Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has caused a global pandemic of COVID-19, resulting in cases of mild to severe respiratory distress and significant mortality. The global outbreak of this novel coronavirus has now infected >20 million people worldwide, with >5 million cases in the United States (11 August 2020). The development of diagnostic and research tools to determine infection and vaccine efficacy is critically needed.
KEYWORDS: SARS-CoV-2, COVID-19, ACE2 blocking assay, serological tests, functional antibodies, SARS-CoV-2 vaccine, SARS-CoV-2 immunity
ABSTRACT
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has caused a global pandemic of COVID-19, resulting in cases of mild to severe respiratory distress and significant mortality. The global outbreak of this novel coronavirus has now infected >20 million people worldwide, with >5 million cases in the United States (11 August 2020). The development of diagnostic and research tools to determine infection and vaccine efficacy is critically needed. We have developed multiple serologic assays using newly designed SARS-CoV-2 reagents for detecting the presence of receptor-binding antibodies in sera. The first assay is surface plasmon resonance (SPR) based and can quantitate both antibody binding to the SARS-CoV-2 spike protein and blocking to the Angiotensin-converting enzyme 2 (ACE2) receptor in a single experiment. The second assay is enzyme-linked immunosorbent assay (ELISA) based and can measure competition and blocking of the ACE2 receptor to the SARS-CoV-2 spike protein with antispike antibodies. The assay is highly versatile, and we demonstrate the broad utility of the assay by measuring antibody functionality of sera from small animals and nonhuman primates immunized with an experimental SARS-CoV-2 vaccine. In addition, we employ the assay to measure receptor blocking of sera from SARS-CoV-2-infected patients. The assay is shown to correlate with pseudovirus neutralization titers. This type of rapid, surrogate neutralization diagnostic can be employed widely to help study SARS-CoV-2 infection and assess the efficacy of vaccines.
INTRODUCTION
The city of Wuhan, China, became the epicenter for a global pandemic in December of 2019 when the first cases of respiratory illnesses were reported and identified as being caused by a novel betacoronavirus now referred to as severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). SARS-CoV-2 is closely related to SARS-CoV-1 and has a high human-to-human transmission rate (1). As of 11 August 2020, over 19,913,080 people have been reported positive for viral infection of SARS-CoV-2, resulting in 732,185 fatalities worldwide (2). SARS-CoV-2 modes of transmission include shedding of the virions in airborne droplets and close contact. Once infected, people develop flu-like symptoms and severe cases can lead to acute respiratory distress syndrome (ARDS), acute respiratory failure, severe pneumonia, pulmonary edema, multiple organ failure, and death. The development of methods and assays to aid in the detection of functional anti-SARS-CoV-2 antibodies is of utmost importance.
Angiotensin-converting enzyme 2 (ACE2) is highly expressed on lung epithelial cells and is the viral receptor for SARS-CoV-1 (3). Recently, ACE2 has also been identified as a receptor utilized by SARS-CoV-2 (4). A 193-amino acid region on the spike protein of SARS-CoV-1 and SARS-CoV-2 termed the receptor binding domain (RBD) was found to interact with the ACE2 receptor and primarily mediates cell entry (5–10). Cryo-electron microscopy was used to determine a high-resolution structure of the prefusion spike protein and to help define the ACE2 interaction site (11, 12). In the SARS-CoV-1 outbreak of 2003, it was reported that high titers of protecting IgG were found in the convalescent-phase serum of patients recovering from infection (13). Early studies show treating COVID-19 patients presenting with severe ARDS with convalescent plasma therapy (CPT) may be beneficial (14, 15). In order to determine if a recovered patient could be reinfected or if their plasma may be useful as a treatment to others, an assessment of functional antibody response is highly valuable. While tests for SARS-CoV-2 seropositivity are being deployed, they do not necessarily correlate with neutralizing antibody responses (16). As there are limited options for rapid diagnostic tests of functional antibody responses, fast and simple functional assays may prove to be a critical assessment tool for discriminating between potential CPT donors.
In parallel with CPT, major academic, industry, and government entities are pushing for therapeutics and vaccines against SARS-CoV-2. Given these urgent needs, there are numerous clinical vaccine trials under way for SARS-CoV-2 in parallel (17, 18). Through preclinical testing, vaccine-induced functional antibody responses are being used to discriminate among potential vaccine candidates. In clinical testing, the humoral responses will need to be analyzed for functionality to assess the immunogenicity of each vaccine. Therefore, rapid assays for detecting functional IgGs in human serum for SARS-CoV-2 are essential to compare vaccine candidates and to understand induced immunity.
Early into this pandemic, diagnostic tools for detecting viral genetic material were developed and utilized in a real-time reverse transcription-PCR assay (19). Serologic assays for detecting anti-SARS-CoV-2 antibodies are now also available (20). However, single-antigen enzyme-linked immunosorbent assays (ELISAs) can suffer from detection limits and numbers of false positives (21) and may not be associated solely with protective responses. Faster and straightforward approaches which detect specific interactions are needed. Neutralization assays that can detect functional immunity have been developed with replicating virus but require handling in specialized biosafety level 3 (BSL-3) facilities, severely limiting the number of samples that can be processed. Pseudovirus neutralization assays run in BSL-2 facilities were quickly developed to detect the functional antibody response in sera (22). While this is a critical tool for determining protective antibody titers, it requires several days for a readout and are not standardized between laboratories. The pseudoviruses produced in these assays are not easily manufactured and take time to express, harvest, and titer. One such approach to help augment the methods listed above is an ELISA employed in a competitive manner to determine levels of ACE2 receptor blocking antibodies in a sample. In addition, recent advances of portable and field-deployable surface plasmon resonance (SPR) devices (23) and the widespread availability of SPR instruments in research laboratories make SPR an additional platform for measuring ACE2 receptor blocking. Here, we describe a competition ELISA and an SPR assay developed to rapidly detect ACE2 receptor blocking antibodies in IgGs and sera of vaccinated mice, guinea pigs, rabbits, and nonhuman primates, as well as human samples from SARS-CoV-2 patients.
MATERIALS AND METHODS
DNA design and plasmid synthesis.
Protein sequences for human Angiotensin-converting enzyme 2 (huACE2) were obtained from UniProt (Q9BYF1). DNA encoding the IgG1 human Fc sequence was added to the C terminus of the huACE2 and cloned by Twist Bioscience into a modified mammalian pVax-1 expression plasmid to generate the recombinant plasmid ACE2-IgHu.
Recombinant protein expression and purification.
Expi293F cells were transfected with the plasmid ACE2-IgHu using polyethyleneimine (PEI)/Opti-MEM. Cell supernatants were harvested 6 days posttransfection by centrifuging (4,000 × g, 25 min) and filtering (0.2-μm Nalgene Rapid-Flow filter). Supernatants were first purified with affinity chromatography using the AKTA pure 25 system and HiTrap MabSelect SuRe protein A column (catalog number 11-0034-94; GE Healthcare) for ACE2-IgHu. The eluate fractions from the affinity purification were pooled, concentrated, and dialyzed into 1× phosphate-buffered saline (PBS) before being further purified by size exclusion chromatography (SEC) using the Superdex 200 10/300 GL column (GE Healthcare). Identified SEC eluate fractions were pooled and concentrated to 1 mg/ml. The molecular weight and homogeneity of the purified sample were confirmed through size exclusion chromatography multiangle light scattering (SEC-MALS) by running the sample in PBS over a Superose 6 increase column, followed by Dawn Heleos II and Optilab T-rex detectors. The data collected were analyzed using the protein conjugated analysis from ASTRA software (Wyatt Technology).
Biophysical characterization of ACE2-IgHu.
SPR experiments were performed using a protein A capture chip on a Biacore 8k instrument. The running buffer was HBS-EP (3 M sodium chloride, 200 mM HEPES, 60 mM EDTA, 1.0% Tween 20) with 1 mg/ml bovine serum albumin (BSA). The SARS-CoV-2 RBD was used as an analyte at concentrations of 100 nM, 10 nM, and 1 nM. The experiment had an association phase for 120 sec and the dissociation phase for 600 sec. The data were fit by a 1:1 Langmuir model.
SARS-CoV-2 spike binding ACE2-IgHu/ACE2-IgMu ELISA.
Ninety-six-well half -area plates (Corning) were coated at room temperature for 8 hours with the 1 μg/ml PolyRab anti-His antibody (Ab) (catalog number PA1-983B; ThermoFisher), followed by overnight blocking with blocking buffer containing 1× PBS, 5% skim milk, 1% fetal bovine serum (FBS), and 0.2% Tween 20. The plates were then incubated with 10 μg/ml of SARS-CoV-2 S1+S2 ECD (40589-V08B1; Sino Biological) at room temperature (RT) for 1–2 h, followed by an addition of either ACE2-IgHu serially diluted 5-fold (starting concentration, 2 μg/ml) or ACE2-IgMu (10108-H05H; Sino Biological) serially diluted 5-fold (starting concentration, 50 μg/ml). Serial dilutions were performed using PBS with 1% FBS and 0.2% Tween and incubated at RT for 1 h. The plates with ACE2-IgHu were then incubated at RT for 1 h with goat anti-human IgG-Fc fragment cross-adsorbed Ab (A80-340P; Bethyl Laboratories) at a 1:10,000 dilution. Similarly, the plates with ACE2-IgMu were incubated with goat anti-mouse IgG H+L horseradish peroxidase (HRP) (A90-116P; Bethyl Laboratories) at 1:20,000 dilution. Next, 3,3',5,5'-tetramethylbenzidine (TMB) substrate (ThermoFisher) was added to both plates and then quenched with 1 M H2SO4. Absorbances at 450 nm and 570 nm were recorded with a BioTek plate reader. Four washes were performed between every incubation using PBS with 0.05% Tween.
Competition ELISA—control (ACE2-IgMu versus ACE2-IgHu).
Ninety-six-well half-area plates (Corning) were coated at room temperature for 8 hours with 1 μg/ml PolyRab anti-His antibody (PA1-983B; ThermoFisher), followed by overnight blocking with blocking buffer containing 1× PBS, 5% skim milk, 1% FBS, and 0.2% Tween 20. The plates were then incubated with 10 μg/ml of SARS-CoV-2 S1+S2 ECD (Sino Biological, 40589-V08B1) at room temperature for 1 to 2 hours. ACE2-IgMu (10108-H05H; Sino Biological) was serially diluted 3-fold (starting concentration, 100 μg/ml) with PBS with 1% FBS and 0.2% Tween and premixed with ACE2-IgHu at constant concentrations (ranging from 0.01 to 10 μg/ml) The premixture was then added to the plate and incubated at RT for 1 hour. The plates were further incubated at room temperature for 1 hour with goat anti-human IgG-Fc fragment cross-adsorbed Ab (A80-340P; Bethyl Laboratories) at a 1:10,000 dilution, followed by addition of the TMB substrate (ThermoFisher), and then quenched with 1 M H2SO4. Absorbances at 450 nm and 570 nm were recorded with a BioTek plate reader. Four washes were performed between every incubation using PBS with 0.05% Tween.
Competition ELISA—mouse.
IgG was purified from sera taken from mice immunized with SARS-CoV-2 spike protein (INO-4800) (24) using the Nab protein G spin kit (89949; ThermoFisher). Ninety-six-well half-area plates (Corning) were coated at room temperature for 8 hours with 1 μg/ml PolyRab anti-His antibody (PA1-983B; ThermoFisher), followed by overnight blocking with blocking buffer containing 1× PBS, 5% skim milk, 1% FBS, and 0.2% Tween 20. The plates were then incubated with 10 μg/ml of SARS-CoV-2 S1+S2 ECD (40589-V08B1; Sino Biological) at room temperature for 1– to 2 hours. Purified IgG was serially diluted 3-fold (starting concentration, 100 μg/ml) with PBS with 1% FBS and 0.2% Tween and premixed with ACE2-IgHu at a constant concentration of 0.1 μg/ml. The premixture was then added to the plate and incubated at RT for 1 hour. The plates were further incubated at room temperature for 1 hour with goat anti-human IgG-Fc fragment cross-adsorbed Ab (A80-340P; Bethyl Laboratories) at a 1:10,000 dilution, followed by ant addition of the TMB substrate (ThermoFisher), and then quenched with 1 M H2SO4. Absorbances at 450 nm and 570 nm were recorded with a BioTek plate reader. Four washes were performed between every incubation using PBS with 0.05% Tween.
Competition ELISA—Guinea pig.
Sera from guinea pigs immunized with INO-4800 (24) were collected and pooled prior to immunization (day 0, n = 5) and 2 weeks postimmunization. Sera from naive guinea pigs (n = 5) given a saline control were also pooled. IgG was purified from these sera pools using the Nab protein A/G spin kit (catalog number 89950; ThermoFisher). Ninety-six-well half-area plates (Corning) were coated at room temperature for 8 hours with 1 μg/ml PolyRab anti-His antibody (PA1-983B; ThermoFisher), followed by overnight blocking with blocking buffer containing 1× PBS, 5% skim milk, 1% FBS, and 0.2% Tween 20. The plates were then incubated with 10 μg/ml of SARS-CoV-2 S1+S2 ECD (40589-V08B1; Sino Biological) at room temperature for 1 to 2 hours. Either purified sera or IgG (day 0, day 14, or naive) was serially diluted 3-fold with PBS with 1% FBS and 0.2% Tween and premixed with ACE2-IgHu at a constant concentration of 0.1 μg/ml. The premixture was then added to the plate and incubated at RT for 1 h. The plates were further incubated at room temperature for 1 h with goat anti-human IgG-Fc fragment cross-adsorbed Ab (A80-340P; Bethyl Laboratories) at a 1:10,000 dilution, followed by an addition of the TMB substrate (ThermoFisher), and then quenched with 1 M H2SO4. Absorbances at 450 nm and 570 nm were recorded with a BioTek plate reader. Four washes were performed between every incubation using PBS with 0.05% Tween.
Competition ELISA—rabbit.
Rabbits were immunized with 1 mg or 2 mg of INO-4800 at day 0 and week 4. Sera from immunized rabbits were collected and pooled prior to immunization (day 0, n = 5) and 2 weeks after the 2nd immunization (week 6, n = 5) (33). Sera from naive rabbits (n = 5) given a saline control were also pooled. IgG was purified from these serum pools using the Nab protein A/G spin kit (89950; Thermo). Ninety-six-well half-area plates (Corning) were coated at room temperature for 8 hours with 1 μg/ml PolyRab anti-His antibody (PA1-983B; ThermoFisher), followed by overnight blocking with blocking buffer containing 1× PBS, 5% skim milk, 1% FBS, and 0.2% Tween 20. The plates were then incubated with 10 μg/ml of SARS-CoV-2 S1+S2 ECD (40589-V08B1; Sino Biological) at room temperature for 1 to 2 hours. Either serum pool or purified IgG (IgGr) from day 0, low dose, high dose, or naive animals was serially diluted 3-fold with PBS with 1% FBS and 0.2% Tween and premixed with ACE2-IgHu at a constant concentration of 0.1 μg/ml. The premixture was then added to the plate and incubated at RT for 1 hour. The plates were further incubated at room temperature for 1 hour with goat anti-human IgG-Fc fragment cross-adsorbed Ab (A80-340P; Bethyl Laboratories) at a 1:10,000 dilution, followed by an addition of TMB substrates (ThermoFisher), and then quenched with 1 M H2SO4. Absorbances at 450 nm and 570 nm were recorded with a BioTek plate reader. Four washes were performed between every incubation using PBS with 0.05% Tween.
Competition ELISA—primates.
Rhesus macaques were immunized with 1 mg of INO-4800 at day 0 and week 4. Sera from immunized nonhuman primates (NHPs) were collected and pooled prior to immunization (day 0, n = 5) and 2 weeks after the 2nd immunization (week 6, n = 5) (33). Sera from SARS-CoV-2 PCR-positive patients treated during acute infection at the Hospital of the University of Pennsylvania emergency room were also collected and compared with sera from heathy human donors purchased from BioChemed in December of 2016, well before the SARS-CoV-2 pandemic. Ninety-six-well half-area plates (Corning) were coated at 4°C overnight with 1 μg/ml of SARS-CoV-2 S1+S2 ECD (40589-V08B1; Sino Biological). Plates were washed 4 times with 1× PBS and 0.05% Tween 20 using a plate washer; and then, they were blocked with 1× PBS, 5% skim milk, and 0.1% Tween 20 at room temperature for 1 hour. After being washed, sera were serially diluted 3-fold in 1× PBS, 5% skim milk, and 0.1% Tween 20 and incubated on the plate at room temperature for 1 hour. Plates were washed and then incubated with a constant concentration of 0.4 μg/ml ACE2-IgMu diluted in 1× PBS, 5% skim milk, and 0.1% Tween 20 at room temperature for 1 hour. After being washed, plates were further incubated at room temperature for 1 hour with goat anti-mouse IgG H+L HRP (A90-116P; Bethyl Laboratories) at a 1:20,000 dilution in 1× PBS, 5% skim milk, and 0.1% Tween 20. Plates were then washed, followed by addition of the TMB substrate (ThermoFisher) and then quenched with 1 M H2SO4. Absorbances at 450 nm and 570 nm were recorded with a BioTek plate reader. Four washes were performed between every incubation using PBS with 0.05% Tween.
SPR assay.
Surface plasmon resonance experiments were conducted on a Biacore 8K instrument. The running and dilution buffers were HBS-EP, 1 mg/ml BSA, and 0.05% Tween. The biotin CAPture reagent was injected over the Series S CAP Sensor chip (28920234; Cytiva Life Sciences) at 2 μl/min for 300 s. SARS-CoV-2 receptor binding domain was biotinylated using the Lightning-Link type-A biotinylation kit (370-0005; Expedeon/Abcam) and injected over the chip for 180 s at 10 μl/min, followed by injection of the sample (ACE2-IgHu) at 8 concentrations of 1,500 nM to 0.7 nM. The association was for 120 s at a flow rate of 30 μl/min, followed by a short dissociation time of 15 s. ACE2-IgHu was injected at a constant 100-nM concentration in each flow cell for 120 s, followed by a 60-s dissociation at a flow rate of 30 μl/min. The difference in response units before injection and after dissociation of the sample application and the constant receptor was calculated for each curve. The regeneration solution was made using 3-parts 8 M guanidine hydrochloride and 1-part 1 M sodium hydroxide.
Statistics.
Statistical analyses were performed using GraphPad Prism 8 software. The data were considered significant if the P value was <0.05. The lines in all graphs represent the mean value, and error bars represent the standard deviation. The area under the concentration-time curve (AUC) values were normalized as a percentage of day 0 AUC. No samples were excluded from the analysis. We were not blind to sample identity before performing each experiment.
SARS-CoV-2 pseudovirus assay.
The SARS-CoV-2 pseudovirus was produced by cotransfection of HEK293T cells with a 1:1 ratio of a DNA plasmid encoding the SARS-CoV-2 S protein (GenScript) and the backbone plasmid pNL4-3.Luc.R-E- (NIH AIDS Reagent) using GeneJammer (Agilent) in 10% FBS-Dulbecco’s modified Eagle medium (DMEM) enriched with 1% penicillin-streptomycin (D10 medium). The supernatant containing pseudovirus was harvested 48 hours posttransfection and enriched with FBS to a 12% total volume, sterifiltered, and stored at −80°C. The pseudovirus titers were determined using a stable ACE2-293T cell line that had previously been generated (24). For the neutralization assay, serially diluted samples were incubated with pseudovirus at room temperature for 90 min and added to 10,000 ACE2-293T cells in 200 μl tosylsulfonyl phenylalanyl chloromethyl ketone (TPCK) media (DMEM supplemented with 1% BSA, 25 mM HEPES, 1 μg/ml of TPCK, and 1× penicillin-streptomycin) in 96-well tissue culture plates, and the plates were incubated at 37°C and 5% CO2 for 72 h. The cells were subsequently harvested and lysed with the BriteLite reagent (PerkinElmer), and luminescence from the plates was recorded with a BioTek plate reader.
Data availability.
The data are available from the corresponding author upon request.
RESULTS
Receptor expression and assay development.
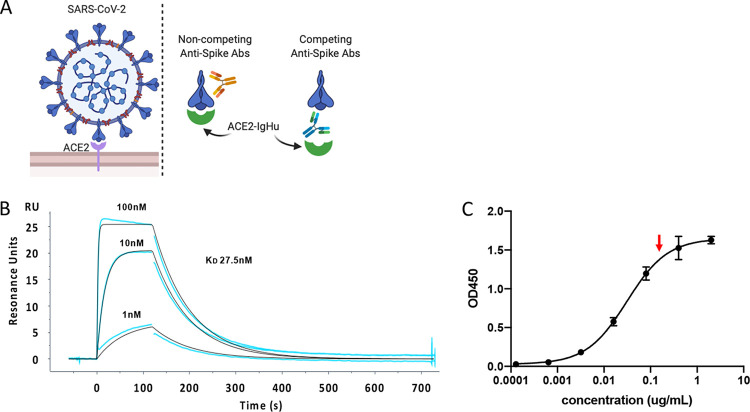
To detect SARS-CoV-2 spike protein binding to ACE2, we designed a soluble variant of the membrane-bound human ACE2 receptor (Fig. 1A). The ectodomain of the receptor was fused to a human IgG1 Fc tag (ACE2-IgHu) for purification and secondary antibody recognition. This protein fusion was expressed in mammalian cells to ensure proper glycosylation and purified on a protein A column. To determine that ACE2-IgHu was dimeric and a homogeneous species in solution, we employed size exclusion chromatography multiangle light scattering (SEC-MALS). The SEC trace shows a single peak (see Fig. S1A in the supplemental material), and the MALS data determined the estimated molecular weight to be 190 kDa for ACE2-IgHu, which is very close to the expected molecular weight of a dimer of ACE2-IgHu at 195 kDa (Fig. S1B). We next sought to confirm the functionality of ACE2-IgHu. Previous studies suggest SARS-CoV-2 binds to ACE2 with an affinity range of 4 to 34 nM (6, 11). We determined that our ACE2-IgHu binds with similar affinity to the receptor binding domain (RBD) of SARS-CoV-2 spike (27.5 nM), as assessed by SPR (Fig. 1B). Next, using enzyme-linked immunosorbent assays (ELISAs), we immobilized the full-length SARS-CoV-2 spike protein (containing both the S1 and S2 subunits) and incubated a dilution series of ACE2-IgHu (Fig. 1C). The binding curves confirmed the high-affinity interaction of the receptor for the spike protein. We further showed similar binding for two independent batches of ACE2, as well as a sample that was frozen and thawed (Fig. S1C). From the binding curve, we hypothesized that a reasonable concentration of ACE2-IgHu needed to see competitive blocking while still binding >90% of immobilized spike protein in the absence of blocking would be around 0.1 to 1 μg/ml (Fig. 1C, red arrow).
FIG 1.
ACE2 receptor expression and affinity. (A) Overview of soluble ACE2 receptor design (ACE2-IgHu). (B) Affinity of SARS CoV-2 receptor binding domain for immobilized ACE2-IgHu assessed by SPR (27 nM) curves are concentrations of RBD X, Y, and Z. (C) Affinity of ACE2-IgHu for immobilized SARS CoV-2 full-length spike protein assessed by ELISA. Optimal concentration of ACE2-IgHu for competition assays (∼EC90, red arrow) requires high signal without excess receptor present.
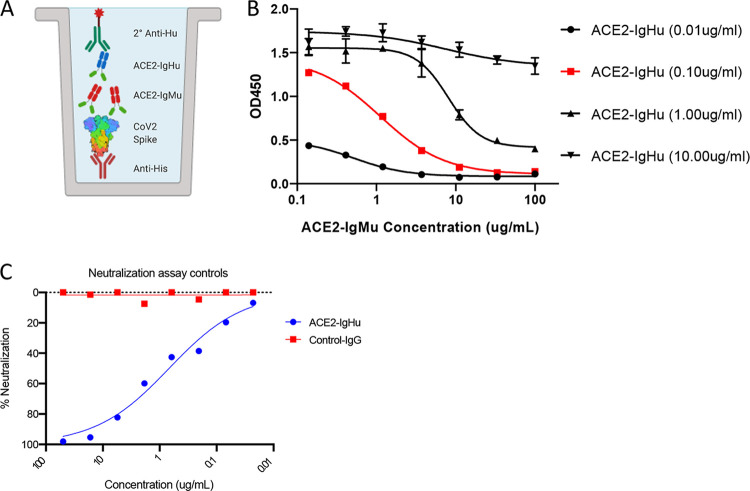
To examine if we could construct a competition assay, we employed ACE2-IgMu (mouse Fc) to act as a competitor to ACE2-IgHu. To match our initial binding ELISA format, the competition ELISA similarly captured a His6×-tagged full-length spike protein by first immobilizing an anti-His polyclonal antibody. A dilution series of the competitor (ACE2-IgMu) was premixed with a constant concentration of the soluble receptor (ACE2-IgHu). A secondary anti-human antibody conjugated to horseradish peroxidase (HRP) determines the amount of ACE2-IgHu present through a TMB colorimetric readout (Fig. 2A). In order to formally determine the optimal concentration of the soluble receptor (ACE2-IgHu), we performed the assay at four concentrations ranging from 0.01 μg/ml to 10 μg/ml (Fig. 2B). The ACE2-IgHu concentration of 0.1 μg/ml (red curve in Fig. 2B) was able to show a complete inhibition curve in the presence of ACE2-IgMu. To further demonstrate functionality of the ACE2-IgHu molecule, we show that ACE2-IgHu could also neutralize SARS-CoV-2 pseudovirus (Fig. 2C).
FIG 2.
ACE2 receptor competition assay development. (A) Competition ELISA schematic displaying immobilized anti-His pAb (red) capturing His6×-tagged SARS-CoV-2 spike protein (rainbow). Premixed ACE2-IgHu (green, blue) at a constant concentration with a dilution series of competitors (green, red) is added, and anti-human HRP (green) determines the amount of ACE2-IgHu remaining in the presence of competitors through a colorimetric readout. (B) Four constant concentrations of ACE2-IgHu were tested with various concentrations of the ACE2-IgMu competitor to establish an optimal ACE2-IgHu concentration which displays a full blocking curve (red, 0.10 μg/ml) from the competitor dilution series while retaining a wide range in signal. (C) Pseudovirus neutralization curves for a control antibody (non-SARS-CoV-2) in red and for ACE2-IgHu in blue.
Animal IgG and serological competition.
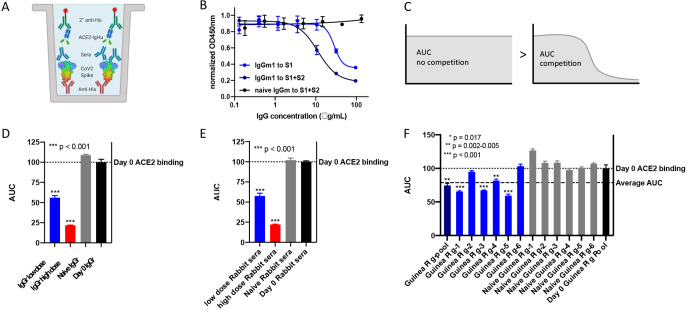
The proof-of-concept competition ELISA displayed a full blocking curve, so we sought to utilize this assay for animals immunized with the SARS-CoV-2 spike protein. The same design for the competition ELISA was used for this assay, replacing the ACE2-IgMu competitor with antibodies induced by vaccination (Fig. 3A). In our previous work, BALB/c mice were immunized with DNA plasmids encoding the SARS-CoV-2 spike protein (24). To examine the activity of antibodies in the sera, IgGs from either naive mice or vaccinated mice 14 days postimmunization were purified using a protein G column. Unlike the ACE2 control which only binds to the receptor binding site (RBS) on the receptor binding domain (RBD) of the spike protein, antibodies from immunized mice can bind to a multitude of epitopes on the spike protein, including epitopes on the S1 subunit (which includes the RBD), S2 subunit or S1-S2 interfaces. While antibodies distal to the RBS should have less effect on ACE2 binding, we hypothesized that such distal antibodies may inhibit ACE2 binding directly by sterically obscuring the RBS or indirectly by causing allosteric conformational shifts in the spike protein. To test this, we immobilized either the full spike protein (S1+S2) or S1 alone to examine the levels of detectable blocking antibodies. A mixture of ACE2-IgHu at a constant concentration of 0.1 μg/ml and a dilution series from a vaccinated mouse IgG (IgGm1) or naive mice IgG (naive IgGm) was incubated on the plate. An anti-human HRP-conjugated antibody was added to determine the ACE2 binding in the presence of IgGm. As Fig. 3B illustrates, there is greater antibody blocking with the full spike protein than with the S1 subunit alone (see Fig. S2A in the supplemental material).
FIG 3.
Animal IgG and serological competition. (A) IgG and serological competition schematic. Anti-His pAb captures SARS-CoV-2 spike protein. Immunized sera or IgG from small animals are used as competitors to block ACE2-IgHu receptor binding when premixed. ACE2-IgHu remaining is determined from an anti-human-HRP colorimetric readout. (B) IgGs present in a vaccinated BALB/c mouse block ACE2-IgHu binding with greater effect when the full-length SARS-CoV-2 S1-S2 spike protein is immobilized versus the S1 subunit by itself. (C) Area under the concentration-time curve (AUC) schematic displaying the larger area for uninhibited ACE2 binding versus the area from curves showing competition with ACE2. (D) AUC of IgGs purified from immunized rabbit sera (IgGr low dose, blue; IgGr high dose, red) versus naive IgGr or day 0 IgGr. (E) AUC of sera from immunized rabbits (low dose rabbit sera, blue; high dose rabbit sera, red) versus naive rabbit sera or day 0 rabbit sera. (F) AUC of sera from immunized guinea pigs at week 2 (dark blue) and individual animals (blue), naive sera (gray), and pooled day 0 sera from all animals (black). The pooled immunized curve displayed a comparable AUC to the average AUC from all individual immunized animals.
To show the utility of this assay in samples from larger mammals, we examined receptor blocking of rabbit sera from SARS-CoV-2 immunization studies. We pooled sera from five rabbits 2 weeks postimmunization in 3 groups (low dose of 1 mg, high dose of 2 mg, and naive), as well as a day 0 pool from all 15 animals. The IgGs from these pools were purified on a protein G column and used as competitors in the competition ELISA. To compare the blocking efficacy, the area under the concentration-time curve (AUC) was calculated for each competition curve. For full, uninhibited ACE2 binding, the AUC will be larger than the AUC for a competitive curve (Fig. 3C). As seen with the mouse IgGs, the pooled vaccinated rabbit IgG displayed statistically significant blocking of ACE2 receptor binding compared with the naive animal pool and the day 0 pool (Fig. 3D, Fig. S2B). The high-dose rabbits reduced ACE2 signal relative to the low-dose group, highlighting the utility of the assay to help discriminate between different vaccine regimens. Up to this point, we have analyzed purified IgGs collected from sera; however, we wanted to validate the use of this assay on serological samples as well. The same rabbit serum pools were used as competitors in the competition ELISA in a dilution series to compare blocking between sera and purified IgG. The rabbit sera displayed statistically significant ACE2 receptor blocking, as we saw in the purified IgG assay (Fig. 3E, Fig. S2C).
Next, we sought to show that we could assess receptor blocking in a third animal model of guinea pigs which were immunized with a SARS-CoV-2 spike-based vaccine (24). We compared a pool of sera collected on day 14 postimmunization to a day 0 pool (Fig. 3F). The immune pool showed significantly lower AUC signal, indicating the immune sera has an ACE2 blocking ability. We then sought to compare how animal serum pools represent the blocking ability of the individual animals. The AUC was calculated for each normalized curve and plotted with the immunized guinea pig pool AUC. Four of the six guinea pigs immunized against SARS-CoV-2 spike showed statistically significant ACE2 blocking, and importantly, the pooled sera was comparable to the average AUC from all six serum samples (Fig. 3F, Fig. S2D). The competition ELISA was used to analyze both the IgGs and the sera from the groups; both groups showed statistically significant blocking of the ACE2 receptor in these assays (Fig. S2E and F).
Primate serological competition.
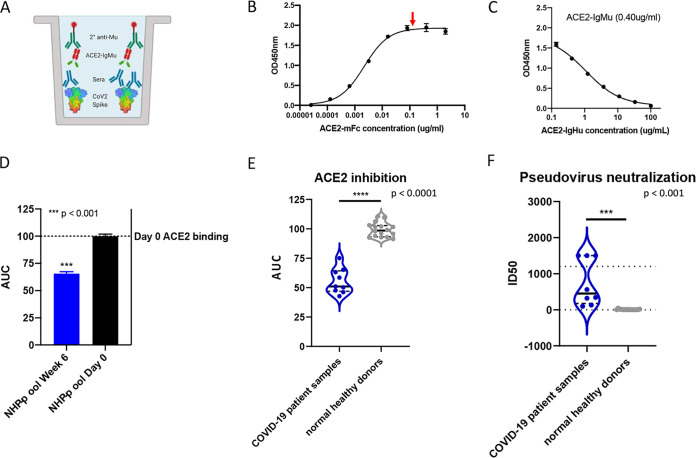
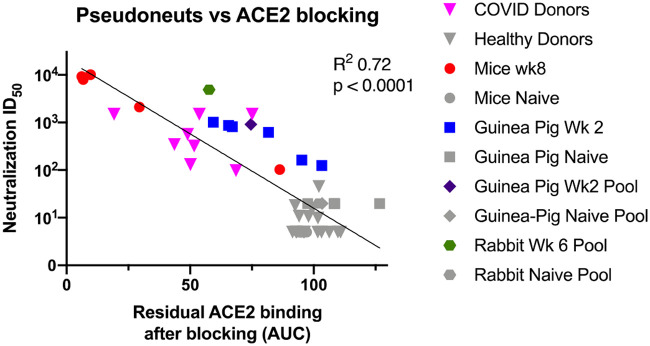
With the competition ELISA demonstrated to be capable of measuring molecular blocking of the ACE2 receptor in small animals with sera and IgG, we next altered the assay for use with primate samples. In the small animal studies, the anti-human secondary antibody was used to detect soluble receptor ACE2-IgHu. However, this secondary antibody would cross-react with antibodies from primates. To remedy this, an ACE2-mouse Fc fusion (ACE2-IgMu) was utilized in place of the ACE2-IgHu and an anti-mouse Fc secondary antibody HRP conjugate was used for detection of ACE2 binding in the presence of primate antibody inhibitors (Fig. 4A). To confirm the function of the replacement ACE2-IgMu, an initial binding ELISA was performed (Fig. 4B). We predicted a similar concentration was needed for optimal competition on the binding curve (Fig. 4B, red arrow) and confirmed this by running a competition ELISA using ACE2-IgHu as the competitor at various ACE2-IgMu constant concentrations (Fig. 4C; see Fig. S3A in the supplemental material). NHP sera from five animals immunized with SARS-CoV-2 spike were pooled, and day 0 sera from the same group of five animals were pooled as a negative control. The NHP immune sera displayed appreciable ACE2 blocking compared with the naive sera (Fig. 4D, Fig. S3B). Next, we examined if we could detect the presence of receptor blocking antibodies in a human intensive care unit (ICU) patient with acute infection. We compared 9 positive COVID-19 patients with 16 naive donor samples taken from 3 years prior to the pandemic. The sera from the COVID-19 patients were able to bind the spike protein and block ACE2 receptor binding with statistical significance, unlike the naive donors (Fig. 4E; see Fig. S4A, B, and C in the supplemental material). To compare healthy and COVID-19 patient samples in an independent assay, we employed a pseudovirus assay we recently developed (24). Briefly, SARS-CoV-2 pseudotyped viruses were generated by transfecting cells with a pNL4-3.Luc.R-E- plasmid and a SARS-CoV-2 spike plasmid. Pseudotype viruses had 50-fold greater relative luminescence units (RLUs) than cells alone. HEK293T cells stably expressing ACE2 were mixed serum and pseudovirus. The serum dilution at which the RLU is 50% signal of control wells is considered the 50% infective dose (ID50). Sera from the healthy donors could not neutralize the virus; yet, sera from COVID-19 patient samples could neutralize the virus (Fig. 4F, Fig. S4D and E). This finding is consistent with the ACE2 blocking data, and we show a correlation between pseudovirus neutralization ID50 and AUC for residual ACE2 blocking across all our data sets (Fig. 5). Thus, we have demonstrated that the ACE2 competition assay can be employed to measure receptor inhibition levels of human samples.
FIG 4.
Primate serological competition. (A) Competition ELISA schematic displaying immobilized His6×-tagged SARS-CoV-2 spike protein (rainbow). Preblocking of the spike protein with primate sera (blue) at various concentrations was added followed by ACE2-IgMu (green, blue) at a constant concentration. Anti-mouse HRP (green) determines the amount of ACE2-IgMu remaining in the presence of competitors through a colorimetric readout. (B) Affinity of ACE2-IgMu for immobilized SARS-CoV-2 S1+S2 full-length spike protein assessed by ELISA. Optimal concentration of ACE2-IgMu for competition assays (red arrow, 0.4 μg/ml) requires high signal without excess receptor present. (C) Optimal ACE2-IgMu concentration which displays a full blocking curve (0.40 μg/ml) from the competitor dilution series (ACE2-IgHu) while retaining a wide range in signal. (D) NHP sera pooled from five vaccinated animals were used as competitors in the primate competition assay. The AUC from vaccinated NHP sera (blue) versus day 0 NHP sera (black). (E) Human sera from nine SARS-CoV-2-positive COVID-19 patients were tested in the primate competition assay and compared with 16 naive human sera collected prepandemic. The AUC of the COVID-19 patient serum (purple) is significantly decreased compared to the prepandemic human serum (gray). The median is shown as a solid black line, and quartiles are shown as dashed black lines. (F) Human sera were analyzed by a pseudovirus neutralization assay. The samples and the coloring are the same as in (E). Statistics include a two-tailed t test with P values indicated.
FIG 5.
ACE2 receptor blocking correlates with pseudovirus neuralization. A symbol represents each of the individual datapoints where we had a paired AUC blocking and pseudovirus ID50 values. The human samples are in triangles, the mice in circles, individual Guinea pigs in squares, Guinea pig pools in diamonds, and rabbit pools in hexagons. SARS-CoV-2 spike-experienced samples are shown in color. Naïve samples and healthy donors are shown in gray. Least-squares fit line is shown with P value and R squared from Prism.
SPR-based assay for ACE2 receptor blocking.
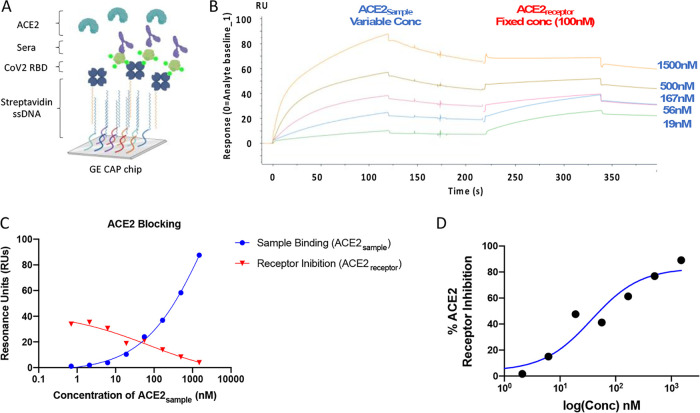
To quantitate blocking of the spike-ACE2 bimolecular interaction in a second, independent experiment, we developed a sensitive surface plasmon resonance (SPR) assay. SPR is a widely used platform that does not require secondary antibodies, and therefore, we could use a single assay format for small animals, NHPs, and humans. In our assay, a CAP sensor chip is used to capture single-stranded DNA coupled to streptavidin. We are then able to capture the biotinylated spike protein to the surface of the sensor chip (Fig. 6A). In SPR, changes in the refractive index occur when molecules interact with the sensor surface or proteins attach to the sensor surface and these changes are reported as resonance units (RUs). Antispike samples in a dilution series can be injected. After a short time, ACE2 receptor at a constant concentration can be injected to measure residual receptor binding. The change in RUs after the second injection is a measure of ACE2 blocking.
FIG 6.
ACE2 receptor competition assay development. (A) Overview of SPR experiment depicting SARS-CoV-2 RBD capture by streptavidin-biotin interaction, sera injected as analyte, and ACE2 injected as second analyte. (B) Sensorgram for ACE2 blocking SPR assay with ACE2-IgHu injected as sample (ACE2sample) as indicated and ACE-IgHu injected as receptor (ACE2receptor) as indicated. Sample responses were referenced to blank injections. Each curve corresponds to a 3-fold dilution of ACE2sample starting at 1,500 nM as indicated on the right, and the ACE2receptor was injected at a constant concentration of 100 nM to all curves. (C) Response in RUs measured at the end of sample (ACE2sample) injection (blue) and receptor (ACE2receptor) injection (red) at each concentration of sample. (D) ACE2 inhibition curve derived from RUs at each concentration.
To demonstrate the feasibility of this assay, we used ACE2-IgHu as both the sample (ACE2sample) and the receptor (ACE2receptor). The sensorgram for this experiment shows ACE2sample injections at various concentrations binding to SARS-CoV-2 RBD between 0 and 125 s (Fig. 6B). At 225 s, we inject ACE2receptor at 100 nM. We observe ACE2receptor binding to SARS-CoV-2 RBD at the lower ACE2sample concentrations but not at the highest ACE2sample concentration. The binding signals of ACE2sample and ACE2receptor intersect close to 100 nM, suggesting the assay is working as expected (Fig. 6C). A measure of % ACE2 receptor inhibition can be calculated to the dose dependence response of the sample (Fig. 6D). While this experiment was done with samples containing a human Fc, this assay can be employed to measure inhibition of the spike:receptor interaction with any sample regardless of species.
DISCUSSION
Diagnostic methods that quickly and accurately detect functional immunity in a SARS-CoV-2 patient or vaccinee are of critical importance during the global pandemic. Serological tests are a central component of our SARS-CoV-2 diagnostic toolbox. Serological tests are often configured to measure the presence of antibodies that recognize SARS-CoV-2 proteins. This may not be a sufficient measurement to determine if a donor’s blood should be used to treat seriously ill patients or to determine if the donor could be reinfected. Instead, a measurement of functional antibodies is required. Furthermore, systematic studies of functional antibody responses and persistence in patients presenting asymptomatic, mild, and severe cases will be critical for understanding humoral responses to SARS-CoV-2. In this study, we developed two new assays which can aid in understanding the specificity and functionality of antibodies in serum. First, we showed that an ELISA-based assay could be employed to measure receptor blocking antibodies in animals vaccinated with the SARS-CoV-2 spike protein. We could measure purified IgG or directly from sera as pools across groups of animals or individual animals. The assay is robust, as exemplified by the ease of adaption to use for five different species. Importantly, the values of ACE2 blocking measured in our assay correlate with pseudovirus neutralization titers, as seen in Fig. 5. Second, we developed an SPR assay for detecting inhibition of the ACE2 receptor binding to the spike protein.
Our assays focus on blocking receptor interactions as a surrogate for virus neutralization. In this study, we examined ACE2 because it appears to be the major receptor for SARS-CoV-2. The assay could be also be used to study antibodies targeting other coronaviruses with similar receptors, such as SARS-CoV-1 or NL63. The Basigin receptor, as known as CD147, has been demonstrated to play a role in SARS-CoV-1 infection (25) and is now hypothesized to function in SARS-CoV-2 invasion of host cells (26). By engineering the ectodomain of CD147 with human and murine Fc tags, these assays could easily be adapted to study antibody inhibition of CD147. In addition, antibodies against other coronaviruses, such as Middle East respiratory syndrome (MERS), which utilize the DPP4 receptor could be studied in our assay format using a similar adaptation.
Neutralizing antibodies (nAbs) can target viral surface proteins to inhibit virus function. Recently, an antibody (B38) was discovered that can neutralize SARS-CoV-2 and has been structurally defined to bind to the same site as ACE2 (27). In addition, three other antibodies (H4, B5, and H2) all showed neutralizing activity, but B5 and H2 did not compete with ACE2 completely, suggesting they bind an alternative neutralizing epitope on the RBD. In another recent study, a nAb, C12.1, binds the RBD spike protein and potently neutralizes and protects Syrian hamsters compared with controls in a passive transfer experiment (28). Recently, a human monoclonal antibody isolated from transgenic mice (47D11) has been shown to neutralize both SARS-CoV-1 and SARS-CoV-2 (29). Probing serological responses for competition and blocking of these protective epitopes could also be easily accomplished through the ELISA and SPR assays reported here.
The Spike-ACE2 interaction is also being considered an important therapeutic target. Indeed, there have been SARS-CoV-1 spike-ACE2 inhibitors developed previously (30). To examine the functionality of these small molecules beyond direct binding to the spike protein, assays such as the one developed here are needed. The SPR instrument is often used for drug discovery, and the SPR assay could be easily adapted to examine blocking capabilities of candidate drugs. The ELISA does not depend on the molecular identity of the competitor, so small molecule or peptide inhibitors could be directly assessed in this assay.
Our study presents a new set of assays for assessing the ability of antibody samples to inhibit SARS-CoV-2 spike interaction with its receptor. A pseudovirus neutralization titer of <30 ID50 for a similar assay has been reported as reasonable a cutoff value for SARS-CoV-2-negative human samples (22). In the comparison of our AUC metric with pseudovirus neutralization in Fig. 5, we see a similar cutoff of 50 ID50 and 85 AUC eliminates all immune negative samples in this study. This AUC cutoff can be used to determine positive inhibition of the ACE2 receptor. Discovering functional monoclonal antibodies can be important for understanding SARS-CoV-2 humoral responses, and serological assays of functional antibodies is just a first step. Indeed, two recent reports observed that only a small portion of monoclonal antibodies sorted from convalescent donors are neutralizing (31, 32). Therefore, it is important to continue to develop and publish assays of various formats to detect functional antibody responses in SARS-CoV-2 spike-exposed people.
In a fast-moving global pandemic, we must quickly bring to bear all our immunological knowledge to create new tools. Receptor-blocking assays can detect functional antibodies in serum samples. Functional antibody assays can be employed widely to help study SARS-CoV-2 infection, help inform on the immune status of the population, and assess the efficacy of vaccines.
Supplementary Material
ACKNOWLEDGMENTS
We thank Matthew Sullivan for providing feedback on the manuscript.
This work was supported by a grant from the Coalition for Epidemic Preparedness Innovations (CEPI).
S.N.W., N.C., and D.W.K. developed the receptor-blocking assays, designed and characterized ACE2-IgHu protein and the SARS-CoV-2 RBD protein, planned and executed the experiments, and analyzed the data. E.L.R. and K.Y.K. optimized, planned, and executed the experiments involving the human samples. E.L.R., E.N.G., A.P., P.T., K.S., J.W., S.R., T.R.F.S., and K.E.B. produced or provided reagents. M.P. and Z.X. created and executed the pseudovirus neutralization assay. E.L.R., M.H., K.M., E.N.G., M.P., Z.X., A.P., and D.B.W. aided in developing parts of the receptor-blocking assay. S.N.W., N.C., and D.W.K. wrote the paper. E.L.R., M.H., K.M., E.N.G., M.P., Z.X., K.S., J.W., S.R., T.R.F.S., K.E.B., A.P., and D.B.W. helped write the paper.
T.R.F.S. and K.S. are employees of Inovio Pharmaceuticals and as such receive salary and benefits, including ownership of stock and stock options, from the company. D.B.W. has received grant funding, participates in industry collaborations, has received speaking honoraria, and has received fees for consulting, including serving on scientific review committees and board services. Remuneration received by D.B.W. includes direct payments or stock or stock options, and in the interest of disclosure, he notes potential conflicts associated with this work with Inovio and possibly others. In addition, D.B.W. has patent pending and patented DNA vaccine technologies owned by Wistar Institute with certain rights licensed to Inovio Pharmaceuticals. We declare no other competing interests.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Li R, Pei S, Chen B, Song Y, Zhang T, Yang W, Shaman J. 2020. Substantial undocumented infection facilitates the rapid dissemination of novel coronavirus (SARS-CoV-2). Science 368:489–493. doi: 10.1126/science.abb3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dong E, Du H, Gardner L. 2020. An interactive Web-based dashboard to track COVID-19 in real time. Lancet Infect Dis 20:533–534. doi: 10.1016/S1473-3099(20)30120-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li W, Moore MJ, Vasilieva N, Sui J, Wong SK, Berne MA, Somasundaran M, Sullivan JL, Luzuriaga K, Greenough TC, Choe H, Farzan M. 2003. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature 426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, Si HR, Zhu Y, Li B, Huang CL, Chen HD, Chen J, Luo Y, Guo H, Jiang RD, Liu MQ, Chen Y, Shen XR, Wang X, Zheng XS, Zhao K, Chen QJ, Deng F, Liu LL, Yan B, Zhan FX, Wang YY, Xiao GF, Shi ZL. 2020. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xiao X, Chakraborti S, Dimitrov AS, Gramatikoff K, Dimitrov DS. 2003. The SARS-CoV S glycoprotein: expression and functional characterization. Biochem Biophys Res Commun 312:1159–1164. doi: 10.1016/j.bbrc.2003.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lan J, Ge J, Yu J, Shan S, Zhou H, Fan S, Zhang Q, Shi X, Wang Q, Zhang L, Wang X. 2020. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature 581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 7.Shang J, Ye G, Shi K, Wan Y, Luo C, Aihara H, Geng Q, Auerbach A, Li F. 2020. Structural basis of receptor recognition by SARS-CoV-2. Nature 581:221–224. doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tai W, He L, Zhang X, Pu J, Voronin D, Jiang S, Zhou Y, Du L. 2020. Characterization of the receptor-binding domain (RBD) of 2019 novel coronavirus: implication for development of RBD protein as a viral attachment inhibitor and vaccine. Cell Mol Immunol 17:613–620. doi: 10.1038/s41423-020-0400-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wan Y, Shang J, Graham R, Baric RS, Li F. 2020. Receptor recognition by the novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS coronavirus. J Virol 94:e00127-20. doi: 10.1128/JVI.00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yan R, Zhang Y, Li Y, Xia L, Guo Y, Zhou Q. 2020. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science 367:1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wrapp D, Wang N, Corbett KS, Goldsmith JA, Hsieh CL, Abiona O, Graham BS, McLellan JS. 2020. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Walls AC, Park YJ, Tortorici MA, Wall A, McGuire AT, Veesler D. 2020. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell 181:281–292.e6. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang J-S, Chen J-T, Liu Y-X, Zhang Z-S, Gao H, Liu Y, Wang X, Ning Y, Liu Y-F, Gao Q, Xu J-G, Qin C, Dong X-P, Yin W-D. 2005. A serological survey on neutralizing antibody titer of SARS convalescent sera. J Med Virol 77:147–150. doi: 10.1002/jmv.20431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shen C, Wang Z, Zhao F, Yang Y, Li J, Yuan J, Wang F, Li D, Yang M, Xing L, Wei J, Xiao H, Yang Y, Qu J, Qing L, Chen L, Xu Z, Peng L, Li Y, Zheng H, Chen F, Huang K, Jiang Y, Liu D, Zhang Z, Liu Y, Liu L. 2020. Treatment of 5 critically ill patients with COVID-19 with convalescent plasma. JAMA 323:1582–1589. doi: 10.1001/jama.2020.4783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Duan K, Liu B, Li C, Zhang H, Yu T, Qu J, Zhou M, Chen L, Meng S, Hu Y, Peng C, Yuan M, Huang J, Wang Z, Yu J, Gao X, Wang D, Yu X, Li L, Zhang J, Wu X, Li B, Xu Y, Chen W, Peng Y, Hu Y, Lin L, Liu X, Huang S, Zhou Z, Zhang L, Wang Y, Zhang Z, Deng K, Xia Z, Gong Q, Zhang W, Zheng X, Liu Y, Yang H, Zhou D, Yu D, Hou J, Shi Z, Chen S, Chen Z, Zhang X, Yang X. 2020. Effectiveness of convalescent plasma therapy in severe COVID-19 patients. Proc Natl Acad Sci U S A 117:9490–9496. doi: 10.1073/pnas.2004168117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harvala H, Robb M, Watkins N, Ijaz S, Dicks S, Patel M, Supasa P, Dejnirattisai W, Liu C, Mongkolsapaya J, Brown A, Bailey D, Vipond R, Grayson N, Temperton N, Bolton J, Fyfe A, Gopal R, Simmonds P, Screaton G, Thompson CP, Brooks T, Zambon M, Miflin G, Roberts D. 2020. Convalescent plasma therapy for the treatment of patients with COVID-19: assessment of methods available for antibody detection and their correlation with neutralising antibody levels. medRxiv 10.1101/2020.05.20.20091694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen WH, Strych U, Hotez PJ, Bottazzi ME. 2020. The SARS-CoV-2 vaccine pipeline: an overview. Curr Trop Med Rep 7:61–64. doi: 10.1007/s40475-020-00201-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Amanat F, Krammer F. 2020. SARS-CoV-2 vaccines: status report. Immunity 52:583–589. doi: 10.1016/j.immuni.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shirato K, Nao N, Katano H, Takayama I, Saito S, Kato F, Katoh H, Sakata M, Nakatsu Y, Mori Y, Kageyama T, Matsuyama S, Takeda M. 2020. Development of genetic diagnostic methods for novel coronavirus 2019 (nCoV-2019) in Japan. Jpn J Infect Dis 73:304–307. doi: 10.7883/yoken.JJID.2020.061. [DOI] [PubMed] [Google Scholar]
- 20.Krammer F, Simon V. 2020. Serology assays to manage COVID-19. Science 368:1060–1061. doi: 10.1126/science.abc1227. [DOI] [PubMed] [Google Scholar]
- 21.Lassaunière R, Frische A, Harboe ZB, Nielsen AC, Fomsgaard A, Krogfelt KA, Jørgensen CS. 2020. Evaluation of nine commercial SARS-CoV-2 immunoassays. medRxiv 10.1101/2020.04.09.20056325. [DOI] [Google Scholar]
- 22.Nie J, Li Q, Wu J, Zhao C, Hao H, Liu H, Zhang L, Nie L, Qin H, Wang M, Lu Q, Li X, Sun Q, Liu J, Fan C, Huang W, Xu M, Wang Y. 2020. Establishment and validation of a pseudovirus neutralization assay for SARS-CoV-2. Emerg Microbes Infect 9:680–686. doi: 10.1080/22221751.2020.1743767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Masson JF. 2020. Portable and field-deployed surface plasmon resonance and plasmonic sensors. Analyst 145:3776–3800. doi: 10.1039/D0AN00316F. [DOI] [PubMed] [Google Scholar]
- 24.Smith TRF, Patel A, Ramos S, Elwood D, Zhu X, Yan J, Gary EN, Walker SN, Schultheis K, Purwar M, Xu Z, Walters J, Bhojnagarwala P, Yang M, Chokkalingam N, Pezzoli P, Parzych E, Reuschel EL, Doan A, Tursi N, Vasquez M, Choi J, Tello-Ruiz E, Maricic I, Bah MA, Wu Y, Amante D, Park DH, Dia Y, Ali AR, Zaidi FI, Generotti A, Kim KY, Herring TA, Reeder S, Andrade VM, Buttigieg K, Zhao G, Wu JM, Li D, Bao L, Liu J, Deng W, Qin C, Brown AS, Khoshnejad M, Wang N, Chu J, Wrapp D, McLellan JS, Muthumani K, Wang B, Carroll MW, Kim JJ, Boyer J, Kulp DW, Humeau LMPF, Weiner DB, Broderick KE. 2020. Immunogenicity of a DNA vaccine candidate for COVID-19. Nat Commun 11:2601. doi: 10.1038/s41467-020-16505-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen Z, Mi L, Xu J, Yu J, Wang X, Jiang J, Xing J, Shang P, Qian A, Li Y, Shaw PX, Wang J, Duan S, Ding J, Fan C, Zhang Y, Yang Y, Yu X, Feng Q, Li B, Yao X, Zhang Z, Li L, Xue X, Zhu P. 2005. Function of HAb18G/CD147 in invasion of host cells by severe acute respiratory syndrome coronavirus. J Infect Dis 191:755–760. doi: 10.1086/427811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang K, Chen W, Zhou Y-S, Lian J-Q, Zhang Z, Du P, Gong L, Zhang Y, Cui H-Y, Geng J-J, Wang B, Sun X-X, Wang C-F, Yang X, Lin P, Deng Y-Q, Wei D, Yang X-M, Zhu Y-M, Zhang K, Zheng Z-H, Miao J-L, Guo T, Shi Y, Zhang J, Fu L, Wang Q-Y, Bian H, Zhu P, Chen Z-N. 2020. SARS-CoV-2 invades host cells via a novel route: CD147-spike protein. bioRxiv 10.1101/2020.03.14.988345. [DOI] [PMC free article] [PubMed]
- 27.Wu Y, Wang F, Shen C, Peng W, Li D, Zhao C, Li Z, Li S, Bi Y, Yang Y, Gong Y, Xiao H, Fan Z, Tan S, Wu G, Tan W, Lu X, Fan C, Wang Q, Liu Y, Zhang C, Qi J, Gao GF, Gao F, Liu L. 2020. A noncompeting pair of human neutralizing antibodies block COVID-19 virus binding to its receptor ACE2. Science 368:1274–1278. doi: 10.1126/science.abc2241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zost SJ, Gilchuk P, Chen R, Case JB, Reidy JX, Trivette A, Nargi RS, Sutton RE, Suryadevara N, Chen EC, Binshtein E, Shrihari S, Ostrowski MA, Chu HY, Didier JE, MacRenaris KW, Jones T, Day S, Myers L, Lee FE-H, Nguyen DC, Sanz I, Martinez DR, Gralinski L, Baric RS, Thackray L, Diamond MS, Carnahan RH, Crowe JE. 2020. Rapid isolation and profiling of a diverse panel of human monoclonal antibodies targeting the SARS-CoV-2 spike protein. bioRxiv 10.1101/2020.05.12.091462. [DOI] [PMC free article] [PubMed]
- 29.Wang C, Li W, Drabek D, Okba NMA, van Haperen R, Osterhaus A, van Kuppeveld FJM, Haagmans BL, Grosveld F, Bosch BJ. 2020. A human monoclonal antibody blocking SARS-CoV-2 infection. Nat Commun 11:2251. doi: 10.1038/s41467-020-16256-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huentelman MJ, Zubcevic J, Hernandez Prada JA, Xiao X, Dimitrov DS, Raizada MK, Ostrov DA. 2004. Structure-based discovery of a novel angiotensin-converting enzyme 2 inhibitor. Hypertension 44:903–906. doi: 10.1161/01.HYP.0000146120.29648.36. [DOI] [PubMed] [Google Scholar]
- 31.Zost SJ, Gilchuk P, Chen RE, Case JB, Reidy JX, Trivette A, Nargi RS, Sutton RE, Suryadevara N, Chen EC, Binshtein E, Shrihari S, Ostrowski M, Chu HY, Didier JE, MacRenaris KW, Jones T, Day S, Myers L, Eun-Hyung Lee F, Nguyen DC, Sanz I, Martinez DR, Rothlauf PW, Bloyet LM, Whelan SPJ, Baric RS, Thackray LB, Diamond MS, Carnahan RH, Crowe JE Jr. 2020. Rapid isolation and profiling of a diverse panel of human monoclonal antibodies targeting the SARS-CoV-2 spike protein. Nat Med doi: 10.1038/s41591-020-0998-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wec AZ, Wrapp D, Herbert AS, Maurer DP, Haslwanter D, Sakharkar M, Jangra RK, Dieterle ME, Lilov A, Huang D, Tse LV, Johnson NV, Hsieh CL, Wang N, Nett JH, Champney E, Burnina I, Brown M, Lin S, Sinclair M, Johnson C, Pudi S, Bortz R III, Wirchnianski AS, Laudermilch E, Florez C, Fels JM, O'Brien CM, Graham BS, Nemazee D, Burton DR, Baric RS, Voss JE, Chandran K, Dye JM, McLellan JS, Walker LM. 2020. Broad neutralization of SARS-related viruses by human monoclonal antibodies. Science 369:731–736. doi: 10.1126/science.abc7424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Patel A, Walters J, Reuschel EL, Schultheis K, Parzych E, Gary EN, Maricic I, Purwar M, Eblimit Z, Walker SN, Guimet D, Bhojnagarwala P, Doan A, Xu Z, Elwood D, Reeder SM, Pessaint L, Kim KY, Cook A, Chokkalingam N, Finneyfrock B, Tello-Ruiz E, Dodson A, Choi J, Generotti A, Harrison J, Tursi NJ, Andrade VM, Dia Y, Zaidi FI, Andersen H, Lewis MG, Muthumani K, Kim JJ, Kulp DW, Humeau LM, Ramos S, Smith TRF, Weiner DB, Broderick KE. 2020. Intradermal-delivered DNA vaccine provides anamnestic protection in a rhesus macaque SARS-CoV-2 challenge model. bioRxiv doi: 10.1101/2020.07.28.225649. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data are available from the corresponding author upon request.