A total of 1,200 serum samples that were tested for SARS-CoV-2 IgG antibody using the Abbott Architect immunoassay targeting the nucleocapsid protein were run in 3 SARS-CoV-2 IgG immunoassays targeting spike proteins (DiaSorin Liaison, Ortho Vitros, and Euroimmun). Consensus-positive and consensus-negative interpretations were defined as qualitative agreement in at least 3 of the 4 assays. Agreement of the 4 individual assays with a consensus-negative interpretation (n = 610) ranged from 96.
KEYWORDS: coronavirus, SARS-CoV-2 IgG, immunoassay
ABSTRACT
A total of 1,200 serum samples that were tested for SARS-CoV-2 IgG antibody using the Abbott Architect immunoassay targeting the nucleocapsid protein were run in 3 SARS-CoV-2 IgG immunoassays targeting spike proteins (DiaSorin Liaison, Ortho Vitros, and Euroimmun). Consensus-positive and consensus-negative interpretations were defined as qualitative agreement in at least 3 of the 4 assays. Agreement of the 4 individual assays with a consensus-negative interpretation (n = 610) ranged from 96.7% to 100%, and agreement with a consensus-positive interpretation (n = 584) ranged from 94.3% to 100%. Laboratory-developed inhibition assays were utilized to evaluate 49 consensus-negative samples that were positive in only one assay; true-positive reactivity was confirmed in only 2 of these 49 (4%) samples. These findings demonstrate very high levels of agreement among 4 SARS-CoV-2 IgG assays authorized for emergency use, regardless of antigen target or assay format. Although false-positive reactivity was identified, its occurrence was rare (no more than 1.7% of samples for a given assay).
INTRODUCTION
As of 14 August 2020, over 21 million cases of coronavirus disease 2019 (COVID-19) caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) have been identified worldwide, with more than 750,000 infection-related deaths (https://coronavirus.jhu.edu). The primary laboratory tool for diagnosing SARS-CoV-2 infection is detection of viral RNA by nucleic acid amplification testing (NAAT) (1). While antibody testing is not currently used to diagnose acute COVID-19 infections, assays for SARS-CoV-2 antibodies are proving important for (i) identifying some acutely infected individuals, particularly those presenting more than 1 to 2 weeks after symptom onset with negative RNA results, (ii) identifying persons who were exposed to the virus (including those with asymptomatic infection) and mounted a measurable antibody response, (iii) evaluating recovered individuals for donation of convalescent-phase plasma, and (iv) assessing prevalence of infection within community settings (https://cdc.gov/coronavirus/2019-ncov/hcp/testing-overview.html). In the future, quantitative SARS-CoV-2 IgG assays may play an important role in assessing vaccine efficacy (2, 3). Whether an IgG antibody response following infection or vaccination translates to protective immunity is still unknown (3).
Many SARS-CoV-2 antibody assays have recently received emergency use authorization (EUA) from the U.S. Food and Drug Administration (https://www.fda.gov/medical-devices/emergency-situations-medical-devices/eua-authorized-serology-test-performance). However, the extent of concordance among the various EUA assays is still being defined; in particular, it is unclear how assays measuring antibodies to the SARS-CoV-2 spike protein compare to assays measuring antibodies to the SARS-CoV-2 nucleocapsid protein (3). Such information is crucial to assisting clinicians and laboratory scientists in evaluating and interpreting results generated by different laboratories that utilize multiple EUA platforms. Here, we compare results obtained using 4 high-throughput SARS-CoV-2 IgG EUA immunoassays; samples positive or negative in an assay measuring IgG to viral nucleocapsid were tested in 3 assays measuring IgG recognizing viral spike protein subunits S1 or both S1 and S2 (S1+S2).
MATERIALS AND METHODS
Samples.
Serum samples with positive (n = 600) or negative (n = 600) SARS-CoV-2 IgG results from the Abbott chemiluminescent microparticle immunoassay (CMIA) utilizing nucleocapsid protein as the antigen target (referred to here as the nucleocapsid-based assay) were selected for further analysis in 3 SARS-CoV-2 IgG assays utilizing spike protein(s) as the antigen target (referred to here as spike-based assays). The 600 deidentified Abbott-positive samples were tested in the Abbott assay at Quest Diagnostics Nichols Institute, Chantilly, VA, and consisted of consecutive positive samples with enough remaining volume for further testing. After testing in the Abbott assay, these samples were frozen and shipped on dry ice to Quest Diagnostics Infections Disease (QDID), San Juan Capistrano, CA; there they were thawed and tested in the other 3 SARS-CoV-2 IgG assays within 24 h of thawing. The 600 deidentified Abbott-negative samples were tested in the Abbott assay at QDID and consisted of consecutive negative samples with enough remaining volume for further testing; the samples were refrigerated a maximum of 48 h before being tested in the other 3 assays. The median age of patients contributing the Abbott-positive samples was 47 years, and 48% were male; the median age of the patients contributing the Abbott-negative samples was 52 years, and 42% were male. Clinical findings and SARS-CoV-2 RNA results were not available for any of the 1,200 patients.
Immunoassays.
In addition to the Abbott CMIA, three SARS-CoV-2 IgG immunoassays (2 chemiluminescent immunoassays [CIAs] and 1 enzyme-linked immunosorbent assay [ELISA]) were evaluated (Table 1). All 4 assays were performed following the instructions for use supplied by the respective manufacturers. For the 3 chemiluminescent assays, undiluted serum samples are placed inside the respective instruments (Table 1), and all assay steps are performed inside the instrument. In contrast, for the Euroimmun ELISA, samples are diluted 1:101 in kit-supplied sample diluent, and the diluted sample is added to reaction wells. Results were classified as positive or negative following the interpretive criteria supplied in the manufacturer’s instructions, with one exception; samples with a Euroimmun equivocal index (0.8 to 1.0) were interpreted as positive for purposes of this study.
TABLE 1.
Information for the 4 SARS-CoV-2 IgG immunoassays used to test study samples
| Assay namea | Manufacturer | Method | Protein (antigen) target | Negative index | Positive index |
|---|---|---|---|---|---|
| Architect SARS-CoV-2 IgG | Abbott Laboratories Inc., Abbott Park, IL | CMIA | Nucleocapsid | <1.4 | ≥1.4 |
| Liaison SARS-CoV-2 S1/S2 IgG | DiaSorin Inc., Stillwater, MN | CIA | Spike 1 + spike 2 | <15.0 | ≥15.0 |
| Vitros anti-SARS-CoV-2 IgG | Ortho Clinical Diagnostics, Raritan, NJ | CIA | Spike 1 | <1.0 | ≥1.0 |
| Anti-SARS-CoV-2 ELISA (IgG) | Euroimmun Inc., Mountain Lakes, NJ | ELISA | Spike 1 | <0.8 | ≥0.8b |
For the CMIA and CIA methods, the assay name begins with the instrument utilized for test performance.
Per the Euroimmun instructions for use, index values of 0.8, 0.9, and 1.0 are considered equivocal; however, for the purposes of this study, equivocal results were interpreted as positive.
Inhibition assays.
Three assays (Abbott, DiaSorin, and Euroimmun) were modified to distinguish true-positive (TP) from false-positive (FP) reactivity based on inhibition of reactivity by soluble SARS-CoV-2 proteins. Preliminary experiments using a sample positive in all 4 assays (presumed to be TP) established the optimal concentrations of inhibitory proteins required for each assay. For the Abbott inhibition assay, serum samples were diluted 1:2 with phosphate-buffered saline (PBS), PBS containing soluble recombinant S1 protein (6 μg/ml; GenScript, Piscataway, NJ), or PBS containing soluble nucleocapsid protein (6 μg/ml; GenScript); thus, the final concentration of S1 protein or nucleocapsid protein in analyzed samples was 3 μg/ml. Similarly, for the DiaSorin inhibition assay, sera were diluted 1:2 with PBS, PBS containing S1 protein (6 μg/ml) and S2 protein (6 μg/ml; RayBiotech, Peachtree Corners, GA), or PBS containing nucleocapsid (12 μg/ml, to match the total S1+S2 concentration of 12 μg/ml). Thus, the final concentrations of analyzed samples for the DiaSorin inhibition assay were 3 μg/ml each for S1 and S2 and 6 μg/ml for nucleocapsid. For the Euroimmun inhibition assay, samples were diluted 1:101 in sample diluent, sample diluent containing S1 protein (3 μg/ml), or sample diluent containing nucleocapsid protein (3 μg/ml). For each inhibition assay, the samples tested included the samples positive only by the comparable routine assay, plus a minimum of 9 samples that were positive in all 4 assays.
Inhibition was calculated as (PBS or diluent index − nucleocapsid or spike index)/PBS or diluent index and expressed as a percentage. The cutoff for distinguishing TP from FP reactivity was defined as the mean percent inhibition value minus 2 standard deviations for the samples positive in all 4 assays that were tested in the inhibition assay; values below this cutoff were considered FP. Cutoff values were 29% for the Abbott inhibition assay, 42% for the DiaSorin inhibition assay, and 61% for the Euroimmun inhibition assay.
Analysis and statistics.
The various reactivity patterns across all 4 SARS-CoV-2 IgG assays were combined into 3 interpretation groups. Consensus-negative was defined as a negative result in at least 3 of 4 assays, and consensus-positive was defined as a positive result in at least 3 of 4 assays; the nonconsensus group included samples positive in 2 of 4 assays. All samples were classified into consensus-negative, consensus-positive, or nonconsensus categories based on the cutoffs listed in Table 1. For each of the 4 assays, positive agreement and negative agreement (and 95% confidence intervals) were calculated to compare the assay’s performance to positive consensus or negative consensus. Samples with a nonconsensus interpretation were not included in positive-agreement/negative-agreement calculations, since there was no majority positive or negative reactivity (i.e., positive in 2 assays and negative in 2 assays). Statistical analyses were conducted using R version 3.6.3, R Foundation for Statistical Computing (https://www.R-project.org).
RESULTS
Consensus interpretations.
Summarized in Table 2 are the results for the 1,200 samples across all 4 SARS-CoV-2 IgG assays. The vast majority of samples in the consensus-negative and consensus-positive groups showed complete agreement across all 4 assays (1,109/1,194 = 93% [95% confidence interval, 91.3% to 94.2%]).
TABLE 2.
Distribution of results across all 4 assays and consensus interpretations
| Interpretation (n) | Individual assay resulta
|
No. of samples | |||
|---|---|---|---|---|---|
| Abbott | DiaSorin | Ortho | Euroimmun | ||
| Consensus-negative (610) | Negative | Negative | Negative | Negative | 561 |
| Negative | Positive | Negative | Negative | 15 | |
| Negative | Negative | Negative | Positive | 20 | |
| Positive | Negative | Negative | Negative | 14 | |
| Consensus-positive (584) | Positive | Positive | Positive | Positive | 548 |
| Positive | Negative | Positive | Positive | 33 | |
| Positive | Positive | Positive | Negative | 1 | |
| Negative | Positive | Positive | Positive | 2 | |
| Nonconsensus (6) | Negative | Positive | Negative | Positive | 2 |
| Positive | Negative | Positive | Negative | 2 | |
| Positive | Negative | Negative | Positive | 2 | |
Discrepant results are in boldface.
Agreement of individual assay results with consensus-negative or -positive interpretations.
Table 3 shows the agreement of results for the 4 individual assays with a consensus-negative or consensus-positive interpretation. High levels of agreement were observed; agreement with a consensus-negative interpretation ranged from 96.7% to 100%, and agreement with a consensus-positive interpretation ranged from 94.3% to 100%. Note that the Ortho assay exhibited the highest agreement (100%) with both the consensus-negative and consensus-positive interpretations.
TABLE 3.
Agreement of individual SARS-CoV-2 IgG assays with negative or positive consensus interpretations
| Assay | Consensus negative (n = 610) |
Consensus positive (n = 584) |
||
|---|---|---|---|---|
| No. agreeing | % Agreement (95% confidence interval) | No. agreeing | Agreement (95% confidence interval) | |
| Abbott | 596 | 97.7 (96.2, 98.6) | 582 | 99.7 (98.8, 99.9) |
| DiaSorin | 595 | 97.5 (96, 98.5) | 551 | 94.3 (92.2, 95.9) |
| Ortho | 610 | 100 (99.4, 100) | 584 | 100 (99.3, 100) |
| Euroimmun | 590 | 96.7 (95, 97.9) | 583 | 99.8 (99, 100) |
Results of inhibition assays.
Within the consensus-negative group were 49 samples (4% of all samples tested) that were positive in only one assay (14 for Abbott, 15 for DiaSorin, and 20 for Euroimmun; no samples were positive by Ortho only). These samples, plus a minimum of 9 samples that were positive in all 4 assays (referred to here as TP samples), were tested in platform-specific inhibition assays designed to discriminate FP from TP reactive. The results are shown in Fig. 1. Of 14 samples positive only by Abbott, 12 exhibited FP reactivity, defined as an inhibition value of <29%. Similarly, all 9 evaluable samples positive only by DiaSorin exhibited FP reactivity (inhibition of <42%), and all 16 evaluable samples positive only by Euroimmun exhibited FP reactivity (inhibition of <61%); 6 DiaSorin-positive only samples and 4 Euroimmun-positive only samples could not be evaluated using the inhibition assay because they were negative when repeated in the routine assay as part of the inhibition protocol. Inhibition by the SARS-CoV-2 protein that was not the target protein in the comparable routine assay (i.e., S1 in the Abbott inhibition assay, nucleocapsid in the DiaSorin inhibition assay, and nucleocapsid in the Euroimmun inhibition assay) was ≤4%, ≤10%, and ≤16%, respectively, for all samples (data not shown). Taken together, these findings demonstrate that, based on inhibition assay results, only 2 of 49 (4%) samples positive in just one assay exhibited TP reactivity (both samples were positive only in the Abbott assay); 96% of samples initially positive in only one of the 4 assays exhibited FP reactivity.
FIG 1.
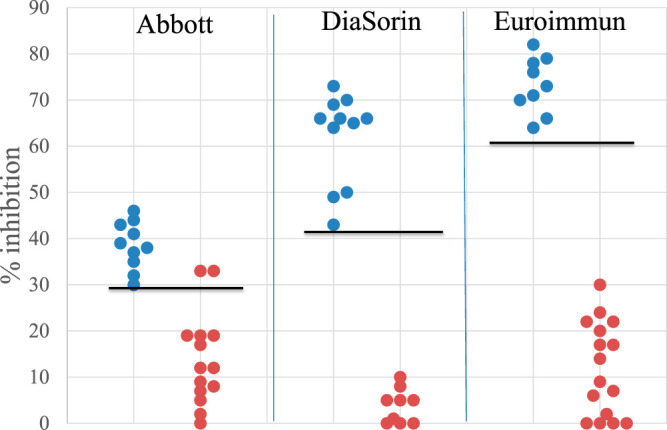
Results from inhibition assays. Blue dots represent samples positive in all 4 assays (TP samples); red dots represent samples that were positive only in the indicated assay. Values for the Abbott assay represent percent inhibition by nucleocapsid, values for the DiaSorin assay represent percent inhibition by S1+S2 proteins, and values for the Euroimmun assay represent percent inhibition by S1 protein. The horizonal lines indicate the assay-specific inhibition value below which reactivity is considered FP.
DISCUSSION
The appropriate interpretation of results from SARS-CoV-2 IgG assays depends on a clear understanding of their performance characteristics and limitations. Robust IgG responses to both the spike protein found on the surface of virus particles and the nucleocapsid protein found inside the virus particle occur following SARS-CoV-2 infection (4–7). However, there are conflicting reports as to the relative sensitivities of spike-based versus nucleocapsid-based IgG assays, particularly during the first 14 days after disease onset. Liu et al. (using ELISAs) and Tang et al. (using the Euroimmun ELISA and the Abbott CIA) found that spike-based IgG assays were slightly more sensitive during this time frame (8, 9), whereas Burbelo et al. (using luciferase immunoprecipitation systems) found that nucleocapsid-based assays were more sensitive (10). By 2 weeks after symptom onset, however, comparable sensitivities are observed (6, 7, 11–14).
Based on these published contradictory results for sensitivity (8–10), plus additional studies presenting contradictory data on the correlation of virus-neutralizing activity with spike antibody levels versus nucleocapsid antibody levels (15, 16), we sought to better understand how spike-based SARS-CoV-2 IgG assays perform compared to nucleocapsid-based assays. We thus selected samples previously tested using the Abbott assay targeting nucleocapsid protein to conduct our comparison study.
Our results show that, when consensus interpretations were defined based on agreement in at least 3 of the 4 assays evaluated, individual assay agreements with the consensus interpretation were very high, ranging from 94% to 100%, regardless of antigen target and assay platform. These findings thus demonstrate highly comparable performance among these 4 SARS-CoV-2 IgG assays. Our findings are consistent with those of other investigators (17–20), who demonstrated good correlation when comparing multiple assays using panels of up to approximately 500 samples. To our knowledge, ours is the first study to assess IgG reactivity in these 4 high-throughput assays using well over 1,000 samples.
A small number of samples within the consensus-negative group (4% of all samples) were positive in only one of the 4 assays. This pattern may represent increased assay sensitivity, decreased specificity, or a combination of the two. To discriminate among these possibilities, inhibition assays were developed and performed. The results showed that all samples positive only in the DiaSorin or Euroimmun assay exhibited FP reactivity; similarly, 12 of 14 samples positive only in the Abbott assay showed FP reactivity. Thus, the vast majority (96%) of samples that tested positive in only 1 of the 4 assays represented decreased assay specificity, rather than increased assay sensitivity. Of note, 13 of the 20 samples with FP reactivity in the Euroimmun assay exhibited index values within the manufacturer’s equivocal range (interpreted as positive for the purpose of this evaluation); however, 12 other Euroimmun-equivocal samples fell within the consensus positive group (also positive by Abbott and Ortho), indicating that some Euroimmun-equivocal samples show TP reactivity. These findings suggest that it may be appropriate to test Euroimmun-equivocal samples using a different SARS-CoV-2 IgG assay, in order to distinguish TP from FP reactivity.
Other investigators (17) evaluating the Euroimmun SARS-CoV-2 IgG assay interpreted Euroimmun-equivocal results as negative rather than as positive, prompting us to question how using that interpretation option might alter our findings. Had samples with an index in the Euroimmun equivocal range been interpreted as negative, the consensus-negative group would increase by 1 sample (from 610 to 611), and the consensus-positive group would decrease by 11 samples (from 584 to 573); most notably, the size of the nonconsensus group would nearly triple, increasing from 6 to 16 samples. The impact of these shifts on consensus-negative percent agreement and consensus-positive percent agreement would be minimal; the Euroimmun consensus-negative agreement would increase from 97% to 99%, and the DiaSorin consensus-positive agreement would increase from 94% to 96%. Thus, when a binary (positive/negative) interpretation was used for Euroimmun assay results, either interpretation of Euroimmun-equivocal results led to the same conclusion: all 4 SARS-COV-2IgG assays evaluated were highly comparable, exhibiting >90% agreement with consensus results.
A recognized limitation of our study is the lack of clinical information for the patients whose serum samples were evaluated. Information on viral RNA results, disease onset, and clinical course would likely help determine if the 2 patients positive only by Abbott but with TP reactivity simply mounted an antibody response to nucleocapsid earlier than a spike-based response, or if these patients for some reason failed to mount an antibody response to spike protein. Likewise, the same questions apply to the 2 patients positive in all 3 spike-based assays but negative in the nucleocapsid-based assay (see Table 2, consensus-positive results). Of note, Kohmer et al. (20) also identified a small number of samples with IgG recognizing only nucleocapsid or spike protein and speculated that there may be individual differences in the antibody response to SARS-CoV-2 proteins. Last, we assume that samples negative in all 4 assays were from uninfected persons and that samples positive in all 4 assays were from infected patients, but without information on viral RNA results, time since infection, and clinical course, we cannot know with certainty.
An additional limitation to the study is possible sample selection bias. As indicated, samples were selected based on the qualitative result in the Abbott SARS-CoV-2 IgG assay. Had we selected samples based on results in a spike-based assay, the levels of agreement among assays might have been different. However, the large number of samples included in the study provided the needed statistical power to demonstrate that the 4 assays yield comparable results.
Last, although data for the inhibition assays showed a clear distinction between TP and FP reactivity, it is unclear why the inhibition values in the Abbott inhibition assay were noticeably lower than the inhibition values in the DiaSorin and Euroimmun assays. The most obvious difference is the target antigen of the assay; the Abbott assay targets nucleocapsid, whereas the DiaSorin and Euroimmun assays target spike protein(s). For all 3 inhibition assays, the soluble inhibitory protein employed was not sourced from the assay manufacturer but rather purchased from a different vendor. Thus, subtle differences in glycosylation and tertiary structure may account for differences in the ability of the soluble protein to inhibit binding of antibodies to the immobilized antigen. Further studies are needed to characterize the relationship between soluble antigen structure and inhibitory activity in SARS-CoV-2 IgG inhibition assays.
In summary, our findings show these 4 SARS-CoV-2 IgG EUA assays exhibit excellent agreement, regardless of the target antigen used or assay format (CMIA/CIA versus ELISA). While variability can occur with any assay, health professionals receiving SARS-CoV-2 IgG results obtained using any of these 4 assays, whether from different laboratories or different platforms within a given laboratory, can be assured that the results are comparable and likely equivalent as a diagnostic adjunct to NAAT.
ACKNOWLEDGMENTS
This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sector.
We thank the Infectious Disease Serology Department staff members at Quest Diagnostics Chantilly and Quest Diagnostics Infectious Disease for expert technical assistance. We thank Ron Kagan for his assistance in retrieving demographic data for the study.
Footnotes
For a commentary on this article, see https://doi.org/10.1128/JCM.02179-20.
REFERENCES
- 1.Rogers AA, Baumann RE, Borillo GA, Kagan RM, Batterman HJ, Galdzicka M, Marlowe EM. 2020. Evaluation of transport media and specimen transport conditions for the detection of SARS-CoV-2 using real time reverse transcription PCR. J Clin Microbiol 58:e00708-20. doi: 10.1128/JCM.00708-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahn DG, Shin HJ, Kim MH, Lee S, Kim HS, Myoung J, Kim BT, Kim SJ. 2020. Current status of epidemiology, diagnosis, therapeutics, and vaccines for novel coronavirus disease 2019 (COVID-19). J Microbiol Biotechnol 30:313–324. doi: 10.4014/jmb.2003.03011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lerner AM, Eisinger RW, Lowy DR, Petersen LR, Humes R, Hepburn M, Cassetti MC. 2020. The COVID-19 Serology Studies workshop: recommendations and challenges. Immunity 53:1–5. doi: 10.1016/j.immuni.2020.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Okba NMA, Müller MA, Li W, Wang C, GeurtsvanKessel CH, Corman VM, Lamers MM, Sikkema RS, de Bruin E, Chandler FD, Yazdanpanah Y, Le Hingrat Q, Descamps D, Houhou-Fidouh N, Reusken CBEM, Bosch BJ, Drosten C, Koopmans MPG, Haagmans BL. 2020. Severe acute respiratory syndrome coronavirus 2-specific antibody responses in coronavirus disease patients. Emerg Infect Dis 26:1478–1488. doi: 10.3201/eid2607.200841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hou H, Wang T, Zhang B, Luo Y, Mao L, Wang F, Wu S, Sun Z. 2020. Detection of IgM and IgG antibodies in patients with coronavirus disease 2019. Clin Transl Immunology 9:e01136. doi: 10.1002/cti2.1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhong L, Chuan J, Gong B, Shuai P, Zhou Y, Zhang Y, Jiang Z, Zhang D, Liu X, Ma S, Huang Y, Lin H, Wang Q, Huang L, Jiang D, Hao F, Tang J, Zheng C, Yu H, Wang Z, Jian Q, Zeng T, Luo M, Zeng F, Zeng F, Liu J, Tian J, Xu Y, Long T, Xu K, Yang X, Liu Y, Shi Y, Jian G, Yang Z. 2020. Detection of serum IgM and IgG for COVID-19 diagnosis. Sci China Life Sci 63:777–780. doi: 10.1007/s11427-020-1688-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xiang F, Wang X, He X, Peng Z, Yang B, Zhang J, Zhou Q, Ye H, Ma Y, Li H, Wei X, Cai P, Ma WL. 2020. Antibody detection and dynamic characteristics in patients with COVID-19. Clin Infect Dis doi: 10.1093/cid/ciaa461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu W, Liu L, Kou G, Zheng Y, Ding Y, Ni W, Wang Q, Tan L, Wu W, Tang S, Xiong Z, Zheng S. 2020. Evaluation of nucleocapsid and spike protein-based enzyme-linked immunosorbent assays for detecting antibodies against SARS-CoV-2. J Clin Microbiol 58:e00461-20. doi: 10.1128/JCM.00461-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tang MS, Hock KG, Logsdon NM, Hayes JE, Gronowski AM, Anderson NW, Farnsworth CW. 2020. Clinical performance of two SARS-CoV-2 serologic assays. Clin Chem 66:1055–1062. doi: 10.1093/clinchem/hvaa120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burbelo PD, Riedo FX, Morishima C, Rawlings S, Smith D, Das S, Strich JR, Chertow DS, Davey RT, Cohen JI. 2020. Sensitivity in detection of antibodies to nucleocapsid and spike proteins of severe acute respiratory syndrome coronavirus 2 in patients with coronavirus disease 2019. J Infect Dis 222:206–213. doi: 10.1093/infdis/jiaa273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sun B, Feng Y, Mo X, Zheng P, Wang Q, Li P, Peng P, Liu X, Chen Z, Huang H, Zhang F, Luo W, Niu X, Hu P, Wang L, Peng H, Huang Z, Feng L, Li F, Zhang F, Li F, Zhong N, Chen L. 2020. Kinetics of SARS-CoV-2 specific IgM and IgG responses in COVID-19 patients. Emerg Microbes Infect 9:940–948. doi: 10.1080/22221751.2020.1762515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Qu J, Wu C, Li X, Zhang G, Jiang Z, Li X, Zhu Q, Liu L. 2020. Profile of immunoglobulin G and IgM antibodies against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Clin Infect Dis doi: 10.1093/cid/ciaa489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beavis KG, Matushek SM, Abeleda APF, Bethel C, Hunt C, Gillen S, Moran A, Tesic V. 2020. Evaluation of the EUROIMMUN anti-SARS-CoV-2 ELISA assay for detection of IgA and IgG antibodies. J Clin Virol 129:104468. doi: 10.1016/j.jcv.2020.104468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bryan A, Pepper G, Wener MH, Fink SL, Morishima C, Chaudhary A, Jerome KR, Mathias PC, Greninger AL. 2020. Performance characteristics of the Abbott Architect SARS-CoV-2 IgG assay and seroprevalence in Boise, Idaho. J Clin Microbiol 58:e00941-20. doi: 10.1128/JCM.00941-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.To KK, Tsang OT, Leung WS, Tam AR, Wu TC, Lung DC, Yip CC, Cai JP, Chan JM, Chik TS, Lau DP, Choi CY, Chen LL, Chan WM, Chan KH, Ip JD, Ng AC, Poon RW, Luo CT, Cheng VC, Chan JF, Hung IF, Chen Z, Chen H, Yuen KY. 2020. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. Lancet Infect Dis 20:565–574. doi: 10.1016/S1473-3099(20)30196-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ni L, Ye F, Cheng ML, Feng Y, Deng YQ, Zhao H, Wei P, Fe J, Gou M, Li X, Sun L, Cao T, Wang P, Zhou C, Zhang R, Liang P, Guo H, Wang X, Qin XF, Chen F, Dong C. 2020. Detection of SARS-CoV-2-specific humoral and cellular immunity in COVID-19 convalescent individuals. Immunity 52:971–977. doi: 10.1016/j.immuni.2020.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Theel ES, Harring J, Hilgart H, Granger D. 2020. Performance characteristics of four high-throughput immunoassays for detection of IgG antibodies against SARS-CoV-2. J Clin Microbiol 58:e01243-20. doi: 10.1128/JCM.01243-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tre-Hardy M, Wilmet A, Beukinga I, Dogne JM, Douxfils J, Blairon L. 2020. Validation of a chemiluminescent assay for specific SARS-CoV-2 antibody. Clin Chem Lab Med 58:1357–1364. doi: 10.1515/cclm-2020-0594. [DOI] [PubMed] [Google Scholar]
- 19.Jaaskelainen AJ, Kuivanen S, Kekalainen E, Ahava MJ, Loginov R, Kallio-Kokko H, Vapalahti O, Jarva H, Kurkela S, Lappalainen M. 2020. Performance of six SARS-CoV-2 immunoassays in comparison with microneutralisation. J Clin Virol 129:104512. doi: 10.1016/j.jcv.2020.104512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kohmer N, Westhaus S, Ruhl C, Ciesek S, Rabenau HF. 2020. Cinical evaluation of six high-throughput SARS-CoV-2 IgG antibody assays. J Clin Virol 129:104480. doi: 10.1016/j.jcv.2020.104480. [DOI] [PMC free article] [PubMed] [Google Scholar]


