The role of mutations in genes associated with phenotypic resistance to bedaquiline (BDQ) and delamanid (DLM) in Mycobacterium tuberculosis complex (MTBc) strains is poorly characterized. A clear understanding of the genetic variants’ role is crucial to guide the development of molecular-based drug susceptibility testing (DST). In this work, we analyzed all mutations in candidate genomic regions associated with BDQ- and DLM-resistant phenotypes using a whole-genome sequencing (WGS) data set from a collection of 4,795 MTBc clinical isolates from six countries with a high burden of tuberculosis (TB).
KEYWORDS: bedaquiline, delamanid, Mycobacterium tuberculosis, resistance to new drugs, whole-genome sequencing
ABSTRACT
The role of mutations in genes associated with phenotypic resistance to bedaquiline (BDQ) and delamanid (DLM) in Mycobacterium tuberculosis complex (MTBc) strains is poorly characterized. A clear understanding of the genetic variants’ role is crucial to guide the development of molecular-based drug susceptibility testing (DST). In this work, we analyzed all mutations in candidate genomic regions associated with BDQ- and DLM-resistant phenotypes using a whole-genome sequencing (WGS) data set from a collection of 4,795 MTBc clinical isolates from six countries with a high burden of tuberculosis (TB). From WGS analysis, we identified 61 and 163 unique mutations in genomic regions potentially involved in BDQ- and DLM-resistant phenotypes, respectively. Importantly, all strains were isolated from patients who likely have never been exposed to these medicines. To characterize the role of mutations, we calculated the free energy variation upon mutations in the available protein structures of Ddn (DLM), Fgd1 (DLM), and Rv0678 (BDQ) and performed MIC assays on a subset of MTBc strains carrying mutations to assess their phenotypic effect. The combination of structural and phenotypic data allowed for cataloguing the mutations clearly associated with resistance to BDQ (n = 4) and DLM (n = 35), only two of which were previously described, as well as about a hundred genetic variants without any correlation with resistance. Significantly, these results show that both BDQ and DLM resistance-related mutations are diverse and distributed across the entire region of each gene target, which is of critical importance for the development of comprehensive molecular diagnostic tools.
INTRODUCTION
The management of drug-resistant tuberculosis (DR-TB) caused by Mycobacterium tuberculosis complex (MTBc) strains poses a serious public health challenge worldwide. The World Health Organization’s (WHO’s) new guidelines recommend the use of bedaquiline (BDQ) for rifampicin-resistant TB (RR-TB), multidrug-resistant TB (MDR-TB) (MTBc strains resistant to at least isoniazid [INH] and rifampicin [RIF]), and extensively drug-resistant TB (XDR-TB) (MDR strains resistant to fluoroquinolones [FQs] and second-line injectable drugs) cases (1). Based on WHO priority groupings of medicines, a delamanid (DLM) compound is recommended when an effective regimen cannot be established using the group A agents BDQ, FQs, and linezolid (LZD) and the group B agents clofazimine (CLF) and cycloserine (CS) (2). Previous studies have shown that MDR/XDR-TB patients treated with BDQ and DLM develop resistance due to fixed mutations in candidate genes, which often appear as previously undescribed novel variants (3–7). Additionally, resistance to BDQ can arise in naive populations of MTBc strains as a consequence of a clofazimine-containing regimen or by random mutations affecting the drug targets (8–14). DLM resistance not associated with exposure has been reported as well (15–19). Moreover, as a member of the nitroimidazooxazines, DLM shares similar resistance mechanisms with a pretomanid (PA-824) compound (20).
Therefore, knowledge of the BDQ and DLM susceptibility status of clinical MTBc isolates before therapy has started and the early detection of emerging resistance in failing regimens are needed to ensure effective treatment of DR-TB.
Here, whole-genome sequencing (WGS)-based approaches, which are rapidly expanding from basic research into routine diagnostic laboratories, provide the advantage of analyzing all resistance targets in a given clinical MTBc strain. However, the routine diagnostic application of WGS requires a much better understanding of the correlation between genotype and phenotype, particularly for new drugs (21, 22).
Currently, the molecular mechanisms leading to resistance to BDQ and DLM are not well described, a fact that jeopardizes the design of a reliable molecular approach to detect resistance.
Mutations in the cellular target of BDQ, the C subunit of ATP synthase encoded by the atpE (rv1305) gene, have been associated with phenotypic resistance (23). In addition, mutations in the transcriptional regulator encoded by the rv0687 gene were frequently associated with an increase of the BDQ MIC due to an upregulation of the MmpL5/MmpS5 efflux pump level, which concurrently leads to cross-resistance to CLF (10, 11, 24). Furthermore, it has been demonstrated that mutations in pepQ (rv2535c) may confer low-level resistance to both CLF and BDQ in clinical isolates (9, 25).
DLM impairs the biosynthesis of mycolic acids and requires activation by the F420-dependent nitroreductase encoded by the ddn gene. The F420 cofactor is synthesized by enzymes encoded by the fbiA, fbiB, fbiC, and fgd1 genes, all of which are involved in DLM off-target mechanisms. Polymorphisms in these genes were shown to be involved in phenotypic DLM resistance (26, 27).
To better describe BDQ and DLM resistance mechanisms, we investigated the genomic regions involved in resistance to BDQ and DLM from 4,795 MTBc isolates collected within a multicountry drug resistance surveillance study and identified variants potentially involved in resistance development (28). All mutations were correlated with strain lineage, DR profile, and country of origin. To combine the genomic data with phenotypes, we performed BDQ and DLM MIC determinations for a subset of these isolates. Finally, for the available three-dimensional (3D) protein structures (Ddn, Fgd1, and Rv0678), we performed an in silico structural analysis in order to estimate the effect of mutations on protein function. This combined information enabled us to provide a first robust catalogue of BDQ and DLM resistance mutations as a basis for the establishment of WGS resistance prediction algorithms for these drugs.
MATERIALS AND METHODS
Study design.
A total of 4,795 MTBc genome sequences (NCBI Sequence Read Archive accession number SRP128089) were considered and investigated in this study. The corresponding MTBc isolates originated from a unique population-based surveillance study across six countries with a high burden of TB or MDR-TB, according to the WHO’s high-burden country list for the period of 2016 to 2020: Azerbaijan (n = 751), Belarus (n = 197), Bangladesh (n = 935), Pakistan (n = 194), South Africa (n = 1,578), and Ukraine (n = 1,140) (28). For our purposes, all sequenced isolates harboring at least one single nucleotide polymorphism (SNP) or insertion/deletion (indel) in at least one of the candidate genomic regions for DLM (n = 5) and/or BDQ (n = 3) resistance were considered for the analysis, excluding synonymous mutations and previously characterized lineage-associated SNPs for which the absence of a correlation with phenotypic DLM resistance was demonstrated (15, 16). For genetic variants detected in more than one isolate, we attempted to replicate results by selecting two isolates from different countries, whenever possible. The flowchart for sample selection, the number of isolates tested, and phenotypic drug susceptibility testing (DST) of selected isolates are reported in Fig. S1 in the supplemental material.
Whole-genome sequencing analysis.
WGS data were generated by both Illumina technology (Illumina, San Diego, CA, USA) and Ion Torrent technology (Thermo Fisher Scientific) as previously described (28). Sequencing data were analyzed using the MTBseq pipeline (version 1.0.2) (29) to identify all variants in the genomes and MTBc lineage. The analysis was performed on the mapped MTBc reads by setting a quality threshold of a mean coverage of at least 20× and an unambiguous base call threshold of ≥70%. A mutation was called only if SNPs and/or indel variants were detected in at least eight reads (both forward and reverse reads) with a minimum Phred quality score of 20 and by considering a mutation frequency of ≥75%. The regions of the MTBc H37Rv reference genome (GenBank accession number NC_000962.3) (30) considered in this study are reported in Table S1 in the supplemental material. The WGS analysis results and distribution of mutations among lineages and countries of isolation are reported in Data Set S1. To verify if promoter variants mapped to potential regulatory regions, promoter sequence prediction analysis was performed by using two online tools, BPROM (http://www.softberry.com/berry.phtml?topic=bprom&group=programs&subgroup=gfindb) and NNPP (http://www.fruitfly.org/seq_tools/promoter.html) (Text S2).
Cluster analysis was performed on the distance matrix generated by the MTBseq pipeline using in-house python scripts (https://github.com/aspitaleri/python). The distance matrix was analyzed using a hierarchical linkage clustering method with a 12-SNP cutoff (31).
MIC assay.
The selected MTBc isolates for genetic variants were subcultured on Lowenstein-Jensen medium and subsequently subjected to MIC testing against BDQ and/or DLM by the resazurin colorimetric microtiter plate assay (REMA) as previously described (16, 32, 33). DLM powder was obtained from Otsuka Pharmaceutical (Tokyo, Japan), and pure BDQ powder was obtained from Janssen Pharmaceutical (Beerse, Belgium). A DLM concentration range of 0.004 to 4 μg/ml and a BDQ concentration range of 0.004 to 2 μg/ml were used, considering the proposed cutoff values of 0.12 μg/ml and 0.06 μg/ml for BDQ and DLM, respectively (34). Based on the MIC results, the isolates were categorized as susceptible (S) (MIC ≤ cutoff), low-level resistant (I) (MIC 1 dilution > cutoff), or resistant (R) (MIC more than 1 dilution > cutoff). All MIC values reported in this work correspond to the MIC100 value that considers any change in color to purple/pink as indicating the presence of viable bacilli (33). For each batch of isolates tested, the H37Rv M. tuberculosis reference strain (M. tuberculosis H37Rv ATCC 27294) was included as a control, and test isolate results of that batch were accepted only if the H37Rv MIC values were within the expected range of ≤0.004 to 0.03 μg/ml for DLM and ≤0.008 to 0.03 μg/ml for BDQ. Further details of REMA protocols are reported in Text S1.
Mutation structural analysis.
We carried out a free energy (ΔΔG) calculation for point mutations in the available protein structures of Ddn (PDB accession number 3R5R), Fgd1 (PDB accession number 3B4Y), and Rv0678 (PDB accession number 4NB5) (35–37) using two endpoint methods, namely, Eris (38) and MAESTRO (39), in order to evaluate the change in protein stability upon mutations. Structural analysis was not performed for stop codons, frameshifts, and SNPs affecting the promoter region; for these variant types, it is not possible to correctly calculate the free energy stability. The stability change, ΔΔG (kilocalories per mole), is computed as the difference between the average stabilities of mutant and wild-type protein structures. Both in silico approaches were used for qualitative cross-validation to evaluate the protein mutation effects, considering 0.34 kcal/mol and 5 kcal/mol as the thresholds for MAESTRO and Eris, respectively.
In addition to folding stability, we calculated the effect of mutations on the complex stabilities of Ddn-F420-H2 and Fgd1-F420-H2 using the DSX pair potential knowledge-based scoring function (40). In the case of the Rv0678 protein, we also performed an in silico analysis to quantify the effect of the mutations on homodimer protein-protein stability. For this purpose, we carried out a molecular mechanics/generalized Born surface area (MM/GBSA) analysis. The calculation was performed using the MMPBSA.py program within the Amber14 suite using the ff14 force field and the GBOBC1 implicit solvent model (41). All in silico results obtained from the analysis are reported in Data Set S2. Further details of in silico analyses are reported in Text S1. Primary protein sequence alignment for the frameshift analysis was performed using the ClustalX algorithm (42). The visualization of mutations on the protein structures was obtained with PyMOL v2.0 (43).
Data availability.
Whole-genome sequencing data for all MTBc isolates were recovered as recalibrated BAM file data from the Sequence Read Archive of the National Center for Biotechnology Information (accession number SRP128089). WGS analysis results of this study, including associated metadata, can be found in Data Set S1 in the supplemental material. In silico results of mutation structural analyses can be found in Data Set S2.
RESULTS
A total of 4,795 whole-genome sequences from MTBc clinical strains were analyzed by considering the candidate genomic regions associated with BDQ resistance (atpE, rv0678, and pepQ) and DLM resistance (ddn, fgd1, fbiA, fbiB, and fbiC). This collection included 731 (17%) MDR and 79 (2%) XDR MTBc strains (see Fig. S1 in the supplemental material). Based on WGS results, we identified totals of 106 and 631 isolates harboring relevant genomic variants potentially involved in BDQ and DLM resistance, respectively. We tested a subset of isolates carrying mutations for phenotypic DST for BDQ (n = 51) and DLM (n = 124) representing 43 and 104 BDQ- and DLM-related variants, respectively (Fig. S1). All genomic variants detected by WGS analysis in candidate genes for BDQ and DLM resistance with the corresponding information on MTBc strain lineage, drug resistance profile, country of isolation, mutation frequency, and MIC results for the tested MTBc isolates are reported in Data Set S1.
Analysis of mutations for BDQ resistance.
The WGS analysis revealed 61 unique mutations in the considered genomic regions associated with BDQ resistance (Fig. S1). The mutation analysis distribution revealed 27 unique mutations in rv0678 (including 7 mutations in the promoter region, 16 nonsynonymous mutations, and 4 indels causing frameshift mutations), 32 unique mutations in the pepQ gene (including 2 upstream mutations, 28 nonsynonymous mutations, and 2 frameshift mutations), and 2 upstream mutations in the atpE gene, while no mutations were found in the atpE coding region (Data Set S1).
Phenotypic testing revealed that isolates carrying four different rv0678 mutations had MIC values above the cutoff of 0.12 μg/ml: two frameshift (fs) mutations in rv0678, G6fs (del_16–17 gg) and the double mutant Q9fs-Y92fs (ins_27 c, ins_274 a), associated with an MIC of 0.5 μg/ml, and two rv0678 nonsynonymous mutations, R96W and M111K, yielding a low level of resistance to BDQ at 0.25 μg/ml (Table 1). The two frameshift mutations associated with BDQ-resistant phenotypes were observed in one MDR isolate and another MDR isolate with concurrent resistance to FQs (corresponding to one new and one previously treated TB case, respectively). Two nonsynonymous mutations associated with low-level resistance were found in two pansusceptible (fully S) MTBc strains (Table 1).
TABLE 1.
rv0678 mutations detected in MTBc strains resistant to BDQg
| Genomic coordinate(s)a | Mutation(s) (amino acid and nucleotide change)h | BDQ MIC (μg/ml) | Countryb | Lineage (Coll lineage no.) | Treatment historyc | DR patternd | Frequencye | Eris ΔΔG (kcal/mol)f |
|---|---|---|---|---|---|---|---|---|
| 779005 | G6fs (Del_16–17 gg) | 0.5 | PAK | Delhi-CAS (3) | New | MDR, FQ R | 1 | NA |
| 779016, 779263i | Q9fs (Ins_27 c), Y92fs (Ins_274 a)i | 0.5 | PAK | EAI (1,1,2) | Retreatment | MDR | 1 | NA |
| 779275 | R96W (cgg/Tgg) | 0.25 | BGD | Delhi-CAS (3) | Retreatment | RIF, INH S | 1 | 10 |
| 779321 | M111K (tag/Aag) | 0.25 | BGD | Haarlem (4,1,2,1) | New | RIF, INH S | 1 | 7.84 |
Genomic position in the H37Rv reference strain (GenBank accession number NC_000962.3).
Country of MTBc isolate origin: Pakistan (PAK) or Bangladesh (BGD).
TB patient treatment history: “New,” new TB case; “Retreatment,” patient with previous TB history and treatment.
Drug resistance pattern of the isolates (see Data Set S1 in the supplemental material).
Frequency of mutation in the WHO database.
ΔΔG values calculated with Eris software are reported where applicable (Data Set 2). NA, not applicable.
All the information is reported for each BDQ resistance-related mutation (MIC ≥ 0.25 μg/ml) and for the MTBc isolate tested for BDQ susceptibility.
Where the original codon and the mutated codon are separated by a slash in parentheses, the uppercase letter in the mutated codon represents the changed nucleotide.
There is a one-to-one correspondence between the pair of entries separated by a comma in the “Genomic coordinate(s)” column and the pair in the “Mutation(s)” column.
The other 39 mutations in the rv0678 and pepQ genes and the corresponding promoter regions were all detected in isolates susceptible to BDQ with MIC values of ≤0.12 μg/ml, including two isolates harboring frameshifts in rv0678: I16fs (ins_46 tcatggaattcg) and A153fs (ins_457 c), showing an MIC of 0.06 μg/ml. (Data Set S1). The protein amino acid sequences obtained from these two frameshift mutations were aligned to the Rv0678 wild-type sequence, highlighting that both wild-type and mutated proteins contain two well-conserved and important regions, from amino acid (aa) positions 34 to 99 (DNA binding domain) and from aa positions 16 to 32 and 101 to 160 (dimerization domains) (Fig. 1). In the case of rv0678 I16fs, the insertion of 12 nucleotides (nt) caused the addition of 4 aa from position 16 of the Rv0678 protein without disrupting the frame of the whole enzyme, while the A153fs mutation caused a change to the last 13 aa of the C terminus of the protein (Fig. 1). This suggests that these frameshifts do not affect protein stability and function resulting in the BDQ-susceptible phenotype.
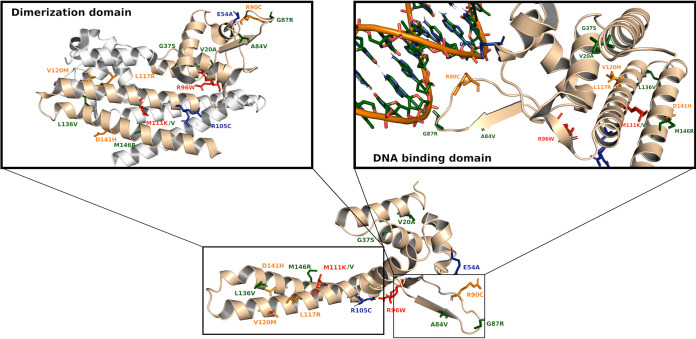
FIG 1.
Amino acid sequence alignment of BDQ-susceptible frameshift mutations in rv0678. The amino acid sequences of the translated proteins with I16fs and A153fs frameshift mutations (associated with a BDQ-susceptible MIC) were aligned with the Rv0678 wild-type amino acid sequence using ClustalX. In both cases, the two insertion mutations cause the addition of new amino acid residues without altering the frame of the Rv0678 protein and functional residues of the protein.
Structural analysis of the effect of mutations in Rv0678 resulting in amino acid changes was performed as described above by using the Eris and MAESTRO computational approaches (Data Set S2). The Rv0678 folding stability calculation suggested that both the R96W and M111K mutations associated with the BDQ-resistant phenotype could alter Rv0678 protein folding/stability (ΔΔG of >5 and >0.34 kcal/mol as determined by Eris and MAESTRO software, respectively). These two mutations are localized in the double-stranded DNA (dsDNA) binding and dimerization domain regions of Rv0678, respectively (Fig. 2). Interestingly, the M111V mutation had a milder effect on protein stability than M111K, which is reflected in the low BDQ MIC value. Both Eris and MAESTRO analyses showed that the other mutations in Rv0678 have a lower estimated effect on protein stability, which seems in accordance with the MIC values for these clinical strains (Data Set S2). As these approaches do not consider the effect of mutations on dimerization protein function, we calculated the protein-protein binding free energy under the MM/GBSA approximation for mutations that localize in the Rv0678 dimerization domain (Fig. 2). The results showed that five mutations (M111K, L117R, V120M, D141H, and M146R) have significant positive ΔΔG (kilocalories per mole) values for homodimer free energy, indicating that they could affect the protein dimerization process (Data Set 2). These data suggest that these mutations could be directly involved in the slight increase in the BDQ MIC for these strains, all with a BDQ MIC of 0.12 μg/ml, except for the MTBc strain harboring the rv0678 M146R variant with a BDQ MIC of 0.06 μg/ml (Fig. 2). The analysis of promoter regions revealed that all mutations observed in the rv0678 and atpE promoters were mapped outside the predicted −10 and −35 regions. For the pepQ promoter, no conserved regulatory element was identified (Text S2). All phenotypically tested strains harboring variants in these promoter regions were susceptible to BDQ (Data Set 1).
FIG 2.
Cartoon representation of Rv0678 protein structures with mutations associated with BDQ-resistant or BDQ-susceptible phenotypes. Shown is a cartoon representation of the monomer present in the X-ray unit cell (PDB accession number 4NB5). The mutations associated with a resistant (red) or susceptible (green) phenotype from BDQ MIC assays are highlighted in sticks. In silico-predicted mutations that could alter dimerization or DNA binding function and are predicted to be associated with the BDQ-susceptible phenotype are shown in orange and blue, respectively. Zoomed-in cartoon representations of the Rv0678 dimerization domain and DNA binding domain are shown.
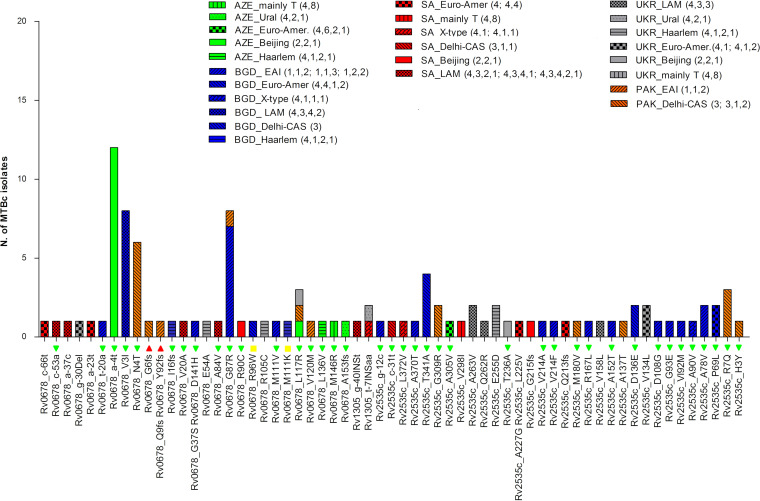
Analysis of the correlation between observed mutations in rv0678, atpE, and pepQ regions and the lineage and country of origin revealed that the majority of mutations (n = 46; 75.4%) occurred only once (Fig. 3). Only four mutations, all in the rv0678 gene, were detected in more than 5 isolates, all of them showing a BDQ-susceptible phenotype: the a-4t mutation in the promoter region of rv0678 was found in 12 Beijing (Coll lineage number 2,2,1) lineage isolates from Azerbaijan, the V3I mutation was found in 8 LAM (4,3,4,2) isolates from Bangladesh, N4T was found in 6 Delhi-CAS (3) isolates from Pakistan, and G87R was found in 8 EAI (1,1,2) isolates from Bangladesh and Pakistan (Fig. 3).
FIG 3.
Lineage and country distributions of MTBc strains with variants in rv0678, atpE (rv1305), and pepQ (rv2535c) genomic regions. The graph reports all mutations found in rv0678, atpE, and pepQ genomic regions and shows their distribution among lineages and countries of isolation. The histograms refer to the number of strains harboring mutations (y axis). The colors of the histograms represent the different countries of isolation, while the patterns inside each bar represent the different lineages. On the x axis, the results of the MIC tests for available MTBc strains are also reported: red triangles are BDQ-resistant strains (MIC > 0.25 μg/ml), yellow boxes are BDQ-low-level-resistant strains (MIC = 0.25 μg/ml), and green triangles are BDQ-susceptible strains (MIC ≤ 0.12 μg/ml).
Analysis of mutations for DLM resistance.
The 631 MTBc isolates identified by WGS as harboring at least one mutation in one of the candidate genes for DLM resistance represented 163 unique DLM-related mutations. The WGS analysis revealed 30 unique mutations in ddn (including 3 nonsense and 2 frameshift mutations), 25 unique mutations in fbiA (including 1 nonsense mutation), 22 unique mutations in fbiB, 62 unique mutations in fbiC (including 2 frameshift mutations), and 24 unique mutations in fgd1 (Data Set S1).
Considering all unique mutations, 20 (12.2%) were combinations of two or three variants in more than one candidate gene. Phenotypic results revealed that out of the 124 isolates tested for DLM, 26 (21%) were resistant to DLM, and 13 (10.5%) showed a low level of resistance (MIC = 0.12 μg/ml), while 85 (68.5%) were DLM susceptible (Data Set S1). The DLM-resistant isolates displayed a total of 32 different mutations (Table 2). Considering the phenotypic drug resistance profiles of the DLM-resistant isolates, only six were MDR-TB strains, five of which were retreatment TB cases and one of which was a new TB case. The analysis of mutation types associated with DLM resistance revealed 3 nonsense mutations leading to truncated proteins, 3 frameshift mutations, and 26 nonsynonymous mutations leading to a single amino acid change (Table 2).
TABLE 2.
ddn, fgd1, fbiA, fbiB, and fbiC mutations detected in MTBc strains resistant to DLMg
| Gene locus | Genomic coordinate(s)a | Mutation(s) (amino acid and nucleotide change)h | DLM MIC (μg/ml) | Countryb | Lineage (Coll lineage no.) | Treatment historyc | DR patternd | Frequencye | Eris ΔΔG (kcal/mol)f |
|---|---|---|---|---|---|---|---|---|---|
| ddn | 3986846 | M1fs (Del_2 t) | 2 | BGD | Eur-Amer. (4,1,2) | New | RIF, INH S | 1 | NA |
| ddn | 3986848 | P2Q (ccg/cAg) | 0.25 | AZE | Mainly T (4,8) | New | RIF, INH S | 1 | NA |
| ddn | 3986885 | S14fs (Del_41 g) | >4 | BGD | Beijing (2,2,1) | Retreatment | MDR | 1 | NA |
| ddn-fbiA | 3986923, 3640445i | W27Stop (tgg/tAg), prom (g-18a)i | >4 | BGD | EAI (1,1,3) | Retreatment | MDR | 1 | NA, NAi |
| ddn | 3986932 | R30H (cgc/cAc) | 1 | BGD | EAI (1,1,3) | Retreatment | RIF, INH S | 1 | NA |
| ddn | 3986944 | G34E (ggg/gAg) | 0.12 | AZE | LAM (4,3,3) | New | RIF, INH S | 9 | NA |
| ddn | 3987015 | Q58Stop (cag/Tag) | >4 | UKR | Beijing (2,2,1) | Retreatment | MDR, FQ R | 3 | NA |
| ddn-fbiA | 3987025, 3641164i | V61G (gtc/gGc), I208V (atc/Gtc)i | 0.12 | SA | Eur-Amer. (4,1,2) | New | RIF, INH S | 3 | 5.33, NAi |
| ddn-fbiA | 3987115, 3640714i | N91T (aac/aCc), V58I (gtc/Atc)i | 0.25 | AZE | Mainly T (4,8) | New | RIF, INH S | 3 | 8.45, NAi |
| ddn | 3987262 | T140I (acc/aTc) | 0.5 | BGD | S type (4,4,1,1) | New | RIF, INH S | 1 | 1.35 |
| fgd1 | 491723 | G314E (gga/gAa) | 0.12 | BGD | Beijing (2,2,1) | New | RIF, INH S | 1 | 0.6 |
| fbiA | 3640546 | K2E (aag/Gag) | 0.12 | AZE | Beijing (2,2,1) | Retreatment | RIF, INH S | 2 | NA |
| fbiA | 3641002 | V154I (gta/Ata) | 0.12 | BGD | Beijing (2,2,1) | Retreatment | MDR, FQ R | 1 | NA |
| fbiA-fbiB | 3641018, 3642877i | P159Q (ccg/cAg), K448R (aag/aGg)i | 0.12 | BGD | Delhi-CAS (3) | Retreatment | RIF, INH S | 1 | NA, NAi |
| fbiA | 3641164 | I208V (atc/Gtc) | 0.25 | BGD | Eur-Amer. (4,1,2) | New | RIF, INH S | 22 | NA |
| fbiA | 3641167 | I209V (atc/Gtc) | 0.5 | BGD | Eur-Amer. (4,5) | New | RIF, INH S | 1 | NA |
| fbiA | 3641403 | C287Stop (tgc/tgA) | 4 | PAK | EAI (1,1,2) | New | RIF, INH S | 1 | NA |
| fbiA | 3641453 | R304Q (cgg/cAg) | 0.25 | PAK | Delhi-CAS (3) | New | MDR | 1 | NA |
| fbiB | 3642195 | G221S (ggc/Agc) | 0.12 | BGD | Beijing (2,2,2) | New | FQ R | 4 | NA |
| fbiB | 3642204 | D224N (gac/Aac) | 0.5 | BGD | Delhi-CAS (3) | New | RIF, INH S | 2 | NA |
| fbiB | 3642351 | G273R (ggc/Cgc) | 0.25 | BGD | X type (4,1,1,3) | New | RIF, INH S | 1 | NA |
| fbiC | 1303241 | Y104C (tat/tGt) | 0.5 | AZE | Beijing (2,2,1) | Retreatment | RIF, INH S | 1 | NA |
| fbiC | 1303265 | G112A (ggc/gCc) | 0.12 | BGD | EAI (1,1,3) | New | RIF, INH S | 2 | NA |
| fbiC | 1303612 | L228F (ctc/Ttc) | 0.12 | BGD | EAI (1,1,3) | New | RIF, INH S | 1 | NA |
| fbiC-fbiB | 1303769, 3642223i | S280L (tcg/tTg), R230Q (cgg/cAg)i | 0.12 | BGD | EAI (1,1,3) | New | RIF, INH S | 1 | NA, NAi |
| fbiC | 1304498 | P523L (cct/cTt) | 0.5 | BGD | EAI (1,1,3) | New | RIF, INH S | 2 | NA |
| fbiC | 1305101 | N724S (aac/aGc) | 0.25 | BGD | Beijing (2,2,1) | New | RIF, INH S | 1 | NA |
| fbiC | 1305215 | S762N (agc/aAc) | 0.12 | BGD | Delhi-CAS (3) | Retreatment | FQ R | 1 | NA |
| fbiC | 1305434 | A835V (gcg/gTg) | 0.5 | BGD | EAI (1,1,3) | New | INH R | 17 | NA |
| fbiC | 1305494 | A855fs (Del 62 nt) | 0.5 | SA | Haarlem (4,1,2,1) | New | RIF, INH S | 28 | NA |
| fbiC-fbiB | 1305494, 3642874i | A855fs (Del 62 nt), L447R (ctg/cGg)i | 0.25 | SA | Mainly T (4,8) | Retreatment | MDR | 1 | NA, NAi |
| fbiC | 1305496 | A856P (gcc/Ccc) | 0.25 | BGD | EAI (1,1,3) | New | RIF, INH S | 1 | NA |
Genomic position in reference to the H37Rv genome (GenBank accession number NC_000962.3).
Country of MTBc isolate origin: Pakistan (PAK), Bangladesh (BGD), Ukraine (UKR), Azerbaijan (AZE), or South Africa (SA).
Patient treatment history: “New,” new TB case; “Retreatment,” patient with previous TB history and treatment.
Drug resistance pattern of the isolates (see Data Set S1 in the supplemental material).
Frequency of mutation in the WHO database.
ΔΔG values calculated using Eris software are reported where applicable (Data Set 2).
All the information is reported for each DLM resistance-related mutation (MIC ≥ 0.12) and for the MTBc isolates tested for DLM susceptibility. prom, promoter; Eur-Amer., Euro-American lineage.
Where the original codon and the mutated codon are separated by a slash in parentheses, the uppercase letter in the mutated codon represents the changed nucleotide.
There is a one-to-one correspondence between the pair of entries separated by a comma in the “Genomic coordinate(s)” column, the pair in the “Mutation(s)” column, and the pair in the “Eris ΔΔG” column.
Overall, the MICs among DLM-resistant isolates ranged from 0.12 μg/ml to ≥4 μg/ml, with the highest MIC values occurring in isolates carrying frameshift or stop codon mutation in the ddn or fbiA gene. The remaining mutations were associated with increased DLM MIC values of between 0.12 and 0.5 μg/ml. Contrary to the high MIC (≥4 μg/ml) observed for the frameshift at codon 14 in the ddn gene, the observed frameshift at the very end of the fbiC gene (codon 855) caused a smaller increase in the MIC, at 0.5 μg/ml (Table 2).
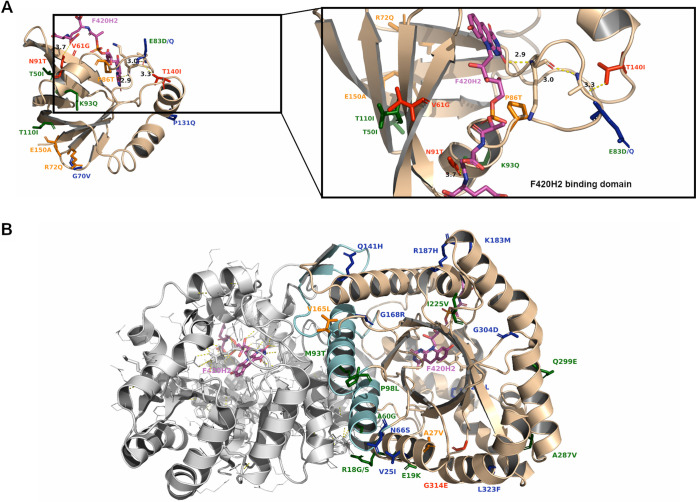
Similar to Rv0678, an in silico evaluation of the effect of mutations on protein stability was performed for the Ddn (PDB accession numbers 3R5L and 3R5R) and Fgd1 (PDB accession numbers 3B4Y and 3C8N) proteins (Data Set S2). The highly conserved Ddn protein catalyzes the reduction of nitroimidazole prodrugs by using the cofactor F420-H2, resulting in the intracellular release of reactive nitrogen species (26). For the on-target Ddn protein, mutations N91T and P86T localize very close to the cofactor binding site (Fig. 4A), and ΔDSX energies resulting from DrugScore analysis indicate that these are the only two mutations that could reduce the binding affinity between Ddn and the cofactor F420-H2 (Data Set S2). Interestingly, the Ddn mutation T140I detected in one isolate with a DLM MIC of 0.5 μg/ml localized 13 to 15 Å away from the cofactor binding site, and its calculated ΔΔG values were highly positive, suggesting that this mutation could act allosterically on decreasing the stability of the Ddn-F420-H2 complex. Indeed, the side chain of the T140 residue is involved in a long hydrogen bond network, including A82-K79-F420-H2, and mutation T140I could also alter this interaction (Fig. 4A). Mutation V61G, which was observed in one isolate with a DLM MIC of 0.12 μg/ml, also showed significantly positive ΔΔG values by both Eris and MAESTRO analyses (Fig. 4A). An analysis of the role of point mutations in Ddn stability without available MIC values revealed significantly positive ΔΔG values for mutations R72Q, P86T, and E150A, suggesting their potential involvement in reducing protein stability (Data Set S2).
FIG 4.
Cartoon representation of Ddn and Fgd1 protein structures with mutations associated with a DLM-resistant or a DLM-susceptible phenotype. (A) Ddn protein bound to the F420 cofactor (PDB accession number 3R5R), with mutations associated with a resistant (red) or susceptible (green) phenotype highlighted in sticks. The in silico-predicted DLM-resistant mutations (orange) and DLM-susceptible mutations (blue) are also reported. F420 is shown as magenta licorice. A zoomed-in cartoon representation of Ddn bound to F420 is shown. The T140-A82-K79-F420-H2 hydrogen bond network is shown as a dashed yellow line, and the A82 and K79 residues are shown as sticks. (B) Fgd1 cartoon representation of homodimer-Fgd1 bound to F420 (PDB accession number 3B4Y). Helices involved in protein dimerization are shown in cyan. Highlighted in sticks are the DLM-resistant (red), DLM-susceptible (green), in silico-predicted DLM-resistant (orange), and in silico-predicted DLM-susceptible (blue) mutations.
The DLM off-target F420-dependent glucose-6-phosphate dehydrogenase (Fgd1) is implicated in DLM redox processes related to nonreplicating persistence by providing the reduced cofactor F420-H2 (35). The reported MIC values did not show a strong effect in the in vitro experiments, probably because all identified mutations are farther than 10 Å away from the cofactor F420 binding site (Fig. 4B). Moreover, the computational Eris approach predicts that two Fgd1 mutations without phenotypic data, A27G and V165L, have potential roles in reducing protein stability (Data Set S2). The Fgd1 mutation G314E, which was observed in a strain with only a moderate increase in the DLM MIC, seemed to poorly correlate with the DLM phenotype, suggesting that other factors could contribute to this small variation in the DLM MIC in MTBc strains.
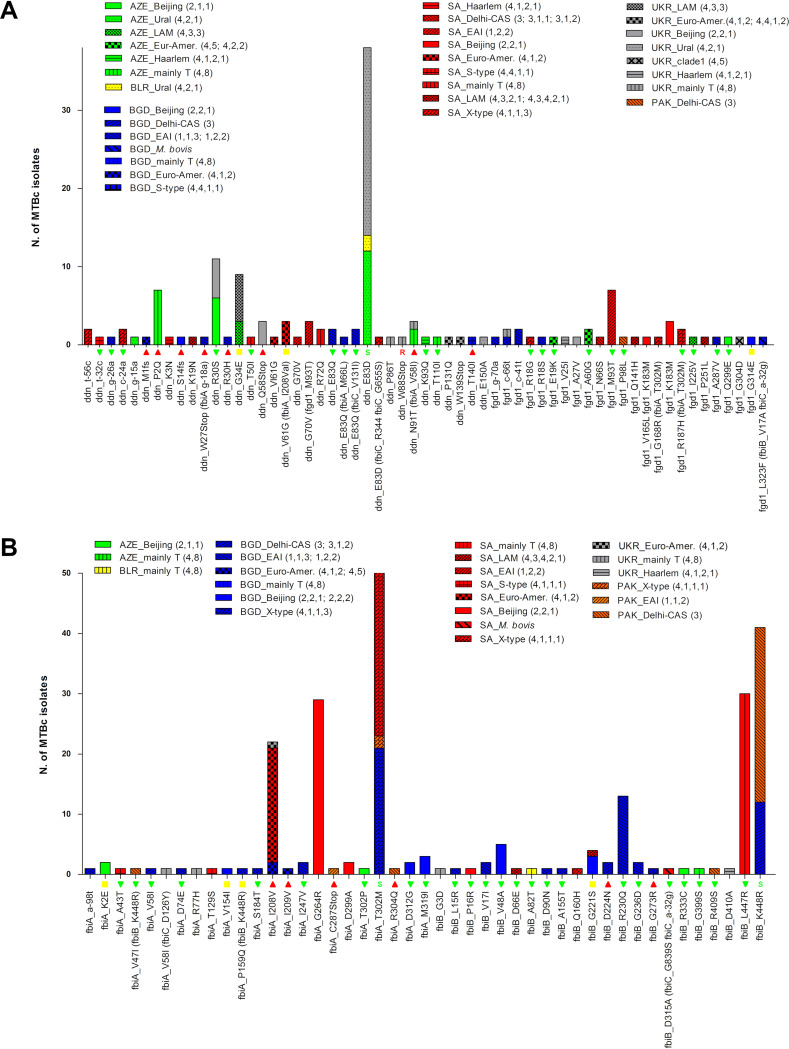
The distribution analysis of mutations across genotypes and countries of isolation showed that ddn mutations detected in isolates with a DLM-resistant phenotype are represented only once or twice. Exceptions are P2Q, which was found in seven mainly T (4,8) isolates (all from Azerbaijan); N91T in ddn in combination with mutation V58I in fbiA in three mainly T (4,8) isolates (two from Azerbaijan and one from Ukraine); and the high-level-resistant stop codon mutation Q58Stop, which was detected in three Beijing (2,2,1) isolates from Ukraine (Fig. 5A). Mutation E83D was the most frequent variant in the ddn gene, observed in 38 Ural (4,2,1,1) isolates from Azerbaijan, Belarus, and Ukraine and in 1 X-type (4,1,1,1) isolate from South Africa (Fig. 5A). The E83D mutation was previously described in one MTBc strain belonging to the Ural (4,2,1,1) lineage with susceptibility to DLM (16). Two DLM-resistant mutations in the fbiA-fbiB region were seen in a single isolate, while the fbiB mutation D224N was found in two Delhi-CAS (3) isolates from Bangladesh. Mutation I208V in fbiA was the most prevalent mutation and was seen in 22 isolates, all belonging to the Euro-American lineage (clade 1; 4,1,2) and isolated in South Africa (n = 19), Bangladesh (n = 2), and Ukraine (n = 1) (Fig. 5B). Four of the seven DLM-resistant fbiC mutation types were seen in single isolates; one was observed in two isolates, while two were more prevalent: the frameshift mutation A855fs (a deletion of 62 nt), which was detected in 30 isolates from South Africa belonging to eight different lineages, and mutation A835V detected in 18 EAI (1,1,3) isolates from Bangladesh only (Fig. 6). Of note, most of the 21 DLM-resistant strains with high MICs (≥0.25 μg/ml) were isolated in Bangladesh (75%), and mutations were detected in ddn (33.5%), fbiC (29%), fbiA (25%), and fbiB (12.5%). Three of them were isolates with a double-mutation pattern (Table 2).
FIG 5.
Lineage and country distributions of MTBc strains with variants in the ddn (rv3547), fgd1 (rv0407), and fbiA-fbiB (rv3261-rv3262) genomic regions. The graph reports all mutations found in the ddn and fgd1 (A) and the fbiA and fbiB (B) genomic regions, showing their distributions among lineages and countries of isolation. The histograms refer to the number of strains harboring mutations (y axis). The colors of the histograms represent the different countries, while the patterns inside each bar represent the different lineages. On the x axis, the results of the MIC tests for available MTBc strains are also reported: red triangles are DLM-resistant strains (MIC ≥ 0.12 μg/ml), yellow boxes are low-level-resistant strains (MIC = 0.12 μg/ml), and green triangles are DLM-susceptible strains (MIC < 0.12 μg/ml). If a mutation was previously described in the literature, it is also reported (“S” for susceptibility and “R” for resistance to DLM).
FIG 6.
Lineage and country distributions of MTBc strains with variants in the fbiC (rv1173) genomic region. In this graph are all mutations found in the fbiC genomic region, showing their distribution among lineages and countries of isolation. See the Fig. 5 legend for details.
Finally, promoter analysis allowed the identification of conserved regulatory regions only in the fbiA-fbiB promoter, but the two detected variants mapped outside these regions (Text S2). Similar to the results from the promoter analysis of BDQ regions, none of the promoter variants in DLM-related genomic regions were observed in MTBc isolates associated with DLM resistance (Data Set 1).
Cluster analysis of the distance matrix was also performed in order to understand if the observation of three or more isolates with mutations associated with DLM resistance were potential clonal clusters. The results showed that the three Beijing (2,2,1) isolates harboring the stop codon mutation Q58Stop in the ddn gene were part of the same transmission chain, at a 12-SNP cutoff (Fig. S2). Furthermore, 10 other clusters of two isolates each were identified in all the other groups harboring mutations associated with DLM resistance: 4 clusters of isolates with the I208V mutation in fbiA; 1 cluster of isolates with the A855fs variant in fbiC; 2 clusters of isolates with P2Q in ddn; 1 cluster of isolates with N91T and V58I mutations in ddn and fbiA, respectively; and 2 clusters of isolates with fbiC A835V (Fig. S2).
DISCUSSION
BDQ and DLM have expanded available treatment options and improved treatment success rates for patients with pulmonary RR-, MDR-, and XDR-TB (44–47), including children with MDR-TB (48, 49). The detection of resistance to BDQ and DLM is critical for ensuring effective treatment and care for DR-TB patients and preventing transmission. Although evidence for the validation and standardization of efficient methods for MICs and the setting of breakpoints for BDQ and DLM continues to expand (22, 34, 50, 51), there is still a notable lack of suitable data on resistance-related genomic variants (52). Moreover, phenotypic methods are too slow to provide an early indication of susceptibility status at the time of treatment initiation. Accurate classification of genetic variants according to their association with drug resistance is therefore essential for the development of interpretable molecular DST for BDQ and DLM and to guide the composition of treatment regimens (7, 53).
In this study, we used a data set containing 4,795 whole-genome sequences of MTBc isolates from different countries with a high burden of either TB or MDR-TB (Azerbaijan, Bangladesh, Belarus, South Africa, Pakistan, and Ukraine) as a unique and accurate source of genetic information for the characterization of genomic variants involved in BDQ and DLM resistance. In particular, this study highlighted the role of genetic variants in BDQ and DLM resistance development by combining the MIC results for MTBc isolates with variants and the results of in silico analysis of available protein structures, paving the way for the construction of an encyclopedia of characterized mutations to be used for molecular DST.
From the entire WGS data set, we identified 61 different BDQ-related variants in rv0678 (44.2%), the atpE promoter (3.2%), and pepQ genomic regions (52.4%) from 106 clinical isolates. Of the 51 MTBc isolates tested for BDQ susceptibility, 4 were associated with a BDQ-resistant phenotype: 2 carrying frameshift mutations in rv0678 associated with high BDQ MICs and 2 with nonsynonymous mutations found associated with low levels of BDQ resistance. To the best of our knowledge, only mutation G6fs (del_16–17 gg) has been previously described in one BDQ-resistant isolate (54). In agreement with the data from the in vitro MIC experiments, the in silico structural analysis of the Rv0678 protein (PDB accession number 4NB5) showed that mutations M111K and R96W have highly positive ΔΔG (kilocalories per mole) values, indicating a potential mutation effect on protein folding stability. Mutations G87R and L117R in rv0678 were previously described in BDQ-susceptible strains (13), confirming the detected low MICs of 0.03 μg/ml and 0.12 μg/ml, respectively (see Data Set S1 in the supplemental material). The role of L117R remains unclear, as another study described this mutation as being associated with both BDQ and CLF resistance (11). Other mutations in rv0678 were described at the same codon position but with a different amino acid change (5, 8, 9, 11, 13). The rv0678 mutations V20F, A84E, and R90P were observed to be linked to increased MICs for CLF and potentially associated with a BDQ-resistant phenotype, while V20G was associated with both BDQ and CLF resistance (5, 9). In this study, the rv0678 mutations V20A and A84V were found in BDQ-susceptible strains (MIC ≤ 0.008 μg/ml), while R90C was associated with a BDQ MIC value of 0.12 μg/ml. Moreover, in silico analysis confirmed that the R90C mutation can have a mild effect on protein stability and a probable role in the small variation of BDQ MICs. Mutations R96Q, M146T, and L136P in rv0678 have been described with increased MIC values for BDQ (8, 11, 13), while in this data set, mutations R96W, M146R, and L136V showed MICs of 0.25 μg/ml (BDQ resistant), 0.06 μg/ml (BDQ susceptible), and 0.03 μg/ml (BDQ susceptible), respectively. Again, the in silico results were in agreement with the phenotypic data, showing that only the R96W mutation has higher positive ΔΔG values than the other two mutations (Data Set S2). Furthermore, in silico MM/GBSA analysis suggested that mutations L117R, V120M, and D141H can affect Rv0678 homodimerization, which could explain the slight increase in the BDQ MIC to 0.12 μg/ml for these MTBc strains, still classified as BDQ susceptible despite being close to the proposed cutoff.
Considering all mutations detected in the five target genes for DLM resistance, a total of 631 isolates (13%) harbored variants potentially involved in DLM resistance. The WGS analysis revealed 163 unique mutations distributed among the ddn (21.4%), fgd1 (14.7%), fbiA (14.1%), fbiB (13.4%), and fbiC (36.1%) genes. Apart from seven previously characterized mutations (16), all the other 156 DLM-related variants have not been previously described, except for N91T in ddn, for which we confirmed its role in DLM resistance (18). Phenotypic results for a subset of isolates showed that 32 different mutations detected in all of the considered genomic regions were associated with DLM resistance. Notably, the in silico mutation analysis revealed that the predicted effect of point mutations on Ddn and Fgd1 protein stability were in agreement with the MIC results (Data Set S2). Indeed, the in silico analysis of the N91T mutation in Ddn suggested its involvement in reducing the binding affinity for the cofactor F420-H2 but also in destabilizing Ddn folding. Mutations V61G and T140I, which were observed in MTBc strains with DLM MICs of 0.12 μg/ml and 0.5 μg/ml, respectively, also showed positive ΔΔG (kilocalories per mole) values, suggesting a potential effect on Ddn stability. Furthermore, these in silico analyses indicate that three mutations in Ddn for which corresponding phenotypic data were not available (R72Q, P86T, and E150A) may have an impact on protein stability and thereby could play a role in the DLM-resistant phenotype.
A concerningly high prevalence of mutations associated with an increased MIC or potentially increasing MICs (by in silico prediction) was detected in clinical isolates from patients who had never been exposed to these drugs, as reported in previous studies (10, 16). By comparing the mutation frequency of the nonessential genes considered in this work (excluding atpE which is essential) with the average frequency of the other 2,813 nonessential genes from the same genomes, we observed similar mutation rates, thus being comparable to the natural genetic variation of M. tuberculosis clinical isolates (data not shown). Furthermore, our data correlate with the previously described in vitro rates of DLM and BDQ spontaneous mutations, higher for DLM, between 10−5 and 10−6, and lower for BDQ, between 10−7 and 10−9 (16, 55).
In addition, data from the correlation analysis between mutations linked to resistant phenotypes and metadata information corroborate previously reported data, suggesting the absence of links between BDQ or DLM resistance and strain lineage or drug resistance profiles of MTBc isolates (10, 16). Globally, considering the DST profiles of BDQ- and DLM-resistant strains, 75% were fully susceptible, 6% were MDR-TB, and the majority of them (68.7%) were from new TB cases.
To complete this set of analyses, we constructed an SNP-based distance matrix to evaluate the relatedness of strains harboring the most frequent DLM-resistant variants, which showed that for these groups, there are no major transmission chains but only small clusters of two to three isolates, meaning that these resistance-associated mutations can arise spontaneously and independently. Conversely, none of the BDQ-resistant variants were detected in clustering groups. Further phylogenetic analysis may be able to elucidate if the mutations observed in these genes arose once or multiple times in the phylogeny (homoplasy) by adding evolutionary evidence of their role in DLM or BDQ resistance (56).
The use of bioinformatics tools like Eris and MAESTRO allowed the evaluation of the potential structural and biophysical effects of putative mutations. In particular, these in silico approaches modeled the effects of mutations to identify a correlation between structural protein features and phenotypic data, as previously described for other M. tuberculosis target proteins (57, 58).
These findings also showed the presence of 125 genetic variants not associated with BDQ and DLM resistance, scattered over the full length of each target gene. Furthermore, considering the absence of fully standardized phenotypic tests, the development of accurate molecular-based DST is wholly dependent on the establishment of a complete database of validated mutations, a scenario which is comparable to the challenges associated with molecular markers of resistance to pyrazinamide and ethionamide (59, 60). A global database containing data from MTBc isolates collected in a large number of settings with the inclusion of different parameters (phenotype, genotype, structure, and free energy analyses) will improve our understanding of the role of mutations in determining the BDQ/DLM susceptibility phenotype. Finally, this database will also be beneficial for studying genetic resistance to other drugs that share a similar genetic basis of resistance, such as clofazimine for BDQ and pretomanid for DLM.
For these reasons, despite some limitations (not all strains were available for MIC determination; there is an absence of standardized methods and breakpoints for the interpretation of BDQ and DLM phenotypes; 3D structures of the FbiA, FbiB, FbiC, and PepQ proteins were unavailable for in silico investigation; and there is a lack of knowledge of other genomic regions potentially involved in BDQ/DLM resistance), this work will be an important source of information for new genome-based sequencing approaches for predicting BDQ and DLM resistance.
Supplementary Material
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.WHO. 2019. Global tuberculosis report, 2019. WHO, Geneva, Switzerland: https://www.who.int/tb/publications/global_report/en/. [Google Scholar]
- 2.WHO. 2019. Consolidated guidelines on drug-resistant tuberculosis treatment. WHO, Geneva, Switzerland: https://www.who.int/tb/publications/2019/consolidated-guidelines-drug-resistant-TB-treatment/en/. [Google Scholar]
- 3.Bloemberg GV, Keller PM, Stucki D, Trauner A, Borrell S, Latshang T, Coscolla M, Rothe T, Hömke R, Ritter C, Feldmann J, Schulthess B, Gagneux S, Böttger EC. 2015. Acquired resistance to bedaquiline and delamanid in therapy for tuberculosis. N Engl J Med 373:1986–1988. doi: 10.1056/NEJMc1505196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hoffmann H, Kohl T, Hofmann-Thiel S, Merker M, Beckert P, Jaton K, Nedialkova L, Sahalchyk E, Rothe T, Keller PM, Niemann S. 2016. Delamanid and bedaquiline resistance in Mycobacterium tuberculosis ancestral Beijing genotype causing extensively drug-resistant tuberculosis in a Tibetan refugee. Am J Respir Crit Care Med 193:337–340. doi: 10.1164/rccm.201502-0372LE. [DOI] [PubMed] [Google Scholar]
- 5.Ghodousi A, Rizvi AH, Baloch AQ, Ghafoor A, Khanzada FM, Qadir M, Borroni E, Trovato A, Tahseen S, Cirillo DM. 2019. Acquisition of cross-resistance to bedaquiline and clofazimine following treatment for tuberculosis in Pakistan. Antimicrob Agents Chemother 63:e00915-19. doi: 10.1128/AAC.00915-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Polsfuss S, Hofmann-Thiel S, Merker M, Krieger D, Niemann S, Rüssmann H, Schönfeld N, Hoffmann H, Kranzer K. 2019. Emergence of low-level delamanid and bedaquiline resistance during extremely drug-resistant tuberculosis treatment. Clin Infect Dis 69:1229–1231. doi: 10.1093/cid/ciz074. [DOI] [PubMed] [Google Scholar]
- 7.Andres S, Merker M, Heyckendorf J, Kalsdorf B, Rumetshofer R, Indra A, Hofmann-Thiel S, Hoffmann H, Lange C, Niemann S, Maurer FP. 2020. Bedaquiline-resistant tuberculosis: dark clouds on the horizon. Am J Respir Crit Care Med 201:1564–1568. doi: 10.1164/rccm.201909-1819LE. [DOI] [PubMed] [Google Scholar]
- 8.Torrea G, Coeck N, Desmaretz C, Van De Parre T, Van Poucke T, Lounis N, de Jong BC, Rigouts L. 2015. Bedaquiline susceptibility testing of Mycobacterium tuberculosis in an automated liquid culture system. J Antimicrob Chemother 70:2300–2305. doi: 10.1093/jac/dkv117. [DOI] [PubMed] [Google Scholar]
- 9.Zhang S, Chen J, Cui P, Shi W, Zhang W, Zhang Y. 2015. Identification of novel mutations associated with clofazimine resistance in Mycobacterium tuberculosis. J Antimicrob Chemother 70:2507–2510. doi: 10.1093/jac/dkv150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Villellas C, Coeck N, Meehan CJ, Lounis N, de Jong B, Rigouts L, Andries K. 2017. Unexpected high prevalence of resistance-associated Rv0678 variants in MDR-TB patients without documented prior use of clofazimine or bedaquiline. J Antimicrob Chemother 72:684–690. doi: 10.1093/jac/dkw502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu J, Wang B, Hu M, Huo F, Guo S, Jing W, Nuermberger E, Lu Y. 2017. Primary clofazimine and bedaquiline resistance among isolates from patients with multidrug-resistant tuberculosis. Antimicrob Agents Chemother 61:e00239-17. doi: 10.1128/AAC.00239-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ismail NA, Omar SV, Joseph L, Govender N, Blows L, Ismail F, Koornhof H, Dreyer AW, Kaniga K, Ndjeka N. 2018. Defining bedaquiline susceptibility, resistance, cross-resistance and associated genetic determinants: a retrospective cohort study. EBioMedicine 28:136–142. doi: 10.1016/j.ebiom.2018.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martinez E, Hennessy D, Jelfs P, Crighton T, Chen SC, Sintchenko V. 2018. Mutations associated with in vitro resistance to bedaquiline in Mycobacterium tuberculosis isolates in Australia. Tuberculosis (Edinb) 111:31–34. doi: 10.1016/j.tube.2018.04.007. [DOI] [PubMed] [Google Scholar]
- 14.Kardan-Yamchi J, Kazemian H, Battaglia S, Abtahi H, Foroushani AR, Hamzelou G, Cirillo DM, Ghodousi A, Feizabadi MM. 2020. Whole genome sequencing results associated with minimum inhibitory concentrations of 14 anti-tuberculosis drugs among rifampicin-resistant isolates of Mycobacterium tuberculosis from Iran. J Clin Med 9:465. doi: 10.3390/jcm9020465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feuerriegel S, Köser CU, Baù D, Rüsch-Gerdes S, Summers DK, Archer JA, Marti-Renom MA, Niemann S. 2011. Impact of fgd1 and ddn diversity in Mycobacterium tuberculosis complex on in vitro susceptibility to PA-824. Antimicrob Agents Chemother 55:5718–5722. doi: 10.1128/AAC.05500-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schena E, Nedialkova L, Borroni E, Battaglia S, Cabibbe AM, Niemann S, Utpatel C, Merker M, Trovato A, Hofmann-Thiel S, Hoffmann H, Cirillo DM. 2016. Delamanid susceptibility testing of Mycobacterium tuberculosis using the resazurin microtitre assay and the BACTEC MGIT 960 system. J Antimicrob Chemother 71:1532–1539. doi: 10.1093/jac/dkw044. [DOI] [PubMed] [Google Scholar]
- 17.Pang Y, Zong Z, Huo F, Jing W, Ma Y, Dong L, Li Y, Zhao L, Fu Y, Huang H. 2017. In vitro drug susceptibility of bedaquiline, delamanid, linezolid, clofazimine, moxifloxacin, and gatifloxacin against extensively drug-resistant tuberculosis in Beijing, China. Antimicrob Agents Chemother 61:e00900-17. doi: 10.1128/AAC.00900-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fujiwara M, Kawasaki M, Hariguchi N, Liu Y, Matsumoto M. 2018. Mechanisms of resistance to delamanid, a drug for Mycobacterium tuberculosis. Tuberculosis (Edinb) 108:186–194. doi: 10.1016/j.tube.2017.12.006. [DOI] [PubMed] [Google Scholar]
- 19.Yang JS, Kim KJ, Choi H, Lee SH. 2018. Delamanid, bedaquiline and linezolid minimum inhibitory concentration distributions and resistance-related gene mutations in multidrug-resistant and extensively drug-resistant tuberculosis in Korea. Ann Lab Med 38:563–568. doi: 10.3343/alm.2018.38.6.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wen S, Jing W, Zhang T, Zong Z, Xue Y, Shang Y, Wang F, Huang H, Chu N, Pang Y. 2019. Comparison of in vitro activity of the nitroimidazoles delamanid and pretomanid against multidrug-resistant and extensively drug-resistant tuberculosis. Eur J Clin Microbiol Infect Dis 38:1293–1296. doi: 10.1007/s10096-019-03551-w. [DOI] [PubMed] [Google Scholar]
- 21.Ellington MJ, Ekelund O, Aarestrup FM, Canton R, Doumith M, Giske C, Grundman H, Hasman H, Holden MTG, Hopkins KL, Iredell J, Kahlmeter G, Köser CU, MacGowan A, Mevius D, Mulvey M, Naas T, Peto T, Rolain JM, Samuelsen Ø, Woodford N. 2017. The role of whole genome sequencing in antimicrobial susceptibility testing of bacteria: report from the EUCAST Subcommittee. Clin Microbiol Infect 23:2–22. doi: 10.1016/j.cmi.2016.11.012. [DOI] [PubMed] [Google Scholar]
- 22.Meehan CJ, Goig GA, Kohl TA, Verboven L, Dippenaar A, Ezewudo M, Farhat MR, Guthrie JL, Laukens K, Miotto P, Ofori-Anyinam B, Dreyer V, Supply P, Suresh A, Utpatel C, van Soolingen D, Zhou Y, Ashton PM, Brites D, Cabibbe AM, de Jong BC, de Vos M, Menardo F, Gagneux S, Gao Q, Heupink TH, Liu Q, Loiseau C, Rigouts L, Rodwell TC, Tagliani E, Walker TM, Warren RM, Zhao Y, Zignol M, Schito M, Gardy J, Cirillo DM, Niemann S, Comas I, Van Rie A. 2019. Whole genome sequencing of Mycobacterium tuberculosis: current standards and open issues. Nat Rev Microbiol 17:533–545. doi: 10.1038/s41579-019-0214-5. [DOI] [PubMed] [Google Scholar]
- 23.Andries K, Verhasselt P, Guillemont J, Göhlmann HW, Neefs JM, Winkler H, Van Gestel J, Timmerman P, Zhu M, Lee E, Williams P, de Chaffoy D, Huitric E, Hoffner S, Cambau E, Truffot-Pernot C, Lounis N, Jarlier V. 2005. A diarylquinoline drug active on the ATP synthase of Mycobacterium tuberculosis. Science 307:223–227. doi: 10.1126/science.1106753. [DOI] [PubMed] [Google Scholar]
- 24.Andries K, Villellas C, Coeck N, Thys K, Gevers T, Vranckx L, Lounis N, de Jong BC, Koul A. 2014. Acquired resistance of Mycobacterium tuberculosis to bedaquiline. PLoS One 9:e102135. doi: 10.1371/journal.pone.0102135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Almeida D, Ioerger T, Tyagi S, Li SY, Mdluli K, Andries K, Grosset J, Sacchettini J, Nuermberger E. 2016. Mutations in pepQ confer low-level resistance to bedaquiline and clofazimine in Mycobacterium tuberculosis. Antimicrob Agents Chemother 60:4590–4599. doi: 10.1128/AAC.00753-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haver HL, Chua A, Ghode P, Lakshminarayana SB, Singhal A, Mathema B, Wintjens R, Bifani P. 2015. Mutations in genes for the F420 biosynthetic pathway and a nitroreductase enzyme are the primary resistance determinants in spontaneous in vitro-selected PA-824-resistant mutants of Mycobacterium tuberculosis. Antimicrob Agents Chemother 59:5316–5323. doi: 10.1128/AAC.00308-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matsumoto M, Hashizume H, Tomishige T, Kawasaki M, Tsubouchi H, Sasaki H, Shimokawa Y, Komatsu M. 2006. OPC-67683, a nitrodihydro-imidazooxazole derivative with promising action against tuberculosis in vitro and in mice. PLoS Med 3:e466. doi: 10.1371/journal.pmed.0030466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zignol M, Cabibbe AM, Dean AS, Glaziou P, Alikhanova N, Ama C, Andres S, Barbova A, Borbe-Reyes A, Chin DP, Cirillo DM, Colvin C, Dadu A, Dreyer A, Driesen M, Gilpin C, Hasan R, Hasan Z, Hoffner S, Hussain A, Ismail N, Kamal SMM, Khanzada FM, Kimerling M, Kohl TA, Mansjö M, Miotto P, Mukadi YD, Mvusi L, Niemann S, Omar SV, Rigouts L, Schito M, Sela I, Seyfaddinova M, Skenders G, Skrahina A, Tahseen S, Wells WA, Zhurilo A, Weyer K, Floyd K, Raviglione MC. 2018. Genetic sequencing for surveillance of drug resistance in tuberculosis in highly endemic countries: a multi-country population-based surveillance study. Lancet Infect Dis 18:675–683. doi: 10.1016/S1473-3099(18)30073-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kohl TA, Utpatel C, Schleusener V, De Filippo MR, Beckert P, Cirillo DM, Niemann S. 2018. MTBseq: a comprehensive pipeline for whole genome sequence analysis of Mycobacterium tuberculosis complex isolates. PeerJ 6:e5895. doi: 10.7717/peerj.5895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lew JM, Kapopoulou A, Jones LM, Cole ST. 2011. TubercuList—10 years after. Tuberculosis (Edinb) 91:1–7. doi: 10.1016/j.tube.2010.09.008. [DOI] [PubMed] [Google Scholar]
- 31.Meehan CJ, Moris P, Kohl TA, Pečerska J, Akter S, Merker M, Utpatel C, Beckert P, Gehre F, Lempens P, Stadler T, Kaswa MK, Kühnert D, Niemann S, de Jong BC. 2018. The relationship between transmission time and clustering methods in Mycobacterium tuberculosis epidemiology. EBioMedicine 37:410–416. doi: 10.1016/j.ebiom.2018.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lopez B, Siqueira de Oliveira R, Pinhata JMW, Chimara E, Pacheco Ascencio E, Puyén Guerra ZM, Wainmayer I, Simboli N, Del Granado M, Palomino JC, Ritacco V, Martin A. 2019. Bedaquiline and linezolid MIC distributions and epidemiological cut-off values for Mycobacterium tuberculosis in the Latin American region. J Antimicrob Chemother 74:373–379. doi: 10.1093/jac/dky414. [DOI] [PubMed] [Google Scholar]
- 33.Palomino JC, Martin A, Camacho M, Guerra H, Swings J, Portaels F. 2002. Resazurin microtiter assay plate: simple and inexpensive method for detection of drug resistance in Mycobacterium tuberculosis. Antimicrob Agents Chemother 46:2720–2722. doi: 10.1128/AAC.46.8.2720-2722.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.WHO. 2018. Technical report on critical concentrations for drug susceptibility testing of medicines used in the treatment of drug-resistant tuberculosis. WHO/CDS/TB/2018.5 WHO, Geneva, Switzerland. [Google Scholar]
- 35.Bashiri G, Squire CJ, Moreland NJ, Baker EN. 2008. Crystal structures of F420-dependent glucose-6-phosphate dehydrogenase FGD1 involved in the activation of the anti-tuberculosis drug candidate PA-824 reveal the basis of coenzyme and substrate binding. J Biol Chem 283:17531–17541. doi: 10.1074/jbc.M801854200. [DOI] [PubMed] [Google Scholar]
- 36.Cellitti SE, Shaffer J, Jones DH, Mukherjee T, Gurumurthy M, Bursulaya B, Boshoff HI, Choi I, Nayyar A, Lee YS, Cherian J, Niyomrattanakit P, Dick T, Manjunatha UH, Barry CE III, Spraggon G, Geierstanger BH. 2012. Structure of Ddn, the deazaflavin-dependent nitroreductase from Mycobacterium tuberculosis involved in bioreductive activation of PA-824. Structure 20:101–112. doi: 10.1016/j.str.2011.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Radhakrishnan A, Kumar N, Wright CC, Chou TH, Tringides ML, Bolla JR, Lei HT, Rajashankar KR, Su CC, Purdy GE, Yu EW. 2014. Crystal structure of the transcriptional regulator Rv0678 of Mycobacterium tuberculosis. J Biol Chem 289:16526–16540. doi: 10.1074/jbc.M113.538959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yin S, Ding F, Dokholyan NV. 2007. Eris: an automated estimator of protein stability. Nat Methods 4:466–467. doi: 10.1038/nmeth0607-466. [DOI] [PubMed] [Google Scholar]
- 39.Laimer J, Hofer H, Fritz M, Wegenkittl S, Lackner P. 2015. MAESTRO—multi agent stability prediction upon point mutations. BMC Bioinformatics 16:116. doi: 10.1186/s12859-015-0548-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Neudert G, Klebe G. 2011. DSX: a knowledge-based scoring function for the assessment of protein-ligand complexes. J Chem Inf Model 51:2731–2745. doi: 10.1021/ci200274q. [DOI] [PubMed] [Google Scholar]
- 41.Miller BR III, McGee TD Jr, Swails JM, Homeyer N, Gohlke H, Roitberg AE. 2012. MMPBSA.py: an efficient program for end-state free energy calculations. J Chem Theory Comput 8:3314–3321. doi: 10.1021/ct300418h. [DOI] [PubMed] [Google Scholar]
- 42.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 43.Schrödinger, LLC. 2015. The PyMOL molecular graphics system, version 2.0. Schrödinger, LLC, New York, NY. [Google Scholar]
- 44.Migliori GB, Pontali E, Sotgiu G, Centis R, D’Ambrosio L, Tiberi S, Tadolini M, Esposito S. 2017. Combined use of delamanid and bedaquiline to treat multidrug-resistant and extensively drug-resistant tuberculosis: a systematic review. Int J Mol Sci 18:341. doi: 10.3390/ijms18020341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ferlazzo G, Mohr E, Laxmeshwar C, Hewison C, Hughes J, Jonckheere S, Khachatryan N, De Avezedo V, Egazaryan L, Shroufi A, Kalon S, Cox H, Furin J, Isaakidis P. 2018. Early safety and efficacy of the combination of bedaquiline and delamanid for the treatment of patients with drug-resistant tuberculosis in Armenia, India, and South Africa: a retrospective cohort study. Lancet Infect Dis 18:536–544. doi: 10.1016/S1473-3099(18)30100-2. [DOI] [PubMed] [Google Scholar]
- 46.Li Y, Sun F, Zhang W. 2019. Bedaquiline and delamanid in the treatment of multidrug-resistant tuberculosis: promising but challenging. Drug Dev Res 80:98–105. doi: 10.1002/ddr.21498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Olayanju O, Esmail A, Limberis J, Dheda K. 2020. A regimen containing bedaquiline and delamanid compared to bedaquiline in patients with drug-resistant tuberculosis. Eur Respir J 55:1901181. doi: 10.1183/13993003.01181-2019. [DOI] [PubMed] [Google Scholar]
- 48.WHO. 2016. The use of delamanid in the treatment of multidrug-resistant tuberculosis in children and adolescents: interim policy guidance. WHO/HTM/TB/2016.14 WHO, Geneva, Switzerland. [PubMed] [Google Scholar]
- 49.Achar J, Hewison C, Cavalheiro AP, Skrahina A, Cajazeiro J, Nargiza P, Herboczek K, Rajabov AS, Hughes J, Ferlazzo G, Seddon JA, du Cros P. 2017. Off-label use of bedaquiline in children and adolescents with multidrug-resistant tuberculosis. Emerg Infect Dis 23:1711–1713. doi: 10.3201/eid2310.170303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rancoita PMV, Cugnata F, Gibertoni Cruz AL, Borroni E, Hoosdally SJ, Walker TM, Grazian C, Davies TJ, Peto TEA, Crook DW, Fowler PW, Cirillo DM, CRyPTIC Consortium. 2018. Validating a 14-drug microtiter plate containing bedaquiline and delamanid for large-scale research susceptibility testing of Mycobacterium tuberculosis. Antimicrob Agents Chemother 62:e00344-18. doi: 10.1128/AAC.00344-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kaniga K, Aono A, Borroni E, Cirillo DM, Desmaretz C, Hasan R, Joseph L, Mitarai S, Shakoor S, Torrea G, Ismail NA, Omar SV. 2020. Validation of bedaquiline phenotypic drug susceptibility testing methods and breakpoints: a multilaboratory, multicountry study. J Clin Microbiol 58:e01677-19. doi: 10.1128/JCM.01677-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Köser CU, Maurer FP, Kranzer K. 2019. ‘Those who cannot remember the past are condemned to repeat it’: drug-susceptibility testing for bedaquiline and delamanid. Int J Infect Dis 80:S32–S35. doi: 10.1016/j.ijid.2019.02.027. [DOI] [PubMed] [Google Scholar]
- 53.Cabibbe AM, Walker TM, Niemann S, Cirillo DM. 2018. Whole genome sequencing of Mycobacterium tuberculosis. Eur Respir J 52:1801163. doi: 10.1183/13993003.01163-2018. [DOI] [PubMed] [Google Scholar]
- 54.Veziris N, Bernard C, Guglielmetti L, Le Du D, Marigot-Outtandy D, Jaspard M, Caumes E, Lerat I, Rioux C, Yazdanpanah Y, Tiotiu A, Lemaitre N, Brossier F, Jarlier V, Robert J, Sougakoff W, Aubry A, CNR MyRMA and the Tuberculosis Consilium of the CNR MyRMA. 2017. Rapid emergence of Mycobacterium tuberculosis bedaquiline resistance: lessons to avoid repeating past errors. Eur Respir J 49:1601719. doi: 10.1183/13993003.01719-2016. [DOI] [PubMed] [Google Scholar]
- 55.Ismail N, Omar SV, Ismail NA, Peters RPH. 2018. Collated data of mutation frequencies and associated genetic variants of bedaquiline, clofazimine and linezolid resistance in Mycobacterium tuberculosis. Data Brief 20:1975–1983. doi: 10.1016/j.dib.2018.09.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tantivitayakul P, Ruangchai W, Juthayothin T, Smittipat N, Disratthakit A, Mahasirimongkol S, Viratyosin W, Tokunaga K, Palittapongarnpim P. 2020. Homoplastic single nucleotide polymorphisms contributed to phenotypic diversity in Mycobacterium tuberculosis. Sci Rep 10:8024. doi: 10.1038/s41598-020-64895-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Portelli S, Phelan JE, Ascher DB, Clark TG, Furnham N. 2018. Understanding molecular consequences of putative drug resistant mutations in Mycobacterium tuberculosis. Sci Rep 8:15356. doi: 10.1038/s41598-018-33370-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lee BM, Harold LK, Almeida DV, Afriat-Jurnou L, Aung HL, Forde BM, Hards K, Pidot SJ, Ahmed FH, Mohamed AE, Taylor MC, West NP, Stinear TP, Greening C, Beatson SA, Nuermberger EL, Cook GM, Jackson CJ. 2020. Predicting nitroimidazole antibiotic resistance mutations in Mycobacterium tuberculosis with protein engineering. PLoS Pathog 16:e1008287. doi: 10.1371/journal.ppat.1008287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vilchèze C, Jacobs WR Jr. 2014. Resistance to isoniazid and ethionamide in Mycobacterium tuberculosis: genes, mutations, and causalities. Microbiol Spectr 2:MGM2-0014-2013. doi: 10.1128/microbiolspec.MGM2-0014-2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Miotto P, Cabibbe AM, Feuerriegel S, Casali N, Drobniewski F, Rodionova Y, Bakonyte D, Stakenas P, Pimkina E, Augustynowicz-Kopeć E, Degano M, Ambrosi A, Hoffner S, Mansjö M, Werngren J, Rüsch-Gerdes S, Niemann S, Cirillo DM. 2014. Mycobacterium tuberculosis pyrazinamide resistance determinants: a multicenter study. mBio 5:e01819-14. doi: 10.1128/mBio.01819-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Whole-genome sequencing data for all MTBc isolates were recovered as recalibrated BAM file data from the Sequence Read Archive of the National Center for Biotechnology Information (accession number SRP128089). WGS analysis results of this study, including associated metadata, can be found in Data Set S1 in the supplemental material. In silico results of mutation structural analyses can be found in Data Set S2.