of Kansas State University.
2.2. Cells
MDCK cells were maintained in Eagle’s minimal essential medium (MEM) (HyClone, Logan, UT) with 5% FBS (HyClone, Logan, UT), 1% 100× antibiotics (Invitrogen, Carlsbad, CA), 1% L-glutamine (Invitrogen, Carlsbad, CA), and 1% MEM vitamin supplement (Invitrogen, Carlsbad, CA). The medium for virus amplification is Eagle’s minimal essential medium (MEM) containing 0.3% BSA (Sigma; St. Louis, MO), 1% 100× antibiotics (Invitrogen, Carlsbad, CA), 1% L-glutamine (Invitrogen, Carlsbad, CA), and 1% MEM vitamin supplement (Invitrogen, Carlsbad, CA).
2.3. Sample collection, screening, and virus isolation
Approximately 5,000 swine samples including nasal swabs and lung tissues, which were collected from diseased pigs in Midwest swine farms or directly obtained from the Kansas state veterinary diagnostic laboratory, were used for screening and isolation of influenza A virus. To detect influenza A virus, the samples were first screened by a real-time RT-PCR targeting the M gene. The lung tissue was processed as 10% homogenate using MEM with 1 μg/mL 100× antibiotics (Invitrogen, Carlsbad, CA). The homogenates were centrifuged at 12,000 rpm for 10 minutes, then the supernatants were collected for the RNA isolation. Nasal swab samples were vortexed, and then samples were centrifuged for 5 minutes at 2,000 rpm to collect the supernatants prior to removal of the swabs from the medium. RNA was extracted using 140 uL of supernatants from the lung homogenates or nasal swab samples. Real-time RT-PCR was performed as previously described (Ma et al., 2010b). The real-time RT-PCR positive samples were filtered using 0.45μm filters to remove any bacteria prior to virus isolation on MDCK cells. If the inoculated samples caused cytopathic effects (CPE) on MDCK cells, the supernatants were collected for further subtyping by sequencing.
2.4. Sequence and phylogenetic analysis
The HA, NA and M genes were first amplified from the isolates using the universal primers as described previously (Hoffmann et al., 2001) and sequenced. If the M gene originated from the A(H1N1)pdm09 after confirmation by analysis, then the full genome of the isolate was sequenced. The subtypes and the origins of the viral segments were determined by sequence BLAST (http://blast.ncbi.nlm.nih.gov) and phylogenetic analysis. The phylogenetic trees of M, and HA genes of selected virus isolates were conducted using Mega software version 4.1. Representative influenza viruses of each subtype were selected as references.
2.5. Viruses
Five genotypes of SIVs with different genetic constellations were isolated and full-genome sequenced, and selected for the pig study. Three endemic viruses include A/swine/Kansas/10–83533/2010 (H3N2), A/swine/Kansas/11–135979/2011 (H1N1) and A/swine/Kansas/11–104259/2011 (H1N2) which are the North American triple reassortant viruses. A/swine/Kansas/11–110529/2011 (H3N2) is a novel reassortant virus (H3N2r) which has NP, M, and NS genes from the A(H1N1)pdm09. A/swine/Kansas/12–156064/2012 (H1N2) which has M gene from the A(H1N1)pdm09 is named H1N2 variant (H1N2v). Other genes of both H1N2v and H3N2r viruses are from North American triple reassortant viruses.
2.6. Pig study
Twelve 4-week-old pigs, which were confirmed to be SIV and porcine reproductive and respiratory syndrome virus negative by testing collected serum and nasal swab samples from each pig using hemagglutination inhibition and real-time RT-PCR assays, were used in this study. A group of five co-housed pigs (pig# M27, M25, M31, M33 and M30) were designated as the infection group, and each pig was intratracheally infected with 105 TCID50 of each virus under anesthesia by intramuscular injection with a mixture of ketamine and xylazine as described previously (Richt et al., 2003; Ma et al., 2015). At one day post infection (dpi), the first group of 4 contact pigs (pig# M28, M29, M32, and M26) were commingled with the original infected animals. The first contact group of pigs were subsequently moved into a new room after 3 days post contact to co-house with the second group of 3 naïve animals (pig# M35, M34, and M36) for 3 days. Clinical signs were monitored daily. Nasal swab samples were collected in original infected pigs at 1, 2, 3 and 4 dpi, and in contact animals starting at 1 day post contact (dpc) until necropsy day. The original 5 infected pigs were euthanized at 4 dpi. The first contact group of 4 pigs were euthanized at 6 dpc, while the second group of 3 pigs were necropsied after 3 dpc with the first group of primary contact animals. During necropsy, gross lung lesions were scored for each pig by a single experienced veterinarian. Bronchioaveolar lavage fluid (BALF) samples were collected by flushing a lung with 50 ml of MEM to study virus replication and transmission. Virus titer of BALF and nasal swab samples was determined on MDCK cells in 96-well plates.
2.7. Library construction and Illumina deep sequencing
The BALF samples were centrifuged at 2,000 rpm for 10 minutes at 4°C and total RNA from collected supernatants was extracted using the QIAamp Viral RNA Mini Kit (Qiagen, CA USA). Isolated RNA was reverse transcribed into cDNA using universal primer (Uni-12). Each segment was then amplified using the pfx polymerase (Invitrogen, CA, USA), and was pooled based on copy numbers to ensure even distribution of all 8 gene segments. The pooled segments were diluted to 0.2 ng/μL, and 1 ng of pooled samples was then used for library preparation and barcoding using the Illumina Nextera XT library preparation kit (Illumina, San Diego, CA, USA) according to manufacturer’s instructions. The barcoded libraries were sequenced on an Illumina MiSeq platform using 2×150 bp paired-end chemistry.
Reads for each sample were parsed into individual files based on barcoded sequences. Parsed reads were loaded into CLC Genomic workbench 7.5 (Qiagen, Carlsbad, CA). The original challenge virus sequence was used as the reference for read mapping, and consensus sequences were extracted for each segment. The consensus sequences of each gene segment were then aligned to a set of references. Pairwise comparison generated from the sequence alignment was used to determine segment identity based on percent identity. Segment identity was called based on the greatest percent identity. A 0.5% cut-off was used to differentiate between closely related genes. To further determine gene identity, a BLAST database was generated with the set of references. Reads were analyzed using an unbiased BLAST search against the reference database using the BioPearl package. The BLAST results were used to generate a heat map in Genesis (Thallinger Lab, Graz, Austria) to determine the origin of each gene segment.
2.8. Plaque assay and identification of viral segment origin
To obtain the single plaques, 10-fold serially diluted BALF samples were used to infect MDCK cells in 6-well plates. One hour post infection, the supernatant was removed from the cells, which were then covered with 0.9% agarose containing 1×MEM, 1% antibiotics and 0.3% BSA. Two days post incubation, the single plaques were picked and amplified on MDCK in 24-well plates. Eight pairs of specific primers were designed based on the conservative regions of each segment of each virus, which can differentiate five viruses used in the pig study (primers available upon request). Eight segments of each viral plaque were amplified using the designed specific primers, sequenced and analyzed to determine the origin of each viral segment.
3. Results
3.1. Characterization of swine influenza virus isolates
From 2010 to 2015, a total of 100 SIVs were isolated from pigs in the Midwestern farms (Table 1), and the full or partial viral genome of these isolates were sequenced. The sequences of partial viral genome included at least the HA, NA, and M genes. Based on the sequence information, these isolates included H1N1 (26 isolates), H1N2 (42 isolates), and H3N2 (32 isolates) (Table 1), and reassortant viruses found in each subtype normally had the Eurasian M gene from the A(H1N1)pdm09 virus (Table 2) as previously reported (Ducatez et al., 2011; Howard et al., 2011; Resende et al., 2017). Sequence analysis showed that 69% (29/42) of H1N2 isolates and 63% (20/32) of H3N2 viruses were reassortants that contain M gene from the A(H1N1)pdm09 (Table 1). In contrast only 35% (9/26) of H1N1 viruses were reassortant viruses between endemic SIVs and A(H1N1)pdm09 viruses, which contained M gene from the A(H1N1)pdm09 and were found in samples collected between 2013 and 2014; the remaining isolates were the traditional North American triple reassortant viruses that are endemic in pigs. Interestingly, most of the reassortant viruses were the H3N2 subtype during 2010–2012, and after 2012 most of the viruses isolated were reassortant H1N1 and H1N2 viruses. Reassortant H1N1 viruses were found after 2013 in our surveillance program.
Table 1:
Virus isolates during 2010–2015
| H3N2 |
H1N1 |
H1N2 |
||||
|---|---|---|---|---|---|---|
| E | R | E | R | E | R | |
| 2010 | 1 | 1 | 5 | 2 | ||
| 2011 | 4 | 7 | 10 | 4 | 1 | |
| 2012 | 2 | 3 | 2 | 9 | ||
| 2013 | 4 | 3 | 3 | 4 | ||
| 2014 | 1 | 8 | 2 | 6 | 2 | 15 |
| 2015 | 1 | |||||
| Total | 12 | 20 | 17 | 9 | 13 | 29 |
E: endemic virus; R: reassortant
Table 2:
Genotype of isolated reassortant viruses
| PB1 | PB2 | PA | HA | NP | NA | M | NS | |
|---|---|---|---|---|---|---|---|---|
| H3N2 | E | E | E | E | P | E | P | P |
| E | E | P | E | E | E | P | E | |
| E | P | P | E | P | E | P | P | |
| H1N2 | E | E | E | E | E | E | P | E |
| P | P | P | E | P | E | P | P | |
| H1N1 | E | E | E | E | E | E | P | E |
| P | P | P | E | E | E | P | E |
E: endemic virus; P: 2009 pandemic H1N1 virus
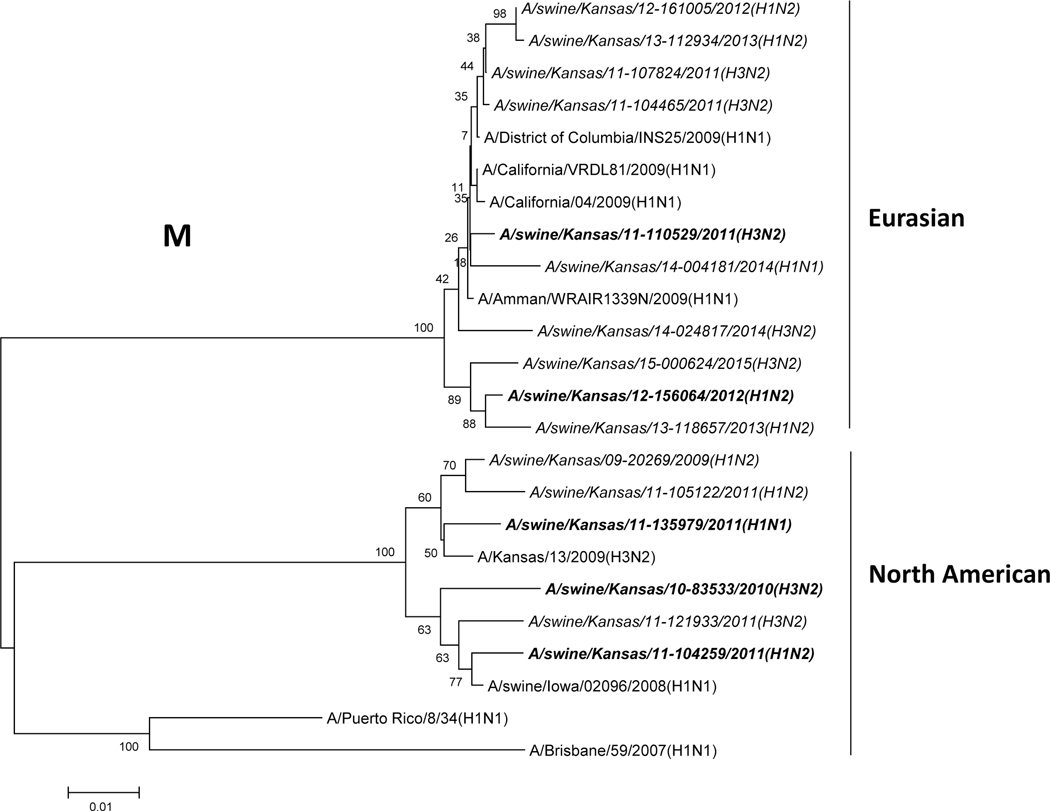
Phylogenetic analysis of the M gene of selected viruses showed that it divided into two lineages (North American and Eurasian swine influenza lineages), but most of them belonged to the Eurasian swine lineage as depicted in Fig. 1A. For the HA of the selected H1 subtype, they belonged to different lineages (Fig. 1B). Three H1 isolates were located in the β lineage, 23 H1 isolates were in the γ lineage and 39 isolates were located in the δ lineage (12 in the δ2 lineage, and 27 in the δ1 lineage). These results indicate that multiple subtypes and genotypes of SIVs exist in Midwestern swine herds.
Figure 1: Phylogenetic trees of M and H1 genes of swine influenza viruses isolated from Midwest swine.
A) Phylogenetic tree of M segment; B) Phylogenetic tree of H1 segment. The trees were generated by software MEGA 4.1 with the distance-based neighbor-joining method. The reliability of the tree was assessed by bootstrap analysis with 1,000 replications. The viruses isolated in this study are in italic, and the isolates used in the animal study are in italic and bold.
Three endemic North American triple reassortant viruses including H1N1, H1N2 and H3N2 viruses and two novel reassortant viruses including H1N2 and H3N2 viruses were selected for further in vivo studies. The reassortant H3N2 virus has NP, M and NS genes from the A(H1N1)pdm09 virus and the remaining 5 genes from endemic H3N2 viruses; the virus has been shown to be pathogenic and transmit efficiently in pigs in our former study (Ma et al., 2015). The reassortant H1N2 virus has the M gene from the A(H1N1)pdm09 virus and the remaining genes from endemic SIVs; this kind of viruses are referred to as H1N2 variant (H1N2v) viruses which have been reported to infect humans in the USA (https://www.cdc.gov/flu/spotlights/h1n2v-cases-mn.htm).
3.2. Virus replication and transmission among pigs
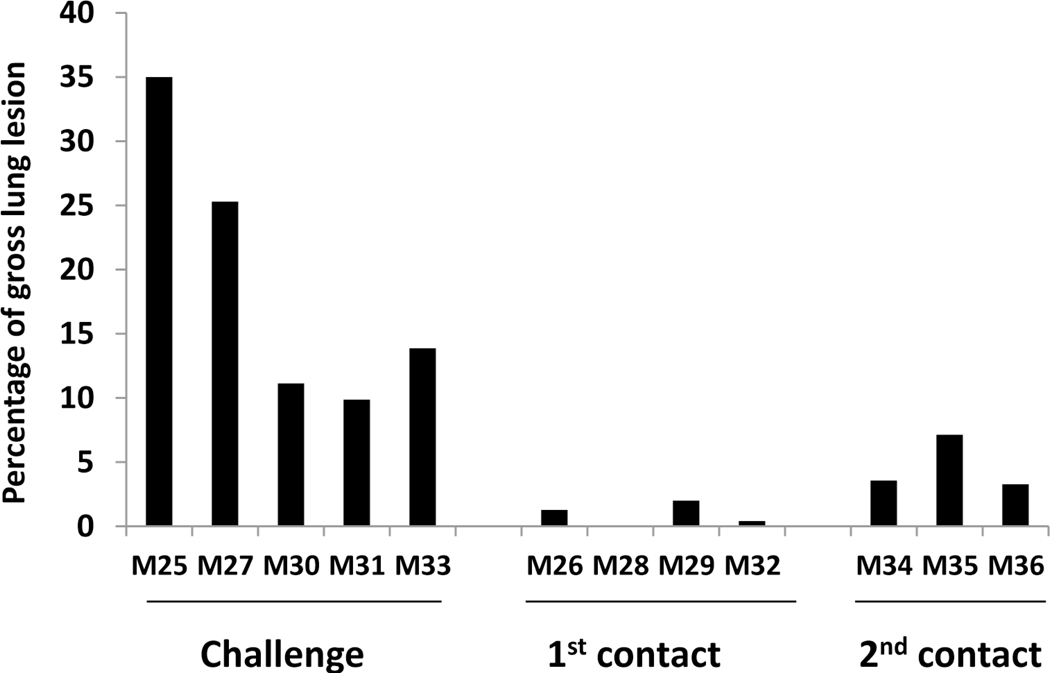
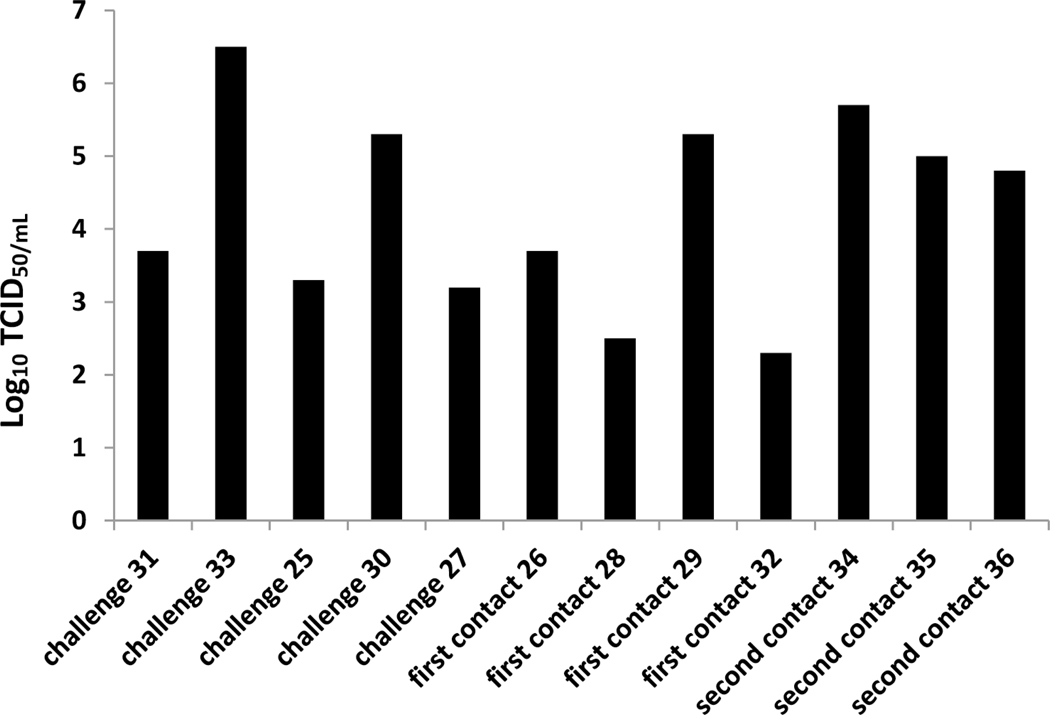
It is very common that multiple genotypes and subtypes of SIVs exist and circulate within the same swine herds. The impacts on viral evolution in the presence of multiple co-circulating genotypes/subtypes of influenza A viruses in swine remains not fully understood. We selected five different genotypes/subtypes of SIVs including H1N1, H3N2, and H1N2 that were isolated from pigs in Midwestern farms, then each one virus was used to infect one individual pig intratracheally respectively. Five pigs infected by each virus were cohoused in one room together, and two contact groups of pigs were introduced to investigate virus transmission and evolution. All pigs did not exhibit obvious respiratory clinical symptoms during the experimental period, but severe pneumonia and lung lesions were observed in each original infected pig although severity was variable (Fig. 2A). Virus was detected in the lung (BALF samples) of each original infected pig with virus titers ranging from 1.5×103 to 5×106 TCID50/mL (Fig. 2B). Minor lung lesions were found in three out of four pigs in the first contact group (Fig. 2A); however, virus was detected in the lung of all four contact pigs with titers ranging from 2×102 to 2×104 TCID50/mL (Fig. 2B). In contrast to the primary contact pigs, all three animals in the secondary contact group showed more severe lung lesions (Fig. 2A) and a higher virus titer was detected in the lungs of each pig ranging from 6×104 to 5×105 TCID50/mL (Fig. 2B).
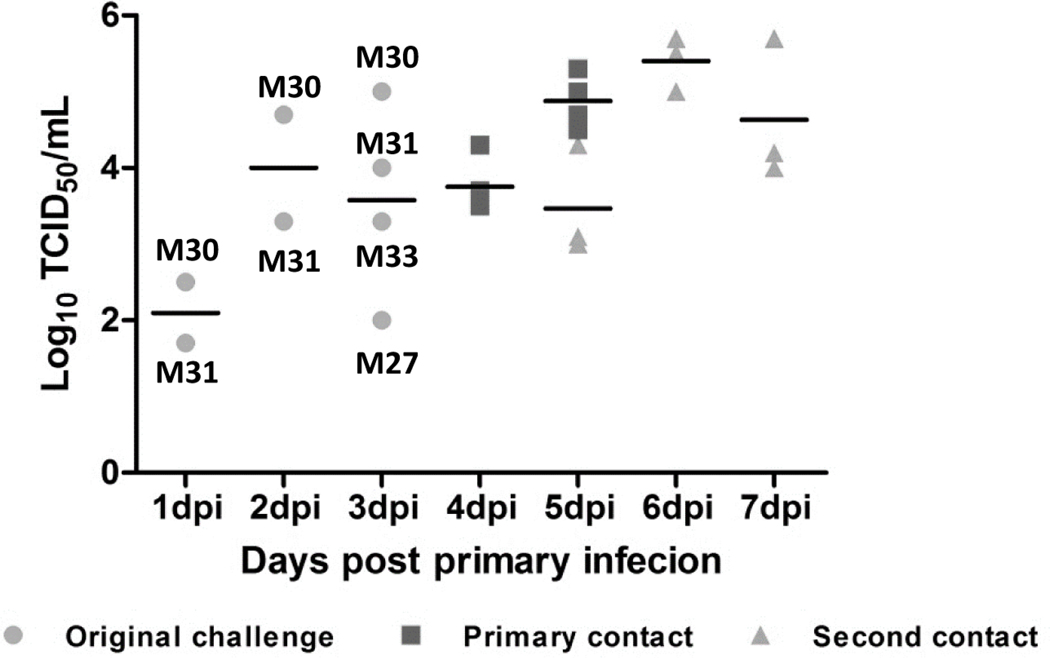
Figure 2: Lung lesions and virus titers in lung and nasal swabs of infected and contact pigs.
A) Lung lesions of original infected and contact pigs. Lung lesions were shown as the average of percentage of lung with lesions. B) Viral titers of bronchial alveoli lavage fluid samples of each pig. C) Viral titers of nasal swab samples collected each day from each pig.
Nasal swabs collected from each pig at different time points were tested to determine viral transmission. In the original infected group, virus was found in nasal swabs collected from both the pig M30 infected with A/swine/Kansas/12–156064/2012 (H1N2) virus and the M31 infected with the A/swine/Kansas/11–110529/2011 (H3N2) virus at all 3 tested time points (1, 2 and 3 dpi) (Fig. 2C); both A/swine/Kansas/10–83533/2010 (H3N2) and A/swine/Kansas/11–104259/2011 (H1N2) were only found in nasal swabs collected from infected pigs at 3 dpi (Fig. 2C). In contrast, no virus was found in nasal swabs collected from the pig M25 infected with the A/swine/Kansas/11–135979/2011 (H1N1) virus (Fig. 2C). These results indicate that the endemic H1N1 virus does not shed efficiently compared to other 4 tested viruses. In the first contact group, all 4 pigs started to shed virus at 3 dpc (equivalent to 4 dpi) and virus shedding was also detected in each pig (Fig. 2C). All 3 pigs in the secondary contact group started to shed virus at 2 days post commingled with the primary contact pigs, virus shedding was detected in each pig until the end of experiment (4 dpc equivalent to 7 dpi) with virus titers averaged at more than 104 TCID50/mL (Fig. 2C).
3.3. H1N2v is predominate and maintained in native pigs after two passages
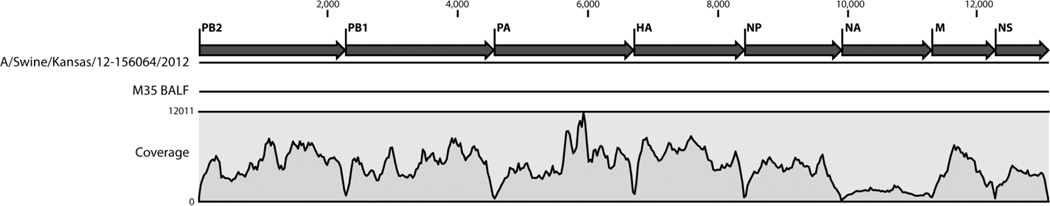
To determine which virus transmits among pigs and which virus will be generated in pigs, Deep sequence was performed for the 12 BALF samples collected from each pig. Approximately 500,000 paired reads for each of the 12 samples were yielded. The high-quality reads for each sample were mapped to a reference sequence and the coverage of mapped reads is shown in Fig. 3. These sequences represent genome cRNA, therefore correspond to the viral genomes. Host cell and ribosomal sequences were subtracted from the dataset, where there was no significant difference between the screening reads and control. The consensus sequences from each sample were aligned to the viral sequences used for infection. Hit rates with each inoculated virus were calculated from the highest to the lowest. The viral sequences with the highest alignment similarity or hit rates higher than 95% were considered majority of the viral mixture. The alignment results indicated that the viral gene segments corresponded to the original viruses in the infection group. In the primary contact group, there was no segment from the endemic H3N2 virus; more viral segments originated from the H3N2r and H1N2v, and only a few segments from both H1N2 and H1N1 viruses. In the secondary contact group, the majority of all the segments were from the H1N2v.
Figure 3: The coverage of each segment in deep sequencing.
The genome coverage of sequencing reads from pig M35 BALF is shown as an example. These were high quality reads that mapped to and covered the whole viral genome. The number reads per sample were more than 500,000.
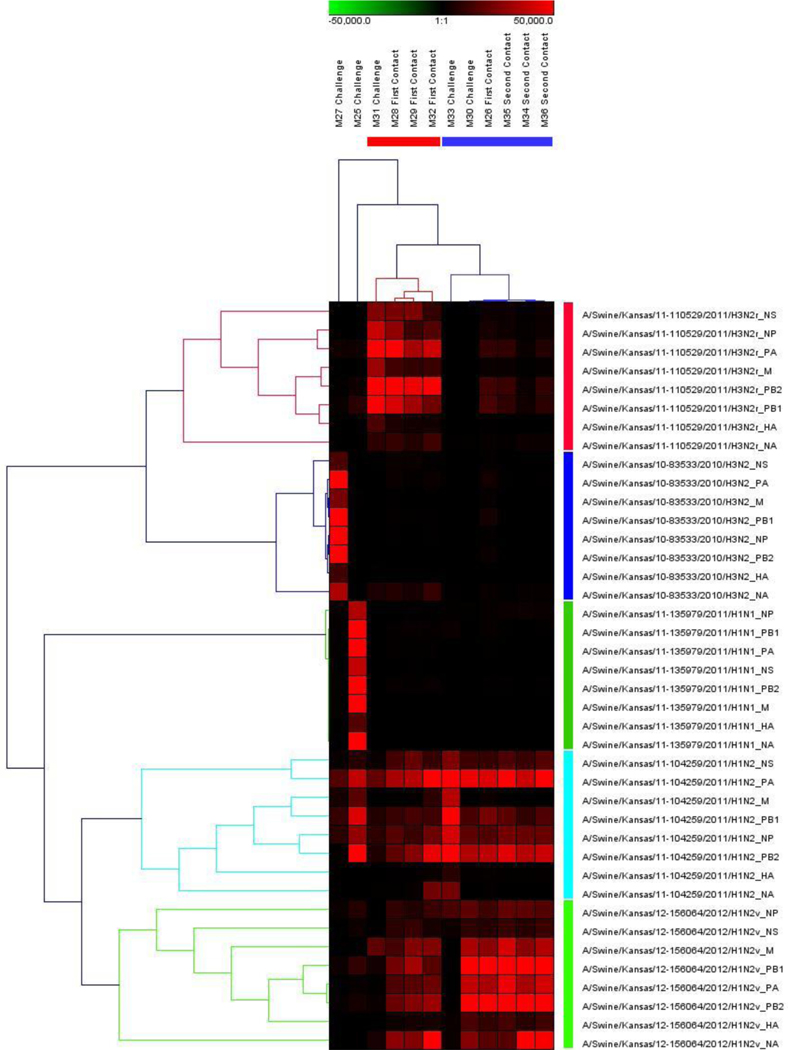
The origin of each segment was interpreted with a heat map as depicted in Fig. 4. The infected pigs contained the original infected virus, except the pigs infected with either H1N2 or H1N2v showed origin from both viruses, as that both endemic H1N2 and reassortant H1N2v viruses share very high homology. Viruses identified in the pigs from the first contact group had a mixture of segments from the H3N2r, H1N2, or H1N2v, while the gene segment of the viruses found in the second contact groups were largely similar to those of both H1N2 and H1N2v viruses. However, both endemic H3N2 and H1N1 viruses didn’t appear in any contact pigs.
Figure 4:
The heat map of the origin of each segment from BALF samples.
The heat map was generated in Genesis to determine the origin of each gene segment. Different intensities of color represent the number of sequencing reads matched with reference sequences.
3.4. Quasispecies of viruses detected in pigs
Plaque assay and subsequent sequences of each gene segment of each single virus was used to determine virus origin detected in BALF samples of each pig. Only the original virus used for infection was found in each of the challenged pigs, based on analysis 2 to 6 single plaques in each BALF sample (Table 3), similar to the deep sequencing results. While more than one phenotype of plaques were observed in the BALF samples collected from the first and second contact groups of pigs, indicating that quasispecies of the viruses might exist in these samples. To determine the gene combinations of each single virus found in two groups of contact pigs, 6 to 12 single plaques were randomly picked from each sample and further analyzed by Sanger sequencing. Results showed that the H1N2v was found in 3 contact pigs and the parental H3N2r virus was also identified in 1 contact pig (M28) in the first contact group (Table 3). Interestingly, both the parental H1N2r and the reassortant H1N2v that has the NS gene from the H3N2r, were identified in 2 primary contact pigs; around 40% of analyzed viruses (M29, 5/12; M32, 4/10) were reassortants in both animals (Table 3). In the second contact group, both the H1N2v and novel reassortant H3N2 viruses were found in pigs. Only the parental H1N2v was found in the pig M35, while both the parental H1N2v and a reassortant H1N2v that has PB2, NA, M and NS from the H3N2r virus, were found in the pig M36 (Table 3). In the pig M34, half (3/6) of analyzed viruses was the parental H1N2v, 2 viruses were a reassortant H1N2v that has the NS gene from the H3N2r virus and one was a novel reassortant H3N2 virus that has the HA gene from the H3N2v and the remaining genes from the H1N2v (Table 3). Noticeably, only one H3N2 virus was identified in the 20 analyzed individual viruses in the second contact group, the remaining were the H1N2v despite several reassortants detected.
Table 3:
Number and subtype of analyzed plaques from each BALF sample of each pig
| Group | Pig number | Total plaque sequenced | Major subtype | Plaques with major subtype | Genomes mixed with |
|---|---|---|---|---|---|
| Original challenge | M25 | 2 | H1N1 | 2/2 | - |
| M30 | 4 | H1N2v | 4/4 | - | |
| M33 | 6 | H1N2 | 6/6 | - | |
| M31 | 2 | H3N2r | 2/2 | - | |
| M27 | 2 | H3N2 | 2/2 | - | |
| Primary contact | M26 | 8 | H1N2v | 8/8 | - |
| M29 | 12 | H1N2v | 8/12 | - | |
| H3N2r | 2/12 | - | |||
| H1N2v | 1/12 | H3N2r PB2 PB1 PA NP NA M NS | |||
| H1N2v | 1/12 | H3N2r PB1 | |||
| M28 | 8 | H3N2r | 8/8 | - | |
| M32 | 10 | H1N2v | 6/10 | ||
| 1/10 | H3N2r PB2 PB1 PA NP M NS | ||||
| 1/10 | H3N2r PB1 NP | ||||
| 2/10 | H3N2r M | ||||
| Secondary contact | M34 | 6 | H1N2v | 3/6 | - |
| 2/6 | H3N2r NS | ||||
| 1/6 | H3N2 HA | ||||
| M35 | 8 | H1N2v | 8/8 | - | |
| M36 | 6 | H1N2v | 5/6 | - | |
| 1/6 | H3N2r PB2, NA, M, NS | ||||
4. Discussion
The segmented feature of influenza virus genome allows reassortment when different influenza viruses infect the same host, which is one of the major mechanisms of influenza virus evolution to generate novel viruses. The generation of novel influenza viruses in swine greatly threatens the pig industry as well as public health. Introduction of A(H1N1)pdm09 into swine results in reassortment with endemic SIVs which expands the genetic diversity in swine (Anderson et al., 2013; Ducatez et al., 2011; Liang et al., 2014; Nelson et al., 2014, 2011). Our surveillance program in swine further demonstrates this fact that multiple reassortant SIVs in addition to endemic H1N1, H3N2, and H1N2 triple reassorant viruses have been detected and isolated in Midwest swine herds since the A(H1N1)pdm09 virus was introduced into swine populations. Interestingly, in contrast to isolated H1 variant viruses, more H3N2 variant viruses were detected in our surveillance program, which have caused human endemic in the US (Greenbaum et al., 2015). We found different reassortment patterns in different subtypes of SIVs in swine. For example, the reassorant H3N2 viruses have 2 (PA and M), 3 (NP, M and NS) or 5 (PA, PB2, NP, M and NS) internal genes from the A(H1N1)pdm09 virus, while the H1N2 reassortant viruses contain 1 (M) or 6 internal genes from the A(H1N1)pdm09 virus. In contrast, the reassortant H1N1 virus with only the A(H1N1)pdm09 virus M gene was found in our surveillance program. These reassortment patterns differ from previously reported patterns (Ducatez et al., 2011; Kitikoon et al., 2011). Noticeably, all these novel reassortant viruses have the M gene from the A(H1N1)pdm09 virus, which is in agreement with other reports (Ducatez et al., 2011; Gao et al., 2017; Kitikoon et al., 2012, 2011). The ‘TRIG’ cassette containing human (PB1), avian (PB2 and PA), and swine (M, NP, and NS) has been shown to support different HA and NA combinations and has changed the dynamics of SIV infection in North America dramatically (Webby et al., 2000). The Eurasian M gene most likely will replace the classical swine M gene to form a novel TRIG cassette that makes virus more viable and promotes virus transmission among pigs (Nelson et al., 2015). Furthermore, some of swine-origin H3N2 and H1 variant viruses with the Eurasian M gene isolated from humans show not only efficient transmission through contact but also efficient respiratory droplet transmission in the ferret model, similar as seasonal human influenza viruses (Pearce et al., 2012; Pulit-Penaloza et al., 2018; Sun et al., 2018). This suggests that these viruses have the potential to replicate and transmit efficiently in mammals including humans (Pearce et al., 2012).
Our results demonstrate that reassortment can occur when two or multiple viruses exist in the same host, but only the fittest virus will be maintained and survive in the host. This result is consistent with our former study in which pigs were co-infected with H1N1 and H3N2 SIVs (Ma et al., 2010a). Based on Hemagglutinin (HA) phylogenetical analysis, HA is divided into six major clusters (α, β, γ1, γ2, δ1, and δ2) phylogenetically (Anderson et al., 2015; Lorusso et al., 2013). Compared to 4 other tested viruses, the H1N2v in δ lineage is more viable and able to outcompete other viruses, which is in agreement with ours and other surveillance results (Rajao et al., 2018; Walia et al., 2018). Importantly, variant viruses, such as H1N2v, have been reported to cause human infections at multiple state fairs across the United States (https://www.cdc.gov/flu/spotlights/h1n2v-cases-mn.htm). Outbreaks of the reassortant H3N2 variant in the US caused more than 400 human cases (Wong et al., 2012) (https://www.cdc.gov/flu/swineflu/variant/h3n2v-cases.htm). Although the H1N2v did not cause as many human cases as the H3N2v did to date, the potential to cross the species barrier to establish in humans cannot be neglected. Moreover, these kind of reassortant viruses were also detected in other countries and resulted in human infections (Resende et al., 2017).
Given a fact that different genotypes/subtypes of influenza A viruses are co-existing in pigs, resulting in generation of novel viruses (Abe et al., 2015; Anderson et al., 2015; Diaz et al., 2017), our study highlighted the importance of SIV co-infection in the field, which increase the genetic diversity of the influenza A viruses that has more incidence to become an endemic or pandemic virus (Salomon and Webster, 2009). Thus, special attentions should be given to coinfections during the SIV surveillances (Vincent et al., 2014).
Taken together, our surveillance data show that multiple reassortant viruses emerged due to introduction of the A(H1N1)pdm09 into swine herds; and in vivo studies reveal that H1N2v is the most viable virus in pigs among the five tested viruses. In conclusion, viral reassortment can be one of major mechanisms to enhance influenza virus fitness and transmission.
Acknowledgements
This work was supported by NIAID funded Centers of Excellence for Influenza Research and Surveillance under contract numbers HHSN 272201400006C and NIAID R21AI121906.
Footnotes
Conflict of Interest
The authors claim no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference
- Abe H, Mine J, Parchariyanon S, Takemae N, Boonpornprasert P, Ubonyaem N, Patcharasinghawut P, Nuansrichay B, Tanikawa T, Tsunekuni R, Saito T, 2015. Co-infection of influenza A viruses of swine contributes to effective shuffling of gene segments in a naturally reared pig. Virology 484, 203–212. 10.1016/j.virol.2015.06.002 [DOI] [PubMed] [Google Scholar]
- Anderson TK, Campbell BA, Nelson MI, Lewis NS, Janas-Martindale A, Killian ML, Vincent AL, 2015. Characterization of co-circulating swine influenza A viruses in North America and the identification of a novel H1 genetic clade with antigenic significance. Virus Res. 201, 24–31. 10.1016/j.virusres.2015.02.009 [DOI] [PubMed] [Google Scholar]
- Anderson TK, Nelson MI, Kitikoon P, Swenson SL, Korslund JA, Vincent AL, 2013. Population dynamics of cocirculating swine influenza A viruses in the United States from 2009 to 2012. Influenza Other Respi. Viruses 7, 42–51. 10.1111/irv.12193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggerstaff M, Reed C, Epperson S, Jhung MA, Gambhir M, Bresee JS, Jernigan DB, Swerdlow DL, Finelli L, 2013. Estimates of the number of human infections with influenza A(H3N2) variant virus, United States, August 2011–April 2012. Clin. Infect. Dis 57, S12–S15. 10.1093/cid/cit273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman AS, Nelson SW, Page SL, Nolting JM, Killian ML, Sreevatsan S, Slemons RD, 2014. Swine-to-human transmission of influenza A(H3N2) virus at agricultural fairs, Ohio, USA, 2012. Emerg. Infect. Dis 20, 1472–1480. 10.3201/eid2009.131082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charoenvisal N, Keawcharoen J, Sreta D, Chaiyawong S, Nonthabenjawan N, Tantawet S, Jittimanee S, Arunorat J, Amonsin A, Thanawongnuwech R, 2013. Genetic characterization of Thai swine influenza viruses after the introduction of pandemic H1N1 2009. Virus Genes 47, 75–85. 10.1007/s11262-013-0927-x [DOI] [PubMed] [Google Scholar]
- Corzo CA, Culhane M, Juleen K, Stigger-Rosser E, Ducatez MF, Webby RJ, Lowe JF, 2013. Active surveillance for influenza a virus among swine, midwestern United States, 2009–2011. Emerg. Infect. Dis 19, 954–960. 10.3201/eid1906.121637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz A, Marthaler D, Culhane M, Sreevatsan S, Alkhamis M, Torremorell M, 2017. Complete Genome Sequencing of Influenza A Viruses within Swine Farrow-to-Wean Farms Reveals the Emergence, Persistence, and Subsidence of Diverse Viral Genotypes. J. Virol 91, e00745–17. 10.1128/JVI.00745-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducatez MF, Hause B, Stigger-Rosser E, Darnell D, Corzo C, Juleen K, Simonson R, Brockwell-Staats C, Rubrum A, Wang D, Webb A, Crumpton JC, Lowe J, Gramer M, Webby RJ, 2011. Multiple reassortment between pandemic (H1N1) 2009 and endemic influenza viruses in pigs, United States. Emerg. Infect. Dis 17, 1624–1629. 10.3201/1709.110338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essere B, Yver M, Gavazzi C, Terrier O, Isel C, Fournier E, Giroux F, Textoris J, Julien T, Socratous C, Rosa-Calatrava M, Lina B, Marquet R, Moules V, 2013. Critical role of segment-specific packaging signals in genetic reassortment of influenza A viruses. Proc. Natl. Acad. Sci 110, E3840–E3848. 10.1073/pnas.1308649110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao S, Anderson TK, Walia RR, Dorman KS, Janas-Martindale A, Vincent AL, 2017. The genomic evolution of H1 influenza a viruses from swine detected in the United States between 2009 and 2016. J. Gen. Virol 98, 2001–2010. 10.1099/jgv.0.000885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenbaum A, Quinn C, Bailer J, Su S, Havers F, Durand LO, Jiang V, Page S, Budd J, Shaw M, Biggerstaff M, De Fijter S, Smith K, Reed C, Epperson S, Brammer L, Feltz D, Sohner K, Ford J, Jain S, Gargiullo P, Weiss E, Burg P, DiOrio M, Fowler B, Finelli L, Jhung MA, 2015. Investigation of an Outbreak of Variant Influenza A(H3N2) Virus Infection Associated with an Agricultural Fair - Ohio, August 2012. J. Infect. Dis 212, 1592–1599. 10.1093/infdis/jiv269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He P, Wang G, Mo Y, Yu Q, Xiao X, Yang W, Zhao W, Guo X, Chen Q, He J, Liang M, Zhu J, Ding Y, Wei Z, Ouyang K, Liu F, Jian H, Huang W, García-Sastre A, Chen Y, 2018. Novel triple-reassortant influenza viruses in pigs, Guangxi, China. Emerg. Microbes Infect 7(1), 85 10.1038/s41426-018-0088-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann E, Stech J, Guan Y, Webster RG, Perez DR, 2001. Universal primer set for the full-length amplification of all influenza A viruses. Arch. Virol 146, 2275–2289. 10.1007/s007050170002 [DOI] [PubMed] [Google Scholar]
- Howard WA, Essen SC, Strugnell BW, Russell C, Barass L, Reid SM, Brown IH, 2011. Reassortant Pandemic (H1N1) 2009 virus in pigs, United Kingdom. Emerg. Infect. Dis 17, 1049–1052. 10.3201/eid1706.101886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T, Couceiro JN, Kelm S, Baum LG, Krauss S, Castrucci MR, Donatelli I, Kida H, Paulson JC, Webster RG, Kawaoka Y, 1998. Molecular basis for the generation in pigs of influenza A viruses with pandemic potential. J. Virol 72, 7367–7373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jhung MA, Epperson S, Biggerstaff M, Allen D, Balish A, Barnes N, Beaudoin A, Berman LS, Bidol S, Blanton L, Blythe D, Brammer L, D’mello T, Danila R, Davis W, De Fijter S, Diorio M, Durand LO, Emery S, Fowler B, Garten R, Grant Y, Greenbaum A, Gubareva L, Havers F, Haupt T, House J, Ibrahim S, Jiang V, Jain S, Jernigan D, Kazmierczak J, Klimov A, Lindstrom S, Longenberger A, Lucas P, Lynfield R, Mcmorrow M, Moll M, Morin C, Ostroff S, Page SL, Park SY, Peters S, Quinn C, Reed C, Richards S, Scheftel J, Simwale O, Shu B, Soyemi K, Stauffer J, Steffens C, Su S, Torso L, Uyeki TM, Vetter S, Villanueva J, Wong KK, Shaw M, Bresee JS, Cox N, Finelli L, 2013. Outbreak of variant influenza A(H3N2) virus in the United States. Clin. Infect. Dis 57, 1703–1712. 10.1093/cid/cit649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitikoon P, Sreta D, Nuntawan Na Ayudhya S, Wongphatcharachai M, Lapkuntod J, Prakairungnamthip D, Bunpapong N, Suradhat S, Thanawongnuwech R, Amonsin A, 2011. Brief report: Molecular characterization of a novel reassorted pandemic H1N1 2009 in Thai pigs. Virus Genes 43, 1–5. 10.1007/s11262-011-0597-5 [DOI] [PubMed] [Google Scholar]
- Kitikoon P, Vincent AL, Gauger PC, Schlink SN, Bayles DO, Gramer MR, Darnell D, Webby RJ, Lager KM, Swenson SL, Klimov A, 2012. Pathogenicity and Transmission in Pigs of the Novel A(H3N2)v Influenza Virus Isolated from Humans and Characterization of Swine H3N2 Viruses Isolated in 2010–2011. J. Virol 86, 6804–6814. 10.1128/JVI.00197-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong W, Wang F, Dong B, Ou C, Meng D, Liu J, Fan ZC, 2015. Novel reassortant influenza viruses between pandemic (H1N1) 2009 and other influenza viruses pose a risk to public health. Microb. Pathog 10.1016/j.micpath.2015.09.002 [DOI] [PubMed] [Google Scholar]
- Liang H, Lam TT-Y, Fan X, Chen X, Zeng Y, Zhou J, Duan L, Tse M, Chan C-H, Li L, Leung T-Y, Yip C-H, Cheung C-L, Zhou B, Smith DK, Poon LL-M, Peiris M, Guan Y, Zhu H, 2014. Expansion of Genotypic Diversity and Establishment of 2009 H1N1 Pandemic-Origin Internal Genes in Pigs in China. J. Virol 88, 10864–10874. 10.1128/JVI.01327-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindstrom S, Garten R, Balish A, Shu B, Emery S, Berman L, Barnes N, Sleeman K, Gubareva L, Villanueva J, Klimov A, 2012. Human infections with novel reassortant influenza A(H3N2)v viruses, United States, 2011. Emerg. Infect. Dis 18, 834–837. 10.3201/eid1805.111922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Ma J, Liu H, Qi W, Anderson J, Henry SC, Hesse RA, Richt JA, Ma W, 2012. Emergence of novel reassortant H3N2 swine influenza viruses with the 2009 pandemic H1N1 genes in the United States. Arch. Virol 157, 555–562. 10.1007/s00705-011-1203-9 [DOI] [PubMed] [Google Scholar]
- Lorusso A, Vincent AL, Gramer MR, Lager KM, Ciacci-Zanella JR, 2013. Contemporary epidemiology of north american lineage triple reassortant influenza a viruses in pigs. Curr. Top. Microbiol. Immunol 370, 113–131. 10.1007/82-2011-196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J, Shen H, Liu Q, Bawa B, Qi W, Duff M, Lang Y, Lee J, Yu H, Bai J, Tong G, Hesse RA, Richt JA, Ma W, 2015. Pathogenicity and Transmissibility of Novel Reassortant H3N2 Influenza Viruses with 2009 Pandemic H1N1 Genes in Pigs. J. Virol 89, 2831–2841. 10.1128/JVI.03355-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma W, Lager KM, Lekcharoensuk P, Ulery ES, Janke BH, Solórzano A, Webby RJ, García-Sastre A, Richt JA, 2010a. Viral reassortment and transmission after co-infection of pigs with classical H1N1 and triple-reassortant H3N2 swine influenza viruses. J. Gen. Virol 91, 2314–2321. 10.1099/vir.0.021402-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma W, Oberst R, Li X, Clouser D, Hesse R, Rowland R, Richt JA, 2010b. Rapid detection of the pandemic 2009 H1N1 virus M gene by real-time and gel-based RT-PCR assays. Influenza Other Respi. Viruses 4, 397–403. 10.1111/j.1750-2659.2010.00180.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall N, Priyamvada L, Ende Z, Steel J, Lowen AC, 2013. Influenza Virus Reassortment Occurs with High Frequency in the Absence of Segment Mismatch. PLoS Pathog. 9 10.1371/journal.ppat.1003421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mena I, Nelson MI, Quezada-Monroy F, Dutta J, Cortes-Fernandez R, Lara-Puente JH, Castro-Peralta F, Cunha LF, Trovao NS, Lozano-Dubernard B, Rambaut A, van Bakel H, Garcia-Sastre A, 2016. Origins of the 2009 H1N1 influenza pandemic in swine in Mexico. Elife 5 2016 10.7554/elife.16777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno A, Di Trani L, Faccini S, Vaccari G, Nigrelli D, Boniotti MB, Falcone E, Boni A, Chiapponi C, Sozzi E, Cordioli P, 2011. Novel H1N2 swine influenza reassortant strain in pigs derived from the pandemic H1N1/2009 virus. Vet. Microbiol 149, 472–477. 10.1016/j.vetmic.2010.12.011 [DOI] [PubMed] [Google Scholar]
- Myers KP, Olsen CW, Gray GC, 2007. Cases of swine influenza in humans: a review of the literature. Clin. Infect. Dis 44, 1084–1088. 10.1086/512813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson MI, Gramer MR, Vincent AL, Holmes EC, 2012. Global transmission of influenza viruses from humans to swine. J. Gen. Virol 93, 2195–2203. 10.1099/vir.0.044974-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson MI, Stratton J, Killian ML, Janas-Martindale A, Vincent AL, 2015. Continual Reintroduction of Human Pandemic H1N1 Influenza A Viruses into Swine in the United States, 2009 to 2014. J. Virol 89, 6218–6226. 10.1128/jvi.00459-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson MI, Lemey P, Tan Y, Vincent A, LamTommy TTY, Detmer S, Viboud C, Suchard MA, Rambaut A, Holmes EC, Gramer M, 2011. Spatial dynamics of human-origin H1 influenza a virus in north american swine. PLoS Pathog. 7 10.1371/journal.ppat.1002077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson MI, Stucker KM, Schobel SA, Trovão NS, Das SR, Dugan VG, Nelson SW, Sreevatsan S, Killian ML, Nolting JM, Wentworth DE, Bowman AS, 2016. Introduction, Evolution, and Dissemination of Influenza A Viruses in Exhibition Swine in the United States during 2009 to 2013. J. Virol 90, 10963–10971. 10.1128/JVI.01457-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson MI, Wentworth DE, Culhane MR, Vincent AL, Viboud C, LaPointe MP, Lin X, Holmes EC, Detmer SE, 2014. Introductions and Evolution of Human-Origin Seasonal Influenza A Viruses in Multinational Swine Populations. J. Virol 88, 10110–10119. 10.1128/JVI.01080-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasma T, Joseph T, 2010. Pandemic (H1N1) 2009 infection in swine herds, Manitoba, Canada. Emerg. Infect. Dis 16, 706–708. 10.3201/eid1604.091636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce MB, Jayaraman A, Pappas C, Belser JA, Zeng H, Gustin KM, Maines TR, Sun X, Raman R, Cox NJ, Sasisekharan R, Katz JM, and Tumpey TM, 2012. Pathogenesis and transmission of swine origin A(H3N2)v influenza viruses in ferrets. Proc. Natl. Acad. Sci 109, 3944–3949. 10.1073/pnas.1119945109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulit-Penaloza JA, Pappas C, Belser JA, Sun X, Brock N, Zeng H, Tumpey TM, Maines TR, 2018. Comparative In Vitro and In Vivo Analysis of H1N1 and H1N2 Variant Influenza Viruses Isolated from Humans between 2011 and 2016. J. Virol 92 10.1128/JVI.01444-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajao DS, Anderson TK, Kitikoon P, Stratton J, Lewis NS, Vincent AL, 2018. Antigenic and genetic evolution of contemporary swine H1 influenza viruses in the United States. Virology 518, 45–54. 10.1016/j.virol.2018.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resende PC, Born PS, Matos AR, Motta FC, Caetano BC, Debur M. do C., Riediger IN, Brown D, Siqueira MM, 2017. Whole-genome characterization of a novel human influenza A(H1N2) virus variant, Brazil. Emerg. Infect. Dis 10.3201/eid2301.161122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richt JA, Janke BH, Webster RG, Lager KM, Webby RJ, Woods RD, 2003. Pathogenic and Antigenic Properties of Phylogenetically Distinct Reassortant H3N2 Swine Influenza Viruses Cocirculating in the United States. J. Clin. Microbiol 41, 3198–3205. 10.1128/jcm.41.7.3198-3205.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomon R, Webster RG, 2009. The Influenza Virus Enigma. Cell. 10.1016/j.cell.2009.01.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer R, Rech RR, Gava D, Cantão ME, da Silva MC, Silveira S, Zanella JRC, 2015. A human-like H1N2 influenza virus detected during an outbreak of acute respiratory disease in swine in Brazil. Arch. Virol 160, 29–38. 10.1007/s00705-014-2223-z [DOI] [PubMed] [Google Scholar]
- Shinde V, Bridges CB, Uyeki TM, Shu B, Balish A, Xu X, Lindstrom S, Gubareva LV, Deyde V, Garten RJ, Harris M, Gerber S, Vagasky S, Smith F, Pascoe N, Martin K, Dufficy D, Ritger K, Conover C, Quinlisk P, Klimov A, Bresee JS, Finelli L, 2009. Triple-Reassortant Swine Influenza A (H1) in Humans in the United States, 2005–2009. N. Engl. J. Med 360, 2616–2625. 10.1056/NEJMoa0903812 [DOI] [PubMed] [Google Scholar]
- Song MS, Lee JH, Pascua PN, Baek YH, Kwon H. il, Park KJ, Choi HW, Shin YK, Song JY, Kim CJ, Choi YK, 2010. Evidence of Human-to-Swine Transmission of the Pandemic (H1N1) 2009 Influenza Virus in South Korea. J. Clin. Microbiol 48, 3204–3211. 10.1128/JCM.00053-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starick E, Lange E, Fereidouni S, Bunzenthal C, Höveler R, Kuczka A, Beilage EG, Hamann HP, Klingelhöfer I, Steinhauer D, Vahlenkamp T, Beerspi-Sup M, Harder T, 2011. Reassorted pandemic (H1N1) 2009 influenza a virus discovered from pigs in germany. J. Gen. Virol 92, 1184–1188. 10.1099/vir.0.028662-0 [DOI] [PubMed] [Google Scholar]
- Sun X, Pulit-Penaloza JA, Belser JA, Pappas C, Pearce MB, Brock N, Zeng H, Creager HM, Zanders N, Jang Y, Tumpey TM, Davis CT, and Maines TR 2018. Pathogenesis and Transmission of Genetically Diverse Swine-Origin H3N2 Variant Influenza A Viruses from Multiple Lineages Isolated in the United States, 2011–2016. J. Virol 92 10.1128/jvi.00665-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijaykrishna D, Poon LLM, Zhu HC, Ma SK, Li OTW, Cheung CL, Smith GJD, Peiris JSM, Guan Y, 2010. Reassortment of pandemic H1N1/2009 influenza a virus in swine. Science (80-. ). 10.1126/science.1189132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent A, Awada L, Brown I, Chen H, Claes F, Dauphin G, Donis R, Culhane M, Hamilton K, Lewis N, Mumford E, Nguyen T, Parchariyanon S, Pasick J, Pavade G, Pereda A, Peiris M, Saito T, Swenson S, Van Reeth K, Webby R, Wong F, Ciacci-Zanella J, 2014. Review of Influenza A Virus in Swine Worldwide: A Call for Increased Surveillance and Research. Zoonoses Public Health. 10.1111/zph.12049 [DOI] [PubMed] [Google Scholar]
- Vincent AL, Ma W, Lager KM, Janke BH, Richt JA, 2008. Swine Influenza Viruses A North American Perspective. Adv. Virus Res 72: 127–154. 10.1016/S0065-3527(08)00403-X [DOI] [PubMed] [Google Scholar]
- Walia RR, Anderson TK, Vincent AL, 2018. Regional patterns of genetic diversity in swine influenza A viruses in the United States from 2010 to 2016. Influenza Other Respi. Viruses 06 April 2. 10.1111/irv.12559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webby RJ, Swenson SL, Krauss SL, Gerrish PJ, Goyal SM, Webster RG, 2000. Evolution of swine H3N2 influenza viruses in the United States. J. Virol 74, 8243–8251. 10.1128/JVI.74.18.8243-8251.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong KK, Greenbaum A, Moll ME, Lando J, Moore EL, Ganatra R, Biggerstaff M, Lam E, Smith EE, Storms AD, Miller JR, Dato V, Nalluswami K, Nambiar A, Silvestri SA, Lute JR, Ostroff S, Hancock K, Branch A, Trock SC, Klimov A, Shu B, Brammer L, Epperson S, Finelli L, Jhung MA, 2012. Outbreak of influenza A (H3N2) variant virus infection among attendees of an agricultural fair, Pennsylvania, USA, 2011. Emerg. Infect. Dis 18, 1937–1944. 10.3201/eid1812.121097 [DOI] [PMC free article] [PubMed] [Google Scholar]