Mycobacterium bovis is the primary cause of bovine tuberculosis (bTB) and infects a wide range of domestic animal and wildlife species and humans. In Germany, bTB still emerges sporadically in cattle herds, free-ranging wildlife, diverse captive animal species, and humans. In order to understand the underlying population structure and estimate the population size fluctuation through time, we analyzed 131 M. bovis strains from animals (n = 38) and humans (n = 93) in Germany from 1999 to 2017 by whole-genome sequencing (WGS), mycobacterial interspersed repetitive-unit–variable-number tandem-repeat (MIRU-VNTR) typing, and spoligotyping.
KEYWORDS: tuberculosis, Mycobacterium bovis, human, animal, transmission, spoligotyping, MIRU-VNTR typing, whole-genome sequencing
ABSTRACT
Mycobacterium bovis is the primary cause of bovine tuberculosis (bTB) and infects a wide range of domestic animal and wildlife species and humans. In Germany, bTB still emerges sporadically in cattle herds, free-ranging wildlife, diverse captive animal species, and humans. In order to understand the underlying population structure and estimate the population size fluctuation through time, we analyzed 131 M. bovis strains from animals (n = 38) and humans (n = 93) in Germany from 1999 to 2017 by whole-genome sequencing (WGS), mycobacterial interspersed repetitive-unit–variable-number tandem-repeat (MIRU-VNTR) typing, and spoligotyping. Based on WGS data analysis, 122 out of the 131 M. bovis strains were classified into 13 major clades, of which 6 contained strains from both human and animal cases and 7 only strains from human cases. Bayesian analyses suggest that the M. bovis population went through two sharp anticlimaxes, one in the middle of the 18th century and another one in the 1950s. WGS-based cluster analysis grouped 46 strains into 13 clusters ranging in size from 2 to 11 members and involving strains from distinct host types, e.g., only cattle and also mixed hosts. Animal strains of four clusters were obtained over a 9-year span, pointing toward autochthonous persistent bTB infection cycles. As expected, WGS had a higher discriminatory power than spoligotyping and MIRU-VNTR typing. In conclusion, our data confirm that WGS and suitable bioinformatics constitute the method of choice to implement prospective molecular epidemiological surveillance of M. bovis. The population of M. bovis in Germany is diverse, with subtle, but existing, interactions between different host groups.
INTRODUCTION
Tuberculosis (TB) is one of the high-priority infectious diseases affecting humans and animals worldwide (1, 2) and is the leading cause of death by a single infectious agent in humans (2). Causative agents for TB are the members of the Mycobacterium tuberculosis complex (MTBC), namely, M. tuberculosis, M. africanum, M. bovis, M. caprae, M. microti, and M. pinnipedii. In addition, M. canetti, M. mungi, and M. orygis have been proposed as separate ecotypes. However, their taxonomic classification is still under debate (3).
M. bovis is the primary cause of bovine TB (bTB) but also affects a wide range of other domestic animal and wildlife species and even humans (4–7). After periods of high prevalence of bTB infection in cattle until the second half of the 20th century, Germany has reached the status of being officially free of bTB. Since 1 July 1996 (decision 97/76/EC), 99.9% of the cattle herds remained officially free of bTB infection and disease for at least six consecutive years (Article 2(d) of Council Directive 64/432/EEC [8–10]). However, bTB is still emerging sporadically in cattle herds (11), free-ranging wildlife, captive animal species (12), and humans (13). Confirmed animal bTB cases are notified through an electronic national disease information system (TSN) and published annually (https://www.fli.de/en/publications/annual-animal-health-reports/). From January 1999 to December 2015, a total of 214 bTB outbreaks in cattle herds were notified in Germany, with about half of the cases caused by either M. bovis or M. caprae. In general, M. caprae is reported mainly in middle European countries, with sporadic cases also in Asia and Peru (14, 15), with cattle and wildlife cases in Germany restricted to an area at the German-Austrian border (16, 17). M. caprae was therefore not included in this study. According to the European Food Safety Authority (EFSA) in 2017, from 2013 to 2017, 43 to 56 bTB cases in humans were diagnosed annually (13). Notification rates for bTB ranged from 0.05 to 0.07 per 100,000 population. M. bovis and the closely related M. caprae make up about 1% of all human TB cases (5,486 cases in 2017; more than 6 per 100,000 population) (13, 18).
As disease transmission dynamics of M. bovis within and between host groups are only partially understood (19), molecular typing methods could offer insights into transmission routes and inform pathogen surveillance (20–22). Classical genotyping methods, including spoligotyping, restriction fragment length polymorphism (RFLP), and mycobacterial interspersed repetitive-unitvariable-number tandem-repeat (MIRU-VNTR) detection, allow analysis of outbreaks, assessment of population structures, and performance of longitudinal molecular epidemiological studies (23–30).
Spoligotyping (24) is based on the analysis of CRISPR-CAS spacer sequences located in a genomic region prone to convergent evolution (20), possibly leading to uncertainty of strain relatedness. Spoligotyping patterns submitted to international databases receive unique identifiers; examples include SITVIT (http://www.pasteur-guadeloupe.fr:8081/SITVIT_ONLINE/ and http://www.pasteur-guadeloupe.fr:8081/SITVIT2/; 31), allowing for MTBC isolates from any host, and the Mycobacterium bovis Spoligotype Database (https://www.mbovis.org/), accepting MTBC strains from animals only. As of October 2018, 39,609 MTBC spoligotypes have been collected in the SITVIT database from more than 121 countries (http://www.pasteur-guadeloupe.fr:8081/SITVIT_ONLINE/). In the Mycobacterium bovis Spoligotype Database, 2,117 patterns are available (last update, April 2020). RFLP is a method with high potential for discrimination for M. tuberculosis but not M. bovis strains due to the small number of analyzed insertion element copies present in the respective genomes. MIRU-VNTR typing possesses a higher discriminatory power, allowing automated high-throughput typing and web-based translation into a digit code identifier (28, 29, 32, 33). The method has high potential to define clusters of related strains but cannot differentiate between closely related strains within outbreaks (34).
Next-generation sequencing (NGS) allows for analysis of the nearly complete genome of a pathogen by whole-genome sequencing (WGS), providing deeper insights into the population structure, pathogen evolution, transmission chains, and biology of bacteria (34–37). WGS analysis facilitates the detection of recent transmission chains and monitoring reemerging of strains after years of nondetection (38–41).
In this study, we used WGS, spoligotyping, and MIRU-VNTR typing to determine the diversity of M. bovis strains isolated from animals and humans in Germany and to define possible transmission chains within and between different host populations over an 18-year period (1999 to 2017). Using Bayesian analyses, we sought insights into the dynamics of strain diversity over the last 800 years in Germany.
MATERIALS AND METHODS
Strain selection and DNA extraction.
In total, 131 M. bovis strains were available for WGS, including the reference strain M. bovis BCG (DSM 43990/ATCC 27289), with 38 strains from the Friedrich-Loeffler-Institut (FLI), Federal Institute for Animal Health, and 93 strains from the National Reference Center (NRC) for Mycobacteria in Borstel, Germany (see Table S1 in the supplemental material). From January 1999 to December 2015 (the study period), a total of 214 bTB outbreaks in cattle herds were notified in Germany by the electronic system implemented by the FLI to monitor bTB outbreaks, with about half of the cases in cattle caused by M. bovis. M. bovis strains from 10 cattle bTB outbreaks, from 5 other domestic animal species, 14 zoo animals, and wild boars were analyzed (Table S2), spanning the period from 1999 to 2015 and covering different regions of the country, including the known hot spot regions in the north and south. At the NRC in Borstel, all German M. bovis strains cultured and archived from 2000 to 2017 were included. The NRC receives samples from all districts in Germany, and while it is not the only laboratory offering specialist mycobacterial diagnostics in Germany, it receives an estimated 50% of all MTBC isolates. At both institutions, strains were cultured according to standard procedures (42–45), and genomic DNA was extracted using the High Pure PCR template preparation kit (Roche Life Science; FLI) and by the cetyltrimethylammonium bromide (CTAB) procedure (NRC), respectively (46).
Classical genotyping.
Spoligotyping of animal strains was performed using a microarray format (Alere Technologies, Jena, Germany) (47). Binary codes were automatically compared with data available through SITVIT and the Mycobacterium bovis Spoligotype Database to identify concordant species and lineages. For human strains, the conventional spoligotyping method was used (24). MIRU-VNTR typing of the strains isolated from animals was performed using conventional PCR and agarose gel electrophoresis (26, 28, 48). For human strains, the automated high-throughput method was used (28). VNTR copy numbers were assessed according to allele calling tables (https://www.miru-vntrplus.org, EU Reference Laboratory for Bovine Tuberculosis [https://www.visavet.es]). The discriminatory power of the method was calculated according to the method of Hunter and Gaston (49) (Tables S3 and S4).
Whole-genome sequencing and data analysis.
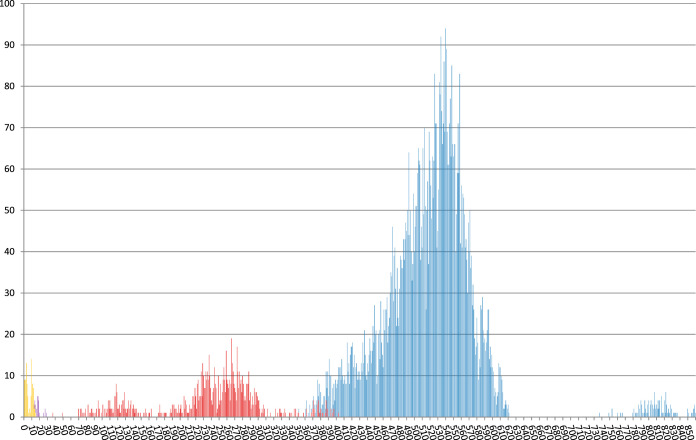
Libraries for WGS were prepared from genomic DNA with a modified Illumina Nextera protocol (50) and run on the Illumina NextSeq NGS platform (Illumina, San Diego, CA). We employed the MTBseq pipeline with default parameters for variant detection and a joint analysis (51), employing a threshold of 12 single nucleotide polymorphisms (SNPs) for cluster detection (52). As deduced from the pairwise SNP distance distribution, we used a cutoff of 350 SNPs to detect major groups (Fig. 1). For all sequenced strains, mean coverage depth was at least 50-fold, and at least 95% of the reference genome fulfilled the MTBseq thresholds for variant detection. From the aligned sequences of concatenated SNP positions produced by MTBseq, we calculated a maximum likelihood tree with FastTree (53) with a general time-reversible (GTR) substitution model, 1,000 resamples, and Gamma20 likelihood optimization to account for rate heterogeneity among sites. The consensus tree was rooted with the “midpoint root” option in FigTree (http://tree.bio.ed.ac.uk/software/figtree), and nodes were arranged in increasing order. The resulting tree was annotated with EvolView software (54). Additionally, we built maximum parsimony trees with the software BioNumerics version 7.5 (Applied Maths, Ghent, Belgium) with default settings.
FIG 1.
Pairwise distance distribution of SNP distances between all sequenced strains (blue) and within WGS d350 groups (red), d30 clusters (purple), and d12 clusters (yellow), with the color indicator for the respective lower thresholds superimposed. The y axis indicates the total number of pairwise distances and the x axis the number of distinct SNPs.
For the coalescent-based analyses, evolutionary rates and tree topologies were analyzed using the GTR and Hasegawa-Kishino-Yano (HKY) substitution models with gamma distributed among-site rate variation with four rate categories (Γ4). The substitution rate was estimated by plotting a regression line that depicts for the sole WGS clusters, in a pairwise manner, the relationship between the elapsed time and the accumulated number of SNPs. Under this model, the slope corresponds to the mutation rate. We tested both a strict molecular clock (which assumes the same evolutionary rates for all branches in the tree) and a relaxed clock that allows different rates among branches. Constant-size, exponential and Bayesian skyline plot models, based on a general, nonparametric prior model that enforces no particular demographic history, were used in BEAST v1.10.4 (55). For each model, two independent chains were conducted for 200 million generations and convergence was assessed by checking the effective sample size values for key parameters using TRACER v1.7.1 (56). We used TRACER V1.7.1 to calculate the log10 Bayes factors in order to compare the models after a burn-in of 10% of the chain. Bayes factors represent the ratio of the marginal likelihood of the models being compared. Approximate marginal likelihoods for each coalescent model were calculated via importance sampling (1,000 bootstraps) using the harmonic mean of the sampled likelihoods. A ratio between 3 and 10 indicates moderate support that one model better fits the data than another, whereas values greater than 10 indicate strong support. For correlation with known clonal complexes, we selected 33 strains representing the known clades contained in a recent publication (15) and performed a joint analysis as described previously.
Data availability.
All WGS data were submitted to the EMBL-EBI ENA SRA archive under accession numbers ERR2212113 to ERR2212125, ERR2815506 to ERR2815614, ERR551004, ERR551009, ERR551191, ERR551252, ERR551427, ERR551917, ERR552138 to ERR552140, ERR552470 to ERR552472, ERR552515, ERR552516, ERR552796, ERR552797, ERR553061, ERR553337, and ERR553338 (see Table S1).
RESULTS
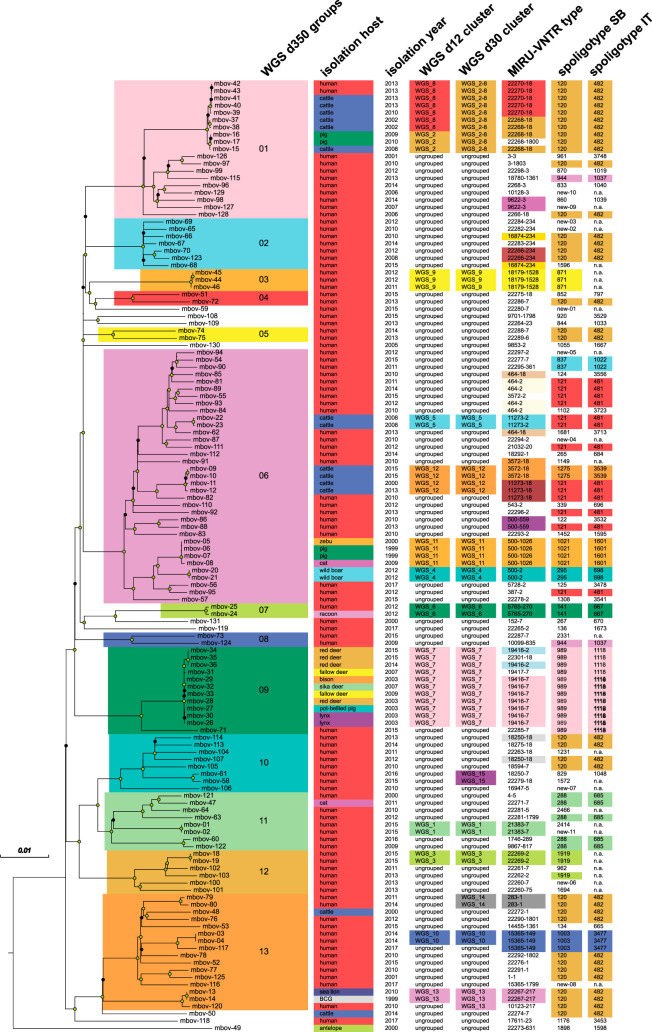
In total, 131 M. bovis strains, 93 of human and 38 of animal origin (Table S1) isolated in Germany from 1999–2017, including 1 M. bovis BCG reference strain, were investigated by spoligotyping, MIRU-VNTR typing, and WGS. WGS data analysis revealed 12,726 variable SNP positions among the genomes analyzed that were used for the calculation of a phylogenetic tree (Fig. 2). Interestingly, the strain mbov-49 was clearly separated from the rest of the study collection. This strain was isolated at the FLI in 2000 from a nilgai antelope (Boselaphus tragocamelus) which died in a German zoo, and the strain was found to be not intrinsically pyrazinamide resistant (57).
FIG 2.
Maximum likelihood tree of 131 M. bovis strains built from 12,726 SNP positions, annotated with host organism, isolation year, WGS cluster, MIRU-VNTR type, and spoligotype from the SITVIT (IT) and mbovis.org (SB) databases. Scale bar indicates the likelihood of per-site substitution and therefore reflects a distance of 127 SNPs bearing reverse mutations. Circles on nodes indicate resampling support of at least 90% (green circles) or at least 70% (black circles).
Overall, the median pairwise distance in distinct SNP positions of the 131 strains was 516 SNPs, and distinct peaks emerged in the frequency distribution between 0 and 30, 70 and 350, 370 and 620, and 780 and 840 distinct SNPs, agreeing with the groups of related strains found by cluster detection with thresholds of 12, 30, and 350 distinct SNPs (d12, d30, and d350) between nearest group members (Fig. 1 and 2). Using the d350 threshold to group strains, we found 13 cladistic groups containing 122/131 strains ranging in size from 2 to 35 members, with, on average, 8 years (2 to 18) between the earliest and latest years of isolation.
Six of the d350 groups contained both human and animal cases, and seven contained only human cases. When comparing d350 groups with the known clonal complexes African 1 and 2 (Af1 and Af2), European 1 and 2 (Eu1 and Eu2), and newly determined Unknown 1 to 8 (15), we could correlate clonal complexes Af1, Eu1, Eu2, and Unknown 2 with d350 groups 08, 07, 06, and 13 (Fig. S1 and Table S6). For clonal complexes Af2, Unknown 1, and Unknown 7, we found only one corresponding strain in our collection (mbov-118, mbov-49, and mbov-119, respectively). Interestingly, three d350 groups (groups 10 to 12) were attributed to clonal complex Unknown 3, and four d350 groups (01, 02, 03, and 04) were attributed to clonal complex Unknown 4. We found no representatives of complexes Unknown 5 and Unknown 6 in our study, as well as correlates of d350 groups 05 and 09 among the collection of known clonal complexes.
Putative transmission clusters.
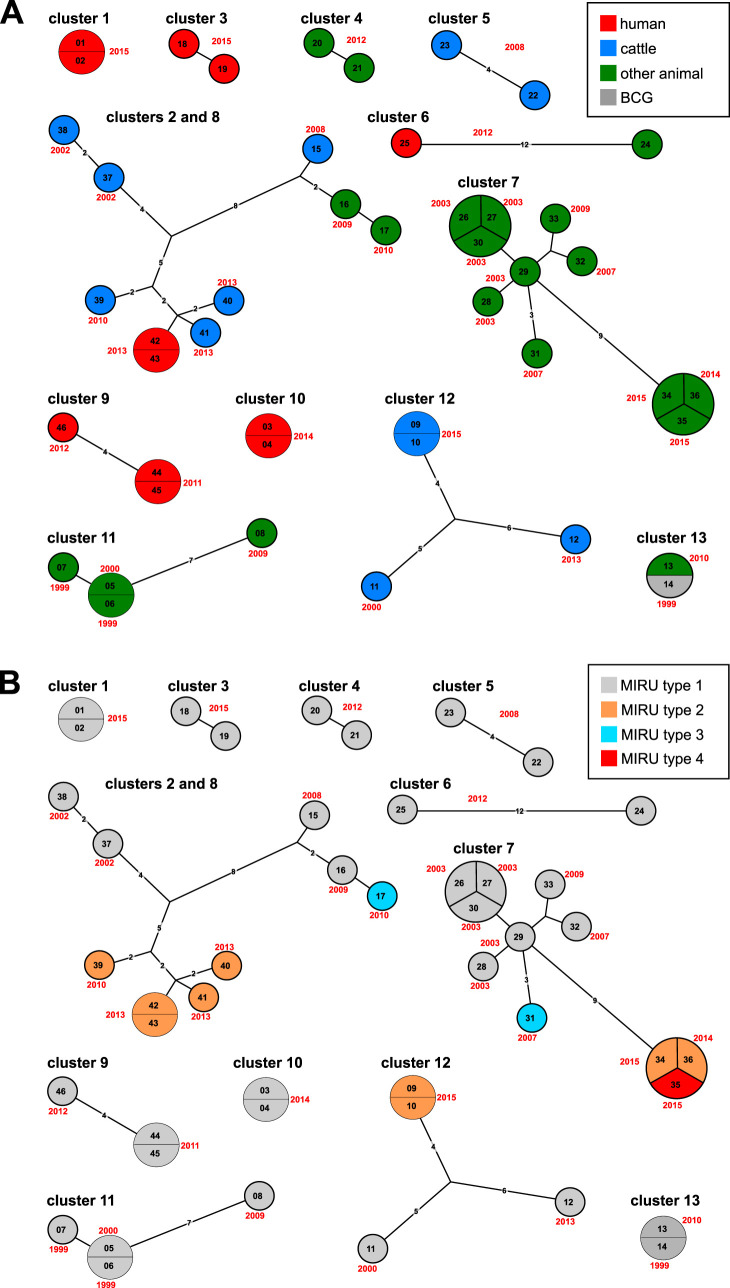
We used a threshold of at most 12 distinct SNP positions to the nearest group member as indication for possible recent transmission (50), which yielded 13 d12 clusters of, all together, 46 strains (Fig. 2 and 3 and Table 1). The d12 clusters ranged in size from 2 to 11 members, spanned up to 15 years, and involved distinct host types, with d12 clusters 5 and 12 comprising only cattle hosts, clusters 4, 7, 11, and 13 only human hosts, and the rest mixed hosts (Table 1). In total, 32 of the 38 animal strains (the pair of M. bovis BCG in d12 cluster 13 not counted) were grouped into WGS d12 clusters. In four of these clusters, animal strains were recovered more than 9 years apart, pointing toward autochthonous persistent bTB infection cycles. In contrast, only 12 out of the 93 human strains were grouped into d12 clusters, with 9 human strains forming four WGS d12 clusters of two and three members, respectively (Table 1). The members of these groups were isolated within at most 2 years from each other. Overall, we found one cluster (cluster 8) with a putative transmission from cattle to humans with respective strains separated by two SNPs and one cluster (cluster 6) of raccoon and human strains separated by 12 SNPs.
FIG 3.
(A) Maximum parsimony trees for the 13 WGS clusters, annotated with host and year of isolation. Numbers on branches indicate number of distinct SNPs; distances of 1 are not indicated. (B) Maximum parsimony trees for the 13 WGS clusters, annotated with MIRU-VNTR types. Numbers on branches indicate number of distinct SNPs; distances of 1 are not indicated.
TABLE 1.
Synopsis of the 13 d12 clusters as deduced from the maximum likelihood tree built from 131 M. bovis strainsa
| Cluster no. | No. of strains | Yr(s) of isolation | Time span (yrs) | Maximum distance by SNPs | Host species | Reference |
|---|---|---|---|---|---|---|
| 1 | 2 | 2015 | 1 | 0 | Human | |
| 2 | 3 | 2008–2010 | 3 | 2 | Cattle, swine | |
| 3 | 2 | 2015 | 1 | 1 | Human | |
| 4 | 2 | 2012 | 1 | 1 | Wild boar | |
| 5 | 2 | 2008 | 1 | 4 | Cattle | 11 |
| 6 | 2 | 2012 | 1 | 12 | Raccoon, human | |
| 7 | 11 | 2003–2015 | 13 | 9 | Different wild animal species | 12 |
| 8 | 7 | 2002–2013 | 12 | 9 | Cattle, human | |
| 9 | 3 | 2011, 2012 | 2 | 4 | Human | |
| 10 | 2 | 2014 | 1 | 0 | Human | |
| 11 | 4 | 1999–2009 | 10 | 7 | Swine, zebu, cat | |
| 12 | 4 | 2000–2015 | 16 | 6 | Cattle | |
| 13 | 2 | 2013 | 1 | 0 | Sea lion (BCG strain) |
The clusters, the number of strains, the years of isolation, spanning time, the maximum distance as indicated by the number of SNPs, and the host organisms are shown.
As the frequency distribution of pairwise SNP distances featured a peak between 0 and 30 SNPs (Fig. 1), we also clustered strains with a threshold of 30 SNPs. This yielded two new clusters of related strains with two members each and an additional member of d12 cluster 13, and d12 clusters 2 and 8 were joined (Fig. 2).
Comparison with classical genotyping.
The 131 strains were differentiated into 45 known spoligotypes and 11 spoligotypes not contained in the established databases (Tables S1 and S5). Five or more strains each fell into four known spoligotypes: SB 120/IT0482 (35 strains), SB 121/IT0481 (13 strains), SB 989/IT1118 (12 strains), and SB 288/IT685 (5 strains). Of these, SB 120 and SB 121 have been reported as predominant spoligotypes circulating among animals around the world (58). Strains of these spoligotypes were present in different branches of the constructed phylogenetic tree and in different MIRU-VNTR and d12 clusters (Fig. 2).
Comparing the composition of the d350 groups in terms of the respective spoligotypes (Fig. 2), we found correlations with the well-established clonal complexes EU1 and EU2 and Af1 and Af2, as well as with the newly determined complexes named Unknown 1 to 8 (15) (Table S7). For example, SB0120, found in d350 groups 01, 02, 04, 05, 10, and 13, was detected in complexes Unknown 2 to 5. This spoligotype has been reported as predominant circulating among animals around the world (58). Seven spoligotypes present in d350 groups 01, 02, 03, and 04 were reported for complex Unknown 4 (15). The 15 spoligotypes found for d350 group 06 corresponded to those for complex Eu2, and the 9 spoligotypes present in d350 groups 10, 11, and 12 were found in clade Unknown 3 (15). Spoligotype SB0989, found in d350 group 09, was reported for singletons not contained in a complex (15).
MIRU-VNTR analysis yielded 92 distinct patterns with 21 strain clusters ranging from two to seven members comprising altogether 62 strains. Using 121 supposedly unrelated strains, the discriminatory power index (HGDI [49]) of each of the 24+ 1-locus MIRU-VNTR loci was determined, with allelic heterogeneity mainly restricted to 2 to 4 repeat copies (Table S3). Allele heterogeneity of >0.5 was found for the loci VNTR 2163a, 2163b, 2165, 2461, and 4052 (Table S4). Overall, MIRU-VNTR types correlated well with both the phylogenetic tree and the d12 clusters. However, 21 strains grouped by MIRU-VNTR were not clustered by d12 analysis, and four d12 clusters encompassed strains with different MIRU-VNTR patterns, with four distinct loci in one and one distinct locus in three of these cases (Fig. 2 and 3).
Mutation rate estimation and demographic inference.
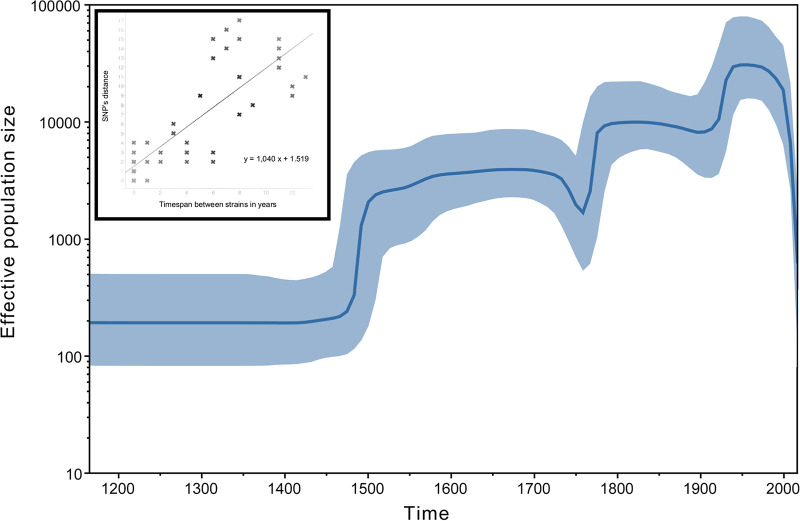
The geographically widespread and phylogenetically diverse nature of our strain collection did not allow implementing a Bayesian tip-dating approach. We therefore focused on the 13 d12 clusters for which the measurably evolving dimension of M. bovis could be captured to infer a realistic estimation of the mutation rate. A positive correlation (r2 = 0.682) was found between the time elapsed between two strains and the number of accumulated SNPs (Fig. 4). The slope was close to 1, corresponding to the acquisition of one SNP every year between two strains and translating to a mutation rate of 1.14 × 10−7 substitution/nucleotide/year.
FIG 4.
Bayesian skyline plot showing the effective population size of the German M. bovis sample through time, estimated from the SNP matrix. According to the coalescence-based approach, the M. bovis population went through three successive expansions followed by a final decline. (Inset) Root-to-tip genetic distances plotted against sampling dates based on 13 WGS clusters. The plot illustrates a positive correlation (r2 = 0.682) of divergence with sampling date and confirms that M. bovis is a measurably evolving population (MEP).
To estimate the effective population size fluctuation through time, three demographic models were compared; the best-fitting evolutionary model was obtained under the Bayesian skyline model with a relaxed clock (Fig. 4). The relaxed-clock model outperforms the constant-clock model (Bayes factor = 40) and the Bayesian skyline was favored to its closest model, constant size (Bayes factor = 14). The time to most recent common ancestor (TMRCA) corresponding to our M. bovis strain collection dated back some 950 years ago (95% highest posterior density [HPD] interval, 836 to 1,062). According to the coalescence-based demographic reconstructions, the German M. bovis population went through three successive expansions: first a 20-fold increase in the late middle age, followed by two mild expansions in the middle of 18th century and the early 20th century (Fig. 4).
DISCUSSION
This investigation provided insights into population structure, persistence, and population size fluctuation of M. bovis strains in Germany over time and the complex interrelations in a multihost pathogen system. In the context of a country declared officially free of bTB for more than 2 decades, special consideration was given to strain persistence, attempting to understand recurrent outbreaks and possible links to human cases, while other publications have mainly concentrated on microevolution of strains in the context of geospatial spreading and transmission dynamics between animal reservoirs (59, 60).
The main limitation of our study is that due to practical limitations related to access to strains, we were not able to collect a fully comprehensive set of M. bovis strains from human and animal cases in Germany. Additionally, due to the restrictions set by data protection regulations, the available metadata for the strains was limited to year and host of isolation. Regrettably, this does not allow an epidemiological analysis of the WGS d12 and d30 clusters. Still, our collection covers a time span from 1999 to 2017 and diverse host species. While we took care to identify and remove duplicate strains from the same host, we cannot fully exclude this possibility for human strains.
We successfully performed WGS for a collection of 93 human and 38 animal M. bovis strains, isolated in Germany from 1999 to 2017. The pairwise distance distribution and the reconstructed phylogenetic tree indicate the presence of 13 d350 groups within the study population. These encompassed the majority of strains (122/131) and represent a snapshot of M. bovis sublineages historically spreading in Germany. Correlating our phylogeny and detected groups with described clonal complexes revealed that our collection contains representatives of the well-known M. bovis complexes Af1, Af2, Eu1, and Eu2, as well as of additional groups defined recently (15). Interestingly, there are at most two strains of complexes Af1, Af2, and Eu1 in our study, and we found no representatives of complexes Unknown 5 and Unknown 6 or correlating complexes for d350 groups 05 and 09. This might indicate a geographically uneven distribution of subgroups and that the M. bovis phylogeny needs to be refined by WGS-based studies with larger, geographically diverse collections.
Using a threshold of 12 distinct SNP positions to identify strains possibly involved in recent transmission events (52), we found that 32 out of the 38 animal strains and 12 out of the 93 human strains grouped into 13 d12 clusters. In four of these clusters, animal strains were recovered more than 9 years apart, pointing toward autochthonous persistent bTB infection cycles. This is further supported by the combination of d12 clusters 2 and 8 into a joint group when clustering with a threshold of 30 SNPs, with the phylogenetic analysis and the number of distinct SNP positions suggesting a relatively recent common source for both clusters. Human strains within clusters were isolated within at most a 1-year difference and with one sole exception had at most one SNP distance, possibly indicating direct transmission.
Despite the imbalance of M. bovis strains included from humans and animals, there seem to be distinct infection dynamics for animals and humans. For cattle and other animals, the majority of strains were found within d12 clusters and several strains were persistently spreading over up to 15 years, pointing toward potential reservoirs of these strains, for example, in the German wildlife population. The mostly unclustered human cases might represent progression to active disease from latently infected individuals as indicated previously (16). In general, human mobility is also higher than mobility of cattle and wild animals. Here, patients having contacts to sources of infection outside Germany may contribute to the detected high diversity of strains isolated from human patients. As reported in 2003 (16), the majority of patients with M. bovis disease in Germany were over 60 years of age, suggesting that they might have acquired the infection at a young age, when the prevalence of bTB in cattle in Germany was much higher than today. Unfortunately, M. bovis strains isolated from cattle before 1999 were not available.
Two of the d12 clusters (6 and 8) contained both animal and human strains, indicating possible recent transmission between humans and animals. The detection of only one human strain contained in a d12 cluster with cattle strains may indicate that the overall risk of human infection with M. bovis is low with respect to consumption of food (milk and meat) or direct contact to indigenous cattle, while transmission can happen in outbreaks settings.
The study results show that WGS is superior in unequivocally detecting genetic relationship between strains and clarify transmission routes compared to spoligotyping and MIRU-VNTR typing. While spoligotyping provides some information of strain relatedness, our results demonstrate that it cannot reliably establish clusters of related strains. MIRU-VNTR typing results correlated well with WGS data. However, MIRU-VNTR typing cannot accurately trace gradual evolution within a transmission cluster. Twenty-one strains clustered by MIRU-VNTR typing were not clustered by d12 analysis, and four d12 clusters encompassed strains with distinct MIRU-VNTR patterns.
We estimated a mutation rate of 1.14 × 10−7 substitution/nucleotide/year for M. bovis. A recent publication on the molecular clock with over 6,000 samples of M. tuberculosis representing the global diversity and covering different epidemiological settings estimated a clock rate between 1 × 10−8 and 5 × 10−7 while stating that sampling times below 15 to 20 years could be insufficient to calibrate a clock rate (61). In another study dealing explicitly with globally distributed M. bovis strains, the clock rate was estimated at between 6.66 × 10−8 and 1.26 × 10−7 (15). Our collection of 131 samples of German M. bovis strains spans a period of 19 years, maybe limiting our ability to estimate the clock rate. However, the rate that we inferred is in full agreement with estimates published for M. tuberculosis outbreaks in Germany (34) and Eurasia (62). Estimates of the effective population size fluctuation through time according to coalescence-based demographic reconstructions suggested that the German M. bovis population went through three successive expansions, first a 20-fold increase in the late middle age, followed by two mild expansions in the mid-18th century and the early 20th century (Fig. 4). These expansions might be due to increasing growth and movement of human and cattle populations as well as increasing growth of human communities and of intensive animal husbandry with time. The population size sharply declined after the 1970s, underlining the absence of ongoing epidemics in Germany and confirming the bTB-free status of the country. Indirectly supporting the data, the Bayesian skyline detected an anticlimax in the period from 1740 to 1760. This observation coincides with the cattle plague outbreak (rinderpest virus [RPV]) that severely impacted the European stocks during that period (63).
In conclusion, in this study for the first time the persistence of infectious cycles of M. bovis in Germany, officially a bTB-free country for more than 10 years, has been clearly demonstrated, pointing toward the challenges controlling this pathogen. As exemplified here, WGS is definitively the method of choice for establishment of an integrated molecular surveillance of M. bovis as well as for outbreak investigations.
Supplementary Material
ACKNOWLEDGMENTS
We thank V. Mohr, F. Boysen, T. Ubben, A. Lüdemann, U. Brommer, and G. Kauth for excellent technical assistance.
Parts of the work have been funded by grants from German Center for Infection Research, Federal Ministry of Education and Research, Germany, from Deutsche Forschungsgemeinschaft (DFG; German Research Foundation) under Germany’s Excellence Strategy—EXC 22167-390884018, and grants from the Leibniz Science Campus EvoLUNG.
All authors provided substantial scientific contributions, read and approved the final manuscript, and agreed to the submission.
None of the authors have any conflicts of interest relevant to this study to disclose.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.World Health Organization. 2018. UN general assembly high-level meeting on ending TB. 26 September 2018, New York. www.who.int/tb/features_archive/UNGA_HLM_ending_TB/en/.
- 2.World Health Organization. 2019. Global tuberculosis report 2019. www.who.int/tb/publications/global_report/en/. Accessed 1 January 2020.
- 3.Riojas MA, McGough KJ, Rider-Riojas CJ, Rastogi N, Hazbón MH. 2018. Phylogenomic analysis of the species of the Mycobacterium tuberculosis complex demonstrates that Mycobacterium africanum, Mycobacterium bovis, Mycobacterium caprae, Mycobacterium microti and Mycobacterium pinnipedii are later heterotypic synonyms of Mycobacterium tuberculosis. Int J Syst Evol Microbiol 68:324–332. doi: 10.1099/ijsem.0.002507. [DOI] [PubMed] [Google Scholar]
- 4.O’Reilly LM, Daborn CJ. 1995. The epidemiology of Mycobacterium bovis infections in animals and man: a review. Tuber Lung Dis 76:1–46. doi: 10.1016/0962-8479(95)90591-X. [DOI] [PubMed] [Google Scholar]
- 5.Palmer MV, Thacker TC, Waters WR, Gortázar C, Corner LA. 2012. Mycobacterium bovis: a model pathogen at the interface of livestock, wildlife, and humans. Vet Med Int 2012:236205. doi: 10.1155/2012/236205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Müller B, Dürr S, Alonso S, Hattendorf J, Laisse CJ, Parsons SD, van Helden PD, Zinsstag J. 2013. Zoonotic Mycobacterium bovis-induced tuberculosis in humans. Emerg Infect Dis 19:899–908. doi: 10.3201/eid1906.120543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barasona JA, Vicente J, Díez-Delgado I, Aznar J, Gortázar C, Torres MJ. 2017. Environmental presence of Mycobacterium tuberculosis complex in aggregation points at the wildlife/livestock interface. Transbound Emerg Dis 64:1148–1158. doi: 10.1111/tbed.12480. [DOI] [PubMed] [Google Scholar]
- 8.Council of the European Union. 1964. Council Directive 64/432/EEC of 26 June 1964 on animal health problems affecting intra-Community trade in bovine animals and swine. https://eur-lex.europa.eu/legal-content/EN/ALL/?uri=CELEX%3A31964L0432.
- 9.European Commission Health & Consumer Protection Directorate-General, Veterinary and International Affairs, Unit G5—Veterinary Programmes. 2013. Working document on eradication of bovine tuberculosis in the EU Accepted by the Bovine tuberculosis subgroup of the task force on monitoring animal disease eradication. SANCO/10067/2013 https://ec.europa.eu/food/sites/food/files/animals/docs/diseases_erad_tb_workingdoc2006_en.pdf.
- 10.Meyn A. 1961. Die Fortschritte der Rindertuberkulosebekämpfung in der Bundesrepublik. Monatsh Tierheilkunde 14:71–78. [Google Scholar]
- 11.Probst C, Freuling C, Moser I, Geue L, Köhler H, Conraths FJ, Hotzel H, Liebler-Tenorio EM, Kramer M. 2011. Bovine tuberculosis: making a case for effective surveillance. Epidemiol Infect 139:105–112. doi: 10.1017/S0950268810000786. [DOI] [PubMed] [Google Scholar]
- 12.Kohl TA, Utpatel C, Niemann S, Moser I. 2018. Mycobacterium bovis persistence in two different captive wild animal populations in Germany: a longitudinal molecular epidemiological study revealing pathogen transmission by whole-genome sequencing. J Clin Microbiol 56:e00302-18. doi: 10.1128/JCM.00302-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.European Food Safety Authority and European Centre for Disease Prevention and Control (EFSA and ECDC). 2018. The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2017. EFSA J 16:5500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Prodinger WM, Brandstätter A, Naumann L, Pacciarini M, Kubica T, Boschiroli ML, Aranaz A, Nagy G, Cvetnic Z, Ocepek M, Skrypnyk A, Erler W, Niemann S, Pavlik I, Moser I. 2005. Characterization of Mycobacterium caprae isolates from Europe by mycobacterial interspersed repetitive unit genotyping. J Clin Microbiol 43:4984–4492. doi: 10.1128/JCM.43.10.4984-4992.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Loiseau C, Menardo F, Aseffa A, Hailu E, Gumi B, Ameni G, Berg S, Rigouts L, Robbe-Austerman S, Zinsstag J, Gagneux S, Brites D. 2020. An African origin for Mycobacterium bovis. Evol Med Public Health 2020:49–59. doi: 10.1093/emph/eoaa005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kubica T, Rüsch-Gerdes S, Niemann S. 2003. Mycobacterium bovis subsp. caprae caused one-third of human M. bovis-associated tuberculosis cases reported in Germany between 1999 and 2001. J Clin Microbiol 41:3070–3077. doi: 10.1128/jcm.41.7.3070-3077.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Domogalla J, Prodinger WM, Blum H, Krebs S, Gellert S, Müller M, Neuendorf E, Sedlmaier F, Büttner M. 2013. Region of difference 4 in alpine Mycobacterium caprae isolates indicates three variants. J Clin Microbiol 51:1381–1388. doi: 10.1128/JCM.02966-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Robert Koch Institut. 2019. Ein historisches Signal für den Kampf gegen Tuberkulose—Deutschland muss das Momentum nutzen. Epidemiologisches Bulletin. https://www.rki.de/DE/Content/Infekt/EpidBull/Archiv/2019/11_12/Art_01.html.
- 19.Brooks-Pollock E, Roberts GO, Keeling MJ. 2014. A dynamic model of bovine tuberculosis spread and control in Great Britain. Nature 511:228–231. doi: 10.1038/nature13529. [DOI] [PubMed] [Google Scholar]
- 20.Comas I, Homolka S, Niemann S, Gagneux S. 2009. Genotyping of genetically monomorphic bacteria. DNA sequencing in Mycobacterium tuberculosis highlights the limitations of current methodologies. PLoS One 4:e7815. doi: 10.1371/journal.pone.0007815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schürch AC, van Soolingen D. 2012. DNA fingerprinting of Mycobacterium tuberculosis: from phage typing to whole-genome sequencing. Infect Genet Evol 12:602–609. doi: 10.1016/j.meegid.2011.08.032. [DOI] [PubMed] [Google Scholar]
- 22.Merker M, Kohl TA, Niemann S, Supply P. 2017. The evolution of strain typing in the Mycobacterium tuberculosis complex. Adv Exp Med Biol 1019:43–78. doi: 10.1007/978-3-319-64371-7_3. [DOI] [PubMed] [Google Scholar]
- 23.van Embden JD, Cave MD, Crawford JT, Dale JW, Eisenach KD, Gicquel B, Hermans P, Martin C, McAdam R, Shinnick TM. 1993. Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J Clin Microbiol 31:406–409. doi: 10.1128/JCM.31.2.406-409.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kamerbeek J, Schouls L, Kolk A, van Agterveld M, van Soolingen D, Kuijper S, Bunschoten A, Molhuizen H, Shaw R, Goyal M, van Embden J. 1997. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J Clin Microbiol 35:907–1014. doi: 10.1128/JCM.35.4.907-914.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cousins DV, Skuce RA, Kazwala RR, van Embden JD. 1998. Towards a standardized approach to DNA fingerprinting of Mycobacterium bovis. International Union against Tuberculosis and Lung Disease, Tuberculosis in Animals Subsection. Int J Tuber Lung Dis 2:471–478. [PubMed] [Google Scholar]
- 26.Frothingham R, Meeker-O'Connell WA. 1998. Genetic diversity in the Mycobacterium tuberculosis complex based on variable numbers of tandem DNA repeats. Microbiology 144:1189–1196. doi: 10.1099/00221287-144-5-1189. [DOI] [PubMed] [Google Scholar]
- 27.Skuce RA, Neill SD. 2001. Molecular epidemiology of Mycobacterium bovis: exploiting molecular data. Tuberculosis (Edinb) 81:169–175. doi: 10.1054/tube.2000.0270. [DOI] [PubMed] [Google Scholar]
- 28.Supply P, Allix C, Lesjean S, Cardoso-Oelemann M, Rüsch-Gerdes S, Willery E, Savine E, de Haas P, van Deutekom H, Roring S, Bifani P, Kurepina N, Kreiswirth B, Sola C, Rastogi N, Vatin V, Gutierrez MC, Fauville M, Niemann S, Skuce R, Kremer K, Locht C, van Soolingen D. 2006. Proposal for standardization of optimized mycobacterial interspersed repetitive unit-variable-number tandem repeat typing of Mycobacterium tuberculosis. J Clin Microbiol 44:4498–4510. doi: 10.1128/JCM.01392-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oelemann MC, Diel R, Vatin V, Haas W, Rüsch-Gerdes S, Locht C, Niemann S, Supply P. 2007. Assessment of an optimized mycobacterial interspersed repetitive-unit–variable-number tandem-repeat typing system combined with spoligotyping for population-based molecular epidemiology studies of tuberculosis. J Clin Microbiol 45:691–697. doi: 10.1128/JCM.01393-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wirth T, Hildebrand F, Allix-Béguec C, Wölbeling F, Kubica T, Kremer K, van Soolingen D, Rüsch-Gerdes S, Locht C, Brisse S, Meyer A, Supply P, Niemann S. 2008. Origin, spread and demography of the Mycobacterium tuberculosis complex. PLoS Pathog 4:e1000160. doi: 10.1371/journal.ppat.1000160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brudey K, Driscoll JR, Rigouts L, Prodinger WM, Gori A, Al-Hajoj SA, Allix C, Aristimuño L, Arora J, Baumanis V, Binder L, Cafrune P, Cataldi A, Cheong S, Diel R, Ellermeier C, Evans JT, Fauville-Dufaux M, Ferdinand S, Garcia de Viedma D, Garzelli C, Gazzola L, Gomes HM, Guttierez MC, Hawkey PM, van Helden PD, Kadival GV, Kreiswirth BN, Kremer K, Kubin M, Kulkarni SP, Liens B, Lillebaek T, Ho ML, Martin C, Martin C, Mokrousov I, Narvskaïa O, Ngeow YF, Naumann L, Niemann S, Parwati I, Rahim Z, Rasolofo-Razanamparany V, Rasolonavalona T, Rossetti ML, Rüsch-Gerdes S, Sajduda A, Samper S, Shemyakin IG, et al. . 2006. Mycobacterium tuberculosis complex genetic diversity: mining the fourth international spoligotyping database (SpolDB4) for classification, population genetics and epidemiology. BMC Microbiol 6:23. doi: 10.1186/1471-2180-6-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Allix-Béguec C, Harmsen D, Weniger T, Supply P, Niemann S. 2008. Evaluation and strategy for use of MIRU-VNTRplus, a multifunctional database for online analysis of genotyping data and phylogenetic identification of Mycobacterium tuberculosis complex isolates. J Clin Microbiol 46:2692–2699. doi: 10.1128/JCM.00540-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weniger T, Krawczyk J, Supply P, Niemann S, Harmsen D. 2010. MIRU-VNTRplus: a web tool for polyphasic genotyping of Mycobacterium tuberculosis complex bacteria. Nucleic Acids Res 38:W326–W331. doi: 10.1093/nar/gkq351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roetzer A, Diel R, Kohl TA, Rückert C, Nübel U, Blom J, Wirth T, Jaenicke S, Schuback S, Rüsch-Gerdes S, Supply P, Kalinowski J, Niemann S. 2013. Whole genome sequencing versus traditional genotyping for investigation of a Mycobacterium tuberculosis outbreak: a longitudinal molecular epidemiological study. PLoS Med 10:e1001387. doi: 10.1371/journal.pmed.1001387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eyre DW, Cule ML, Wilson DJ, Griffiths D, Vaughan A, O’Connor L, Ip CLC, Golubchik T, Batty EM, Finney JM, Wyllie DH, Didelot X, Piazza P, Bowden R, Dingle KE, Harding RM, Crook DW, Wilcox MH, Peto TEA, Walker AS. 2013. Diverse sources of C. difficile infection identified on whole-genome sequencing. N Engl J Med 369:1195–1205. doi: 10.1056/NEJMoa1216064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Harrison EM, Paterson GK, Holden MT, Larsen J, Stegger M, Larsen AR, Petersen A, Skov RL, Christensen JM, Bak Zeuthen A, Heltberg O, Harris SR, Zadoks RN, Parkhill J, Peacock SJ, Holmes MA. 2013. Whole genome sequencing identifies zoonotic transmission of MRSA isolates with the novel mecA homologue mecC. EMBO Mol Med 5:509–515. doi: 10.1002/emmm.201202413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Niemann S, Merker M, Kohl T, Supply P. 2016. Impact of genetic diversity on the biology of Mycobacterium tuberculosis complex strains. Microbiol Spectr 4:TBTB2-0022-2016. doi: 10.1128/microbiolspec.TBTB2-0022-2016. [DOI] [PubMed] [Google Scholar]
- 38.Biek R, Pybus OG, Lloyd-Smith JO, Didelot X. 2015. Measurably evolving pathogens in the genomic era. Trends Ecol Evol 30:306–313. doi: 10.1016/j.tree.2015.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kao RR, Price-Carter M, Robbe-Austerman S. 2016. Use of genomics to track bovine tuberculosis transmission. Rev Sci Tech 35:241–258. doi: 10.20506/rst.35.1.2430. [DOI] [PubMed] [Google Scholar]
- 40.Bjorn-Mortensen K, Soborg B, Koch A, Ladefoged K, Merker M, Lillebaek T, Andersen AB, Niemann S, Kohl TA. 2016. Tracing Mycobacterium tuberculosis transmission by whole genome sequencing in a high incidence setting: a retrospective population-based study in East Greenland. Sci Rep 6:33180. doi: 10.1038/srep33180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bjorn-Mortensen K, Lillebaek T, Koch A, Soborg B, Ladefoged K, Sørensen HC, Kohl TA, Niemann S, Andersen AB. 2017. Extent of transmission captured by contact tracing in a tuberculosis high endemic setting. Eur Respir J 49:1601851. doi: 10.1183/13993003.01851-2016. [DOI] [PubMed] [Google Scholar]
- 42.Deutsches Institut für Normung. 2011. Medical microbiology—diagnosis of tuberculosis—part 3: detection of mycobacteria by culture methods, p 338–356. In DIN-Taschenbuch 222—Medizinische Mikrobiologie und Immunologie—Diagnostische Verfahren. Beuth Verlag, Berlin, Germany: (In German and English; DIN 58943–3: 2011–03.) [Google Scholar]
- 43.Rodriguez JG, Mejia GA, Del Portillo P, Patarroyo ME, Murillo LA. 1995. Species-specific identification of Mycobacterium bovis by PCR. Microbiology 141:2131–2138. doi: 10.1099/13500872-141-9-2131. [DOI] [PubMed] [Google Scholar]
- 44.Moser I, Prodinger WM, Hotzel H, Greenwald R, Lyashchenko KP, Bakker D, Gomis D, Seidler T, Ellenberger C, Hetzel U, Wuennemann K, Moisson P. 2008. Mycobacterium pinnipedii: transmission from South American sea lion (Otaria byronia) to Bactrian camel (Camelus bactrianus bactrianus) and Malayan tapirs (Tapirus indicus). Vet Microbiol 127:399–406. doi: 10.1016/j.vetmic.2007.08.028. [DOI] [PubMed] [Google Scholar]
- 45.Berg S, Firdessa R, Habtamu M, Gadisa E, Mengistu A, Yamuah L, Ameni G, Vordermeier M, Robertson BD, Smith NH, Engers H, Young D, Hewinson RG, Aseffa A, Gordon SV. 2009. The burden of mycobacterial disease in Ethiopian cattle: implications for public health. PLoS One 4:e5068. doi: 10.1371/journal.pone.0005068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.van Soolingen D, Hermans PW, de Haas PE, Soll DR, van Embden JD. 1991. Occurrence and stability of insertion sequences in Mycobacterium tuberculosis complex strains: evaluation of an insertion sequence dependent DNA polymorphism as a tool in the epidemiology of tuberculosis. J Clin Microbiol 29:2578–2586. doi: 10.1128/JCM.29.11.2578-2586.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ruettger A, Nieter J, Skrypnyk A, Engelmann I, Ziegler A, Moser I, Monecke S, Ehricht R, Sachse K. 2012. Rapid spoligotyping of Mycobacterium tuberculosis complex bacteria by use of a microarray system with automatic data processing and assignment. J Clin Microbiol 50:2492–2495. doi: 10.1128/JCM.00442-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Skuce RA, McCorry TP, McCarroll JF, Roring SM, Scott AN, Brittain D, Hughes SL, Hewinson RG, Neill SD. 2002. Discrimination of Mycobacterium tuberculosis complex bacteria using novel VNTR-PCR targets. Microbiology 148:519–528. doi: 10.1099/00221287-148-2-519. [DOI] [PubMed] [Google Scholar]
- 49.Hunter PR, Gaston MA. 1988. Numerical index of the discriminatory ability of typing systems: an application of Simpson’s index of diversity. J Clin Microbiol 26:2465–2466. doi: 10.1128/JCM.26.11.2465-2466.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Baym M, Kryazhimskiy S, Lieberman TD, Chung H, Desai MM, Kishony R. 2015. Inexpensive multiplexed library preparation for megabase-sized genomes. PLoS One 10:e0128036. doi: 10.1371/journal.pone.0128036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kohl TA, Utpatel C, Schleusener V, De Filippo MR, Beckert P, Cirillo DM, Niemann S. 2018. MTBseq: a comprehensive pipeline for whole genome sequence analysis of Mycobacterium tuberculosis complex isolates. PeerJ 6:e5895. doi: 10.7717/peerj.5895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Walker TM, Ip CL, Harrell RH, Evans JT, Kapatai G, Dedicoat MJ, Eyre DW, Wilson DJ, Hawkey PM, Crook DW, Parkhill J, Harris D, Walker AS, Bowden R, Monk P, Smith EG, Peto TE. 2013. Whole-genome sequencing to delineate Mycobacterium tuberculosis outbreaks: a retrospective observational study. Lancet Infect Dis 13:137–146. doi: 10.1016/S1473-3099(12)70277-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Price MN, Dehal PS, Arkin AP. 2010. FastTree 2—approximately maximum-likelihood trees for large alignments. PLoS One 5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.He Z, Zhang H, Gao S, Lercher MJ, Chen WH, Hu S. 2016. Evolview v2: an online visualization and management tool for customized and annotated phylogenetic trees. Nucleic Acids Res 44:W236–W241. doi: 10.1093/nar/gkw370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Suchard MA, Lemey P, Baele G, Ayres DL, Drummond AJ, Rambaut A. 2018. Bayesian phylogenetic and phylodynamic data integration using BEAST 1.10. Virus Evol 4:vey016. doi: 10.1093/ve/vey016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rambaut A, Drummond AJ, Xie D, Baele G, Suchard MA. 2018. Posterior summarization in Bayesian phylogenetics using Tracer 1.7. Syst Biol 67:901–904. doi: 10.1093/sysbio/syy032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Loiseau C, Brites D, Moser I, Coll F, Pourcel C, Robbe-Austerman S, Escuyer V, Musser KA, Peacock SJ, Feuerriegel S, Kohl TA, Niemann S, Gagneux S, Köser CU. 2019. Revised interpretation of the Hain Lifescience GenoType MTBC to differentiate Mycobacterium canettii and members of the M. tuberculosis complex. Antimicrob Agents Chemother 63:e00159-19. doi: 10.1128/AAC.00159-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ghavidel M, Mansury D, Nourian K, Ghazvini K. 2018. The most common spoligotype of Mycobacterium bovis isolated in the world and the recommended loci for VNTR typing; a systematic review. Microb Pathog 118:310–315. doi: 10.1016/j.micpath.2018.03.036. [DOI] [PubMed] [Google Scholar]
- 59.Trewby H, Wright D, Breadon EL, Lycett SJ, Mallon TR, McCormick C, Johnson P, Orton RJ, Allen AR, Galbraith J, Herzyk P, Skuce RA, Biek R, Kao RR. 2016. Use of bacterial whole-genome sequencing to investigate local persistence and spread in bovine tuberculosis. Epidemics 14:26–35. doi: 10.1016/j.epidem.2015.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Biek R, O’Hare A, Wright D, Mallon T, McCormick C, Orton RJ, McDowell S, Trewby H, Skuce RA, Kao RR. 2012. Whole genome sequencing reveals local transmission patterns of Mycobacterium bovis in sympatric cattle and badger populations. PLoS Pathog 8:e1003008. doi: 10.1371/journal.ppat.1003008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Menardo F, Duchêne S, Brites D, Gagneux S. 2019. The molecular clock of Mycobacterium tuberculosis. PLoS Pathog 15:e1008067. doi: 10.1371/journal.ppat.1008067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Merker M, Blin C, Mona S, Duforet-Frebourg N, Lecher S, Willery E, Blum MG, Rüsch-Gerdes S, Mokrousov I, Aleksic E, Allix-Béguec C, Antierens A, Augustynowicz-Kopeć E, Ballif M, Barletta F, Beck HP, Barry CE III, Bonnet M, Borroni E, Campos-Herrero I, Cirillo D, Cox H, Crowe S, Crudu V, Diel R, Drobniewski F, Fauville-Dufaux M, Gagneux S, Ghebremichael S, Hanekom M, Hoffner S, Jiao WW, Kalon S, Kohl TA, Kontsevaya I, Lillebæk T, Maeda S, Nikolayevskyy V, Rasmussen M, Rastogi N, Samper S, Sanchez-Padilla E, Savic B, Shamputa IC, Shen A, Sng LH, Stakenas P, Toit K, Varaine F, Vukovic D, Wahl C, Warren R, Supply P, Niemann S, Wirth T. 2015. Evolutionary history and global spread of the Mycobacterium tuberculosis Beijing lineage. Nat Genet 47:242–249. doi: 10.1038/ng.3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Broad J. 1983. Cattle plague in eighteenth-century England. Agric Hist Rev 31:104–115. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All WGS data were submitted to the EMBL-EBI ENA SRA archive under accession numbers ERR2212113 to ERR2212125, ERR2815506 to ERR2815614, ERR551004, ERR551009, ERR551191, ERR551252, ERR551427, ERR551917, ERR552138 to ERR552140, ERR552470 to ERR552472, ERR552515, ERR552516, ERR552796, ERR552797, ERR553061, ERR553337, and ERR553338 (see Table S1).