Entamoeba histolytica infection is an increasingly common sexually transmitted infection in Japan. Currently, stool ova and parasite examination (O&P) is the only approved diagnostic method. Here, we assessed the utility of the commercially available rapid antigen detection test (Quik Chek) for E. histolytica. A multicenter cross-sectional study was conducted. Stool samples that had been submitted for O&P were included. The samples were subjected to both Quik Chek and PCR, and the Quik Chek results were assessed in comparison with PCR as the reference standard.
KEYWORDS: parasitology, amebiasis, diagnosis, cyst, Entamoeba histolytica, parasitology
ABSTRACT
Entamoeba histolytica infection is an increasingly common sexually transmitted infection in Japan. Currently, stool ova and parasite examination (O&P) is the only approved diagnostic method. Here, we assessed the utility of the commercially available rapid antigen detection test (Quik Chek) for E. histolytica. A multicenter cross-sectional study was conducted. Stool samples that had been submitted for O&P were included. The samples were subjected to both Quik Chek and PCR, and the Quik Chek results were assessed in comparison with PCR as the reference standard. E. histolytica infection was confirmed in 5.8% (38/657) of the samples and comprised 20 diarrheal and 18 nondiarrheal cases. The overall sensitivity and specificity of Quik Chek were 44.7% (95% confidence interval, 30.1 to 60.3) and 99.8% (99.1 to 100), respectively. The sensitivity of Quik Chek was higher for diarrheal cases (60.0%) than for nondiarrheal cases (27.8%). Furthermore, the combined use of Quik Chek with O&P increased the sensitivity (78.9%), especially for diarrheal cases (up to 90%). The E. histolytica burden assessed by quantitative PCR was similar between Quik Chek-positive and -negative samples. The Quik Chek assay sensitivity was lower for cyst-containing stools than for trophozoite-containing stools, although it was shown that cultured E. histolytica clinical strains from Quik Chek-negative cyst-containing stools exhibited antigenicity in vitro. The present study confirmed the high specificity of Quik Chek for E. histolytica infection. Combined use with O&P increased the sensitivity of detection, facilitating the use of Quik Chek in point-of-care settings in nonendemic situations.
INTRODUCTION
Entamoeba histolytica, the causative agent of invasive amebiasis, is the second most common parasitic cause of mortality worldwide (1). Over the past 2 decades, invasive amebiasis has become prevalent not only in developing countries, where food and water are frequently contaminated by feces, but also in several developed countries in Asia and Europe (2–5). In these areas, the pathogen spreads as a sexually transmitted infection, especially among men who have sex with men and within the HIV-infected population (2, 5, 6). Furthermore, recent data indicate that this pathogen is also spreading among HIV-uninfected men and women in Japan (7–9).
In Japan, stool ova and parasite examination (O&P) is the only approved diagnostic method for E. histolytica infection as of 25 August 2020. O&P is a low-cost and rapid diagnostic tool for enteric parasite infection; however, in most developed countries, it is not recommended for the diagnosis of E. histolytica (10, 11). This is because (i) O&P cannot distinguish nonpathogenic Entamoeba spp., such as E. dispar and E. moshkovskii, from potentially pathogenic E. histolytica, and (ii) the sensitivity and specificity of O&P are highly dependent on local health care settings, such as the skill of the technician and the timing of the examination, as O&P should be carried out by a well-trained laboratory technician immediately (within 1 h) after sampling. Moreover, enteric parasite infections are not common in regions with good hygiene, resulting in many developed countries having fewer opportunities for the training of health care professionals in the O&P technique. The PCR is the most reliable diagnostic tool for the detection of E. histolytica; however, this procedure, which includes the extraction of DNA from various types of clinical samples, remains too technically complex for point-of-care use and too expensive for the routine clinical diagnosis of E. histolytica infection (10, 11). On the other hand, E. histolytica antigen detection tests are easy to perform and relatively inexpensive. For example, the E. histolytica Quik Chek test is an FDA-approved rapid test based on immunochromatography, and the utility of this test has been proven in several studies performed in developing countries (12–14). However, reports from developed countries are limited (15).
In the present study, we assessed the utility of Quik Chek through a multicenter cross-sectional study in Japan, and we sought to determine the best use of this technique to improve the diagnosis of amebiasis in clinical settings.
MATERIALS AND METHODS
Study design and sampling.
This multicenter cross-sectional study was carried out between October 2018 and December 2019. Clinical specimens were prospectively collected from patients with suspected enteric parasite infections at five regional core hospitals in Japan, including the National Center for Global Health and Medicine (primary facility, Tokyo), Hokkaido University Hospital (Hokkaido), Tokyo Metropolitan Cancer and Infectious Diseases Center Komagome Hospital (Tokyo), The Institute of Medical Science at The University of Tokyo (Tokyo), and Kumamoto University Hospital (Kumamoto). All stool samples, which had been submitted for O&P for diagnostic purposes, were recruited for the present analysis after anonymization. Clinical information other than the O&P diagnosis and stool form as scored with the Bristol stool scale were not handled in the present study. Thereafter, the collected specimens were examined with the Quik Chek assay (E. histolytica Quik Chek; Techlab, Blacksburg, VA, USA) independently from O&P at each institution. The remainder of the specimens was stored at −20°C and transferred to the National Institute of Infectious Diseases (Tokyo) for PCR detection of E. histolytica and other protozoa. This study was approved by the ethics committee of each facility as well as the National Center for Global Health and Medicine Center (approval no. NCGM-G-002516-00). This study was implemented in accordance with the provisions of the Declaration of Helsinki.
DNA extraction from stool samples.
Stool specimens (approximately 0.2 g) were weighed and subjected to DNA extraction using a QIAamp Fast DNA Stool minikit (Qiagen, Hilden, Germany). DNA extraction was performed according to the manufacturer’s instructions and a previous report (16). The DNA was eluted in 100 μl of elution buffer (Qiagen) and stored at −80°C until further analysis.
Conventional and quantitative PCR.
A single-round conventional PCR (cPCR) assay for the detection of three Entamoeba species (E. histolytica, E. dispar, and E. moshkovskii) was carried out. The primer set was designed based on signature sequences in the small-subunit rRNA of each species, of which, the utility was confirmed in a previous study (17). The primer set consisted of the same forward primer (EntaF, 5′-ATGCACGAGAGCGAAAGCAT-3′) in combination with three reverse primers, one for each of the three species (EhR, 5′-GATCTAGAAACAATGCTTCTCT-3′; EdR, 5′-CACCACTTACTACC-3′; EmR, 5′-CACCACCACTTACTATCCCTACC-3′). Entamoeba species were differentiated based on the sizes of the PCR products (a 166-bp PCR product for E. histolytica, a 752-bp PCR product for E. dispar, and a 580-bp PCR product for E. moshkovskii). Finally, the results were confirmed by DNA sequencing. For cases in which E. histolytica infection was confirmed by cPCR, quantitative PCR (qPCR) was additionally performed using a 6-carboxyfluorescein (FAM)-conjugated probe (TCATT+GAATGAATTGGCCATTT) and an Entamoeba primer set (Ehd-88R, 5′-GCGGACGGCTCATTATAACA-3′, and EM-RT-F2, 5′-GTCCTCGATACTACCAAC-3′) (18). The pathogen burden of E. histolytica was presented as the quantity of trophozoite DNA per milligram stool relative to standard reference samples from an axenically cultured experimental strain (HM-1:IMSS).
Antigen detection test using the E. histolytica Quik Chek assay.
For the antigen detection test, we used the E. histolytica Quik Chek (Techlab, Blacksburg, VA, USA) assay according to the package insert. In brief, all reagents and specimens were brought to room temperature before testing. Next, 25 μl of liquid stool or a 2-mm-diameter portion of solid stool homogenized with 500 μl of diluent was premixed with a drop of the conjugate. All of the diluted sample (∼500 μl) was added to a sample well of the test membrane in the device and incubated for 15 min. Finally, 300 μl of wash buffer, followed by two drops of the substrate, was added directly to the reaction window.
E. histolytica culture from clinical specimens.
Cultivation of E. histolytica was attempted for all cases diagnosed as having E. histolytica infection at the National Center for Global Health and Medicine, within the ambit of the written informed consent obtained for another study (approval no. NCGM-G-001566-02). Isolation of axenic E. histolytica was performed in accordance with the protocols previously published by Clark and Diamond (19). In brief, stool samples were first inoculated into xenic culture media (e.g., BR medium [R medium precultured with Escherichia coli] followed by Robinson medium), either directly for trophozoite-containing diarrheal stools or after initial treatment with 0.1 N hydrochloric acid for 10 min for cyst-containing formed stools. Thereafter, E. histolytica was cultivated in the trophozoite form in xenic culture media for several weeks or months. After obtaining stable growth in xenic culture media, monoaxenic culture was achieved by washing xenically cultured E. histolytica in phosphate-buffered saline and placing it into a rich medium containing Crithidia fasciculata and antibiotics (e.g., 8,000 U penicillin G, 0.04 g streptomycin, 12,000 U polymyxin B, plus 0.2 ml of antibiotic antimycotic 100× solution [Sigma-Aldrich, Merck KGaA, Germany] in 4 ml of medium). After cultivation in the monoxenic medium for several weeks or months, E. histolytica was finally cultured in Crithidia-free medium. The axenic clinical strains were then maintained in YIMDHA-S medium.
Statistical analyses.
In the present study, cases were defined as being positive for E. histolytica infection where the identification of E. histolytica was confirmed by PCR in stool samples. The sensitivity and specificity of O&P or the antigen detection test were calculated with reference to the PCR data. Comparisons of the qualitative data were carried out with the chi-square test, and analysis of variance (ANOVA) was used for comparisons of quantitative data. Statistical significance was defined as a two-sided P value of <0.05. All statistical analyses were performed using GraphPad Prism 7.0 (GraphPad Software, Inc., San Diego, CA, USA).
RESULTS
Study subjects.
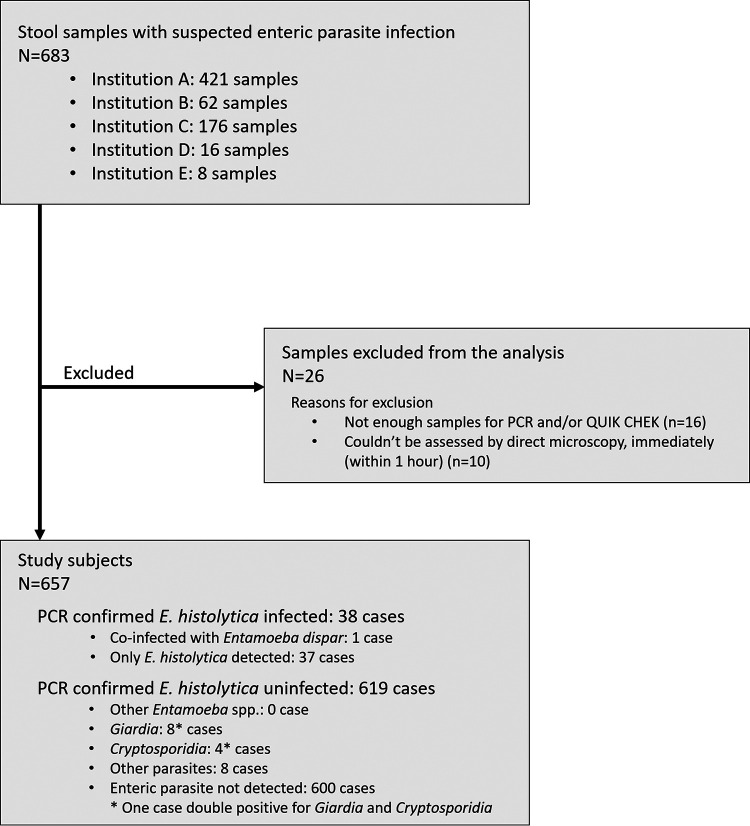
In total, 683 stool samples were collected during the study period (Fig. 1; see also Fig. S1 in the supplemental material). Of these, 26 were excluded from the analyses, either because there was insufficient sample remaining after O&P (16 samples) or O&P could not be performed immediately after sampling due to the unavailability of a skilled laboratory technician (10 samples). Therefore, 657 samples were included for further analysis to assess the utility of Quik Chek. E. histolytica PCR identified 5.8% (38/657) of samples as positive, including 20 diarrheal cases (Bristol score 6 or 7) and 18 nondiarrheal cases (Bristol score 5 or lower). During O&P, trophozoite forms were observed more frequently in diarrheal stools, whereas the cystic forms were more frequently seen in formed stools (Fig. 2A). Coinfection with E. dispar was reported in one of the E. histolytica-positive diarrheal cases. On the other hand, among the 619 E. histolytica-negative samples, enteric parasites were detected in 19 cases, and these included eight cases of Giardia and four cases of Cryptosporidium. However, nonpathogenic Entamoeba spp. were not reported in the E. histolytica-negative samples, although recently discovered E. bangladeshi was not assessed in the present study (Fig. 1). These results suggest that E. histolytica infection is the most common enteric parasite in Japan, which emphasizes the need for well-constructed diagnostic systems for E. histolytica infection.
FIG 1.
Workflow of the clinical specimen collection. Institutions A, B, C, D, and E are National Center for Global Health and Medicine (primary facility, Tokyo), The Institute of Medical Science at The University of Tokyo (Tokyo), Tokyo Metropolitan Cancer and Infectious Diseases Center at Komagome Hospital (Tokyo), Kumamoto University Hospital (Kumamoto), and Hokkaido University Hospital (Hokkaido), respectively.
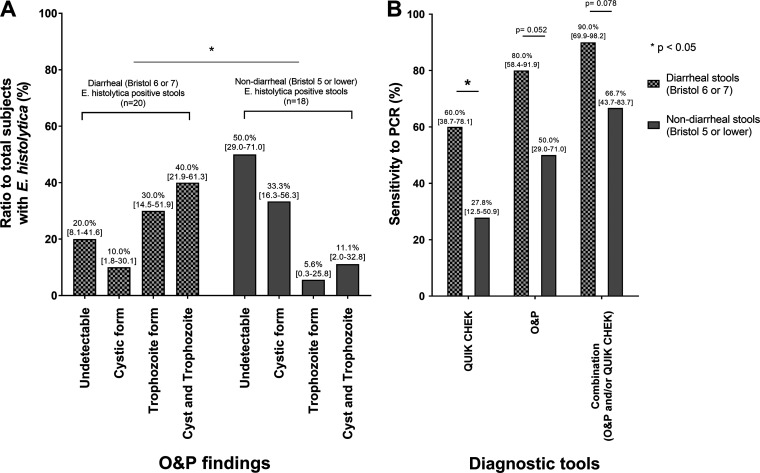
FIG 2.
O&P findings and diagnostic sensitivities among diarrheal and nondiarrheal cases. (A) Morphologic diagnosis by stool ova and parasite examination (O&P) of the 657 samples included in this study identified 38 that were positive for E. histolytica. The number of cases in each subgroup of the O&P findings is expressed as a ratio to the total number of subjects in each of the diarrheal stool (n = 20) and formed stool (n = 18) groups. (B) The sensitivity of each diagnostic tool was compared between diarrheal and nondiarrheal stool samples. When using a combination of O&P and Quik Chek, a sample was deemed positive when either O&P or the antigen detection test was positive. O&P, ova and parasite examination; Quik Chek, rapid antigen detection test (E. histolytica Quik Chek).
Diagnostic value of E. histolytica antigen testing and O&P.
Next, we investigated the sensitivity and specificity of Quik Chek and O&P using the PCR results as a reference standard (Table 1). The sensitivity of Quik Chek was 44.7% (95% confidence interval, 30.1% to 60.3%), which was significantly lower than that of O&P (65.8% [49.9% to 78.8%]). On the other hand, the specificity of Quik Chek was 99.8% (99.1% to 100%). No cross-reactivity was seen with the other protozoa infections. Interestingly, the specificity of O&P was equivalent to that of Quik Chek in this study population, probably because nonpathogenic Entamoeba spp. are rarely seen in Japan. When a combination of O&P and Quik Chek was applied to the diagnosis of E. histolytica infection, and positive samples were defined as having either a positive O&P or Quik Chek result, the sensitivity and specificity were 78.9% (63.7% to 88.9%) and 99.7% (98.8% to 99.9%), respectively (Table 1). For the diarrheal cases, in particular, the combined use of O&P and Quik Chek increased the sensitivity to 90% (Fig. 2B).
TABLE 1.
Sensitivity and specificity of each method for the diagnosis of E. histolytica infection with reference to PCR-confirmed cases
| Result | Performance relative to PCR (no. of samples) |
Sensitivity (% [95% CIa ]) | Specificity (% [95% CI]) | ||
|---|---|---|---|---|---|
| Positive | Negative | Total | |||
| E. histolytica Quik Chek | |||||
| Positive | 17 | 1 | 18 | 44.7 (30.1–60.3) | |
| Negative | 21 | 618 | 639 | 99.8 (99.1–100) | |
| Total | 38 | 619 | |||
| O&Pb | |||||
| Positive | 25 | 1 | 26 | 65.7 (49.9–78.8) | |
| Negative | 13 | 618 | 631 | 99.8 (99.1–100) | |
| Total | 38 | 619 | |||
| Combination of O&P and Quik Chekc | |||||
| Positive | 30 | 2 | 32 | 78.9 (63.7–88.9) | |
| Negative | 8 | 617 | 625 | 99.7 (98.8–99.9) | |
| Total | 38 | 619 | |||
CI, confidence interval.
O&P, ova and parasite examination.
Judged as positive when either O&P or Quik Chek produced a positive result.
Effects of pathogen forms and burden on the sensitivity of the antigen detection test.
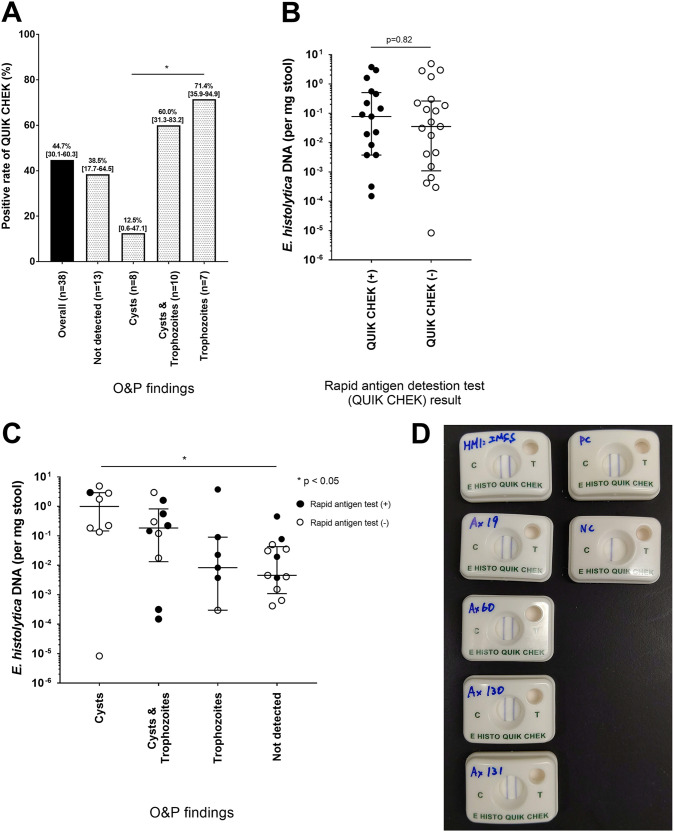
Next, to assess the factors affecting the sensitivity of Quik Chek, we compared the sensitivities of Quik Chek according to the O&P findings. The targeted antigen for this immunochromatography kit is a surface adhesin (Gal/GalNAc lectin) that is highly expressed on the surface of trophozoites (13, 14). As expected, the sensitivity of Quik Chek was only 12.5% for stools containing cysts alone, whereas it was relatively high (70.6%) for trophozoite-containing stools (Fig. 3A). Interestingly, it was shown that Quik Chek identified E. histolytica infection in 38.5% (17.7% to 64.5%) of stool samples with negative O&P results.
FIG 3.
Factors potentially influencing the diagnostic test outcome. (A) The sensitivity of the rapid antigen detection test (Quik Chek) was compared in accordance with the O&P findings. (B) The pathogen burden in the stool samples was determined by quantitative PCR (qPCR) and compared between the positive and negative stool samples based on the antigen detection test. (C) The pathogen burden quantified by qPCR was compared in accordance with the O&P findings. (D) Quik Chek was applied to in vitro cultured clinical strains using a concentration of 106 trophozoites/ml. HM-1:IMSS, laboratory reference strain; Ax19, clinical strain from Quik Chek-positive liver abscess sample; Ax60, Ax130, and Ax131, clinical strains from Quik Chek-negative stool samples; PC, positive control; NC, negative control; O&P, ova and parasite examination; Quik Chek, rapid antigen detection test (E. histolytica Quik Chek).
The pathogen burden assessed by quantitative PCR for E. histolytica was similar between Quik Chek-positive and -negative stool samples (Fig. 3B). Furthermore, the cyst-containing stools showed a relatively higher pathogen burden than the trophozoite-containing or O&P-negative stool samples (Fig. 3C). We sought to check the potential antigenicity of E. histolytica in the Quik Chek-negative cases by in vitro culture. Three strains (Ax60, Ax130, and Ax131) were successfully isolated by in vitro passage of the trophozoites from cyst-containing Quik Chek-negative stool samples. Positive Quik Chek results were then obtained using these cultured strains (Fig. 3D). Thus, taken together, the sensitivity of this rapid antigen detection test is more dependent on the form of E. histolytica in the stool samples than on the pathogen burden or genetic properties of each strain.
DISCUSSION
In the present study, we evaluated the utility of a commercial rapid antigen detection test based on immunochromatography (E. histolytica Quik Chek) in clinical settings in Japan. Notably, Quik Chek showed high specificity (99.8%). On the other hand, the overall sensitivity of Quik Chek in detecting PCR-confirmed cases was 44.7%, although it was shown that Quik Chek exhibited relatively higher sensitivity (60.0%) when looking at diarrheal cases alone. Unexpectedly, in the present study, the sensitivity of Quik Chek was lower than that of O&P, not only for nondiarrheal E. histolytica-positive cases but also diarrheal cases. This was probably because O&P was carried out at core hospitals in Japan by highly trained technicians, resulting in the relatively high sensitivity of O&P. However, in 10 cases, stool samples had to be excluded from O&P analysis because of the unavailability of a technician at the time of sampling. Furthermore, it was shown that 38.5% of the stool samples diagnosed as negative by O&P were diagnosed as positive by Quik Chek. Importantly, it was shown that the combined use of O&P with Quik Chek resulted in increased sensitivity overall. Therefore, Quik Chek could play an important role in the diagnosis of invasive amebiasis in point-of-care settings in Japan.
Here, the sensitivity of Quik Chek was found to be particularly low for cyst-containing stools, which are typically nondiarrheal samples. This was as expected, because the antigen targeted by this assay is an adhesin (Gal/GalNAc lectin) on the surface of the trophozoite form of E. histolytica (11).
The potency of antigen presentation in trophozoite isolates was not assessed in previous studies. In the present study, it was confirmed that Quik Chek showed positive results when using in vitro cultured strains derived from cyst-containing antigen-negative stool samples. Thus, the sensitivity of Quik Chek is more dependent on adhesin expression (a phenotypic property of the pathogen) in different stool environments than on the genetic properties of each strain. This indicates that this antigen detection test may be widely applicable to local clinical strains, although more data should be collected from other geographical areas to confirm this. In addition, factors affecting lectin expression in the gut environment, such as the gut microbiome, should be investigated in future studies.
There were several limitations to the present study. First, this study was carried out using anonymized stool samples that were previously submitted for suspected enteric parasitic infections. Hence, the patients’ other clinical data were not available. Factors affecting the sensitivity and specificity, such as antibiotic treatment history and travel history to an area of endemicity, could not be assessed. Moreover, other etiologies of intestinal infectious/noninfectious diseases could not be ruled out. Second, O&P in this study was carried out by highly trained technicians at core hospitals in Japan. The sensitivity and specificity of O&P are highly influenced by technical skill, the level of which differs between health care settings. Local health care settings should, therefore, be taken into consideration before applying our study results.
In conclusion, the present study used a multicenter cross-sectional study design in Japan that confirmed the high specificity of Quik Chek for E. histolytica infection. The combined use of Quik Chek with O&P increased the sensitivity of diagnosis, which may facilitate the detection of E. histolytic infection in point-of-care settings in nonendemic situations.
Supplementary Material
ACKNOWLEDGMENTS
We thank all the medical staff who participated in the patients’ care and assisted with sample collection at each study site. We also thank Natasha Beeton-Kempen, from Edanz Group, for editing a draft of the manuscript.
We declare no conflict of interest.
This work was supported by a Health Labor Sciences research grant (grant number H30-AIDS-general-010), Research Program on Emerging and Re-emerging Infectious Diseases of Japan from the Japan Agency for Medical Research and Development (AMED) (grant number JP20fk0108138), and a grant from the National Center for Global Health and Medicine (grant number 19A2016).
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Lozano R, Naghavi M, Foreman K, Lim S, Shibuya K, Aboyans V, Abraham J, Adair T, Aggarwal R, Ahn SY, Alvarado M, Anderson HR, Anderson LM, Andrews KG, Atkinson C, Baddour LM, Barker-Collo S, Bartels DH, Bell ML, Benjamin EJ, Bennett D, Bhalla K, Bikbov B, Bin Abdulhak A, Birbeck G, Blyth F, Bolliger I, Boufous S, Bucello C, Burch M, Burney P, Carapetis J, Chen H, Chou D, Chugh SS, Coffeng LE, Colan SD, Colquhoun S, Colson KE, Condon J, Connor MD, Cooper LT, Corriere M, Cortinovis M, de Vaccaro KC, Couser W, Cowie BC, Criqui MH, Cross M, Dabhadkar KC, et al. 2012. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet 380:2095–2128. doi: 10.1016/S0140-6736(12)61728-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nagata N, Shimbo T, Akiyama J, Nakashima R, Nishimura S, Yada T, Watanabe K, Oka S, Uemura N. 2012. Risk factors for intestinal invasive amebiasis in Japan, 2003–2009. Emerg Infect Dis 18:717–724. doi: 10.3201/eid1805.111275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hung CC, Chang SY, Ji DD. 2012. Entamoeba histolytica infection in men who have sex with men. Lancet Infect Dis 12:729–736. doi: 10.1016/S1473-3099(12)70147-0. [DOI] [PubMed] [Google Scholar]
- 4.James R, Barratt J, Marriott D, Harkness J, Stark D. 2010. Seroprevalence of Entamoeba histolytica infection among men who have sex with men in Sydney, Australia. Am J Trop Med Hyg 83:914–916. doi: 10.4269/ajtmh.2010.10-0231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roure S, Valerio L, Soldevila L, Salvador F, Fernandez-Rivas G, Sulleiro E, Manosa M, Sopena N, Mate JL, Clotet B. 2019. Approach to amoebic colitis: epidemiological, clinical and diagnostic considerations in a non-endemic context (Barcelona, 2007–2017). PLoS One 14:e0212791. doi: 10.1371/journal.pone.0212791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Watanabe K, Gatanaga H, Escueta-de Cadiz A, Tanuma J, Nozaki T, Oka S. 2011. Amebiasis in HIV-1-infected Japanese men: clinical features and response to therapy. PLoS Negl Trop Dis 5:e1318. doi: 10.1371/journal.pntd.0001318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yanagawa Y, Nagata N, Watanabe K, Tsukada K, Teruya K, Kikuchi Y, Gatanaga H, Akiyama J, Uemura N, Oka S. 2016. Increases in Entamoeba histolytica antibody-positive rates in human immunodeficiency virus-infected and noninfected patients in Japan: a 10-year hospital-based study of 3,514 patients. Am J Trop Med Hyg 95:604–609. doi: 10.4269/ajtmh.16-0134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ishikane M, Arima Y, Kanayama A, Takahashi T, Yamagishi T, Yahata Y, Matsui T, Sunagawa T, Nozaki T, Oishi K. 2016. Epidemiology of domestically acquired amebiasis in Japan, 2000–2013. Am J Trop Med Hyg 94:1008–1014. doi: 10.4269/ajtmh.15-0560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yanagawa Y, Nagashima M, Gatanaga H, Kikuchi Y, Oka S, Yokoyama K, Shinkai T, Sadamasu K, Watanabe K. 2020. Seroprevalence of Entamoeba histolytica at a voluntary counselling and testing centre in Tokyo: a cross-sectional study. BMJ Open 10:e031605. doi: 10.1136/bmjopen-2019-031605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bennett JE, Dolin R, Blaser MJ. 2019. Mandell, Douglas, and Bennett's principles and practice of infectious diseases, ninth edition Elsevier/Saunders, Philadelphia, PA. [Google Scholar]
- 11.Saidin S, Othman N, Noordin R. 2019. Update on laboratory diagnosis of amoebiasis. Eur J Clin Microbiol Infect Dis 38:15–38. doi: 10.1007/s10096-018-3379-3. [DOI] [PubMed] [Google Scholar]
- 12.Verkerke HP, Hanbury B, Siddique A, Samie A, Haque R, Herbein J, Petri WA Jr. 2015. Multisite clinical evaluation of a rapid test for Entamoeba histolytica in stool. J Clin Microbiol 53:493–497. doi: 10.1128/JCM.02836-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Korpe PS, Stott BR, Nazib F, Kabir M, Haque R, Herbein JF, Petri WA Jr. 2012. Evaluation of a rapid point-of-care fecal antigen detection test for Entamoeba histolytica. Am J Trop Med Hyg 86:980–981. doi: 10.4269/ajtmh.2012.11-0661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leo M, Haque R, Kabir M, Roy S, Lahlou RM, Mondal D, Tannich E, Petri WA Jr. 2006. Evaluation of Entamoeba histolytica antigen and antibody point-of-care tests for the rapid diagnosis of amebiasis. J Clin Microbiol 44:4569–4571. doi: 10.1128/JCM.01979-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Calderaro A, Piergianni M, Piccolo G, Rossi S, Montecchini S, Buttrini M, Arcangeletti MC, Medici MC, Chezzi C, De Conto F. 2016. Diagnostic performances of antigen detection compared to conventional and nucleic acid detection of Entamoeba histolytica in a non-endemic setting. New Microbiol 39:153–155. [PubMed] [Google Scholar]
- 16.Liu J, Kabir F, Manneh J, Lertsethtakarn P, Begum S, Gratz J, Becker SM, Operario DJ, Taniuchi M, Janaki L, Platts-Mills JA, Haverstick DM, Kabir M, Sobuz SU, Nakjarung K, Sakpaisal P, Silapong S, Bodhidatta L, Qureshi S, Kalam A, Saidi Q, Swai N, Mujaga B, Maro A, Kwambana B, Dione M, Antonio M, Kibiki G, Mason CJ, Haque R, Iqbal N, Zaidi AKM, Houpt ER. 2014. Development and assessment of molecular diagnostic tests for 15 enteropathogens causing childhood diarrhoea: a multicentre study. Lancet Infect Dis 14:716–724. doi: 10.1016/S1473-3099(14)70808-4. [DOI] [PubMed] [Google Scholar]
- 17.Hamzah Z, Petmitr S, Mungthin M, Leelayoova S, Chavalitshewinkoon-Petmitr P. 2006. Differential detection of Entamoeba histolytica, Entamoeba dispar, and Entamoeba moshkovskii by a single-round PCR assay. J Clin Microbiol 44:3196–3200. doi: 10.1128/JCM.00778-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ngobeni R, Samie A, Moonah S, Watanabe K, Petri WA Jr, Gilchrist C. 2017. Entamoeba species in South Africa: correlations with the host microbiome, parasite burdens, and first description of Entamoeba bangladeshi outside of Asia. J Infect Dis 216:1592–1600. doi: 10.1093/infdis/jix535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clark CG, Diamond LS. 2002. Methods for cultivation of luminal parasitic protists of clinical importance. Clin Microbiol Rev 15:329–341. doi: 10.1128/cmr.15.3.329-341.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.