Abstract
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) poses a huge threat to public health. Viral nucleic acid testing is the diagnostic gold standard and can play an important role in the prevention and control of this infection. In this study, bacteriophage MS2 virus-like particles encapsulating specific RNA sequences of SARS-CoV-2 and other coronaviruses were prepared by genetic engineering. The assessment panel, consisting of four positive samples with concentrations of 2.8, 3.5, 4.2, and 4.9 log10 copies/mL and five negative samples with other human coronaviruses, was prepared and distributed to evaluate the accuracy of routine viral RNA detection. Results of 931 panels from 844 laboratories were collected. The overall percentage agreement, positive percentage agreement (PPA), and negative percentage agreement, defined as the percentage of agreement between the correct results and total results submitted for all, positive, and negative samples were 96.8% (8109/8379), 93.9% (3497/3724), and 99.1% (4612/4655), respectively. For samples with concentrations of 4.9 and 4.2 log10 copies/mL, the PPAs were >95%. However, for 3.5 and 2.8 log10 copies/mL, the PPAs were 94.6% (881/931) and 84.9% (790/931), respectively. For all negative samples, the negative percentage agreement values were >95%. Thus, most laboratories can reliably detect SARS-CoV-2. However, further improvement and optimization are required to ensure the accuracy of detection in panel members with lower concentrations of viral RNA.
In December 2019, patients exhibited unexplained pneumonia in Wuhan, Hubei Province, China.1 After virus isolation and genome sequencing were performed, a new coronavirus was discovered and named severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2).2 The resultant disease was named coronavirus disease 2019 [COVID-19; World Health Organization (WHO), https://www.who.int/emergencies/diseases/novel-coronavirus-2019/technical-guidance/naming-the-coronavirus-disease-(covid-2019)-and-the-virus-that-causes-it, last accessed October 15, 2020].3 SARS-CoV-2 is an enveloped, positive-sense, single-stranded RNA virus, and its genome is composed of two flanking untranslated regions and a long open reading frame (ORF). The long ORF consists of a replicase gene and structural genes, including the spike (S), envelope (E), membrane (M), and nucleocapsid (N) genes.4 Epidemiologic studies have shown that SARS-CoV-2 has strong human-to-human transmission capability,5 and common symptoms of COVID-19 include fever, dry cough, fatigue, and dyspnea, whereas severe infection leads to respiratory failure and even death.2 To date, SARS-CoV-2 has resulted in >14,300,000 confirmed cases and has caused 600,000 deaths throughout the world (WHO, https://www.who.int/emergencies/diseases/novel-coronavirus-2019, last accessed July 21, 2020); thus, early diagnosis, timely isolation, and therapy for infected individuals are critical to prevent the spread of the pandemic.
Nucleic acid detection is the gold standard for the diagnosis of SARS-CoV-2 infection, and the most commonly used method for this detection is real-time RT-PCR. Since the outbreak of COVID-19, the WHO, the China Centers for Disease Control and Prevention, and the US Food and Drug Administration (FDA) have issued several testing protocols for molecular diagnosis of SARS-CoV-2 infection and suggested the selection of target genes for nucleic acid detection (WHO, https://www.who.int/publications-detail/laboratory-testing-for-2019-novel-coronavirus-in-suspected-human-cases-20200117, last accessed April 27, 2020; and US Food and Drug Administration, https://www.fda.gov/medical-devices/coronavirus-disease-2019-covid-19-emergency-use-authorizations-medical-devices/vitro-diagnostics-euas, last accessed July 21, 2020). Generally, the conserved E gene serves as the first-line screening tool, and the specific ORF1ab, N, and S genes are used for confirmation (WHO, https://www.who.int/docs/default-source/coronaviruse/protocol-v2-1.pdf, last accessed January 17, 2020).6 On the basis of these recommendations, a series of assays have been approved by the US FDA Emergency Use Authorization and China's National Medical Products Administration (NMPA), and these assays have been used in response to COVID-19 infection emergencies. The assays vary in their composition of reagents and specific targets involved, and the analytical performance claimed in the corresponding instructions by manufacturers has not been thoroughly validated. In clinical applications, more factors, such as differences in nucleic acid extraction methods, real-time RT-PCR processes, interpretation of results, personnel, and equipment, lead to variations in testing results among different laboratories. Therefore, large-scale quality evaluation of SARS-CoV-2 nucleic acid testing is urgently needed to help laboratories conduct quality management and improvements. At present, SARS-CoV-2 proficiency test surveys have been undertaken by the College of American Pathologists, the Society for Promoting Quality Assurance in Medical Laboratories, and Quality Control for Molecular Diagnostics, including laboratories in the United States and European Union/European Economic Area countries (College of American Pathologists, https://www.cap.org/laboratory-improvement/proficiency-testing/sars-cov-2-proficiency-testing-programs, last accessed July 31, 2020; Society for Promoting Quality Assurance in Medical Laboratories, https://www.instand-ev.de/fileadmin/uploads/Begleithefte/Coronavirus_EN.pdf, last accessed July 31, 2020; and Quality Control for Molecular Diagnostics, http://www.qcmd.org/index.php?pageId=49&pageVersion=EN, last accessed July 31, 2020).
In this study, an external quality assessment (EQA) was conducted to evaluate the performance and status of SARS-CoV-2 nucleic acid detection. The analytical sensitivity and specificity of each assay were evaluated, as well as the ability to detect positive and negative samples in clinical practice. Moreover, the possible causes of these false-negative and false-positive results were assessed. Finally, several practical suggestions were provided to improve laboratory capabilities and increase assay reliability.
Materials and Methods
Preparation of Simulated Samples
Armored RNA enveloping the specific sequence of SARS-CoV-2 was used as a positive sample. A preliminary survey examining the target sequences of the NMPA-approved assays (as of March 26, 2020) and laboratory-developed tests (LDTs) was conducted when clinical laboratories were enrolled in EQA to ensure that the virus-like particles (VLPs) were suitable for all of the assays used in participating laboratories. ORF1ab, N, and E genes were selected on the basis of the assays used by participants, including the sequences recommended by the WHO and the Centers for Disease Control and Prevention of China (WHO, https://www.who.int/docs/default-source/coronaviruse/protocol-v2-1.pdf, last accessed January 17, 2020; and WHO, https://www.who.int/docs/default-source/coronaviruse/whoinhouseassays.pdf, last accessed November 23, 2020), as well as some specific regions in the three genes. None of these assays targeted sequences in the S gene. Thus, the RNA sequence enveloped was 2908 bp in length, consisting of a part of the ORF1ab, N, and E genes, including all of the target sequences selected by the assays. Target segments of nucleotide sequences from the SARS-CoV-2 genome [BetaCoV/Wuhan/IVDC-HB-01/2019|EPI_ISL_402119 (Global Initiative on Sharing All Influenza Data, http://platform.gisaid.org/epi3/frontend, registration required, last accessed January 17, 2020)] were synthesized by Sangon Biotech Co, Ltd (Shanghai, China). A specific 19-bp stem-loop operator sequence was added at both the 5′ and 3′ ends. Two ends of the synthetic sequence were flanked by KpnI and PacI restriction digestion sites, respectively, such that after enzymatic digestion and gel purification, the synthetic sequence was subcloned into the KpnI/PacI site of the pACYC-MS2 vector.7 , 8 Expression and purification of MS2 VLPs were performed according to previously published protocols in the study laboratory.7, 8, 9 In addition, VLPs containing sequences corresponding to the same ORF1ab, N, and E gene target regions of SARS-CoV-2 were constructed on the basis of the sequences of six other coronaviruses capable of infecting humans and were prepared by the same method. Genome sequences of other coronaviruses were downloaded from GenBank [available from https://www.ncbi.nlm.nih.gov/genbank, accession numbers SARS-coronavirus (CoV), NC_004718; NL63-CoV, JX504050.1; 229E-CoV, NC_002645; HKU1-CoV, NC_006577.2; OC43-CoV, NC_006213.1; Middle East respiratory syndrome-CoV, NC_019843].
MS2 VLP Concentration Determination
First, the VLPs were diluted 10,000-fold with phosphate-buffered saline (PBS) solution, and 140 μL of the dilution was subsequently used for RNA extraction with the QIAamp Viral RNA Mini Kit (Qiagen, Hilden, Germany). Universal primers and 2 μL of extracted RNA were used to synthesize cDNA using the Qiagen OneStep RT-PCR Kit. According to the manufacturer's instructions, 1 μL of cDNA was needed to prepare a digital PCR system, and subsequently, the reaction was performed with ddPCR Supermix for Probes (Bio-Rad, Hercules, CA) on a Bio-Rad QX200 instrument. The primers and probes used are shown in Table 1 . The results were analyzed using the supporting QuantaLife software version 1.7.4 (Bio-Rad) by providing the copies of cDNA in the digital PCR, which can be used to deduce the initial VLP concentration.
Table 1.
Primers and Probes for Digital PCR
| VLPs | Primers and probes | Amplicon size, bp | |
|---|---|---|---|
| SARS-CoV-2 | Forward primer | 5′-CACATTGGCACCCGCAATC-3′ | 128 |
| Reverse primer | 5′-GAGGAACGAGAAGAGGCTTG-3′ | ||
| Probe | 5’-[FAM]-ACTTCCTCAAGGAACAACATTGCCA-[BHQ1]-3′ | ||
| SARS-CoV | Forward primer | 5′-TACGTCCAAATACCTACCACT-3′ | 181 |
| Reverse primer | 5′-GCTGCACTTACACCGCAAA-3′ | ||
| Probe | 5’-[FAM]-TTGTGACCAACTCCGCGAAC-[BHQ1]-3′ | ||
| 229E-CoV | Forward primer | 5′-ATGAAGCTGATTACCGTTG-3′ | 125 |
| Reverse primer | 5′-AGCCTGTAATGTAGAACCA-3′ | ||
| Probe | 5’-[FAM]-CTTGTACACACCTCCGACTG-[BHQ1]-3′ | ||
| HKU1-CoV | Forward primer | 5′-CTGAGGCTACTATTAACCAAG-3′ | 148 |
| Reverse primer | 5′-AGAATAGGATCTTTTATACCCAA-3′ | ||
| Probe | 5’-[FAM]-CCTCAGTTTGTATTTATTGCCGTG-[BHQ1]-3′ | ||
| OC43-CoV | Forward primer | 5′-AAGCCAGTTACTTTGCCTA-3′ | 245 |
| Reverse primer | 5′-CCAAAATATGTAGCGACACT-3′ | ||
| Probe | 5’-[FAM]-AATAGTACCAGCAGGCAACCA-[BHQ1]-3′ | ||
| NL63-CoV | Forward primer | 5′-TGTCATAGCACCATCAACC-3′ | 150 |
| Reverse primer | 5′-GAAACCTTAATACGCAACAC-3′ | ||
| Probe | 5’-[FAM]-CATTGTAACACCGACAACTCCC-[BHQ1]-3′ | ||
| MERS-CoV | Forward primer | 5′-CTCGCTGTATAATGTCACAC-3′ | 89 |
| Reverse primer | 5′-CACTATGGAAGTTATCGCCTC-3′ | ||
| Probe | 5’-[FAM]-CTTACGCCATCACGAGCAGT-[BHQ1]-3′ | ||
BHQ1, Black Hole Quencher 1; CoV, coronavirus; FAM, carboxyfluorescein; MERS, Middle East respiratory syndrome; SARS, severe acute respiratory syndrome; VLP, virus-like particle.
Organization of EQA
A coded panel consisting of four positive samples and five negative samples was prepared. Detailed information about the coding panel is shown in Table 2 . The SARS-CoV-2 VLPs were appropriately diluted with PBS to obtain the sample with the highest concentration (4.9 log10 copies/mL), which was similar to the actual viral load in clinical samples,10 , 11 and subsequently, this sample was serially diluted fivefold to prepare positive samples with other concentrations. PBS was used as a negative sample or dilution buffer to mimic the properties of respiratory secretions. Similarly, VLP-encapsulated sequences of other coronaviruses were diluted as negative samples to test whether their presence could lead to erroneous test results. All samples were dispensed in 0.5-mL aliquots and stored at −20°C until distribution. Before delivering samples to the participants, the panels were tested using four commercial assays [Sansure Biotech Inc. (Sansure), Changsha, China; Daan Gene Co, Ltd, of Sun Yat-sen University (Daan), Guangzhou, China; BGI Bio-tech Co, Ltd (BGI), Wuhan, China; and BioGerm Medical Co, Ltd (BioGerm), Shanghai, China] to confirm whether each sample was positive or negative. Sample panels were distributed to each participating laboratory via dry ice delivery. The laboratories participating in the EQA were required to assay the panels using routine molecular procedures and report the results. Nucleic acid extraction and detection methods, instrument and reagent details, manufacturers, raw detection data, and qualitative results were all required to be submitted.
Table 2.
Composition and the Overall Results of the External Quality Assessment Panel
| Classification | Sample no. | Contents | Concentration, log10 copies/mL | Correct results, N | Total results, N | Agreement, % |
|---|---|---|---|---|---|---|
| Positive | 2020-B | SARS-CoV-2 | 2.8 | 790 | 931 | 84.9 |
| 2020-C | SARS-CoV-2 | 3.5 | 881 | 931 | 94.6 | |
| 2020-H | SARS-CoV-2 | 4.2 | 920 | 931 | 98.8 | |
| 2020-I | SARS-CoV-2 | 4.9 | 906 | 931 | 97.3 | |
| Overall | 3497 | 3724 | 93.9 | |||
| Negative | 2020-A | SARS | 6.5 | 913 | 931 | 98.1 |
| 2020-D | PBS | 926 | 931 | 99.5 | ||
| 2020-E | NL63 | 6.2 | 928 | 931 | 99.7 | |
| 2020-F | HKU1 | 6.0 | 925 | 931 | 99.4 | |
| 229E | 6.4 | |||||
| 2020-G | MERS | 4.0 | 920 | 931 | 98.8 | |
| OC43 | 6.0 | |||||
| Overall | 4612 | 4655 | 99.1 | |||
| Total | 8109 | 8379 | 96.8 |
MERS, Middle East respiratory syndrome; PBS, phosphate-buffered saline; SARS, severe acute respiratory syndrome; SARS-CoV-2, SARS coronavirus 2.
Statistical Analysis
Because of the current critical nature of accurately detecting SARS-CoV-2 during the pandemic, all of the samples were required to be detected correctly in the EQA. The results were classified into two categories as follows: competent (100% correct responses) or improvable (≥1 incorrect result). All data were analyzed using Excel (Microsoft, Redmond, WA) and SPSS 19.0 (IBM, New York, NY), and the variances among different groups were compared using the Pearson χ2 test or the Fisher exact test. P < 0.05 was considered significant.
Results
Evaluation of the Sample Panel
The armored RNA digested by DNase I and RNase A was subjected to 1% agarose gel electrophoresis, and the single band that was observed indicated the construction of stable simulated samples (Supplemental Figure S1). Real-time RT-PCR was performed on nucleic acids extracted from VLPs, and the sequencing results of amplified products provided further evidence of the successful packaging of target segments (data not shown). The concentrations of all types of VLPs were not <10.0 log10 copies/mL. Each sample of the panel was assessed using Sansure, Daan, BGI, and BioGerm assays before distribution. All of the samples containing SARS-CoV-2 VLPs were detected to be positive by the four assays, whereas other samples containing PBS and other coronaviruses were all negative.
Participants and Methods
In total, 931 results were submitted from 844 participants because 87 laboratories returned data generated by two different assays. Of all returns, 97.5% (908/931) were based on real-time RT-PCR, whereas reverse transcription–loop-mediated isothermal amplification, high-throughput sequencing, time-of-flight mass spectrometry, and microarray analysis accounted for 0.54% (5/931), 1.1% (10/931), 0.32% (3/931), and 0.54% (5/931), respectively. Various nucleic acid extraction methods were applied by the laboratories. Among these methods, manual column-based, manual magnetic bead-based, automated column-based, and automated magnetic bead-based methods accounted for 21.3% (198/931), 15.3% (142/931), 1.5% (14/931), and 51.7% (481/931), respectively. In addition, 8.8% (82/931) of laboratories used rapid lysis extraction, which destroys the virus directly and releases nucleic acids without isolation and purification. Only 36.6% (341/931) of the laboratories applied unmodified extraction reagents, same with those in the manufacturers' instructions for use (IFU), whereas other laboratories used modified extraction reagents, which were different from those in the manufacturers’ IFU.
Percentage Agreements of Overall Results
Of the 931 completed data sets, the performances were found to be competent in 748 (80.3%) analyses. The accuracy of the laboratory results was evaluated by calculating the overall percentage agreement, positive percentage agreement (PPA), and negative percentage agreement (NPA), which are defined herein as the percentage of agreement between the intended correct results and total results submitted for all, positive, and negative samples, respectively. The overall percentage agreement, PPA, and NPA were 96.8% (8109/8379), 93.9% (3497/3724), and 99.1% (4612/4655), respectively (Table 2). The PPAs were >97% for samples with concentrations of 4.9 and 4.2 log10 copies/mL but were 94.6% (881/931) for 3.5 log10 copies/mL and 84.9% (790/931) for 2.8 log10 copies/mL. The NPAs of PBS, NL63, and HKU1/229E were all >99%, whereas they were 98.1% (913/931) for SARS and 98.8% (920/931) for Middle East respiratory syndrome/OC43 (Table 2).
Percentage Agreements of the Results for Positive Samples
Detection Method
The laboratories participating in this EQA used a total of five detection methods, and the PPAs of each method are shown in Table 3 . Real-time RT-PCR showed PPAs >95% for samples with 4.9, 4.2, and 3.5 log10 copies/mL and 85.1% (773/908) for 2.8 log10 copies/mL. Reverse transcription–loop-mediated isothermal amplification, high-throughput sequencing, time-of-flight mass spectrometry, and microarray techniques all demonstrated high PPAs for samples with 4.9 and 4.2 log10 copies/mL. However, for 3.5 and 2.8 log10 copies/mL, the PPAs varied considerably.
Table 3.
The Results of Four Positive Samples Submitted by the Laboratories Using Different Detection Methods
| Method | Data sets, N | Agreement, % (n/total) |
|||
|---|---|---|---|---|---|
| 2020-I (4.9 log10 copies/mL) | 2020-H (4.2 log10 copies/mL) | 2020-C (3.5 log10 copies/mL) | 2020-B (2.8 log10 copies/mL) | ||
| Real-time RT-PCR | 908 | 97.4 (884/908) | 98.8 (897/908) | 95.2 (864/908) | 85.1 (773/908) |
| RT-LAMP | 5 | 100.0 (5/5) | 100.0 (5/5) | 100.0 (5/5) | 60.0 (3/5) |
| High-throughput sequencing | 10 | 90.0 (9/10) | 100.0 (10/10) | 80.0 (8/10) | 90.0 (9/10) |
| Time-of-flight mass spectrometry | 3 | 100.0 (3/3) | 100.0 (3/3) | 100.0 (3/3) | 100.0 (3/3) |
| Mircoarray | 5 | 100.0 (5/5) | 100.0 (5/5) | 20.0 (1/5) | 40.0 (2/5) |
RT-LAMP, reverse transcription–loop-mediated isothermal amplification.
Nucleic Acid Extraction
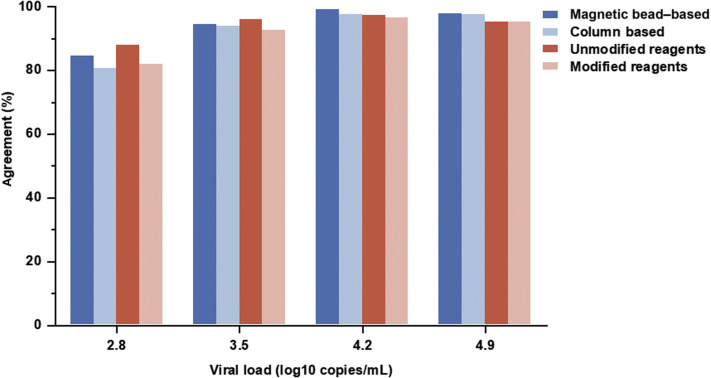
For samples with 4.9 and 4.2 log10 copies/mL, the PPAs of magnetic bead–based and column-based nucleic acid extraction methods were >95%; however, for 3.5 and 2.8 log10 copies/mL, the rates of both decreased (Figure 1 ). For each positive sample, the percentage agreement of the laboratories using magnetic bead–based extraction method was higher than that of the laboratories using column-based extraction method (Figure 1). The overall PPA of the laboratories using unmodified extraction reagents in the manufacturers’ IFU (94.1%, 1284/1364) was higher than that of the laboratories using modified extraction reagents (91.7%, 2163/2360), and the situation was similar for positive samples at each concentration (Figure 1).
Figure 1.
The results of four positive samples submitted by the laboratories using different nucleic acid extraction methods and extraction reagents.
Detection Assays
Among the 931 received data sets, 823 employed nine assays approved by the NMPA and the others applied LDT assays (Supplemental Tables S1 and S2). The PPA of NMPA-approved assays (94.3%, 3106/3292) was higher than that of LDTs (90.5%, 391/432), which revealed a statistical difference in performance between them (P = 0.002). The NMPA-approved assays employed by >20 laboratories were analyzed. For samples with 4.9 and 4.2 log10 copies/mL, the PPAs of the six assays were all >95%. However, for 3.5 and 2.8 log10 copies/mL, the rates were variable (Table 4 ). Sansure and BGI assays produced PPAs >95% for both samples, and Daan obtained 97.3% (218/224) only in detecting samples with 3.5 log10 copies/mL, whereas the remaining assays all displayed rates of <95%. Assays manufactured by XABT Bio-tech Co, Ltd (XABT; Beijing, China), BioGerm, and ZJ Bio-tech Co, Ltd (ZJ; Shanghai, China) only demonstrated percentage agreements of 75% (15/20), 76.9% (80/104), and 74.1% (123/166), respectively, in detecting the sample with 2.8 log10 copies/mL (Table 4). Two laboratories failed to identify any positive samples. Both of them used the modified extraction reagents and employed the Daan kit and the BioGerm kit for detection, respectively. Both laboratories used plasmids as positive controls and detected positive controls with Ct values of approximately 29 and <24, respectively.
Table 4.
The Results of Four Positive Samples Submitted by the Laboratories Using the Six Most Frequently Used NMPA Assays
| Assays | 2020-I (4.9 log10 copies/mL) |
2020-H (4.2 log10 copies/mL) |
2020-C (3.5 log10 copies/mL) |
2020-B (2.8 log10 copies/mL) |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PPA, % (n/total) | Average Ct value | CV, % | PPA, % (n/total) | Average Ct value | CV, % | PPA, % (n/total) | Average Ct value | CV, % | PPA, % (n/total) | Average Ct value | CV, % | ||
| BGI | Overall result | 97.1 (33/34) | NA | NA | 100.0 (34/34) | NA | NA | 100.0 (34/34) | NA | NA | 97.1 (33/34) | NA | NA |
| ORF1ab | 97.1 (33/34) | 31 | 4.8 | 100.0 (34/34) | 33 | 4.8 | 100.0 (34/34) | 34.6 | 5.2 | 97.1 (33/34) | 36.2 | 6.1 | |
| Sansure | Overall result | 97.3 (251/258) | NA | NA | 100.0 (258/258) | NA | NA | 100.0 (258/258) | NA | NA | 96.9 (250/258) | NA | NA |
| ORF1ab | 81.4 (210/258) | 34.8 | 9.1 | 78.3 (202/258) | 36 | 8.9 | 57.0 (147/258) | 36.9 | 6 | 43.8 (113/258) | 37.4 | 5.3 | |
| N gene | 96.5 (249/258) | 32.2 | 9.9 | 99.2 (256/258) | 33.2 | 9.6 | 98.8 (255/258) | 35.5 | 6.5 | 96.1 (248/258) | 36.4 | 6.6 | |
| Daan | Overall result | 96.4 (216/224) | NA | NA | 99.1 (222/224) | NA | NA | 97.3 (218/224) | NA | NA | 86.6 (194/224) | NA | NA |
| ORF1ab | 93.8 (210/224) | 35.2 | 6.8 | 93.3 (209/224) | 36.6 | 5.7 | 77.2 (173/224) | 38.1 | 4.2 | 46.0 (103/224) | 38.2 | 4.4 | |
| N gene | 93.8 (210/224) | 33.8 | 6.2 | 96.9 (217/224) | 35 | 10.2 | 93.3 (209/224) | 36.9 | 5.1 | 73.2 (164/224) | 37.6 | 4.8 | |
| BioGerm | Overall result | 97.1 (101/104) | NA | NA | 97.1 (101/104) | NA | NA | 88.5 (92/104) | NA | NA | 76.9 (80/104) | NA | NA |
| ORF1ab | 90.4 (94/104) | 34.1 | 4.7 | 79.8 (83/104) | 35.4 | 6.8 | 49.0 (51/104) | 36 | 3.9 | 29.8 (31/104) | 36.2 | 4.7 | |
| N gene | 97.1 (101/104) | 32.7 | 5.2 | 99.0 (103/104) | 34.3 | 4.1 | 91.4 (95/104) | 35.8 | 3.6 | 81.7 (85/104) | 36.4 | 3.3 | |
| ZJ | Overall result | 98.2 (163/166) | NA | NA | 98.2 (163/166) | NA | NA | 92.8 (154/166) | NA | NA | 74.1 (123/166) | NA | NA |
| ORF1ab | 98.2 (163/166) | 34.8 | 5.7 | 97.0 (161/166) | 36 | 4.4 | 88.6 (147/166) | 37 | 4.6 | 70.5 (117/166) | 37.5 | 4.8 | |
| N gene | 21.1 (35/166) | 36.4 | 5.8 | 22.9 (38/166) | 37.7 | 5.8 | 18.1 (30/166) | 38.6 | 5 | 11.4 (19/166) | 38.5 | 6.5 | |
| E gene | 95.8 (159/166) | 33.4 | 6.6 | 95.8 (159/166) | 34.6 | 5.5 | 86.1 (143/166) | 35.7 | 6.2 | 49.4 (82/166) | 35.9 | 6.7 | |
| XABT | Overall result | 100.0 (20/20) | NA | NA | 100.0 (20/20) | NA | NA | 90.0 (18/20) | NA | NA | 75.0 (15/20) | NA | NA |
| ORF1ab | 100 (20/20) | 35.8 | 7.2 | 65.0 (13/20) | 36.4 | 4.4 | 35.0 (7/20) | 35.8 | 4.7 | 20.0 (4/20) | 37.6 | 5.3 | |
| N gene | 100 (20/20) | 32.1 | 9.3 | 100 (20/20) | 34.1 | 6.7 | 85.0 (17/20) | 35 | 6.3 | 75.0 (15/20) | 35.9 | 7 | |
| E gene | 60.0 (12/20) | 33.2 | 3.9 | 65.0 (13/20) | 34 | 8.2 | 66.7 (8/20) | 36.2 | 3.3 | 45.0 (9/20) | 36.4 | 5.8 | |
BGI, BGI Bio-tech Co, Ltd, Wuhan, China; BioGerm, BioGerm Medical Co, Ltd, Shanghai, China; CV, CV for Ct values of positive results in each target gene; Daan, Daan Gene Co, Ltd, of Sun Yat-sen University, Guangzhou, China; NA, not applicable; NMPA, National Medical Products Administration; PPA, positive percentage agreement; Sansure, Sansure Biotech Inc., Changsha, China; XABT, XABT Bio-tech Co, Ltd, Beijing, China; ZJ, ZJ Bio-tech Co, Ltd, Shanghai, China.
The claimed limits of detection (LODs) of BGI, XABT, Sansure, Daan, BioGerm, and ZJ were 100, 200, 200, 500, 1000, and 1000 copies/mL (2.0, 2.3, 2.3, 2.7, 3, and 3 log10 copies/mL), respectively. In general, assays with LODs ≤200 copies/mL performed better than those with LODs >200 copies/mL. However, there were also several exceptions. For example, XABT showed a PPA of 90.0% (18/20) in detecting the sample with 3.5 log10 copies/mL, which was lower than that of Daan (97.3%, 218/224) and Sansure (100%, 258/258). Because the claimed LODs of these six assays are different, ranging from 2.0 to 3.0 log10 copies/mL, the PPAs of the samples with the same multiples of their individual LODs were compared to evaluate the robustness of these assays, which is defined as the ability to proceed optimally despite slight variation in conditions, such as differences in extraction reagents and expertise of personnel. For samples with nucleic acid concentrations ≥6.4× the LOD, all assays could achieve percentage agreements of >95%, except XABT assays (Supplemental Table S3). However, for samples with nucleic acid concentrations equal to 3.2× the LOD, the percentage agreements of the Sansure, ZJ, BioGerm, and XABT assays differed greatly, ranging from 75.0% to 96.9% (Supplemental Table S3).
The six assays mentioned previously in this study were classified according to the number of detection targets, and the PPAs among them were calculated. In total, assays detecting single gene targets obtained a percentage agreement of 96.3% (131/136), whereas those with dual and triple targeted regions showed PPAs of 95.6% (2241/2344) and 90.9% (676/744), respectively. Regarding multitarget assays, the PPAs of different targeted genes also exhibited differences. For Sansure, in descending order of concentration, the PPAs were 96.5% (249/258), 99.2% (256/258), 98.8% (255/258), and 96.1% (248/258) for the N gene but were 81.4% (210/258), 78.3% (202/258), 57.0% (147/258), and 43.8% (113/258) for ORF1ab (Table 4). For 3.5 and 2.8 log10 copies/mL, the PPAs of the ORF1ab gene of most multitarget assays did not exceed 50% (Table 4). The sensitivity of the N gene was generally higher than that of ORF1ab, with lower sample concentrations yielding greater differences. In addition, the multitarget assays by Sansure, Daan, BioGerm, and XABT all tended to obtain lower Ct values than those with the ORF1ab gene when detecting the N or E gene. Because the cutoff values of the target regions are the same in an assay, a lower Ct value represents a better ability to detect this target. This phenomenon was observed for all concentrations of positive samples, and only the average Ct values for ORF1ab for 2.8 log10 copies/mL determined with BioGerm and for 4.2 log10 copies/mL determined with XABT were slightly lower. However, the ZJ assay was an exception. The Ct values obtained by detecting the N gene at each concentration were greater than those with ORF1ab and exhibited poor PPAs.
Percentage Agreements of the Results for Negative Samples
The NPAs of SARS, PBS, NL63, HKU1/229E, and Middle East respiratory syndrome/OC43 were 98.1% (913/931), 99.5% (926/931), 99.7% (928/931), 99.4% (925/931), and 98.8% (920/931), respectively. The six NMPA-approved assays mentioned previously were associated with NPAs >95% for all negative samples, except for SARS (Table 5 ). Only ZJ showed false-positive results in detecting SARS samples (7.2%, 12/166). Noticeably, 8.3% (1/12) of the erroneous results from the laboratories using ZJ assays had a single positive gene target, 41.7% (5/12) had two positive gene targets, and 50% (6/12) showed all three gene targets as positive.
Table 5.
The Results of Five Negative Samples Submitted by the Laboratories Using the Six Most Frequently Used NMPA Assays
| Manufacturers | Laboratories reporting, N | 2020-A (SARS), % (n/total) | 2020-D (PBS), % (n/total) | 2020-E (NL63), % (n/total) | 2020-F (HUK1/229E), % (n/total) | 2020-G (MERS/OC43), % (n/total) |
|---|---|---|---|---|---|---|
| BGI | 34 | 100 (34/34) | 97.1 (33/34) | 100 (34/34) | 97.1 (33/34) | 100 (34/34) |
| Sansure | 258 | 100 (258/258) | 100 (258/258) | 100 (258/258) | 100 (258/258) | 98.8 (255/258) |
| Daan | 224 | 100 (224/224) | 100 (224/224) | 100 (224/224) | 100 (224/224) | 99.6 (223/224) |
| BioGerm | 104 | 100 (104/104) | 100 (104/104) | 100 (104/104) | 100 (104/104) | 100 (104/104) |
| ZJ | 166 | 92.8 (154/166) | 100 (166/166) | 99.4 (165/166) | 99.4 (165/166) | 97.6 (162/166) |
| XABT | 20 | 100 (20/20) | 100 (20/20) | 100 (20/20) | 100 (20/20) | 100 (20/20) |
BGI, BGI Bio-tech Co, Ltd, Wuhan, China; BioGerm, BioGerm Medical Co, Ltd, Shanghai, China; Daan, Daan Gene Co, Ltd, of Sun Yat-sen University, Guangzhou, China; MERS, Middle East respiratory syndrome; NMPA, National Medical Products Administration; PBS, phosphate-buffered saline; Sansure, Sansure Biotech Inc., Changsha, China; SARS, severe acute respiratory syndrome; XABT, XABT Bio-tech Co, Ltd, Beijing, China; ZJ, ZJ Bio-tech Co, Ltd, Shanghai, China.
Discussion
Ideally, quality control materials should be obtained from clinical samples after inactivation, but this approach is not feasible because of the limited number of clinical samples and potential infection.12 Armored RNA is similar in structure to natural viruses, resistant to RNase A, and unable to replicate by itself both in vivo and in vitro.13 Therefore, armored RNA is stable for storage and transport, is not pathogenic, and is able to participate in the whole nucleic acid extraction process. In previous studies related to external quality assessment for nucleic acid detection, VLPs have been used as a substitute for infectious pathogens.8 , 9 , 14 In this study, bacteriophage VLPs were prepared as simulated samples for SARS-CoV-2 and other coronaviruses. Subsequently, an EQA on the proficiency of SARS-CoV-2 nucleic acid detection in laboratories was conducted.
In this study, the viral load of simulated positive samples ranged from 2.8 to 4.9 log10 copies/mL. The SARS-CoV-2 virus replicates actively in the early stage of infection and reaches a peak concentration (approximately 4.0 to 7.0 log10 copies/mL) in the respiratory tract early after the onset of symptoms. Then, the viral load gradually decreases to <4.0 log10 copies/mL in the late stage of infection.15 The viral loads of respiratory samples ranged from 2.8 to 11.1 log10 copies/mL, showing a median of 4.9 log10 copies/mL in throat swabs and 5.9 log10 copies/mL in sputum. The lowest detected viral load was 2.8 log10 copies/mL in the late stages of infection.10 Another study showed that the average viral load of respiratory samples was 4.6 log10 copies/mL, and the minimum viral load in the later stage of the disease was 3.2 log10 copies/mL.11 A dynamic analysis of sputum samples from 44 confirmed patients showed that the viral load was 3.1 to 4.7 log10 copies/mL in the early and late stages.16 Therefore, the EQA positive samples used in this study are in keeping with the characteristics of clinical samples and could reflect the infection status at various stages of infection.
The results for positive samples showed that clinical laboratories could reliably detect SARS-CoV-2 nucleic acids with PPA of 93.9% (3497/3724). The results showed that the PPAs of 2020-I (4.9 log10 copies/mL) and 2020-H (4.2 log10 copies/mL) were 97.3% and 98.8%, respectively. Most of the false-negative results were obtained from the weakly positive sample (2020-B, 2.8 log10 copies/mL). False-negative results are a problem for nucleic acid testing to diagnose SARS-CoV-2, as they not only lead to diagnostic confusion but also reduce the credibility of nucleic acid detection. Two laboratories missed all positive samples. Both of these laboratories used modified extraction methods and employed the Daan kit and the BioGerm kit for detection, respectively. The two assays select human RNase P gene as endogenous internal control. Endogenous internal controls might be negative in improperly collected, stored, or possessed samples; thus, they are helpful to monitor sample collection, storage, and extraction process. However, endogenous internal controls are not always effective to monitor the detection of viral RNA, when the amount of virus is much less than human cells. The detection of fluorescence probe for viral RNA might be not as efficient as that for internal controls when the equipment is not well maintained. It is essential for laboratories to incorporate quality control materials containing virus or simulated virus with low concentration to find the errors in the whole analytical process. In this study, the two laboratories used high-positive plasmids as quality control, which were not able to monitor nucleic acid extraction process and the detection of low positive samples. The false-negative results might be due to problems in the test systems, such as extraction process, competency of staff, and performance of equipment.
Nucleic acid extraction is one of the most critical steps to ensure the reliability of molecular diagnosis.17 The percentage of the laboratories using automated assays in the magnetic bead–based extraction method was 77.2% (481/623), which was considerably higher than the percentage (6.6%, 14/212) of the laboratories using automated assays in the column-based extraction method. This might lead to a higher percentage agreement of the laboratories using magnetic bead–based extraction method. Automated extraction has some advantages, such as easy operation, reduced working time, and high-throughput capability. Most important, automated extraction systems are often able to recover a more consistent yield of nucleic acids, avoiding differences caused by operators.18 A study comparing the efficiency of two automated methods and two manual methods for extracting enterovirus RNA showed that the automated extraction method has similar or better results than the manual method.19 In the current study, it was also observed that 36.6% (341/931) of laboratories used unmodified extraction reagents in the manufacturers’ IFU, which showed a higher PPA than those using modified extraction reagents. When extraction reagents different from those in the IFU were introduced into the testing process, the entire integrated test system was modified. The potential risk of inefficient SARS-CoV-2 RNA isolation and purification might exist because the efficiency of pathogen lysis and nucleic acid binding might be different, considering the various components of kits provided by manufacturers.20 According to MM03, Molecular Diagnostic Methods for Infectious Diseases. 3rd ed, issued by the Clinical and Laboratory Standards Institute, modified FDA-cleared or approved assays required full validation.21 Therefore, it is suggested that the laboratories use unmodified extraction reagents or validate the whole system thoroughly before the modified extraction reagents are applied.
Overall, the main nucleic acid detection method of SARS-CoV-2 in China is real-time RT-PCR, with an application rate of 97.4% (884/908) and a PPA of 94.1% (3418/3632). Likewise, 89.9% (62/69) of the nucleic acid assays approved by the US FDA Emergency Use Authorization are based on real-time RT-PCR, whereas a minority are based on isothermal amplification (US Food and Drug Administration, https://www.fda.gov/medical-devices/coronavirus-disease-2019-covid-19-emergency-use-authorizations-medical-devices/vitro-diagnostics-euas, last accessed July 21, 2020). The PPAs of time-of-flight mass spectrometry are 100% in all four positive samples. However, it cannot be concluded that time-of-flight mass spectrometry is more sensitive than other methods because only three laboratories used this method. Xiu et al22 also showed that the LOD of mass spectrometry to detect coronavirus was 10 to 100 copies per reaction, similar to that of real-time RT-PCR. The high accuracy might be due to a specific assay, because all three laboratories used Bioyong assay (Beijing, China). For RT-PCR method, the groups using Hybribio (Chaozhou, China), Huirui (Shanghai, China), Rdbio (Shanghai, China), Szprk (Shenzhen, China), and Amplly (Xiamen, China) LDTs also detected all of the positive results correctly (Supplemental Table S2). For assays involved in this study, the overall PPA based on four positive samples of NMPA-approved assays was slightly higher than that for LDTs (94.3% versus 90.5%); in addition, the performance of the six most frequently used NMPA-approved assays varied for different samples, especially for the weakly positive samples. The capability of assays to detect weakly positive samples is an essential analytical performance metric evaluated for each assay. For example, the LOD of cobas SARS-CoV-2 (Roche Diagnostics, Basel, Switzerland), approved by the US FDA Emergency Use Authorization, is 0.009 tissue culture infectious dose-50 per mL (ORF1ab) and 0.003 tissue culture infectious dose-50 per mL (E), and this assay reached a 100% PPA when detecting samples with nucleic acid concentrations of 1.5× and 4× the LOD in clinical evaluation.15 , 23 The LOD of the Abbott RealTime SARS-CoV-2 assay (Abbott Molecular, Inc., Des Plaines, IL) was 2.0 log10 copies/mL, and the PPA was 100% when detecting samples with nucleic acid concentrations of 1 to 2× the LOD in clinical evaluation.23 In this study, most assays did not achieve a 95% detection rate for samples with 3.5 and 2.8 log10 copies/mL, except for the Sansure, BGI, and Daan assays used in clinical practice (Table 4). The varying abilities of assays to detect weakly positive samples should be due to the difference in the claimed LODs of these assays. However, the assays with the same claimed LODs also performed differently (Supplemental Table S3). The claimed LODs of Sansure and XABT are both 2.3 log10 copies/mL. When detecting samples with nucleic acid concentrations of 2.8 log10 copies/mL (3.2× the LOD), the PPAs of the laboratories using Sansure assays were 96.9%, considerably higher than the percentage agreements (75.0%) of the laboratories using XABT assays. The PPA of ZJ and BioGerm for the sample with a concentration of 2.8 log10 copies/mL was only 74.1% and 76.9%, respectively, because the claimed LODs of ZJ and BioGerm were both 3.0 log10 copies/mL. However, for the samples with concentrations of 3.2× the LOD, the PPAs of ZJ and BioGerm assays were 92.8% and 88.5%, respectively, considerably higher than that of the XABT assay. Sansure, ZJ, and BioGerm assays appeared to have better robustness than the XABT assay, which is important for the assays to be applied more consistently in different laboratories.
Currently, real-time RT-PCR assays approved by the US FDA Emergency Use Authorization and NMPA mainly target the ORF1ab, N, E, and S genes.15 In this study, the targets of assays involved the ORF1ab, N, and E genes. Among the six assays, BGI is the only assay with a single target (namely, ORF1ab) with a high PPA and reproducibility. In theory, assays with multiple target regions are able to detect each target of a positive sample. However, because of the primer design and the competition of primer set in the multiplex reaction, it is normal for an assay to have different amplification efficiencies for each target, which causes differences in the sensitivity for each target.24 , 25 Multiple pairs of primers in a PCR analysis will also increase the probability of mispairing and nonspecific amplification products (primer dimers).26 , 27 In addition, enzymes, primers, the optimal salt ion concentration, and cycle numbers required for primers based on different genes might be different. Thus, when targets with higher sensitivity are positive, regions with lower sensitivity might be undetected, especially in weakly positive samples. In this study, inconsistent PPAs in the ORF1ab, N, and E genes occurred in all multitarget assays (Table 4). For the assays that report positive results based on positive amplification of both ORF1ab and N genes, such as the BioGerm assay, a less sensitive target will result in a final false negative. Therefore, it is essential to perform validation and evaluate the performance of individual target in the multitarget real-time RT-PCR when the assay is developed.
For the SARS-CoV sample, 1.9% (18/931) of the false-positive results were reported. The Ct values of these results ranged from 26 to 32, indicating the high concentration of samples, and these laboratories detected all other SARS-CoV-2 negative samples correctly. Therefore, it is speculated that cross-contamination during operation or residual contamination from amplification was not the primary reason for this finding. Furthermore, 66.7% (12/18) of the false-positive results were detected by laboratories using the ZJ assay. Among the 12 false results, 50% (6/12) were reported because of positive results on N genes (or combined E gene). The E gene and N gene of SARS-CoV-2 serve as the first-line screening tool for the ZJ assay, and this finding might have been due to cross-reaction of the selected primers with SARS-CoV and the high similarity of the targeted regions with that of other coronaviruses. However, the instructions of ZJ specify that a positive result is only premised on a positive result based on the ORF1ab gene. Therefore, the six false-positive results were caused by mistakes in interpretation. The other six false positives involved positive results based on three targets, and the related assays were clustered into several specific kit lots, which suggests that the lot-to-lot reproducibility of ZJ assays was not acceptable.
In summary, an EQA was organized to evaluate the current accuracy of SARS-CoV-2 molecular diagnosis in clinical laboratories. The results showed that SARS-CoV-2 can be reliably detected for most participants. For the assays used by a large number of laboratories, the PPAs and NPAs in the current study showed their analytical sensitivity and cross-reactivity with other human coronaviruses, which is valuable for selecting the real-time RT-PCR assay kit. It should be emphasized that other performance characteristics, such as precision, the effect of interfering substances, and clinical evaluation, are also crucial for the assays. False negatives were the main cause of errors. Inefficient nucleic acid extraction, poor performance of detection assays, and unstandardized personnel operations should all be taken seriously. Laboratories should perform verification studies before routine implementation of tests and monitor reliability throughout the entire process through daily quality management. Future studies will include more samples with low concentrations to monitor false-negative results because of the importance for clinical laboratories to monitor the ability to detect weakly positive samples.
Footnotes
Supported by AIDS and Hepatitis, and Other Major Infectious Disease Control and Prevention Program of China grant 2018ZX10102001 (Rui.Z.).
Z.W., Y.C., J.Y., and Y.H. contributed equally to this work.
Disclosures: None declared.
Supplemental material for this article can be found at http://doi.org/10.1016/j.jmoldx.2020.10.008.
Supplemental Data
Supplemental Figure S1.
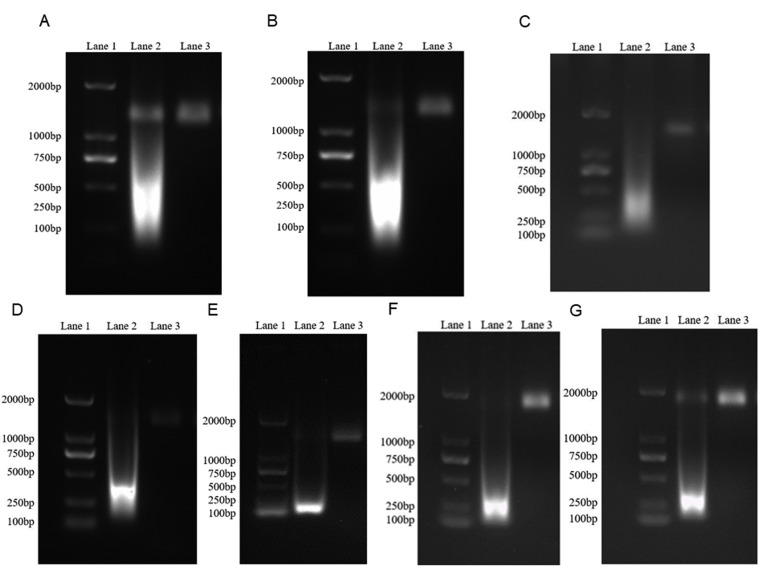
Verification of virus-like particles (VLPs) encapsulating specific RNA fragments of human coronaviruses. After incubation with RNase A and DNase I at 37°C for 40 minutes, the single band over 1% agarose gel electrophoresis near 2 kb proved the successful construction of VLPs. A: Results of electrophoresis of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) VLPs. Lane 1, 2000-bp DNA marker; lane 2, SARS-CoV-2 VLPs without incubation with RNase A and DNase I; lane 3, SARS-CoV-2 VLPs incubated with RNase A and DNase I. B: Results of electrophoresis of SARS VLPs. Lane 1, 2000-bp DNA marker; lane 2, SARS-CoV VLPs not incubated with RNase A and DNase I; lane 3, SARS-CoV VLPs incubated with RNase A and DNase I. C: Results of electrophoresis of 229E VLPs. Lane 1, 2000-bp DNA marker; lane 2, 229E VLPs without incubation with RNase A and DNase I; lane 3, 229E VLPs incubated with RNase A and DNase I. D: Results of electrophoresis of HKU1 VLPs. Lane 1, 2000-bp DNA marker; lane 2, HKU1 VLPs without incubation with RNase A and DNase I; lane 3, HKU1 VLPs incubated with RNase A and DNase I. E: Results of electrophoresis of OC43 VLPs. Lane 1, 2000-bp DNA marker; lane 2, OC43 VLPs without incubation with RNase A and DNase I; lane 3, OC43 VLPs after incubation with RNase A and DNase I. F: Results of electrophoresis of NL63 VLPs. Lane 1, 2000-bp DNA marker; lane 2, NL63 VLPs without incubation with RNase A and DNase I; lane 3, NL63 VLPs digested by RNase A and DNase I. G: Results of electrophoresis of Middle East respiratory syndrome (MERS) VLPs. Lane 1, 2000-bp DNA marker; lane 2, MERS VLPs without incubation with RNase A and DNase I; lane 3, MERS VLPs incubated with RNase A and DNase I.
References
- 1.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., Zhang L., Fan G., Xu J., Gu X., Cheng Z., Yu T., Xia J., Wei Y., Wu W., Xie X., Yin W., Li H., Liu M., Xiao Y., Gao H., Guo L., Xie J., Wang G., Jiang R., Gao Z., Jin Q., Wang J., Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ren L.L., Wang Y.M., Wu Z.Q., Xiang Z.C., Guo L., Xu T. Identification of a novel coronavirus causing severe pneumonia in human: a descriptive study. Chin Med J (Engl) 2020;133:1015–1024. doi: 10.1097/CM9.0000000000000722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Coronaviridae Study Group of the International Committee on Taxonomy of Viruses The species severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat Microbiol. 2020;5:536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chan J.F., Kok K.H., Zhu Z., Chu H., To K.K., Yuan S., Yuen K. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg Microbes Infect. 2020;9:221–236. doi: 10.1080/22221751.2020.1719902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rothe C., Schunk M., Sothmann P., Bretzel G., Froeschl G., Wallrauch C., Zimmer T., Thiel V., Janke C., Guggemos W., Seilmaier M., Drosten C., Vollmar P., Zwirglmaier K., Zange S., Wölfel R., Hoelscher M. Transmission of 2019-nCoV infection from an asymptomatic contact in Germany. N Engl J Med. 2020;382:970–971. doi: 10.1056/NEJMc2001468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Corman V.M., Landt O., Kaiser M., Molenkamp R., Meijer A., Chu D.K., Bleicker T., Brünink S., Schneider J., Schmidt M.L., Mulders D.G., Haagmans B.L., van der Veer B., van den Brink S., Wijsman L., Goderski G., Romette J.L., Ellis J., Zambon M., Peiris M., Goossens H., Reusken C., Koopmans M.P., Drosten C. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Euro Surveill. 2020;25:2000045. doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhan S., Li J., Xu R., Wang L., Zhang K., Zhang R. Armored long RNA controls or standards for branched DNA assay for detection of human immunodeficiency virus type 1. J Clin Microbiol. 2009;47:2571–2576. doi: 10.1128/JCM.00232-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang L., Hao M., Zhang K., Zhang R., Lin G., Jia T., Zhang D., Chang L., Xie J., Li J. External quality assessment for the molecular detection of MERS-CoV in China. J Clin Virol. 2016;75:5–9. doi: 10.1016/j.jcv.2015.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sun Y., Jia T., Sun Y., Han Y., Wang L., Zhang R., Zhang K., Lin G., Xie J., Li J. External quality assessment for avian influenza A (H7N9) virus detection using armored RNA. J Clin Microbiol. 2013;51:4055–4059. doi: 10.1128/JCM.02018-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pan Y., Zhang D., Yang P., Poon L.L.M., Wang Q. Viral load of SARS-CoV-2 in clinical samples. Lancet Infect Dis. 2020;20:411–412. doi: 10.1016/S1473-3099(20)30113-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chan J.F., Yip C.C., To K.K., Tang T.H., Wong S.C., Leung K.H., Fung A.Y., Ng A.C., Zou Z., Tsoi H.W., Choi G.K., Tam A.R., Cheng V.C., Chan K.H., Tsang O.T., Yuen K. Improved molecular diagnosis of COVID-19 by the novel, highly sensitive and specific COVID-19-rdrp/hel real-time reverse transcription-polymerase chain reaction assay validated in vitro and with clinical specimens. J Clin Microbiol. 2020;58 doi: 10.1128/JCM.00310-20. e00310-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clinical and Laboratory Standards Institute . ed 2. Clinical and Laboratory Standards Institute; Wayne, PA: 2013. Design of Molecular Proficiency Testing/External Quality Assessment: Approved Guideline. CLSI document MM14-A2 (ISBN 1-56238-876-6) [Google Scholar]
- 13.Fu Y., Li J. A novel delivery platform based on bacteriophage MS2 virus-like particles. Virus Res. 2016;211:9–16. doi: 10.1016/j.virusres.2015.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang G., Sun Y., Zhang K., Jia T., Hao M., Zhang D., Chang L., Zhang L., Zhang R., Lin G., Peng R., Li J. External quality assessment of molecular detection of Ebola virus in China. PLoS One. 2015;10:e0132659. doi: 10.1371/journal.pone.0132659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shi J., Han D., Zhang R., Li J., Zhang R. Molecular and serological assays for SARS-CoV-2: insights from genome and clinical characteristics. Clin Chem. 2020;66:1030–1046. doi: 10.1093/clinchem/hvaa122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu F., Yan L., Wang N., Yang S., Wang L., Tang Y., Gao G., Wang S., Ma C., Xie R., Wang F., Tan C., Zhu L., Guo Y., Zhang F. Quantitative detection and viral load analysis of SARS-CoV-2 in infected patients. Clin Infect Dis. 2020;71:793–798. doi: 10.1093/cid/ciaa345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ali N., Rampazzo R.C.P., Costa A.D.T., Krieger M.A. Current nucleic acid extraction methods and their implications to point-of-care diagnostics. Biomed Res Int. 2017;2017:1–13. doi: 10.1155/2017/9306564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shin J.H. In: Advanced Techniques in Diagnostic Microbiology. Tang Y.W., Stratton C., editors. Springer US; Boston, MA: 2013. pp. 209–225. [Google Scholar]
- 19.Knepp J.H., Geahr M.A., Forman M.S., Valsamakis A. Comparison of automated and manual nucleic acid extraction methods for detection of enterovirus RNA. J Clin Microbiol. 2003;41:3532–3536. doi: 10.1128/JCM.41.8.3532-3536.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Obersteller S., Neubauer H., Hagen R.M., Frickmann H. Comparison of five commercial nucleic acid extraction kits for the PCR-based detection of Burkholderia Pseudomallei DNA in formalin-fixed, paraffin-embedded tissues. Eur J Microbiol Immunol (Bp) 2016;6:244–252. doi: 10.1556/1886.2016.00028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Clinical and Laboratory Standards Institute . Molecular Diagnostic Methods for Infectious Diseases. ed 3. Clinical and Laboratory Standards Institute; Wayne, PA: 2015. CLSI document MM03 (ISBN 1-56238-998-X) [Google Scholar]
- 22.Xiu L., Zhang C., Wu Z., Peng J. Establishment and application of a universal coronavirus screening method using MALDI-TOF mass spectrometry. Front Microbiol. 2017;8:1510. doi: 10.3389/fmicb.2017.01510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chauhan D.S., Prasad R., Srivastava R., Jaggi M., Chauhan S.C., Yallapu M.M. Comprehensive review on current interventions, diagnostics, and nanotechnology perspectives against SARS-CoV-2. Bioconjug Chem. 2020;31:2021–2045. doi: 10.1021/acs.bioconjchem.0c00323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Berwouts S., Christensen T.M., Brandon J., Bejjani B.A., Barton D.E., Dequeker E. Multiallelic synthetic quality control material: lessons learned from the cystic fibrosis external quality assessment scheme. Genet Test Mol Biomarkers. 2011;15:579–586. doi: 10.1089/gtmb.2011.0015. [DOI] [PubMed] [Google Scholar]
- 25.Weidmann M., Armbruster K., Hufert F.T. Challenges in designing a Taqman-based multiplex assay for the simultaneous detection of Herpes simplex virus types 1 and 2 and Varicella-zoster virus. J Clin Virol. 2008;42:326–334. doi: 10.1016/j.jcv.2008.03.005. [DOI] [PubMed] [Google Scholar]
- 26.Garcia E.P., Dowding L.A., Stanton L.W., Slepnev V.I. Scalable transcriptional analysis routine--multiplexed quantitative real-time polymerase chain reaction platform for gene expression analysis and molecular diagnostics. J Mol Diagn. 2005;7:444–454. doi: 10.1016/S1525-1578(10)60575-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brownie J., Shawcross S., Theaker J., Whitcombe D., Ferrie R., Newton C., Little S. The elimination of primer-dimer accumulation in PCR. Nucleic Acids Res. 1997;25:3235–3241. doi: 10.1093/nar/25.16.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.