Background.
Approximately 15% of kidney transplant recipients (KTRs) develop BK viremia (BKV), with 1%–10% developing BK virus-associated nephropathy (BKVAN), which histologically resembles rejection. The Diagnosing Acute Rejection in Kidney Transplant Recipients (DART) study showed that donor-derived cell-free DNA (dd-cfDNA) levels <1% have a negative predictive value of 85% for active allograft rejection. Using data from this study, we evaluated the association of dd-cfDNA with plasma BK viral loads and biopsy findings to determine if dd-cfDNA can distinguish asymptomatic BKV from BKVAN.
Methods.
Data on dd-cfDNA, plasma BK viral loads, and biopsy findings from patients from the DART study were retrospectively examined. BKV was defined as 500–10 000 copies/mL. Presumptive BKVAN was defined as BK >10 000 copies/mL.
Results.
Of 102 participants with biopsies, 10 patients with BKV and BKVAN had paired dd-cfDNA, and viral loads available for analysis. Patients diagnosed with BKV and BKVAN had a median dd-cfDNA of 0.58% (IQR 0.43–1.15) and 3.38% (IQR 2.3–4.56, P = 0.001), respectively. dd-cfDNA titers correlated with BK PCR viral loads (R = 0.874, P = 0.01) and the presence of histologic evidence of BKVAN (100% sensitivity, 50% specificity). Five of 7 patients with BKVAN, but only 2 of 7 with BKV, had biopsies meeting Banff criteria for T-cell–mediated rejection. Median dd-cfDNA in nonrejection patients was 0.43% versus 2.84% in rejection patients (P = 0.001).
Conclusion.
Higher dd-cfDNA titers were associated with higher BK viral loads, biopsy-diagnosed BVAN, as well histologic changes meeting Banff criteria for as T-cell–mediated rejection. dd-cfDNA may be a useful noninvasive test to assess for progression of BKV to BKVAN.
Between 10% and 30% of kidney transplant recipients (KTRs) develop BK viremia (BKV); with 1%–10% developing BK virus-associated nephropathy (BKVAN). BK virus appears in the urine before occurrence of viremia by median of 4 weeks,1 followed by BKV, which precedes BKVAN by a median of 8 weeks. The polymerase chain reaction (PCR) threshold that best predicts BKVAN is a viral load >10 000 copies/mL.2,3 However, this remains controversial, likely due to variation in quantification methods and discordance in biopsy reads.4,5 Thus, the definitive diagnosis of BKVAN currently relies on biopsy confirmation and is managed through reduction of immunosuppression.6
The Diagnosing Acute Rejection in Kidney Transplant Recipients (DART) study (ClinicalTrials.gov Identifier: NCT02424227) showed that donor-derived cell-free DNA (dd-cfDNA; AlloSure CareDx, Brisbane, CA) levels <1% have a negative predictive value of approximately 85% for active allograft rejection.7 Whether dd-cfDNA has a role in discriminating BKV from BKVAN or acute rejection is unknown. To answer these questions, we retrospectively assessed dd-cfDNA, plasma BK viral loads, and biopsy findings from patients from the DART study.
MATERIALS AND METHODS
Study Design and Data Collection
The parent DART study has been previously described.7 Data on dd-cfDNA, plasma BK viral loads, and biopsy findings from patients from the DART study were retrospectively examined (WIRB protocol number 20150504). Institutional review board committee approval was obtained for this study. Data were anonymized and stored in secure encrypted electronic location.
BKV was defined as BK 500–10 000 copies/mL of plasma. Presumptive BKVAN was defined as PCR >10 000 copies/mL. Patients with the presence of BKV and BKVAN were included in the study. Histologic BKVAN was defined by positive staining for large T-antigen of simian virus 40 (SV40) or the presence of characteristic intranuclear BKV inclusions (Table 1).
TABLE 1.
Rank ordered dd-cfDNA by percent, BK diagnosis, and concurrent pathology reports
| AlloSure % | Patient | BK PCR copies | BK diagnosis | Pathology report from concurrent biopsy | Rejection |
|---|---|---|---|---|---|
| 0.29 | 1 | 2100 | BK viremia | 2 globally sclerosed glomeruli, cd68(0-3), CD3(0), mononuclear infiltrate, 45% cortical scarring, SV40 positive, C4d negative, trichrome interstitial fibrosis and tubular atrophy | No acute rejection |
| 0.43 | 4 | 1500 | BK viremia | No sclerosed glomeruli, CD68(0-1) CD3(0-4), mononuclear infiltrate, severe tubulitis (ti2_, iatr3, I, t1, moderate IF/TA, arterial intimal fibrosis | No acute rejection |
| 0.43 | 5 | 4800 | BK viremia | No sclerosed glomeruli, CD68(0-1) CD3(0-4), no significant infiltrate, no tubulitis (ti, iatr3, i, t, tatr, ptc), mild sclerosis no arterial inflammation. C4d negative | No acute rejection |
| 0.58 | 7 | 7500 | BK viremia | 26 glomeruli/8 sclerosed, 5 with hyaline deposit, focal fibrosis −30% IF/TA, no tubulitis or capillaritis, C4d negative, SV40 negative, C3 2+ t0, v0, i1, g0, ci2, ct2, cg1b, mm, cv2, ah3, ptc0 | No acute rejection |
| 0.68 | 5 | 7700 | BK viremia | 22 glomeruli, non sclerosed. cd68(0-3), CD3(0-7) (g_cg_mm), minimal mononuclear infiltrate, multifocal IF/TA(15%) (ti_, iatr3, i_, t, tatr, ptc), no capillaritis/tubulitis. Arterioles mild hyalinosis (v_cv_ahl) SVC4d negative, decoy cells in urine | No rejection |
| 1.15 | 3 | 9800 | BK Viremia | Minimal inflammatory infiltrate, Mild IF/TA | Borderline T-cell–mediated rejection |
| 1.22 | 6 | 10 000 | BK viremia | No formal path report | Grade IA rejection |
| 1.77 | 7 | 37 100 | BKVAN | 4 unremarkable glomeruli. CD68 0, CD3 0-1 (g_cg_, mm), moderate inflammatory infiltrate. No capillaritis but multifocal tubulitis (ti3, iatr3, i3, t2, tatr3, ptc). Mild IF/TA. SV40 positive in 75% of biopsy. C4d negative, DSA DQ2 positive | No rejection |
| 2.3 | 5 | 68 100 | BKVAN | 6 glomeruli, CD68(1-8), CD3(0-2) (g_, cg_, mm_), dense mononuclear infiltrate, plasma cell stain for IgM, SV40 focally positive, moderate tubuliits, no capillaritis, mild IF/TA (ti1, iatr1, i1, t3, tatr_, ptc_)C4d negative. Urine with decoy cells | Grade IA ACR rejection |
| 2.31 | 8 | 150 300 | BKVAN | 37 glomeruli, none sclerosed, moderate infiltrate, C4d negative. severe IF/TA, extensive tubulitis, no arteritis or capillaritis t2, i3, g2 | Patient went to have grade IA rejection |
| 3.38 | 9 | 500 000 | BKVAN | 8 glomeruli, CD68(0-6), CD3(1-4) (g_, cg_, mm_)prominent inflammatory infiltrate, severe tubulitis, no capillaritis (ti3, iatr2, i2, t3, tatr3, ptc_)moderate focal IF/TA, scarring in 25% of cortex (ci, ct1) SV40positive, C4d negative | T-cell mediated rejection, type IB |
| 3.56 | 10 | 289 000 | BKVAN | No formal path report— Moderate IF/TA, t2, i2 | Rejection IA |
| 4.54 | 2 | 384 000 | BKVAN | No formal path report—Moderate IF/TA | Grade IIA rejection |
| 4.65 | 3 | 22 000 000 | BKVAN | 37 glomeruli/1 sclerosed, intimal arteritis, arteriolar hyaline deposit, focal fibrosis -moderate IF/TA, severe tubulitis,C4d negative, SV40 negative (i2-3, t3, ci1, ct2, cv0, cg0, ah2, ptc0) | Banff IB |
ah, arteriolar hyalinosis; aah, hyaline arteriolar thickening; BKVAN, BK virus–associated nephropathy; ci, interstitial fibrosis; ct, tubular atrophy; cv, vascular fibrous intimal thickening; dd-cfDNA, donor-derived cell-free DNA; i, interstitial inflammation; i-IFTA, inflammation in the area of IF/TA; iatr, inflammation in areas of tubular atrophy; IF/TA, interstitial fibrosis and tubular atrophy; g, glomerulitis; mm, mesangial matrix expansion; ptc, peritubular capillaritis; SV40, simian virus 40; t, tubilitis; tatr, tubulitis in areas of tubular atrophy; ti, total inflammation; v, initimal arteritis.
Statistical Analysis
Linear regression models, Wilcoxon rank sum tests, and parametric ANOVA were used to compare groups. Pearson’s correlation-coefficient was used for correlation analyses. All analyses were performed using Stata (version 12.1; College Station, TX).
RESULTS
Of 102 trial participants with biopsies, 10 patients with BKV or BKVAN also had paired dd-cfDNA available for analysis. Patients diagnosed with BKV (median 7500 copies/mL, IQR 3450–8750/mL) had a median dd-cfDNA of 0.58% (IQR 0.43–1.15), while those with BKVAN (median 289 000 copies/mL, IQR 109 200–442 000 copies/mL) had a median dd-cfDNA of 3.38% (IQR 2.3–4.56, P = 0.001, Figure 1A).
FIGURE 1.
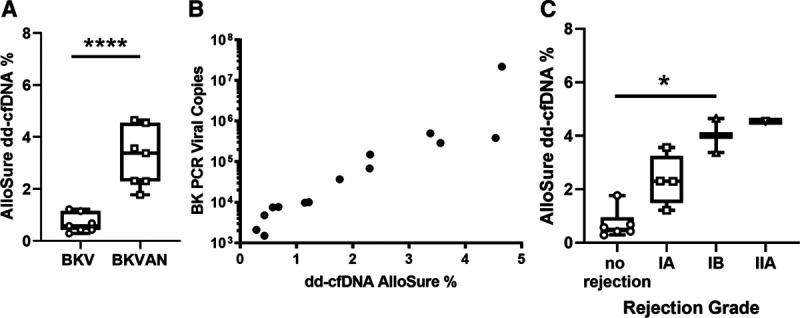
Correlation of dd-cfDNA Allosure levels with BK PCR viral copy number and biopsy diagnosis. A, Patient stratification by BKV vs BKVAN. B, BK PCR viral copies from BKVAN patients vs dd-cfDNA AlloSure levels. C, dd-cfDNA levels of biopsies meeting Banff criteria ≥Banff TCMR1A. BKV, BK viremia; BKVAN, BK virus–associated nephropathy; dd-cfDNA, donor-derived cell-free DNA.
Seven biopsies, all from different patients, met histologic criteria for BKVAN, with diagnostic “ground glass” type intranuclear inclusions in tubular epithelial cells (Table 1). In 3 biopsies, this was confirmed by immunohistochemical staining for SV40. Of 14 total biopsies, a formal biopsy report was not available for 3, but had an interpretation from the principal investigator. Three patients progressed from BKV to BKVAN, with concomitant rise in BK viral loads and dd-cfDNA.
BKV PCR titers correlated with dd-cfDNA results, and biopsy findings with 100% sensitivity and 50% specificity. There was a strong positive correlation of dd-cfDNA and BK viral load (Pearson R = 0.874, P = 0.01, Figure 1B).
Five of 7 patients with BKVAN, but only 2 of 7 patients with BKV, had biopsies meeting Banff criteria for T-cell–mediated rejection (TCMR). One patient with BKV had borderline TCMR and 1 had Banff grade 1A rejection. In the BKVAN cohort, 2 patients each met Banff criteria for grade 1A and 1B rejection, respectively, with one other having grade 2A rejection.
Median dd-cfDNA in nonrejection (including borderline) patients was 0.43%, versus 2.84% in those patients meeting a diagnosis of TCMR per Banff criteria (P = 0.001, Figure 1C). There was no significant difference in BK PCR load in biopsies meeting Banff criteria for TCMR ≥Banff 1A (P = 0.45).
DISCUSSION
BK viral reactivation in the transplanted kidney first manifests as viruria, preceding viremia by a median of 4 weeks. 1 Thereafter, BKV precedes BKVAN by a median of 8 weeks, with a better positive predictive value for the development of BKVAN than viruria, especially if the viral load is >10 000 copies/mL.2,3 However, there are no standardized BK PCR assays, and paucity of international standards preclude interlaboratory comparison.5,8,9 Diagnosis of BKVAN relies on kidney biopsy, which is also associated with considerable limitations. As recently demonstrated by Nankivell et al,10 even with kidney biopsy, detection rates are also dependent on deeper tissue sampling targeted at the medulla and SV40 staining is positive only in 48%, along can frequently be missed on microscopy. This phenomenon likely explains discordant BKVAN positive and BKVAN negative cores have been identified in nearly 30% cases. 11 Thus, owing to the focal nature of BK viral replication in the kidney, biopsy for BKVAN has limited sensitivity and a false negative rate of 10%–30%, especially in cases of biopsies done early after the onset of viremia and lacking medullary tissue.11,12
Circulating dd-cfDNA is a marker of allograft injury and rejection.7,13 Progression of BKV is associated with direct viral-induced cell necrosis and indirect tubular cell damage in morphologically noninfected tubules.14 In our study, dd-cfDNA predicted progression of BKV to BKVAN, with higher levels also associated with de novo BKVAN (median dd-cfDNA in BKV and BKVAN was 0.58% and 3.38%, respectively; Figure 1A).
Another pertinent finding was that levels of dd-cfDNA correlated with BK viral loads (Pearson R = 0.874, P = 0.01, Figure 1B) and had excellent sensitivity and specificity for BKVAN (100% sensitivity and 50% specificity). These findings support the utility of dd-cfDNA as a surrogate for predicting progression from BKV to BKVAN. It has been suggested previously that cfDNA could be utilized for broadly screening for infectious complications after transplantation. A correlation has been reported between cfDNA- and CMV-induced graft injuries in lung transplant recipients. 15
Five of the patients in our study with BKVAN met histologic Banff criteria for TCMR. Median dd-cfDNA in nonrejection patients was 0.43%, versus 2.84% in patients meeting a diagnosis of TCMR per Banff criteria (P = 0.001, Figure 1C). The tubulointerstitial nephritis typical of BKVAN can mimic rejection.6 Histologic acute cellular rejection can occur simultaneously with BKVAN and be present in more than half of patients with sustained BKV.16,17 There also is a bidirectional relationship between viral infection and acute rejection, and viral infection can trigger acute rejection or vice versa.17 Methods to distinguish these identities are not definitive, and reliance on clinical judgment continues.18,19 In comparison to the DART study, patients with IA TCMR had higher levels of dd-cfDNA in our cohort, which is likely driven by the presence of concurrent BKVAN.
This study is the first to ascertain the utility of dd-cfDNA in BKVAN, by examining dd-cfDNA, BK viral loads, and histology simultaneously. Additionally, we also present tracked data from the time of BKV detection to BKVAN progression to demonstrate the clinical applicability of dd-cfDNA.
Our study has several limitations. The study is retrospective and has a small sample size. Three of the biopsies did not have a formal pathology report available, and in 4 cases of BKVAN, the diagnosis was made solely by histology (characteristic intranuclear inclusions) without confirmation by immunohistochemical staining for SV40. However, limitations of biopsy sampling and associated SV40 staining are being increasingly recognized.10 The DART study was not able to distinguish borderline and 1A rejections using a dd-cfDNA threshold of 1%. Recent studies using a dd-cfDNA threshold of 0.7% were able to identify Banff Borderline and 1A rejections.20 Finally, there are no long-term follow-up data available on the patient cohort in this study.
In conclusion, dd-cfDNA positively correlated with BK viral loads, biopsy-diagnosed BVAN, and TCMR. dd-cfDNA may be useful in assessing the clinical course of BKV and BKVAN, distinguishing pathogenic and nonpathogenic BKV, along with detecting and tracking concomitant allograft injury. Additionally, it may aid in more precise diagnosis of BKVAN in those patients with borderline viral loads and confounding biopsy results. Rejection or BKVAN is unlikely when the dd-cfDNA level is below 0.7%.20
Footnotes
Published online 23 October, 2020.
Clinicaltrials.gov The Diagnosing Acute Rejection in Kidney Transplant Recipients (DART) study (ClinicalTrials.gov Identifier: NCT02424227).
All authors participated in research design, in the writing of the paper, in the performance of the research, and in data analysis.
D.B. has received honoraria for speaking, consulting fees and research support to Johns Hopkins from CareDx. S.K. is supported in part by a Fellowship Grant to Johns Hopkins School of Medicine. J.B. has received research support to the University of Maryland from CareDx. M.H. has received honoraria for speaking and consulting fees from CareDx.
REFERENCES
- 1.Hirsch HH, Brennan DC, Drachenberg CB, et al. Polyomavirus-associated nephropathy in renal transplantation: interdisciplinary analyses and recommendations. Transplantation. 2005; 79:1277–1286 [DOI] [PubMed] [Google Scholar]
- 2.Hirsch HH, Knowles W, Dickenmann M, et al. Prospective study of polyomavirus type BK replication and nephropathy in renal-transplant recipients. N Engl J Med. 2002; 347:488–496 [DOI] [PubMed] [Google Scholar]
- 3.Randhawa P, Ho A, Shapiro R, et al. Correlates of quantitative measurement of BK polyomavirus (BKV) DNA with clinical course of BKV infection in renal transplant patients. J Clin Microbiol. 2004; 42:1176–1180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pinto GG, Poloni JAT, Paskulin DD, et al. Quantitative detection of BK virus in kidney transplant recipients: a prospective validation study. J Bras Nefrol. 2018; 40:59–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Solis M, Meddeb M, Sueur C, et al. ; French BKV Study Group. Sequence variation in amplification target genes and standards influences interlaboratory comparison of BK virus DNA load measurement. J Clin Microbiol. 2015; 53:3842–3852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bohl DL, Brennan DC. BK virus nephropathy and kidney transplantation. Clin J Am Soc Nephrol. 2007; 2Suppl 1S36–S46 [DOI] [PubMed] [Google Scholar]
- 7.Bloom RD, Bromberg JS, Poggio ED, et al. ; Circulating Donor-Derived Cell-Free DNA in Blood for Diagnosing Active Rejection in Kidney Transplant Recipients (DART) Study Investigators. Cell-Free DNA and active rejection in kidney allografts. J Am Soc Nephrol. 2017; 28:2221–2232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hassan S, Mittal C, Amer S, et al. Currently recommended BK virus (BKV) plasma viral load cutoff of ≥4 log10/mL underestimates the diagnosis of BKV-associated nephropathy: a single transplant center experience. Transplant Infectious Disease. 2014; 16:55–60 [DOI] [PubMed] [Google Scholar]
- 9.Hoffman NG, Cook L, Atienza EE, et al. Marked variability of BK virus load measurement using quantitative real-time PCR among commonly used assays. J Clin Microbiol. 2008; 46:2671–2680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nankivell BJ, Renthawa J, Shingde M, et al. The importance of kidney medullary tissue for the accurate diagnosis of BK virus allograft nephropathy. Clin J Am Soc Nephrol. 2020; 15:1015–1023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Drachenberg CB, Papadimitriou JC, Hirsch HH, et al. Histological patterns of polyomavirus nephropathy: correlation with graft outcome and viral load. Am J Transplant. 2004; 4:2082–2092 [DOI] [PubMed] [Google Scholar]
- 12.Pang XL, Doucette K, LeBlanc B, et al. Monitoring of polyomavirus BK virus viruria and viremia in renal allograft recipients by use of a quantitative real-time PCR assay: one-year prospective study. J Clin Microbiol. 2007; 45:3568–3573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Celec P, Vlková B, Lauková L, et al. Cell-free DNA: the role in pathophysiology and as a biomarker in kidney diseases. Expert Rev Mol Med. 2018; 20:e1. [DOI] [PubMed] [Google Scholar]
- 14.Drachenberg CB, Papadimitriou JC, Wali R, et al. BK polyoma virus allograft nephropathy: ultrastructural features from viral cell entry to lysis. Am J Transplant. 2003; 3:1383–1392 [DOI] [PubMed] [Google Scholar]
- 15.De Vlaminck I, Martin L, Kertesz M, et al. Noninvasive monitoring of infection and rejection after lung transplantation. Proc Natl Acad Sci U S A. 2015; 112:13336–13341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nickeleit V, Hirsch HH, Zeiler M, et al. BK-virus nephropathy in renal transplants—tubular necrosis, MHC-class II expression and rejection in a puzzling game. Nephrol Dial Transplant. 2000; 15:324–332 [DOI] [PubMed] [Google Scholar]
- 17.Masutani K, Shapiro R, Basu A, et al. Putative episodes of T-cell-mediated rejection in patients with sustained BK viruria but no viremia. Transplantation. 2012; 94:43–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ahuja M, Cohen EP, Dayer AM, et al. Polyoma virus infection after renal transplantation. Use of immunostaining as a guide to diagnosis. Transplantation. 2001; 71:896–899 [DOI] [PubMed] [Google Scholar]
- 19.Collins AB, Schneeberger EE, Pascual MA, et al. Complement activation in acute humoral renal allograft rejection: diagnostic significance of C4d deposits in peritubular capillaries. J Am Soc Nephrol. 1999; 10:2208–2214 [DOI] [PubMed] [Google Scholar]
- 20.Stites E, Kumar D, Olaitan O, et al. High levels of dd-cfDNA identify patients with TCMR 1A and borderline allograft rejection at elevated risk of graft injury. Am J Transplant. 2020; 20:2491–2498 [DOI] [PMC free article] [PubMed] [Google Scholar]


