The ongoing coronavirus disease 2019 (COVID-19) pandemic has resulted in shortages of nasopharyngeal swabs (NPS) and viral transport media, necessitating the search for alternate diagnostic specimens, such as saliva. We directly compared matched saliva and NPS specimens from symptomatic patients suspected of having COVID-19. An enhanced saliva specimen (i.e., strong sniff, elicited cough, and collection of saliva/secretions) was collected without transport medium prior to collection of NPS from 224 patients with symptoms deemed consistent with COVID-19.
KEYWORDS: COVID, SARS-CoV-2, saliva, specimen
ABSTRACT
The ongoing coronavirus disease 2019 (COVID-19) pandemic has resulted in shortages of nasopharyngeal swabs (NPS) and viral transport media, necessitating the search for alternate diagnostic specimens, such as saliva. We directly compared matched saliva and NPS specimens from symptomatic patients suspected of having COVID-19. An enhanced saliva specimen (i.e., strong sniff, elicited cough, and collection of saliva/secretions) was collected without transport medium prior to collection of NPS from 224 patients with symptoms deemed consistent with COVID-19. Both specimens were tested with the CDC 2019 nCoV real-time RT-PCR diagnostic panel (4 February 2020 version), with the NPS result used as the reference standard. For the 216 patients included in the final analysis, there was 100% positive agreement (38/38 positive specimens) and 99.4% negative agreement (177/178 negative specimens). The one discrepant specimen had the presence of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) confirmed in the saliva specimen using an alternate FDA EUA assay. The overall mean difference in cycle threshold (CT) values for the positive NPS and saliva specimens was −3.61 (95% confidence interval [CI], −5.78 to −1.44; P = 0.002). An enhanced saliva specimen performed as well as NPS for the qualitative detection of SARS-CoV-2 in symptomatic patients, although the overall mean viral load in saliva was lower.
INTRODUCTION
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has rapidly spread from person to person, crossing international boundaries at an unprecedented rate (1). The ensuing pandemic has uncovered a number of deficits in the ability of societies to respond rapidly to an infectious disease public health emergency (2). Inadequate testing equipment and supplies, shortages of ventilators, insufficient personal protective equipment, exhaustion of viral/universal transport medium supplies, and inadequate nasopharyngeal swab (NPS) inventories have necessitated innovation and adaptation in health care delivery (2). Specifically, the shortage of NPSs, the standard specimen collection device for the diagnosis of respiratory viral infections, has necessitated the investigation and employment of nasal midturbinate swabs, anterior nares swabs, and even saliva as alternate specimens for the diagnosis of coronavirus disease 2019 (COVID-19) infection (3–5). Several studies have investigated the use of saliva as a specimen for testing for SARS-CoV-2, with mixed results (3, 5–11). We therefore undertook a direct NPS-to-saliva specimen comparison to provide further clarification regarding the use of saliva as a potential specimen for testing.
MATERIALS AND METHODS
We undertook a comparison of saliva as a clinical specimen for SARS-CoV-2 testing with the standard NPS specimen, following the approval of the Institutional Review Board of the Cleveland Clinic. The age and self-declared race and sex were collected for each patient.
Specimen collection.
Patients with symptoms consistent with COVID-19, as determined by a licensed health care provider and reviewed by a second health care provider, were directed to a specimen collection location. Upon registration, the patients were asked if they would be willing to voluntarily submit a saliva specimen for specimen validation purposes and were provided with a patient information sheet explaining the rationale of the study and the additional specimen that would be required. If the patient agreed to submit a saliva specimen for comparative purposes, they were provided with a specimen collection cup and given instructions (see the supplemental material) on how to provide the specimen while they waited in line in their car for NPS collection. The patient was instructed to “snuff” (i.e., sniff strongly) to gather any nasal secretion/mucus into the oropharynx, to cough to produce any phlegm, and then to submit these secretions and additional saliva until the specimen reached the premarked fill line on the sterile specimen collection container (also called a urine cup). We requested 3 ml of saliva/naso-oropharyngeal secretions (referred to here as “enhanced saliva”) for this study but accepted whatever volume the patient could provide. After enhanced-saliva collection, an NPS was collected in a standard manner by a trained health care professional. The NPS was placed in 0.9% normal saline, which is a validated, standard-of-care specimen transport medium for SARS-CoV-2 testing (12). Both specimens were simultaneously submitted and processed for SARS-CoV-2 testing by a reverse transcription-PCR (RT-PCR) method. The specimens were refrigerated at the collection site and transported to the laboratory in insulated coolers with ice packs.
SARS-CoV-2 testing.
All specimens were tested on the day they were collected. The saliva specimen, if viscous, was rendered more liquid using 0.5% dithiothreitol. After this, both the saliva and the NPS in transport medium were independently processed for SARS-CoV-2 testing, as described elsewhere (13). In brief, 200 μl of specimen was rendered noninfectious using bacterial lysis buffer (Roche Diagnostics, Indianapolis, IN). Then, 200 μl of inactivated specimen was processed for nucleic acid extraction using the MagNA Pure system (Roche Diagnostics, Indianapolis, IN), which yielded 50 μl of eluate.
Five microliters of eluate was added to 15 μl of PCR master mix for each of the PCR wells for each of the four components of the CDC 2019 nCoV real-time RT-PCR diagnostic panel (4 February 2020 version) (14). ABI 7500 Fast Dx instruments were used for real-time PCR. The four components consist of primers/probes designed to detect three unique targets on the nucleocapsid (N) gene of the SARS-CoV-2 genome and a human gene (RNase P) amplification control. The amplification and detection of all three N gene targets were required to characterize a specimen as positive. If amplification of only one or two of the N gene targets occurred, then the result was characterized as indeterminate. Any specimen characterized as indeterminate was documented but excluded from further analysis. If the N gene targets were not detected, then detection of the RNase P gene was required to characterize a specimen as negative. Specimens with no amplification of the N gene targets and no amplification of the RNase P gene target were characterized as reflecting amplification control failure and excluded from further analysis.
Statistical analysis.
The NPS result was used as the reference standard to calculate the positive and negative percent agreement (PPA and NPA) for the matched enhanced saliva specimen. The mean cycle threshold (CT) value was determined from the three SARS-CoV-2 genetic loci targeted by the CDC assay (i.e., N1, N2, and N3) for each of the positive specimens. The matched CT values from the NPS and enhanced saliva specimens were recorded and correlated with one another using the Pearson correlation coefficient. The overall mean CT values were determined for positive NPS specimens and positive saliva specimens and compared. Additionally, the CT values of the human RNase P gene amplification control from the NPS specimens were compared with those from the corresponding saliva specimens as a surrogate for cellularity of the specimens.
RESULTS
Two hundred twenty-four patients with symptoms deemed consistent with COVID-19 submitted an enhanced saliva specimen 5 to 10 min prior to having a matched NPS collected by a trained medical professional. After testing and for simplicity, 8 specimens were excluded from the final comparative analysis, due to internal amplification control failure (1) or an indeterminate test result in one of the specimens (7). Five (71%) of the indeterminate results occurred with saliva specimens and two (29%) with NPS specimens. Of the remaining 216 paired specimens, there were 38 NPS specimens that were positive for SARS-CoV-2, all of which had corresponding positive saliva specimens (100% PPA). Of the remaining 178 NPS specimens that were negative for SARS-CoV-2, 177 had corresponding negative saliva specimens (99.4% NPA). The one discordant specimen with negative NPS results and positive saliva results had the presence of SARS-CoV-2 in the saliva specimen confirmed using the Aptima SARS-CoV-2 transcription mediated amplification assay (Panther System) (Hologic, Inc., San Diego, CA). The CT values from RT-PCRs from the saliva from this discordant specimen were 34.4 cycles for N1, 34.7 cycles for N2, and 34.6 cycles for N3, which indicates a low viral load.
The findings from the seven specimens that were excluded from analysis because the result from either the NPS specimen or saliva specimen was indeterminate are further described. Four of the seven had a negative result from the NPS specimen and an indeterminate because one of the three PCRs of the CDC assay was positive in the saliva specimen. Two of the remaining saliva specimens were positive, whereas the corresponding NPS specimens were indeterminate due to the amplification of one or two of the N gene targets. Conversely, one NPS specimen was positive, whereas the corresponding saliva specimen demonstrated amplification of only 2 of the 3 N gene targets. Therefore, if all positive results were included in the final analysis, an additional positive would be added for saliva specimens over NPS specimens. The specimen with the amplification control failure was an NPS specimen.
The mean (SD) CT value of the RNase P amplification control of the NPS specimens was 25.1 (2.2) cycles, whereas the mean (SD) CT value of this target for the saliva specimens was 23.0 (1.8). The mean difference in CT values was 2.1 (95% confidence interval [CI], 1.8 to 2.4; P < 0.001), which suggests that the saliva specimens had a greater cellularity than the NPS specimens.
The ages of the patients presenting for testing ranged from 18 to 82 years old, with a mean age of 44 years. The demographic characteristics of the patients with laboratory-confirmed SARS-CoV-2 infection were as follows. The average age was 47 years old. Twenty-five patient were female, and 14 were male. The self-declared races were black (n = 19), white (n = 13), multiracial (n = 3), and one each of Hispanic, other, declined, and unavailable.
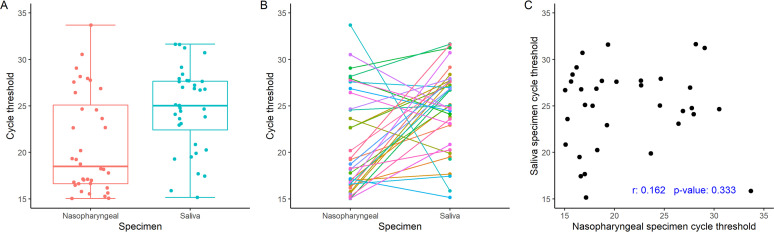
Although cycle threshold values were distributed widely overall, those for NPS specimens were lower than those for saliva specimens (Fig. 1A). The overall mean (SD) CT value for the positive NPS specimens was 20.55 (5.36) cycles, whereas the corresponding overall mean (SD) CT value for enhanced saliva specimens was 24.16 (4.80) cycles, for a mean difference in CT value for paired NPS and saliva specimens of −3.61 (95% CI, −5.78 to −1.44; P = 0.002). Seventy-one percent of the saliva specimens had higher CT values (i.e., lower viral loads) than the matched NPS specimens (Fig. 1B). There was little correlation between the CT values (i.e., viral loads) of the positive saliva and NPS specimens (Pearson correlation coefficient, r = 0.162; P = 0.333) (Fig. 1C). The three RT-PCRs that targeted SARS-CoV-2 were highly correlated one to another within a specimen type (Fig. S1).
FIG 1.
(A) The median CT for saliva specimens is significantly higher overall than that for NPS specimens. (B) Individual CT values for matched NPS and saliva specimens. Seventy-one percent of NPS specimens had lower CT values (i.e., higher viral loads) than matched saliva specimens. (C) Poor correlation between CT values for NPS and matched saliva specimens.
DISCUSSION
The national shortage of supplies, which includes NPS and viral transport medium, has necessitated innovative approaches and the evaluation of alternate specimen types for the laboratory-based diagnosis of COVID-19 (2, 15). Saliva is an attractive specimen type, since it eliminates the need for a swab and transport medium and can potentially decrease the interaction time between health care providers and potentially infected individuals, which could affect personal protective equipment usage and exposure risk (9). Although several groups have investigated the use of saliva for the detection of SARS-CoV-2, the results have been inconsistent with regard to the superiority of this specimen type. We therefore undertook a study to evaluate and further clarify the use of saliva as an alternate specimen type for the detection of SARS-CoV-2 by comparing saliva samples with matched NPSs.
The well-conducted study by Wyllie et al. (11) examined first morning saliva specimens and demonstrated a higher SARS-CoV-2 viral load in saliva compared with matched NPS in hospitalized patients. This study also reported less temporal variability between specimens and more consistent self-sampling, which led this group to conclude that first morning saliva was a superior specimen to NPS (11). The finding by Wyllie et al. of a greater viral load in first morning saliva specimens than in NPS specimens was in contrast to the findings reported in the current study, which used a midday enhanced saliva specimen as the NPS comparator.
The conclusions of Wyllie et al. were supported by the work of Kojima and colleagues, who studied supervised and unsupervised oral fluid collection for the detection of SARS-CoV-2, with NPS as a comparator (3). They reported that supervised oral fluid collection identified the most infected patients (90%), followed by NPS (79%), and finally unsupervised oral fluid collection (66%). The reason for the poor performance of the unsupervised oral fluid specimen was that some of the infected patients failed to elicit a cough, as instructed, which was informative for our study design. To et al. studied the temporal profiles of posterior oropharyngeal specimens (sputa) and found that they were highest after the first week of onset of symptoms and higher in older individuals (16).
The model of collecting and evaluating a first morning saliva collection did not fit with our approach to outpatient testing. We therefore designed a study to compare a self-collected saliva specimen from symptomatic patients in an outpatient testing center (i.e., drive through) that was obtained just prior to the routine NPS collection. Testing symptomatic patients when they presented to the testing center (i.e., regardless of the time of day) was more consistent with our practice than requesting a first morning saliva specimen. Based on the report by Kojima et al. (3), we sought to obtain an enhanced saliva specimen. Our instruction sheet requested that the patient snuff (i.e., obtain nasal/nasopharyngeal secretions using a strong sniff), elicit a cough to introduce lung and tracheal secretions into the oropharynx, and finally expel the mouth contents and continue to collect saliva until 3 ml was obtained. Although we did not record the compliance of the patients with these instructions, we did demonstrate a 100% PPA for the detection of SARS-CoV-2 between the NPS and the enhanced saliva specimens.
Saliva, however, has not been found to be an adequate specimen compared to NPS in all studies. Chen et al. reported an only 84.5% PPA when NPS was compared with oropharyngeal saliva specimens (6). In this study, 10.3% of patients with laboratory-confirmed COVID-19 were identified by NPS only, whereas 5.2% were identified by saliva only, and the remaining 84.5% were identified using both specimen types. Similarly, Fernández-Pittol and colleagues (17) reported a sensitivity of saliva of only 83.8%, using a heat-inactivation protocol prior to testing. This group also reported that the cohort that yielded the sensitivity of 83.8% included individuals with more remote disease, which has been shown by others to have decreased viral loads in upper respiratory specimens (18, 19). When only individuals with symptoms of <9 days duration were included in the analysis, then the sensitivity of saliva in this study increased to 90%, which is consistent with findings of others (5, 7); the study was limited to adults. Additional studies are needed to determine the acceptability of saliva as an alternate specimen type in children.
The cycle threshold (CT) has been used appropriately throughout this pandemic as a surrogate for viral load (13, 20). Although the enhanced saliva specimens were adequate to confirm the diagnosis of COVID-19 in this study (100% PPA), we, like others, detected lower viral loads (i.e., higher CT values) in saliva specimens than in the corresponding NPS specimens (6, 9, 10). The CT values for NPS were lower (i.e., the viral loads were higher) than those for saliva in 71% of the specimens (Fig. 1A and B). The lower viral load in saliva may also explain the higher rate of indeterminate results (i.e., failure to amplify all three N gene targets in the CDC assay) for saliva specimens (5/7) compared with) NPS specimens (2/7).
In conclusion, our studies support the use of saliva as an alternate diagnostic specimen type for the laboratory-based diagnosis of COVID-19 in symptomatic individuals. The lower viral load in an enhanced saliva specimen suggests that false negatives would be more likely to occur with saliva specimens than with NPS specimens in individuals with lower viral loads. This finding may have implications if enhanced saliva specimens are used for asymptomatic screening (e.g., presurgical screening), as viral loads may be lower than in cases of symptomatic COVID-19, but the consequences of missing individuals with such low viral loads should be insignificant.
Supplementary Material
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Wang P, Anderson N, Pan Y, Poon L, Charlton C, Zelyas N, Persing D, Rhoads D, Babcock H. 2020. The SARS-CoV-2 outbreak: diagnosis, infection prevention, and public perception. Clin Chem Epub ahead of print. . doi: 10.1093/clinchem/hvaa080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kaul KL. 2020. Laboratories and pandemic preparedness: a framework for collaboration and oversight. J Mol Diagn 22:841–843. doi: 10.1016/j.jmoldx.2020.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kojima N, Turner F, Slepnev V, Bacelar A, Deming L, Kodeboyina S, Klausner JD. 2020. Self-collected oral fluid and nasal swabs demonstrate comparable sensitivity to clinician collected nasopharyngeal swabs for Covid-19 detection. medRxiv doi: 10.1101/2020.04.11.20062372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.LeBlanc JJ, Heinstein C, MacDonald J, Pettipas J, Hatchette TF, Patriquin G. 2020. A combined oropharyngeal/nares swab is a suitable alternative to nasopharyngeal swabs for the detection of SARS-CoV-2. J Clin Virol 128:104442. doi: 10.1016/j.jcv.2020.104442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheuk S, Wong Y, Tse H, Siu HK, Kwong TS, Chu MY, Yau FYS, Cheung IYY, Tse CWS, Poon KC, Cheung KC, Wu TC, Chan JWM, Cheuk W, Lung DC. 2020. Posterior oropharyngeal saliva for the detection of SARS-CoV-2. Clin Infect Dis Epub ahead of print. doi: 10.1093/cid/ciaa797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen JH, Yip CC, Poon RW, Chan KH, Cheng VC, Hung IF, Chan JF, Yuen KY, To KK. 2020. Evaluating the use of posterior oropharyngeal saliva in a point-of-care assay for the detection of SARS-CoV-2. Emerg Microbes Infect 9:1356–1359. doi: 10.1080/22221751.2020.1775133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jamal AJ, Mozafarihashjin M, Coomes E, Powis J, Li AX, Paterson A, Anceva-Sami S, Barati S, Crowl G, Faheem A, Farooqi L, Khan S, Prost K, Poutanen S, Taylor M, Yip L, Zhong XZ, McGeer AJ, Mubareka S, Toronto Invasive Bacterial Diseases Network COVID-19 Investigators. 2020. Sensitivity of nasopharyngeal swabs and saliva for the detection of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Clin Infect Dis Epub ahead of print. doi: 10.1093/cid/ciaa848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Leung EC, Chow VC, Lee MK, Lai RW. 2020. Deep throat saliva as an alternative diagnostic specimen type for the detection of SARS-CoV-2. J Med Virol Epub ahead of print. doi: 10.1002/jmv.26258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McCormick-Baw C, Morgan K, Gaffney D, Cazares Y, Jaworski K, Byrd A, Molberg K, Cavuoti D. 2020. Saliva as an alternate specimen source for detection of SARS-CoV-2 in symptomatic patients using Cepheid Xpert Xpress SARS-CoV-2. J Clin Microbiol 58:e01109-20. doi: 10.1128/JCM.01109-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pasomsub E, Watcharananan SP, Boonyawat K, Janchompoo P, Wongtabtim G, Suksuwan W, Sungkanuparph S, Phuphuakrat A. 2020. Saliva sample as a non-invasive specimen for the diagnosis of coronavirus disease 2019: a cross-sectional study. Clin Microbiol Infect Epub ahead of print. doi: 10.1016/j.cmi.2020.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wyllie AL, Fournier J, Casanovas-Massana A, Campbell M, Tokuyama M, Vijayakumar P, Geng B, Muenker MC, Moore AJ, Vogels CBF, Petrone ME, Ott IM, Lu P, Lu-Culligan A, Klein J, Venkataraman A, Earnest R, Simonov M, Datta R, Handoko R, Naushad N, Sewanan LR, Valdez J, White EB, Lapidus S, Kalinich CC, Jiang X, Kim DJ, Kudo E, Linehan M, Mao T, Moriyama M, Oh JE, Park A, Silva J, Song E, Takahashi T, Taura M, Weizman O-E, Wong P, Yang Y, Bermejo S, Odio C, Omer SB, Dela Cruz CS, Farhadian S, Martinello RA, Iwasaki A, Grubaugh ND, Ko AI. 2020. Saliva is more sensitive for SARS-CoV-2 detection in COVID-19 patients than nasopharyngeal swabs. medRxiv doi: 10.1101/2020.04.16.20067835. [DOI] [Google Scholar]
- 12.Rodino KG, Espy MJ, Buckwalter SP, Walchak RC, Germer JJ, Fernholz E, Boerger A, Schuetz AN, Yao JD, Binnicker MJ. 2020. Evaluation of saline, phosphate-buffered saline, and minimum essential medium as potential alternatives to viral transport media for SARS-CoV-2 testing. J Clin Microbiol 58:e00590-20. doi: 10.1128/JCM.00590-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shrestha NK, Marco Canosa F, Nowacki AS, Procop GW, Vogel S, Fraser TG, Erzurum SC, Terpeluk P, Gordon SM. 2020. Distribution of transmission potential during non-severe COVID-19 illness. Clin Infect Dis Epub ahead of print. doi: 10.1093/cid/ciaa886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lu X, Wang L, Sakthivel SK, Whitaker B, Murray J, Kamili S, Lynch B, Malapati L, Burke SA, Harcourt J, Tamin A, Thornburg NJ, Villanueva JM, Lindstrom S. 2020. US CDC real-time reverse transcription PCR panel for detection of severe acute respiratory syndrome coronavirus 2. Emerg Infect Dis 26:1654–1665. doi: 10.3201/eid2608.201246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cox JL, Koepsell SA. 2020. 3D-printing to address COVID-19 testing supply shortages. Lab Med 51:e45–e46. doi: 10.1093/labmed/lmaa031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.To KK, Tsang OT, Leung WS, Tam AR, Wu TC, Lung DC, Yip CC, Cai JP, Chan JM, Chik TS, Lau DP, Choi CY, Chen LL, Chan WM, Chan KH, Ip JD, Ng AC, Poon RW, Luo CT, Cheng VC, Chan JF, Hung IF, Chen Z, Chen H, Yuen KY. 2020. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. Lancet Infect Dis 20:565–574. doi: 10.1016/S1473-3099(20)30196-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fernández-Pittol M, Hurtado JC, Moreno-García E, Rubio E, Navarro M, Valiente M, Peiró A, Capón A, Seijas N, Martínez MJ, Casals-Pascual C, Vila J. 2020. Assessment of the use and quick preparation of saliva for rapid microbiological diagnosis of COVID-19. bioRxiv 10.1101/2020.06.25.172734. [DOI]
- 18.Kucirka LM, Lauer SA, Laeyendecker O, Boon D, Lessler J. 2020. Variation in false-negative rate of reverse transcriptase polymerase chain reaction-based SARS-CoV-2 tests by time since exposure. Ann Intern Med 173:262–267. doi: 10.7326/M20-1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu J, Liu X, Liu J, Liao H, Long S, Zhou N, Wu P. 2020. Coronavirus disease 2019 test results after clinical recovery and hospital discharge among patients in China. JAMA Netw Open 3:e209759. doi: 10.1001/jamanetworkopen.2020.9759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zou L, Ruan F, Huang M, Liang L, Huang H, Hong Z, Yu J, Kang M, Song Y, Xia J, Guo Q, Song T, He J, Yen HL, Peiris M, Wu J. 2020. SARS-CoV-2 viral load in upper respiratory specimens of infected patients. N Engl J Med 382:1177–1179. doi: 10.1056/NEJMc2001737. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.