We prospectively compared health care worker-collected nasopharyngeal swabs (NPS) to self-collected anterior nasal swabs (ANS) and straight saliva for the diagnosis of coronavirus disease 2019 (COVID-19) in 354 patients. The percent positive agreement between NPS and ANS or saliva was 86.3% (95% confidence interval [CI], 76.7 to 92.9%) and 93.8% (95% CI, 86.0 to 97.9%), respectively. The percent negative agreement was 99.6% (95% CI, 98.0 to 100.0%) for NPS versus ANS and 97.8% (95% CI, 95.3 to 99.
KEYWORDS: SARS-Cov-2, alternative specimen, anterior nasal swab, nasopharyngeal swab, saliva
ABSTRACT
We prospectively compared health care worker-collected nasopharyngeal swabs (NPS) to self-collected anterior nasal swabs (ANS) and straight saliva for the diagnosis of coronavirus disease 2019 (COVID-19) in 354 patients. The percent positive agreement between NPS and ANS or saliva was 86.3% (95% confidence interval [CI], 76.7 to 92.9%) and 93.8% (95% CI, 86.0 to 97.9%), respectively. The percent negative agreement was 99.6% (95% CI, 98.0 to 100.0%) for NPS versus ANS and 97.8% (95% CI, 95.3 to 99.2%) for NPS versus saliva. More cases were detected by the use of NPS (n = 80) and saliva (n = 81) than by the use of ANS (n = 70), but no single specimen type detected all severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infections.
INTRODUCTION
Rapid and accurate diagnostic tests are essential for controlling the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) pandemic. The Centers for Disease Control and Prevention (CDC) currently recommends collecting and testing an upper respiratory tract specimen for initial SARS-CoV-2 diagnostic testing (1), but the most sensitive specimen type has not been defined. Nasopharyngeal swabs (NPS) have historically been considered the reference specimen type used for respiratory virus detection. In addition, anterior nasal swabs (ANS) are routinely used for influenza virus nucleic acid amplification testing (NAAT). Recurrent shortages of swabs and personal protective equipment (PPE), however, have prompted evaluation of alternatives to NPS, including the use of patient self-collected ANS and saliva.
The advantages of ANS and saliva are the minimally invasive nature of sampling and the potential for patient self-collection, which may reduce health care worker exposure to infectious aerosols. Saliva also has the added benefit of being a swab-free specimen type known to contain high concentrations of SARS-CoV-2 RNA (2–4). Surprisingly few studies have assessed the performance of patient self-collected ANS for SARS-CoV-2 testing (5, 6). Small sample sizes and the use of selected cases limit the available evidence for ANS. More performance data exist for saliva than for ANS (7), but the published studies vary substantially in the way in which the specimens were obtained. Many saliva collection protocols require patients to cough before pooling saliva in their mouth (2, 3, 8); entail avoidance of food, water, or tooth brushing prior to testing (9); and/or rely on RNA stabilization reagents as a part of the collection device. Forced cough, if performed in the presence of a health care worker, necessitates the need for PPE, and restrictions on eating and drinking are not feasible in most health care settings. Furthermore, RNA stabilizers increase the cost of testing, are vulnerable to supply shortages, are not compatible with all NAAT chemistries, and can be potentially toxic to use. Larger studies that compare the performance of patient self-collected ANS and straight saliva to health care worker-collected NPS for SARS-CoV-2 detection are needed. Therefore, we performed a prospective comparative study to evaluate the performance of patient self-collected ANS and saliva versus that of health care provider-collected NPS for SARS-CoV-2 diagnostic testing.
MATERIALS AND METHODS
Study subjects.
Adult patients presenting to a drive-through test center with symptoms suggestive of coronavirus disease 2019 (COVID-19) were included. Criteria for testing included the presence of at least one of the following: fever, cough, shortness of breath, sore throat, malaise, chills, and/or a decreased sense of smell or taste. After obtaining consent, the subjects were instructed to swab both nostrils, pool saliva in their mouth without coughing, and then repeatedly spit a minimum of 1 ml saliva into a sterile empty tube in the presence of a health care worker. Detailed instructions for the patient self-collection procedures are included in the supplemental material.
The NPS was collected last in the sampling sequence and was collected by a technique matching the Infectious Diseases Society of America (IDSA) and CDC guidelines for the collection of samples for SARS-CoV-2 nucleic acid amplification testing (10). The University of Utah Institutional Review Board approved all study procedures.
Specimen collection and processing.
Flocked minitip and foam swabs (Puritan Medical Products) were used for the nasopharyngeal and nasal swab collections, respectively. Swabs placed in 3 ml of sterile 1× phosphate-buffered saline (ARUP Laboratories) and straight saliva collected in a sterile empty 50-ml Falcon tube (without prealiquoted stabilization medium) were transported to the clinical laboratory at 4○C. The study samples were stored refrigerated and tested within 5 days of receipt in the clinical laboratory, which is within our validated stability parameters for each specimen type. Saliva was then diluted 1:1 in ARUP Laboratories transport medium (ATM) at the time of testing. Mixing was performed directly in an Hologic Aptima lysis tube by gently inverting the tube three times to ensure homogenization, prior to testing on the instrument.
SARS-CoV-2 detection.
All specimens were analyzed using an Hologic Aptima SARS-CoV-2 transcription-mediated amplification (TMA) assay (Hologic Inc.), which is approved by the U.S. Food and Drug Administration (FDA) under an emergency use authorization for NPS and ANS. Samples producing an invalid TMA result were repeat tested using the original specimen and a 1:1 dilution in ATM. Discrepant NAAT results across specimens collected from the same patient triggered repeat testing using an Hologic Panther Fusion (Hologic Inc.) real-time reverse transcription-PCR (RT-PCR) platform to assess the cycle threshold (CT) value as a surrogate measure of the RNA concentration. Per the manufacturer’s package insert, a CT value of ≤42 by PCR is considered a positive result.
Statistical methods.
The standard-of-care NPS results obtained by TMA were used as the benchmark for assessments of test agreement. GraphPad Quick Calcs software was used to calculate kappa coefficients (κ) and proportions (P values) by the chi-square test. Percent positive or negative agreement for categorical variables was calculated in Microsoft Excel software using the Analyse-it software package.
RESULTS
A total of 1,104 specimens were collected from 368 unique patients between 29 May and 25 June 2020. The average age of the study participants was 35 years (range, 18 to 75 years), 47% were female, and 53% were male. Saliva samples from 12 patients (3.3%) generated invalid TMA results due to automated sample processing errors or internal control failure, and an additional 2 patients did not provide an adequate saliva volume for testing. Repeat testing in response to invalid results did not resolve sample failures. Data for patients with missing saliva data (n = 14) were excluded from the primary analysis.
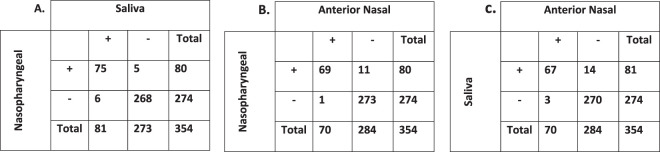
Figure 1 and Table 1 contain summaries of all TMA results. There was nearly perfect qualitative agreement across sample types (for NPS versus saliva, κ = 0.912 [95% confidence interval [CI], 0.86 to 0.96]; for NPS versus ANS, κ = 0.889 [95% CI, 0.84 to 0.95]). In all, 66 (18.6%) patients had SARS-CoV-2 detected in all 3 specimen types, 13 (3.7%) had SARS-CoV-2 detected in 2 specimen types, 7 (2.0%) had SARS-CoV-2 detected in 1 specimen type, and 268 (75.7%) had completely negative test results. Of the 13 patients for whom two of the three specimen collection types were positive, 9 (69.2%) had SARS-CoV-2 detected in NPS and saliva, 3 (23%) had SARS-CoV-2 detected in NPS and ANS, and a single patient (7.7%) had SARS-CoV-2 detected in saliva and ANS. The 7 patients for whom single specimens were positive included 2 (28.6%) in whom infections were detected in NPS only and 5 (71.4%) in whom infections were detected in saliva only. Positivity rates were higher for NPS (22.5%; 80/354) and saliva (22.9%; 81/354) than for ANS (19.7%; 70/354) alone, but this did not reach statistical significance (P = 0.408 for the NPS-versus-ANS comparison). The greatest case detection rate combined NPS sampling with saliva sampling (23.6%; 86/354).
FIG 1.
Qualitative result comparisons across all specimen types. The numbers represent the numbers of patients. Abbreviations: +, positive; −, negative.
TABLE 1.
Percent agreement between nasopharyngeal swabs and alternative specimen types
| Agreement | % agreement (95% CI) |
|
|---|---|---|
| Saliva vs NPS | ANS vs NPS | |
| Positive | 93.8 (86.0–97.9) | 86.3 (76.7–92.9) |
| Negative | 97.8 (95.3–99.2) | 99.6% (98.0–100.0) |
An adequate residual sample volume was available for 15 of 20 discordant specimen sets to perform repeat PCR testing. Figure 2 and Table S1 in the supplemental material display the CT values across discordant specimen sets. The average CT values for NPS-positive-only or saliva-positive-only specimens were 27.0 (range, 19.7 to 32.7) and 28.2 (range, 18.3 to 37.5), respectively. Similar CT value ranges (22.0 to 35.7) were seen in the NPS-positive/ANS-negative specimens, with the average CT value being 28.3. Interestingly, 3 specimens (1 saliva specimen and 2 ANS specimens) initially reported to be negative by TMA had low levels of viral RNA detected by RT-PCR upon repeat testing (average CT value, 35.7; range, 33.4 to 37.3).
FIG 2.
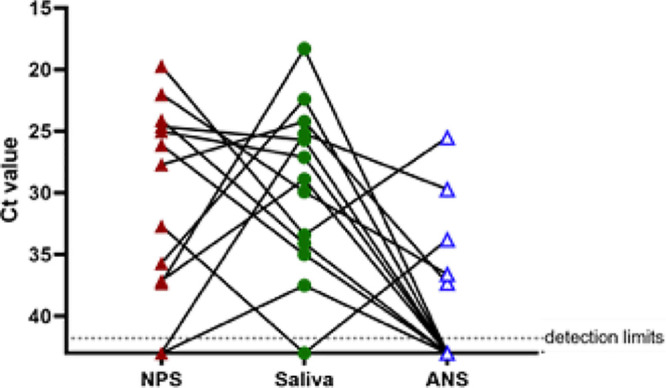
RT-PCR cycle threshold (CT) values for discordant NPS, saliva, and ANS specimen sets. A three-way comparison of CT values is shown, with solid lines linking RT-PCR results across specimen types.
DISCUSSION
Sensitive detection of SARS-CoV-2 RNA is critical for patient management decisions, hospital infection prevention, and curbing the ongoing public health emergency. The selection and adequate collection of clinical specimens play essential roles in diagnostic test performance, and this holds true for sensitive NAAT methods. Both the CDC (1) and IDSA (10) endorse the use of NPS or ANS (either health care worker or patient collected) for the diagnosis of COVID-19. However, few data from comparisons of the performance of different sample types collected from the same patient at the same time and by the use of FDA-authorized NAAT platforms exist.
This study represents one of the largest prospective comparisons of specimen types to date and demonstrates excellent agreement between provider-collected NPS and patient self-collected saliva and ANS. The majority (91.9%) of patients with positive results had SARS-CoV-2 nucleic acid detected in at least two specimen types concurrently. NPS and saliva samples had the greatest positivity rates overall. Given that all participants had a strong clinical suspicion for COVID-19 and molecular testing in general has a very high specificity, it is likely that the results for the NPS or saliva specimens that were the only specimens positive are true positives, but the lack of an accepted external reference standard precludes calculations of clinical sensitivity and specificity. Even though there was excellent qualitative agreement across specimen types, relying on ANS alone could have missed infection in 10 or 11 patients compared with relying on NPS or saliva alone, respectively. Missed COVID-19 cases have major clinical implications affecting isolation decisions for symptomatic patients and are a lost opportunity for contact tracing.
No single sample type detected all potential COVID-19 cases, and discrepant results were not always explained by high CT values (i.e., low RNA concentrations near the limit of detection of the test). There are several potential explanations for false-negative results. First, inadequate swab collection technique is possible. We did not include a host genomic marker to ensure the presence of respiratory epithelial cells on the swab, nor did we compare patient self-collected ANS to health care provider-collected ANS. Previous respiratory virus studies, however, suggest that the performance of patient self-collected ANS is equivalent to that of provider-collected ANS (11). We also did not evaluate the impact of swab type on SARS-CoV-2 detection. This study relied on foam nasal swabs and flocked nasopharyngeal swabs, so the results may not be generalizable to other swab types. Additionally, the level of viral replication in the nasopharynx or posterior oropharynx/salivary glands may vary over the course of infection. We did not collect information on the duration or type of symptoms at the time of specimen collection, which is an additional limitation of the study. Lastly, in an attempt to exclude the possibility of RNA degradation in straight saliva as a potential explanation for false-negative results, we performed stability studies at ambient and refrigerated temperatures for up to 5 days and saw no reduced TMA or PCR signal (data not shown).
In conclusion, NPS and saliva were clinically superior to ANS alone for the detection of SARS-CoV-2 in symptomatic patients. These observations, along with other recent reports (9, 12), suggest that straight saliva is an acceptable specimen type for symptomatic patients, especially if swab or PPE supplies are limited. However, not all patients could provide an adequate volume of saliva, and saliva is a complex matrix that requires clinical laboratories to validate this specimen type on their respective NAAT platforms. Saliva processing also required an additional pipetting step to dilute the specimen in ATM prior to testing. Additional processing has work flow and ergonomic implications for the clinical laboratory. Despite sample dilution, an increased rate of indeterminate or invalid results was observed for saliva (3.3% for saliva versus 0% for swabs in saline). This could be related to issues of sample viscosity affecting the automated pipetting and/or internal control inhibition. Repeat testing of the original specimen (diluted 1:1 in ATM) did not resolve invalid results and, therefore, is not recommended. We did not test whether a higher dilution factor (e.g., 1:2 or 1:3) with proportionally more ATM would reduce the invalid result rate without losing sensitivity. Regardless of the approach, repeat testing and recreation of the dilution series increase the time to results, the amount of labor required, and the overall cost of testing.
Combination testing with simultaneous sample collection from multiple anatomic sites may increase SARS-CoV-2 detection rates slightly, but multisite testing could be impractical, given the current swab and reagent shortages. Requiring two separate NAAT reactions would also increase costs. Given ongoing supply limitations, validating multiple specimen types provides redundancy and allows clinical laboratories options for testing. Ultimately, the availability of materials and staffing and cost considerations will influence what testing can be offered by individual laboratories.
Supplementary Material
ACKNOWLEDGMENT
This study was supported by the ARUP Institute for Clinical and Experimental Pathology.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Centers for Disease Control and Prevention. 2020. Interim guidelines for collecting, handling, and testing clinical specimens for COVID-19. CDC, Atlanta, GA: https://www.cdc.gov/coronavirus/2019-ncov/lab/guidelines-clinical-specimens.html. Accessed 14 July 2020. [Google Scholar]
- 2.To KK, Tsang OT, Chik-Yan Yip C, Chan KH, Wu TC, Chan JMC, Leung WS, Chik TS, Choi CY, Kandamby DH, Lung DC, Tam AR, Poon RW, Fung AY, Hung IF, Cheng VC, Chan JF, Yuen KY. 2020. Consistent detection of 2019 novel coronavirus in saliva. Clin Infect Dis 71:841–843. doi: 10.1093/cid/ciaa149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.To KK, Tsang OT, Leung WS, Tam AR, Wu TC, Lung DC, Yip CC, Cai JP, Chan JM, Chik TS, Lau DP, Choi CY, Chen LL, Chan WM, Chan KH, Ip JD, Ng AC, Poon RW, Luo CT, Cheng VC, Chan JF, Hung IF, Chen Z, Chen H, Yuen KY. 2020. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. Lancet Infect Dis 20:565–574. doi: 10.1016/S1473-3099(20)30196-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yoon JG, Yoon J, Song JY, Yoon SY, Lim CS, Seong H, Noh JY, Cheong HJ, Kim WJ. 2020. Clinical significance of a high SARS-CoV-2 viral load in the saliva. J Korean Med Sci 35:e195. doi: 10.3346/jkms.2020.35.e195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Altamirano J, Govindarajan P, Blomkalns AL, Kushner LE, Stevens BA, Pinsky BA, Maldonado Y. 2020. Assessment of sensitivity and specificity of patient-collected lower nasal specimens for sudden acute respiratory syndrome coronavirus 2 testing. JAMA Netw Open 3:e2012005. doi: 10.1001/jamanetworkopen.2020.12005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kojima N, Slepnev V, Bacelar A, Deming L, Kodeboyina S, Klausner JD. 2020. Self-collected oral fluid and nasal swabs demonstrate comparable sensitivity to clinician collected nasopharyngeal swabs for Covid-19 detection. medRxiv doi: 10.1101/2020.04.11.20062372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Khurshid Z, Zohaib S, Joshi C, Moin SF, Zafar MS, Speicher DJ. 2020. Saliva as a non-invasive sample for the detection of SARS-CoV-2: a systematic review. medRxiv https://www.medrxiv.org/content/10.1101/2020.05.09.20096354v1. [Google Scholar]
- 8.Chen JH, Yip CC, Poon RW, Chan KH, Cheng VC, Hung IF, Chan JF, Yuen KY, To KK. 2020. Evaluating the use of posterior oropharyngeal saliva in a point-of-care assay for the detection of SARS-CoV-2. Emerg Microbes Infect 9:1356–1359. doi: 10.1080/22221751.2020.1775133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wyllie AL, Casanovas-Massana A, Campbell M, Tokuyama M, Vijayakumar P, Geng B, Muenker MC, Moore AJ, Vogels CBF, Petrone ME, Ott IM, Lu P, Lu-Culligan A, Klein J, Venkataraman A, Earnest R, Simonov M, Datta R, Handoko R, Naushad N, Sewanan LR, Valdez J, White EB, Lapidus S, Kalinich CC, Jiang X, Kim DJ, Kudo E, Linehan M, Mao T, Moriyama M, Oh JE, Park A, Silva J, Song E, Takahashi T, Taura M, Weizman O-E, Wong P, Yang Y, Bermejo S, Odio C, Omer SB, Dela Cruz CS, Farhadian S, Martinello RA, Iwasaki A, Grubaugh ND, Ko AI. 2020. Saliva is more sensitive for SARS-CoV-2 detection in COVID-19 patients than nasopharyngeal swabs. medRxiv doi: 10.1101/2020.04.16.20067835. [DOI] [Google Scholar]
- 10.Hanson KE, Caliendo AM, Arias CA, Englund JA, Lee MJ, Loeb M, Patel R, El Alayli A, Kalot MA, Falck-Ytter Y, Lavergne V, Morgan RL, Murad MH, Sultan S, Bhimraj A, Mustafa RA. 16 June 2020. Infectious Diseases Society of America guidelines on the diagnosis of COVID-19. Clin Infect Dis doi: 10.1093/cid/ciaa760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Akmatov MK, Gatzemeier A, Schughart K, Pessler F. 2012. Equivalence of self- and staff-collected nasal swabs for the detection of viral respiratory pathogens. PLoS One 7:e48508. doi: 10.1371/journal.pone.0048508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Williams E, Bond K, Zhang B, Putland M, Williamson DA. 2020. Saliva as a noninvasive specimen for detection of SARS-CoV-2. J Clin Microbiol 58:e00776-20. doi: 10.1128/JCM.00776-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.