Abstract
The learning health system is a conceptual model for continuous learning and knowledge generation rooted in the daily practice of medicine. While companies such as Google and Amazon use dynamic learning systems that learn iteratively through every customer interaction, this efficiency has not materialized on a comparable scale in health systems. An ideal learning health system would learn from every patient interaction to benefit the care for the next patient. Notable advances include the greater use of data generated in the course of clinical care, Common Data Models, and advanced analytics. However, many remaining barriers limit the most effective use of large and growing health care data assets. In this review, we explore the accomplishments, opportunities, and barriers to realizing the learning health system.
Keywords: learning health system, electronic health data, medical records, medical algorithms
INTRODUCTION
Companies such as Google and Amazon exemplify successful learning systems, where the system learns from every customer interaction and iteratively improves to provide a better user experience or a more refined product recommendation for the next customer. Despite its immense promise, this efficiency in processing and analyzing big data to improve outcomes has not been realized in health care.1 Even today, most high-quality clinical evidence takes years to generate and disseminate.2 Nevertheless, health systems generate voluminous amounts of health care data that provide ample learning opportunities.3 The McKinsey Global Institute estimated that applying big-data analytics could generate up to $100 billion in value annually across US health care systems through broad improvements, including clinical trial efficiency, safety monitoring, payment innovation, administrative efficiency, and disease prevention.4
However, the opportunities are largely untapped despite the current technology that is well within the reach of effectively leveraging such data.5 These data continue to exist in silos, and the limited interoperability between major electronic health record systems slows the effective use of enormous data in generating new knowledge.6 The COVID-19 pandemic has put into bright relief the inability of health systems to rapidly learn from experience. Few health systems were well positioned to glean insights from the volumes of patients admitted with COVID-19 or to collaborate across systems.7 Agile responses were restricted to those who had the information systems in place that could be directed toward producing the needed knowledge at a rapid pace.8
In 2007, The Institute of Medicine proposed the concept of the learning health system, a conceptual model for continuous learning and knowledge generation embedded in the daily practice of medicine.9 It set a framework for health systems to improve outcomes with efficient utilization of data, similar to what is being achieved in the information technology sector (Figure 1). After more than a decade since it was proposed and even after some accomplishments, the learning health system remains to be fully actualized.
Figure 1.
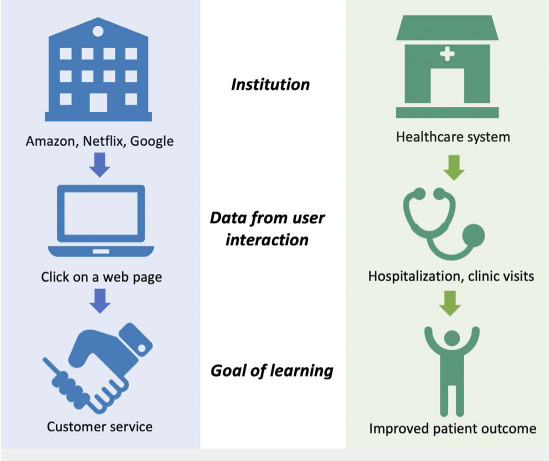
This figure shows similarities between the learning system successfully realized in companies and aspired in health care. Amazon improves the system via every web page click to increase profit. This efficiency has not materialized in health care to leverage the information gathered in every patient interaction to improve patient outcome.
WHAT IS AN IDEAL LEARNING HEALTH SYSTEM?
An ideal learning health system rapidly turns electronic health data into actionable knowledge in almost real time, contributing to a collective wisdom that enables each patient to benefit from the experiences of prior patients. Electronic health data include a variety of sources such as text data from provider notes, image data, laboratory values, diagnostic codes, continuously measured vital signs, data from personal digital devices (eg, wearables), and data captured in devices such as mechanical ventilators and cardiopulmonary bypass machines. In the era of digital medicine, effective use of the data requires rapid data capture, efficient data processing and analysis, and interpretable and actionable data output.
These processes should be automated and optimized such that the generation of data in electronic health data systems can seamlessly translate into an output delivered to the bedside in near real time. These outputs can provide value by conveying information about risk and patient response—for example, the probability of the patient experiencing readmission in the next 30 days, or the likelihood that the patient would benefit from a certain medical product(s), diagnostic test strategy, surgery, or other health care intervention. These systems can also help with diagnoses by leveraging pattern recognition. Ideally, such outputs would be available at the point of care to maximize the use of time-sensitive information and to inform timely clinical decision making for individual patients. In addition, an ideal system would automate the iterative learning process to rapidly refine algorithms. Although the infrastructure and technology to achieve each of these steps already exist, such platforms have not materialized to a large extent. As the Institute of Medicine's report emphasized, data utility and a digital platform form key components to realizing the ideal learning health system.10
SCOPE OF THE PROBLEM
In addition to predictive analytics, the learning health system can feed discovery, comparative effectiveness, and experimental research designs. The current approach in clinical research is slow, expensive, and rigid. For example, a clinical trial takes 5 years on average from the time of design to publication in a peer-reviewed journal,2 and the median estimated cost of trials testing therapeutic agents that were approved by the US Food and Drug Administration (FDA) between 2015 and 2016 was $19 million.11 Also, despite the value of adaptive trials, they are hard to implement largely because of the slow accumulation of data. In the time it takes to recruit subjects and collect data, the drug and/or intervention of interest may become obsolete or be used for slightly different clinical indications, whereby the study loses its relevance. Another problem is that trial inclusion criteria may be so limited that the resulting data is not considered inclusive enough to inform optimal therapy in real-world situations.12 Additionally, the estimation of average effect size in a clinical trial cohort may not necessarily inform whether or not a particular patient with differing demographics or clinical characteristics would actually benefit from the therapy.
There are similar issues of delay, cost, and complexity when using data to assess health care quality. For example, several states publicly report center- and provider-level outcomes for percutaneous coronary interventions and coronary artery bypass graft surgery, but performance in a given year is published 3 years later.13 The Society of Thoracic Surgeons Adult Cardiac Surgery Database distributes center- and surgeon-level quarterly reports, but this process is also delayed by the manual data abstraction process with the lag of at least several months.14 Therefore, data collection and reporting mechanisms limit timely response to the reported outcomes.
WHAT HAS BEEN DONE?
Electronic Health Record Data
Electronic health record data may provide an opportunity to generate high-quality evidence that reflects real-world practice and behaviors faster and with less effort. In addition, investing in an effective data platform may allow use of data that have traditionally been underutilized—such as information streams from mechanical ventilators, continuously monitored vital signs and telemetry, and cardiopulmonary bypass—to better assess and guide clinical care.5 These data streams can feed observational and experimental research in ways that enable faster, more accurate data acquisition.
To use health system data in research, it must be stored at the institutional level and easy to retrieve and process. Current electronic health record systems may house the bulk of the data, but continuous physiologic data and data captured in cardiopulmonary bypass machines and mechanical ventilators are often stored elsewhere in a separate local database. Effective storage of this massive data from multiple sources has been accelerated by distributed storage and cloud computing, allowing for parallel processing of computationally expensive tasks.15 Also, electronic health records are often not labeled in ways that make the information easy to aggregate and analyze; however, machine learning approaches can help with consistent and accurate data mapping without requiring substantial resources.
One advance in fostering the interoperability of data platforms is the Common Data Model (CDM), which provides a shared data language that applications can use. This type of model has a set of standardized data schema that represent commonly used concepts. An example is the Observational Medical Outcomes Partnership (OMOP) CDM that was developed by the Observational Health Data Sciences and Informatics (OHDSI) group.16 With OMOP, health systems can generate evidence using standardized tools. Fast Healthcare Interoperability Resources (FHIR) is a standard describing data formats and an application program interface that is used to unify multiple CDMs.17
One challenge with data platforms is that many exist within proprietary systems that constrain choices. However, they may be enabled by an open-source software framework for distributed storage and scalable, parallel data processing, which allows effective storage of massive data from multiple sources.18 Using this type of platform, our group developed an integrated data lake to store large health care datasets that can be accessed in near real time and transformed into CDMs fit for purpose.19 One feature of this platform is the ability to store and analyze longitudinal physiologic monitoring data such as vital signs and ventilator data. Another key feature is the ability to map, organize, and analyze data in almost real time. The data lake can leverage an analytic layer that provides exceptional computing capability. We are also in the process of integrating genomic data. In cases such as the COVID-19 pandemic, where efficient use of rapidly evolving data is needed, the versatility of this agile analytics platform allowed us to rapidly shift our focus to COVID-19–related topics, providing near real-time analysis of the data necessary to support operational intelligence, clinical care delivery, and advanced discovery.20 In such cases where a rapid and complex diagnosis is critical, computed phenotypes of patients based on almost real-time clinical data are also valuable.21
The rapidly increasing volume of data often makes it impractical to store all relevant data at a single, central location. This problem gave rise to the concept of “federated optimization,” in which analytical algorithms are tuned locally (ie, at each hospital), and the pooled results are used to update and improve the central model.16,22 A successful example of this is the OHDSI collaboration, which consists of an international data network with 11 data sources, using OMOP.16 This federated approach may bypass many issues encountered in the data centralization approach taken by large clinical registries. For example, the Society of Thoracic Surgeons database and the National Cardiovascular Data Registry depend on each hospital to collect data according to a standardized form and submit the data to a central repository. While this approach bypasses the interoperability issues between institutional electronic health records, the standardized collection of patient-level data is resource intensive and, by its nature, reductionist because information is lost when translating into case report forms. Additionally, there is an increasing concern about data security, even in de-identified data, because triangulation of de-identified individuals becomes easier with a large volume of ancillary data.23 This centralized approach may also create an asymmetry in benefit between the local sites and the central data manager, as the local sites bear the burden of data collection while benefits derived from the data may be harvested by the data manager.22 Therefore, federated models of data utilization may become an effective complement for the learning health system.
Another emerging approach is to support each patient's agency over their data. Bringing together an individual patient's data is a challenge when it is often scattered across multiple health care systems. Interoperability between electronic health record vendors has progressed slowly and remains limited despite the Health Information Technology for Economic and Clinical Health Act of 2009. Consequently, it is difficult to understand the health course of a patient being treated at multiple health care systems. Studying simple outcomes such as medication use and readmission over the long term remains a major challenge outside of payer-specific claims databases such as Medicare datasets,24 which is a limited population and often lack granularity such as laboratory and imaging data.
User-mediated health information exchange enables people to assemble their own longitudinal record across venues, ultimately producing an integrated record of the patient experience that feeds into research and clinical care.25 In this approach, a digital platform facilitates access to patients' own medical records through each hospital system's patient record portal, allowing patients to centralize all of their health records in one place, even data from pharmacies, payors, and wearable devices (Figure 2). Patients may then choose to share the data for specific clinical uses or research projects. By empowering patients to be the hub of their own health care data, this approach may foster research partnerships, bypass the interoperability issues across different electronic health record vendors, and benefit patients by providing a platform to keep track of their health records. From a research perspective, with a patient's permission, this may help create a repository of more comprehensive longitudinal health care data, unlike most current clinical databases that are tied to providers, institutions, or payors. Such platforms can enable patients to partner with their clinicians or with researchers to share their data, improve the data about their care, and drive research forward.
Figure 2.
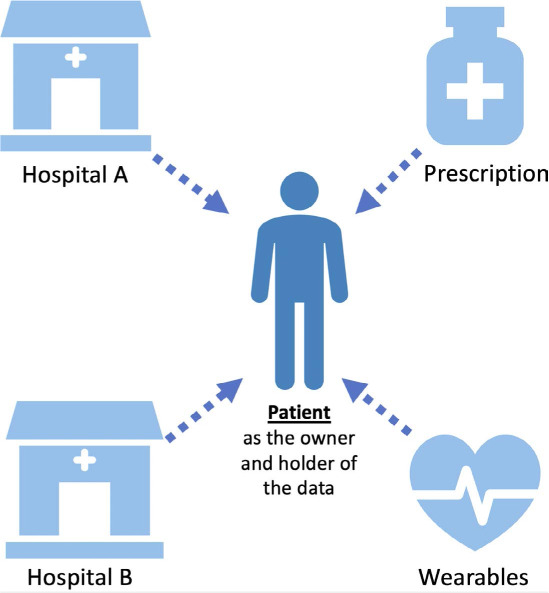
Model for patient as the holder of health care data. Hospitals often have interoperability issues to automatically share patients' health records (red cross). Empowering patients to centralize their health care data could form a meaningful research partnership and create efficient ways to help the next patient.
Advanced Analytics
The promise of advanced analytics applied to high-dimensional health care data is just on the horizon. Such approaches could help with prediction, pattern recognition, causal inference, computational phenotyping, and treatment matching. Diagnosis has perhaps been the most successful application of these methods. Machine and deep-learning approaches have successfully leveraged various data from unconventional sources to demonstrate the algorithms' diagnostic capacity, which is comparable or even superior to that of clinicians.26,27 Although large-scale implementation of such algorithms and their impact on improving patient outcomes remain largely unexamined, these examples represent a major step forward in realizing a next-generation learning health system.
The use of sensors is also showing promise, although it has not yet been fully realized. For example, Yan and colleagues have twice shown that high-throughput screening for atrial fibrillation may be possible by using video recordings of patients' faces in lieu of an electrocardiogram or even physical contact with the patient.26,28 The algorithm they developed using deep convoluted neural networks successfully discriminated 20 individuals who were experiencing atrial fibrillation from 24 individuals in sinus rhythm, with a sensitivity of 94%, specificity of 98%, positive predictive value of 98%, and negative predictive value of 94%.26 Other such examples include use of a screening electrocardiogram for the automated diagnosis of hyperkalemia27 and left ventricular dysfunction29 and identification of malignancies from various imaging sources.30,31
Some of the algorithms have already gained the approval of regulatory agencies. For example, deep learning algorithms demonstrated excellent performance in diagnosing diabetic retinopathy using retinal fundus photographs,32 and the FDA approved this software (IDx-DR) for clinical use.33 As more such software gains FDA approval, researchers are beginning to study how its implementation in the clinical arena may impact outcomes and costs of caring for these patient populations. Early evidence suggests that the use of advanced analytics may improve tailoring of treatment through improved phenotyping of diseases. Bhargava and colleages, for example, found that a machine learning algorithm detected critical variations in the stromal morphology of prostate cancer in black and white patients that prognosticated cancer recurrence.34 There are many other publications touting analytic approaches, and it is likely that we will soon see many such algorithms integrated into practice.
WHAT NEEDS TO BE DONE?
Implementation, Value Validation, and Algorithm Regulation
There are abundant reports of the use of big data and advanced analytical techniques demonstrating superior predictive performance compared with a conventional approach. Even so, there is little evidence regarding their implementation in clinical practice and validation that they add value in the real-world clinical setting. Several trial results evaluating the efficacy of analytical algorithms in improving clinical outcomes are much awaited. Moreover, defining the lifecycle of novel algorithms and determining how they should be regulated by governing agencies remain active areas of interest.35,36 Algorithms turned into software for clinical use instigated recognition of “Software as a Medical Device” (SaMD)—a term defined by the International Medical Device Regulators Forum (IMDRF) as “software intended to be used for one or more medical purposes that perform these purposes without being part of a hardware medical device.”37 This effort by the IMDRF to provide a harmonized approach for regulating how SaMD will be used for clinical evaluation is critical and timely to address unique challenges of such software that are not well regulated in the existing framework for devices. Nevertheless, we are still early in optimizing our approach to software oversight.
Additionally, predictive analytics that depend on historical data must be cautiously evaluated for potential bias that resides in the data,38 eg, sex- and race-based differences in outcomes and quality of care that exist in cardiovascular disease. Using algorithms trained on historical data that harbor potential sources of human biases can unintentionally propagate this problem.
Finally, we must remain cognizant that the current paradigm of treatment decisions is based on a group-level average of treatment effects. This approach may obscure individuals who, based on specific phenotypes, are actually harmed by a treatment that is beneficial to the group when the outcome is averaged.
Data Privacy and Access
Improvement in the way data are being used must be accompanied by the disruption of the current health care data market, which allows hospitals and companies to monetize patient health care data. Poor interoperability between electronic health record systems developed by different manufacturers promotes the way these data are treated as proprietary assets by limiting access. Although patient-directed centralization of health records through a digital platform may be a partial solution, a system-wide solution is needed to allow for a more efficient way of liberalizing health care data trapped in silos. At this juncture, the limited interoperability and modifiability of such electronic health record platforms, even within health systems, represents one of the largest barriers towards a rapidly adaptive learning health system.
Additionally, integration of multiple data sources must be done in a secure way with patients' permission. Ownership of patients' health care data is becoming increasingly controversial with a significant market interest.39,40 For example, there is the famous Dinerstein v Google case, in which a patient sued Google and the University of Chicago for turning over thousands of patient data that could triangulate a unique individual even without the identifiers that the Health Insurance Portability and Accountability Act (HIPAA) currently regulates.41 This case illustrated the inability of HIPAA to address contemporary privacy issues that are unique to the enormous scale of data. The current law is poorly equipped to answer the simple question of who owns health care data42 and urgently requires modernization to provide accountability and transparency to the data-sharing process.
Clearly defining and regulating access to health care data are critical because the medical record request form in about half of all hospitals does not provide an option for patients to acquire the entire medical record that they are entitled to by federal and state law.43 Efforts are underway to promote patient data agency by giving patients the option to share their data for research purposes, and this may be a way to drive people-powered generation of knowledge.25,44
The 21st Century Cures Act by the US-based Office of the National Coordinator for Health Information Technology (ONC) was created in 2016 to promote interoperability and patient access to health care records.45 This act was notable in that it prohibited information blocking, defined as “a practice by a health care provider, health IT developer, health information exchange, or health information network that…is likely to interfere with, prevent, or materially discourage access, exchange, or use of electronic health information.” The act also specified examples of information blocking, including (1) imposing formal or informal restriction on access, exchange, or use of electronic health information, and (2) discouraging efforts to develop or use interoperable technologies or services by exercising influence over customers, users, or other persons.46 The recent rules from ONC and the Centers for Medicare & Medicaid Services make tangible the aspirations of the act and will further advance this agenda. Thus, while the full promise of interoperability is yet to be delivered, a learning health system must strive towards it and ensure that health care organizations abide by these rules.
CONCLUSIONS
The digital era presents unprecedented opportunities to improve health care, and many promises are on the cusp of being actualized. Significant advances include the use of advanced predictive analytics that leverage unconventional data source; creation of data science platforms that enable centralization and analysis of rich health care data; awareness and discussions surrounding data security, privacy, and ownership; and regulation of advanced analytical algorithms. Remaining challenges include large-scale clinical implementation of analytical algorithms and demonstration of clinical benefit, improving interoperability of data storage mechanisms, and promotion of value and quality through disruption of the current health care data market.
KEY POINTS
The Learning Health System is a conceptual model that deserves re-examination in the context of the COVID-19 pandemic, which highlighted the gap between health systems that could and could not rapidly turn data into knowledge to respond quickly.
Advances made towards actualizing the Learning Health System include greater use of data generated in the course of clinical care, Common Data Models, and advanced analytics.
Remaining challenges include broader implementation of systems to efficiently capture and store various types of health care data; implementation, validation of value, and regulation of analytical algorithms; and improving compliance with regulations surrounding data privacy and access issues.
Footnotes
Conflict of Interest Disclosure:
Dr. Krumholz is an advisor for UnitedHealth Group, IBM Watson Health, Element Science, Aetna, Siegfried and Jensen LLP, Arnold & Porter Kaye Scholer LLP, Martin/Baughman law firm, and the National Center for Cardiovascular Diseases in Beijing (through Yale University); receives grant funding from the US Centers for Medicare & Medicaid Services (CMS), Medtronic, Johnson & Johnson, and the US Food and Drug Administration (FDA); and has financial relationships with HugoHealth and Refactor Health. Dr. Schulz was an investigator for a research agreement, through Yale University, from the Shenzhen Center for Health Information; collaborates with the National Center for Cardiovascular Diseases (Beijing); is a consultant to HugoHealth and Interpace Diagnostics Group; and is a cofounder of Refractor Health. Dr. Lin works under contract with CMS. Dr. Ross has received research support from Medtronic, Inc., the FDA, CMS, the Blue Cross Blue Shield Association, Johnson & Johnson, the Medical Device Innovation Consortium (as part of the National Evaluation System for Health Technology), the Agency for Healthcare Research and Quality, the National Heart, Lung and Blood Institute, and the Laura and John Arnold Foundation.
REFERENCES
- 1.Krumholz HM. Big Data and New Knowledge in Medicine: The Thinking, Training, and Tools Needed for a Learning Health System. Health Aff (Millwood) 2014 Jul;33(7):1163–70. doi: 10.1377/hlthaff.2014.0053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ioannidis JP. Effect of the Statistical Significance of Results on the Time to Completion and Publication of Randomized Efficacy Trials. JAMA. 1998 Jan 28;279(4):281–6. doi: 10.1001/jama.279.4.281. [DOI] [PubMed] [Google Scholar]
- 3.EMC.com [Internet] International Data Corporation. The digital universe of opportunities: rich data and the increasing value of the internet of things. Round Rock, TX: Dell Inc.; c2020. 2014. Apr, [cited 2020 Jun 9]. Available from: https://www.emc.com/leadership/digital-universe/2014iview/index.htm. [Google Scholar]
- 4.McKinsey & Company [Internet]. Cattell J, Chilukuri S, Levy M. How big data can revolutionize pharmaceutical R&D. New York, NY: McKinsey & Company; ©1996–2020. 2013 Apr 1 [cited 2020 Jun 9]. Available from: https://www.mckinsey.com/industries/pharmaceuticals-and-medical-products/our-insights/how-big-data-can-revolutionize-pharmaceutical-r-and-d. [Google Scholar]
- 5.Mori M, Schulz WL, Geirsson A, Krumholz HM. Tapping Into Underutilized Healthcare Data in Clinical Research. Ann Surg. 2019 Aug;270(2):227–9. doi: 10.1097/SLA.0000000000003329. [DOI] [PubMed] [Google Scholar]
- 6.Adler-Milstein J. Moving past the EHR interoperability blame game. NEJM Catalyst. 2017 Jul 18;3(4) [Google Scholar]
- 7.Dong E, Du H, Gardner L. An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect Dis. 2020 May;20(5):533–534. doi: 10.1016/S1473-3099(20)30120-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schulz WL, Durant TJS, Torre CJ, Hsiao AL, Krumholz HM. Agile Health Care Analytics: Enabling Real-Time Disease Surveillance With a Computational Health Platform. J Med Internet Res. 2020 May 28;22(5):e18707. doi: 10.2196/18707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Olson L, Aisner D, McGinnis MJ, editors. The Learning Healthcare System: Workshop Summary (2007) Washington, DC: The National Academies Press; c2007. p. 374. p. [PubMed] [Google Scholar]
- 10.Institute of Medicine. The learning health system and its innovation collaboratives update report. Washington, DC: National Academies Press; 2011. [Google Scholar]
- 11.Moore TJ, Zhang H, Anderson G, Alexander GC. Estimated Costs of Pivotal Trials for Novel Therapeutic Agents Approved by the US Food and Drug Administration, 2015–2016. JAMA Intern Med. 2018 Nov 1;178(11):1451–7. doi: 10.1001/jamainternmed.2018.3931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kennedy-Martin T, Curtis S, Faries D, Robinson S, Johnston J. A Literature Review on the Representativeness of Randomized Controlled Trial Samples and Implications for the External Validity of Trial Results. Trials. 2015 Nov 3;16:495. doi: 10.1186/s13063-015-1023-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mori M, Shahian DM, Suter LG, Geirsson A, Lin Z, Krumholz HM. Relevance of Cardiac Surgery Outcome Reporting 3 Years Later in a New York and California Statewide Analysis. JAMA Surg. 2020 Mar 4;155(5):442–4. doi: 10.1001/jamasurg.2019.6367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.The Society of Thoracic Surgeons [Internet]. STS National Database harvest schedule and information. Chicago, IL: The Society of Thoracic Surgeons; c2020. 2020 [cited 2020 Jun 9]. Available from: https://www.sts.org/registries-research-center/sts-national-database/harvest-schedule-and-information. [Google Scholar]
- 15.Li S, Li X. International Conference on Cyberspace Technology (CCT 2013) 2013. Nov 23, Distributed storage based on cloud computing; pp. 196–8. Beijing, China. 2013 Nov. [Google Scholar]
- 16.Hripcsak G, Ryan PB, Duke JD et al. Characterizing Treatment Pathways at Scale Using the OHDSI Network. Proc Natl Acad Sci U S A. 2016 Jul 5;113(27):7329–36. doi: 10.1073/pnas.1510502113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Braunstein ML. 2015 International Conference on Collaboration Technologies and Systems (CTS) 2015. Jun 1–5, Patient—Physician collaboration on FHIR (Fast Healthcare Interoperability Resources) pp. 501–3. Atlanta, GA. 2015. p. [Google Scholar]
- 18.Dean J, Ghemawat S. MapReduce: simplified data processing on large clusters. Commun ACM. 2008 Jan;51(1):107–113. [Google Scholar]
- 19.McPadden J, Durant TJ, Bunch DR et al. Health Care and Precision Medicine Research: Analysis of a Scalable Data Science Platform. J Med Internet Res. 2019 Apr 9;21(4):e13043. doi: 10.2196/13043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Medium [Internet]. Schulz W. Agile analytics & operational intelligence for COVID-19. San Francisco, CA: A Medium Corporation; 2020. 2020 Mar 22 [cited 2020 Jun 9]. Available from: https://medium.com/@wade.schulz/agile-analytics-operational-intelligence-for-covid-19-61c7f5401e59. [Google Scholar]
- 21.MedRxiv [Internet]. Schulz WL, Young PH, Coppi A, et al. Temporal relationship of computed and structured diagnoses in electronic health record data. 2020 Jan 2; doi: 10.1186/s12911-021-01416-x. [cited 2020 Jun 9]. Available from: https://www.medrxiv.org/content/10.1101/2019.12.28.19015628v1. [DOI] [PMC free article] [PubMed]
- 22.arXiv [Internet]. Konečný J, McMahan HB, Ramage D. Federated optimization: distributed optimization beyond the datacenter. New York, NY: Cornell University; c2020. 2015 Nov 11 [cited 2020 Jun 9]. Available from: https://arxiv.org/abs/1511.03575. [Google Scholar]
- 23.Rocher L, Hendrickx JM, de Montjoye YA. Estimating the Success of Re-Identifications in Incomplete Datasets Using Generative Models. Nat Commun. 2019 Jul 23;10(1):3069. doi: 10.1038/s41467-019-10933-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jacob JA. On the Road to Interoperability, Public and Private Organizations Work to Connect Health Care Data. JAMA. 2015 Sep 22–29;314(12):1213–5. doi: 10.1001/jama.2015.5930. [DOI] [PubMed] [Google Scholar]
- 25.Dhruva SS, Ross JS, Akar JG et al. Aggregating Multiple Real-World Data Sources Using a Patient-Centered Health-Data-Sharing Platform. NPJ Digit Med. 2020 Apr 20;3:60. doi: 10.1038/s41746-020-0265-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yan BP, Lai WHS, Chan CKY et al. High-Throughput, Contact-Free Detection of Atrial Fibrillation From Video With Deep Learning. JAMA Cardiol. 2019 Nov 27;5(1):105–7. doi: 10.1001/jamacardio.2019.4004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Galloway CD, Valys AV, Shreibati JB et al. Development and Validation of a Deep-Learning Model to Screen for Hyperkalemia From the Electrocardiogram. JAMA Cardiol. 2019 May 1;4(5):428–36. doi: 10.1001/jamacardio.2019.0640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yan BP, Lai WHS, Chan CKY et al. Contact-Free Screening of Atrial Fibrillation by a Smartphone Using Facial Pulsatile Photoplethysmographic Signals. J Am Heart Assoc. 2018 Apr 5;7(8):e008585. doi: 10.1161/JAHA.118.008585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Attia ZI, Kapa S, Lopez-Jimenez F et al. Screening for Cardiac Contractile Dysfunction Using an Artificial Intelligence-Enabled Electrocardiogram. Nat Med. 2019 Jan;25(1):70–4. doi: 10.1038/s41591-018-0240-2. [DOI] [PubMed] [Google Scholar]
- 30.Shen L, Margolies LR, Rothstein JH, Fluder E, McBride R, Sieh W. Deep Learning to Improve Breast Cancer Detection on Screening Mammography. Sci Rep. 2019 Aug 29;9(1):12495. doi: 10.1038/s41598-019-48995-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Munir K, Elahi H, Ayub A, Frezza F, Rizzi A. Cancer Diagnosis Using Deep Learning: A Bibliographic Review. Cancers (Basel) 2019 Aug 23;11(9):1235. doi: 10.3390/cancers11091235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gulshan V, Peng L, Coram M et al. Development and Validation of a Deep Learning Algorithm for Detection of Diabetic Retinopathy in Retinal Fundus Photographs. JAMA. 2016 Dec 13;316(22):2402–10. doi: 10.1001/jama.2016.17216. [DOI] [PubMed] [Google Scholar]
- 33.U. S. Food & Drug Administration [Internet] FDA permits marketing of artificial intelligence-based device to detect certain diabetes-related eye problems. Silver Spring, MD: U.S. Food and Drug Administration; c2020. 2018. Apr 11, [cited 2020 Jun 9]. Available from: https://www.fda.gov/news-events/press-announcements/fda-permits-marketing-artificial-intelligence-based-device-detect-certain-diabetes-related-eye. [Google Scholar]
- 34.Bhargava HK, Leo P, Elliott R et al. Computationally Derived Image Signature of Stromal Morphology Is Prognostic of Prostate Cancer Recurrence Following Prostatectomy in African American Patients. Clin Cancer Res. 2020 Apr 15;26(8):1915–23. doi: 10.1158/1078-0432.CCR-19-2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schulz WL, Durant TJS, Krumholz HM. Validation and Regulation of Clinical Artificial Intelligence. Clin Chem. 2019 Oct 1;65(10):1336–7. doi: 10.1373/clinchem.2019.308304. [DOI] [PubMed] [Google Scholar]
- 36.Hwang TJ, Kesselheim AS, Vokinger KN. Lifecycle Regulation of Artificial Intelligence- And Machine Learning-Based Software Devices in Medicine. JAMA. 2019 Nov 22; doi: 10.1001/jama.2019.16842. [DOI] [PubMed] [Google Scholar]
- 37.U. S. Food & Drug Administration [Internet] Software as a Medical Device (SaMD) Silver Spring, MD: U.S. Food and Drug Administration; c2020. 2018 Dec 4 [cited 2020 Jun 9]. Available from: https://www.fda.gov/medical-devices/digital-health/software-medical-device-samd. [Google Scholar]
- 38.Char DS, Shah NH, Magnus D. Implementing Machine Learning in Health Care - Addressing Ethical Challenges. N Engl J Med. 2018 Mar 15;378(11):981–3. doi: 10.1056/NEJMp1714229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mikk KA, Sleeper HA, Topol EJ. The Pathway to Patient Data Ownership and Better Health. JAMA. 2017 Oct 17;318(15):1433–4. doi: 10.1001/jama.2017.12145. [DOI] [PubMed] [Google Scholar]
- 40.Wachter RM, Cassel CK. Sharing Health Care Data With Digital Giants: Overcoming Obstacles and Reaping Benefits While Protecting Patients. JAMA. 2020 Jan 16; doi: 10.1001/jama.2019.21215. [DOI] [PubMed] [Google Scholar]
- 41.Cohen IG, Mello MM. Big Data, Big Tech, and Protecting Patient Privacy. JAMA. 2019 Aug 9; doi: 10.1001/jama.2019.11365. [DOI] [PubMed] [Google Scholar]
- 42.Cassel C, Bindman A. Risk, Benefit, and Fairness in a Big Data World. JAMA. 2019 Jul 9;322(2):105–6. doi: 10.1001/jama.2019.9523. [DOI] [PubMed] [Google Scholar]
- 43.Lye CT, Forman HP, Gao R et al. Assessment of US Hospital Compliance With Regulations for Patients' Requests for Medical Records. JAMA Netw Open. 2018 Oct 5;1(6):e183014. doi: 10.1001/jamanetworkopen.2018.3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Evans BJ, Krumholz HM. People-powered Data Collaboratives: Fueling Data Science With the Health-Related Experiences of Individuals. J Am Med Inform Assoc. 2019 Feb 1;26(2):159–61. doi: 10.1093/jamia/ocy159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lye CT, Forman HP, Daniel JG, Krumholz HM. The 21st Century Cures Act and Electronic Health Records One Year Later: Will Patients See the Benefits? J Am Med Inform Assoc. 2018 Sep 1;25(9):1218–20. doi: 10.1093/jamia/ocy065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.HealthIT.gov [Internet] Information Blocking. Washington, DC: U.S. Department of Health and Human Services; c2020. 2020 May 22 [cited 2020 Jun 9]. Available from: https://www.healthit.gov/topic/information-blocking. [Google Scholar]


