Abstract
Objectives:
Coronavirus disease 2019 continues to spread rapidly with high mortality. We performed metabolomics profiling of critically ill coronavirus disease 2019 patients to understand better the underlying pathologic processes and pathways, and to identify potential diagnostic/prognostic biomarkers.
Design:
Blood was collected at predetermined ICU days to measure the plasma concentrations of 162 metabolites using both direct injection-liquid chromatography-tandem mass spectrometry and proton nuclear magnetic resonance.
Setting:
Tertiary-care ICU and academic laboratory.
Subjects:
Patients admitted to the ICU suspected of being infected with severe acute respiratory syndrome coronavirus 2, using standardized hospital screening methodologies, had blood samples collected until either testing was confirmed negative on ICU day 3 (coronavirus disease 2019 negative) or until ICU day 10 if the patient tested positive (coronavirus disease 2019 positive).
Interventions:
None.
Measurements and Main Results:
Age- and sex-matched healthy controls and ICU patients that were either coronavirus disease 2019 positive or coronavirus disease 2019 negative were enrolled. Cohorts were well balanced with the exception that coronavirus disease 2019 positive patients suffered bilateral pneumonia more frequently than coronavirus disease 2019 negative patients. Mortality rate for coronavirus disease 2019 positive ICU patients was 40%. Feature selection identified the top-performing metabolites for identifying coronavirus disease 2019 positive patients from healthy control subjects and was dominated by increased kynurenine and decreased arginine, sarcosine, and lysophosphatidylcholines. Arginine/kynurenine ratio alone provided 100% classification accuracy between coronavirus disease 2019 positive patients and healthy control subjects (p = 0.0002). When comparing the metabolomes between coronavirus disease 2019 positive and coronavirus disease 2019 negative patients, kynurenine was the dominant metabolite and the arginine/kynurenine ratio provided 98% classification accuracy (p = 0.005). Feature selection identified creatinine as the top metabolite for predicting coronavirus disease 2019-associated mortality on both ICU days 1 and 3, and both creatinine and creatinine/arginine ratio accurately predicted coronavirus disease 2019-associated death with 100% accuracy (p = 0.01).
Conclusions:
Metabolomics profiling with feature classification easily distinguished both healthy control subjects and coronavirus disease 2019 negative patients from coronavirus disease 2019 positive patients. Arginine/kynurenine ratio accurately identified coronavirus disease 2019 status, whereas creatinine/arginine ratio accurately predicted coronavirus disease 2019-associated death. Administration of tryptophan (kynurenine precursor), arginine, sarcosine, and/or lysophosphatidylcholines may be considered as potential adjunctive therapies.
Keywords: biomarker, coronavirus disease 2019, diagnoses, intensive care unit, metabolomics, prognoses
Coronavirus disease 2019 (COVID19) is caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), which continues to spread rapidly worldwide (1, 2). Diagnosis of COVID19 typically requires polymerase chain reaction for SARS-CoV-2 genes or immunoassay for SARS-CoV-2 antigens. COVID19 primarily affects lungs, but dysfunction of other organs, such as heart and kidneys, has also been reported (3–6). The severity of COVID19 may involve the excessive release of inflammatory mediators (7–9) together with microvascular thrombi formation secondary to endothelial injury/activation and glycocalyx degradation (10). Critically ill COVID19 patients are admitted to the ICU, where the mortality rate is reported to be 31–40% with standardized ICU care (11, 12). Although a number of protein mediators have been identified that predict COVID19-associated death (9, 12), a further characterization of COVID19-associated processes and pathways is essential for the identification of novel diagnostic/prognostic biomarkers and for improving COVID19 patient outcomes.
Metabolomics measures a person’s metabolite profile (chemicals with an molecular weight < 1,500 Da), including amino acids, organic acids, biogenic amines, acylcarnitines, glycerophospholipids, sphingolipids, sugars, and many other compounds (13). Metabolites fall downstream of genetic, transcriptomic, proteomic, and environmental events, thus providing a cohesive measure of a subject’s recent phenotype. Two complementary analytical methods for metabolomics are proton nuclear magnetic resonance (1H NMR; μM range) spectroscopy and mass spectrometry (MS; nM-pM range). Previous studies have demonstrated the diagnostic and prognostic potentials of metabolomics profiling in selecting patient populations (e.g., traumatic brain injury [14]).
Metabolomics profiling of critically ill COVID19 patients over the first 10 days of their ICU stay was the overall aim of this exploratory study, thereby identifying potential metabolite candidates and/or combinations as diagnostic/prognostic biomarkers. Our specific objectives were: 1) to determine/compare the metabolomes between COVID19 positive (+) ICU patients and either healthy control subjects or COVID19 negative (–) ICU patients, 2) to determine specific metabolites that most accurately differentiated COVID19+ from either healthy control subjects or COVID19– ICU patients, and 3) to determine whether specific metabolites can predict COVID19 outcome shortly after ICU admission.
MATERIALS AND METHODS
This study was approved by the Western University, Human Research Ethics Board (HREB). Given the unprecedented pandemic situation and the restricted hospital access for substitute decision makers, waived consent was approved for a short, defined period of time (Research Ethics Board [REB] ID# 1670; issued March 20, 2020). In keeping with the Society for Critical Care Medicine statement on “Waiver of Informed Consent in Emergency Situations” (15), the following criteria were considered relevant for HREB approval of waived consent: the subjects were admitted to the ICU with a life-threatening condition; the subjects had impaired decisional capacity; the research staff encountered significant obstacles and delays when attempting to contact the absent substitute decision makers; the study risk was minimal; the research knowledge gained on this new, lethal disease offered an eventual chance of benefit; and community consultation had been implemented. Given the pandemic circumstances and the waived consent model applied, no further attempts were made to contact the surviving patients and/or substitute decision makers. The last patient enrolled under waived consent was May 1, 2020.
Study Participants and Clinical Data
We enrolled consecutive patients who were admitted to our level 3 academic ICUs at the London Health Sciences Centre (London, ON, Canada) and were suspected of having COVID19 based on standard hospital screening procedures (16). Blood sampling began on ICU admission for up to 3 days in COVID19– patients or up to 7 days in COVID19+ patients, with an additional blood draw occurring on day 10 for COVID19+ patients who have not been discharged. COVID19 status was confirmed as part of standard hospital testing by nasopharyngeal swab detection of two SARS-CoV-2 viral genes on polymerase chain reaction (17). Patient baseline characteristics were recorded at admission and included age, sex, comorbidities, medications, hematologic labs, creatinine, arterial-partial-pressure-to-inspired-oxygen ratio, and chest x-ray findings. We calculated Multiple Organ Dysfunction Score (18) and Sequential Organ Failure Assessment score (19) for both COVID19+ and COVID19– patient groups to enable objective comparison of their illness severity. Both patient groups were characterized as having confirmed or suspected sepsis diagnosis using Sepsis 3.0 criteria (19). We also recorded clinical interventions received during the observation period including use of antibiotics, antiviral agents, systemic corticosteroids, vasoactive medications, VTE prophylaxis, antiplatelet or anticoagulation treatment, renal replacement therapy, high-flow oxygen therapy, and mechanical ventilation (invasive and noninvasive). Final participant groups were constructed by age- and sex-matching COVID19+ patients with COVID19– patients and healthy controls without disease, acute illness, or prescription medications that were previously banked in the Translational Research Centre, London, ON, Canada (REB ID# 16986E; reissued March 10, 2020; “Repository of control biological specimens from healthy volunteers for future research purposes”; Directed by Dr. D. D. Fraser; https://translationalresearchcentre.com/) (20, 21).
Blood Draws
Standard operating procedures were used to ensure all samples were treated rapidly and equally. Blood was obtained from critically ill ICU patients via indwelling catheters in the morning and placed immediately on ice. If a venipuncture was required, research blood draws were coordinated with a clinically indicated blood draw. In keeping with accepted research phlebotomy protocols for adult patients, blood draws did not exceed maximal volumes (22). Once transferred to a negative pressure hood, blood was centrifuged and plasma was isolated, aliquoted at 250 μL, and frozen at –80°C. All samples remained frozen until use and freeze/thaw cycles were avoided.
Direct Injection-LC-Mass Spectrometry/Mass Spectrometry
A targeted quantitative metabolomics approach was used to analyze the samples using a combination of direct injection (DI) MS with a reverse-phase LC-MS/MS custom assay. This custom assay, in combination with an ABSciex 4000 QTrap (Applied Biosystems, Foster City, CA/MDS Sciex, Foster City, CA) mass spectrometer, can be used for the targeted identification and quantification of up to 150 different endogenous metabolites including amino acids, acylcarnitines, biogenic amines and derivatives, uremic toxins, glycerophospholipids, sphingolipids, and sugars (23, 24). The method combines the derivatization and extraction of analytes, and the selective mass-spectrometric detection using multiple reaction monitoring pairs. Isotope-labeled internal standards and other internal standards were used for metabolite quantification. The custom assay contained a 96-deep-well plate with a filter plate attached with sealing tape, and reagents and solvents used to prepare the plate assay. The first 14 wells were used for one blank, three zero samples, seven standards, and three quality control samples. For all metabolites except organic acid, samples were thawed on ice and subsequently vortexed and centrifuged at 13,000× g; 10 µL of each sample was then loaded onto the center of the filter on the upper 96-well plate and dried in a stream of nitrogen. Subsequently, phenyl-isothiocyanate was added for derivatization. After incubation, the filter spots were dried again using an evaporator. Extraction of the metabolites was then achieved by adding 300 µL of extraction solvent. The extracts were obtained by centrifugation into the lower 96-deep-well plate, followed by a dilution step with the MS running solvent (0.2% formic acid in water, 0.2% formic acid in acetonitrile for biogenic amines and amino acids, and 0.02% formic acid in methanol for all other classes of metabolites).
For organic acid analysis, 150 µL of ice-cold methanol and 10 µL of isotope-labeled internal standard mixture were added to 50 µL of serum sample for overnight protein precipitation at –20°C, followed by centrifugation at 13,000× g for 20 minutes. A total of 50 µL of supernatant was loaded into the center of wells of a 96-deep-well plate, followed by the addition of 3-nitrophenylhydrazine reagent. After incubation for 2 hours, butylated hydroxytoluene stabilizer (2 mg/mL) and water were added before LC-MS injection.
Mass spectrometric analysis was performed on an ABSciex 4000 Qtrap tandem MS instrument (Applied Biosystems/MDS Analytical Technologies, Foster City, CA) equipped with an Agilent 1260 series UHPLC system (Agilent Technologies, Palo Alto, CA). The samples were delivered to the mass spectrometer by an LC method followed by a DI method. Data analysis was done using Analyst 1.6.2 (Foster City, CA).
Proton Nuclear Magnetic Resonance
Plasma samples contain a significant concentration of large-molecular-weight proteins and lipoproteins, which affects the identification of the small-molecular-weight metabolites by NMR spectroscopy. A deproteinization step, involving ultrafiltration as previously described (25), was therefore introduced in the protocol to remove plasma proteins. Prior to filtration, 3-kDa cutoff centrifugal filter units (Amicon Microcon YM-3, Burlington, MA) were rinsed five times each with 0.5 mL of H2O and centrifuged (9,400× g for 10 min) to remove residual glycerol bound to the filter membranes. Aliquots of each plasma sample were then transferred into the centrifuge filter devices and spun (9,400× g for 20 min) to remove macromolecules (primarily protein and lipoproteins) from the sample. The filtrates were checked visually for any evidence that the membrane was compromised, and for these samples, the filtration process was repeated with a different filter and the filtrate inspected again. The subsequent filtrates were collected and the volumes were recorded. If the total volume of the sample was under 250 µL, an appropriate amount from a 150-mM KH2PO4 buffer (pH 7) was added until the total volume of the sample was 173.5 µL. Any sample that had to have buffer added to bring the solution volume to 173.5 μL was annotated with the dilution factor and metabolite concentrations were corrected in the subsequent analysis. Subsequently, 46.5 µL of a standard buffer solution (54% D2O:46% 1.75-mM KH2PO4 pH 7.0 v/v containing sodium trimethylsilylpropanesulfonate (DSS) [5.84-mM 2,2-dimethyl-2-silcepentane-5-sulphonate, 5.84-mM 2-chloropyrimidine-5 carboxylate, and 0.1% NaN3 in H2O]) was added to the sample.
The plasma sample (250 µL) was then transferred to a 3-mm SampleJet NMR tube for subsequent spectral analysis. All 1H-NMR spectra were collected on a 700-MHz Avance III (Bruker, Billerica, MA) spectrometer equipped with a 5-mm hydrogen, carbon, nitrogen Z-gradient pulsed-field gradient cryoprobe. 1H-NMR spectra were acquired at 25°C using the first transient of the Nuclear Overhauser Effect Spectroscopy (NOESY) presaturation pulse sequence (NOESY1DPR), chosen for its high degree of quantitative accuracy (26). All free induction decays were zero-filled to 250 K data points. The singlet produced by the DSS methyl groups was used as an internal standard for chemical shift referencing (set to 0 ppm). For quantification, all 1H-NMR spectra were processed and analyzed using an in-house version of the magnetic resonance for metabolomics (MAGMET)-automated analysis software package using a custom metabolite library. MAGMET allows for qualitative and quantitative analyses of an NMR spectrum by automatically fitting spectral signatures from an internal database to the spectrum. Each spectrum was further inspected by an NMR spectroscopist to minimize compound misidentification and misquantification. Typically, all of visible peaks were assigned. Most of the visible peaks were annotated with a compound name. It has been previously shown that this fitting procedure provides absolute concentration accuracy of 90% or better (27).
Population Statistics
Medians (interquartile ranges [IQRs]) and frequency (%) were used to report ICU patient baseline characteristics for continuous and categorical variables, respectively; continuous variables were compared using Mann-Whitney U tests (or Kruskal-Wallis tests, as appropriate), and categorical variables were compared using Fisher exact chi-square, with p values of less than 0.05 considered statistically significant. Receiver operating characteristic (ROC) curves were conducted to determine sensitivity and specificity of individual metabolite ratios for predicting a binary outcome. Area-under-the-curve (AUC) was calculated as an aggregate measure of metabolite ratio performance across all possible classification thresholds. All analyses were conducted using SPSS version 26 (IBM, Armonk, NY).
Machine Learning
COVID19 analyte data were visualized with a nonlinear dimensionality reduction on the full data matrix using the t-distributed stochastic nearest neighbor embedding (t-SNE) algorithm (28). t-SNE assumes that the “optimal” representation of the data lies on a manifold with complex geometry, but with low dimension, embedded in the full-dimensional space of the raw data. For feature selection, the raw data for each subject were ingested within each feature, across subjects. A random forest classifier was trained on the variables to predict COVID19 status or COVID19 outcome (“scikit-learn” module for Python 3.8.5 Open Source). A random forest is a set of decision trees, and consequently, we were able to interrogate this collection of trees to identify the features that have the highest predictive value. Feature selection was not performed in preprocessing. During training, the random forest classifier performed an implicit feature selection; the top features are those that appear highest ranked in the most trees. To reduce overfitting, the number of trees and maximum depth of each tree was limited (29); thus, COVID19 status was determined using a six-fold cross validation with a random forest of 10 trees, whereas patient outcome was determined using a three-fold cross validation with a random forest of 10 trees and a maximum depth of 6. To remain conservative and to limit the risk of overfitting further, no hyperparameters were tuned or optimized by design and intent. Furthermore, to validate the results and ensure no overfitting occurred, a simple linear support vector machine classifier was used to compare the predication accuracies with excellent concordance.
RESULTS
We investigated 10 COVID19+ patients (median years of age = 61.0, IQR = 54.8–67.0), 10 age- and sex-matched COVID19– patients (median years of age = 58.0, IQR = 52.5–63.0), and 10 age- and sex-matched healthy controls (median years of age = 57.5, IQR = 52.8–62.8; p = 0.686). Baseline demographic characteristics, comorbidities, laboratory values, and chest x-ray findings are reported in Table 1. The COVID19– patients had significantly higher unilateral pneumonia, whereas COVD19+ patients were more likely to have bilateral pneumonia. Sepsis was “confirmed” by infectious pathogen identification in only 20% of COVID19– patients, whereas sepsis was “suspected” in the remaining 80% (19). A mortality rate of 40% was determined for COVID19+ patients.
Table 1.
Subject Demographics and Clinical Data
| Variable | Healthy Control Subjects | COVID19– Patients | COVID19+ Patients | p |
|---|---|---|---|---|
| n | 10 | 10 | 10 | 1.000 |
| Age, yr | 57.5 (52.8–62.8) | 58.0 (52.5–63.0) | 61.0 (54.8–67.0) | 0.686 |
| Sex | 7 Female:3 Male | 7 Female:3 Male | 7 Female:3 Male | 1.000 |
| Multiple Organ Dysfunction Score | 6.0 (3.8–8.0) | 4.0 (2.5–7.3) | 0.251 | |
| Sequential Organ Failure Assessment score | 7.5 (4.8–11.0) | 4.5 (2.8–9.3) | 0.160 | |
| Comorbidities | ||||
| Hypertension | 8 (80) | 6 (60) | 0.628 | |
| Diabetes | 4 (40) | 3 (30) | 1.000 | |
| Chronic kidney disease | 1 (10) | 2 (20) | 1.000 | |
| Cancer | 1 (10) | 2 (20) | 1.000 | |
| Chronic obstructive pulmonary disease | 1 (10) | 0 (0) | 1.000 | |
| Baseline medications | ||||
| Antiplatelet agents | 6 (60) | 2 (20) | 0.170 | |
| Anticoagulants | 1 (10) | 0 (0) | 1.000 | |
| Baseline labs | ||||
| WBC | 15.3 (11.1–23.0) | 8.5 (6.3–16.1) | 0.064 | |
| Neutrophils | 12.2 (8.1–15.2) | 7.7 (5.7–13.3) | 0.197 | |
| Lymphocytes | 1.6 (0.5–2.3) | 0.7 (0.6–1.0) | 0.141 | |
| Platelets | 184 (159–245) | 206 (109–294) | 0.623 | |
| Hemoglobin | 130 (104–142) | 122 (102–136) | 0.364 | |
| Creatinine | 80 (54–147) | 107 (55–288) | 0.571 | |
| Chest x-ray findings | ||||
| Bilateral pneumonia | 1 (10) | 9 (90) | 0.001* | |
| Unilateral pneumonia | 5 (50) | 0 (0) | 0.033* | |
| Interstitial infiltrates | 1 (10) | 1 (10) | 1.000 | |
| Normal | 3 (30) | 0 (0) | 0.211 | |
| Arterial-partial-pressure-to-inspired-oxygen ratio | 172 (132–304) | 124 (69–202) | 0.153 | |
| Sepsis diagnosis | ||||
| Suspected | 8 (80) | 0 (0) | 0.001* | |
| Confirmed | 2 (20) | 10 (100) | 0.001* | |
| Interventions during study | ||||
| Antibiotics | 10 (100) | 10 (100) | 1.000 | |
| Antivirals | 0 (0) | 3 (30) | 0.211 | |
| Steroids | 3 (30) | 2 (20) | 1.000 | |
| Vasoactive medications | 6 (60) | 7 (70) | 1.000 | |
| VTE prophylaxis | 10 (100) | 10 (100) | 1.000 | |
| New antiplatelets | 0 (0) | 1 (10) | 1.000 | |
| New anticoagulation | 2 (20) | 1 (10) | 1.000 | |
| Renal replacement therapy | 1 (10) | 2 (20) | 1.000 | |
| High-flow nasal cannula | 2 (20) | 5 (50) | 0.350 | |
| Noninvasive MV | 8 (80) | 6 (60) | 0.628 | |
| Invasive MV | 8 (80) | 7 (70) | 1.000 | |
| Patient outcome | ||||
| Venous thromboembolism/ischemic stroke | 2 (20) | 1 (10) | 1.000 | |
| Survived | 10 (100) | 6 (60) | 0.087 | |
MV = mechanical ventilation.
Continuous data are presented as medians (interquartile ranges) and categorical data are presented as n (%). VTE prophylaxis represents the number of patients receiving venous thromboembolism prophylaxis with regular- or low-molecular heparin; new antiplatelets represents the number of patients who were started on aspirin or clopidogrel during ICU stay; new anticoagulation represents the number of patients who were started on therapeutic anticoagulation with regular- or low-molecular heparin, or novel oral anticoagulants
We measured a total of 183 plasma metabolites using both DI-LC-MS/MS and 1H NMR. In the event of metabolite repeats measured with both techniques (21 metabolites), the 1H NMR metabolite repeat measurements were deleted from the combined metabolite database, yielding a final number of 162 metabolites analyzed.
Figure 1A shows a t-SNE plot illustrating that the ICU day 1 COVID19+ patient metabolome was distinct and easily separable from age- and sex-matched healthy control subjects. In fact, classification accuracy was 100% when comparing the two metabolomes. We then identified the top eight metabolites underlying these differences between the cohorts, which are shown in Figure 1B with their associated % importance. In the COVID19+ cohort, relative to the healthy control subjects, kynurenine increased 5.1-fold whereas arginine decreased 0.5-fold, sarcosine decreased 0.6-fold, and lysophosphatidylcholines (LysoPCs) all decreased 0.3-fold on average. The least number of metabolites that were required to maintain a 100% classification accuracy between the cohorts was then determined, with only arginine (cutoff ≤ 52.8 μM) and kynurenine (cutoff ≥ 3.1 μM) required. The excellent predictive ability of an arginine/kynurenine ratio for discriminating a COVID19 patient from a healthy control subject (cutoff ≤ 15.7) is shown with ROC analysis in Figure 1C (AUC = 1.00; p = 0.0002).
Figure 1.
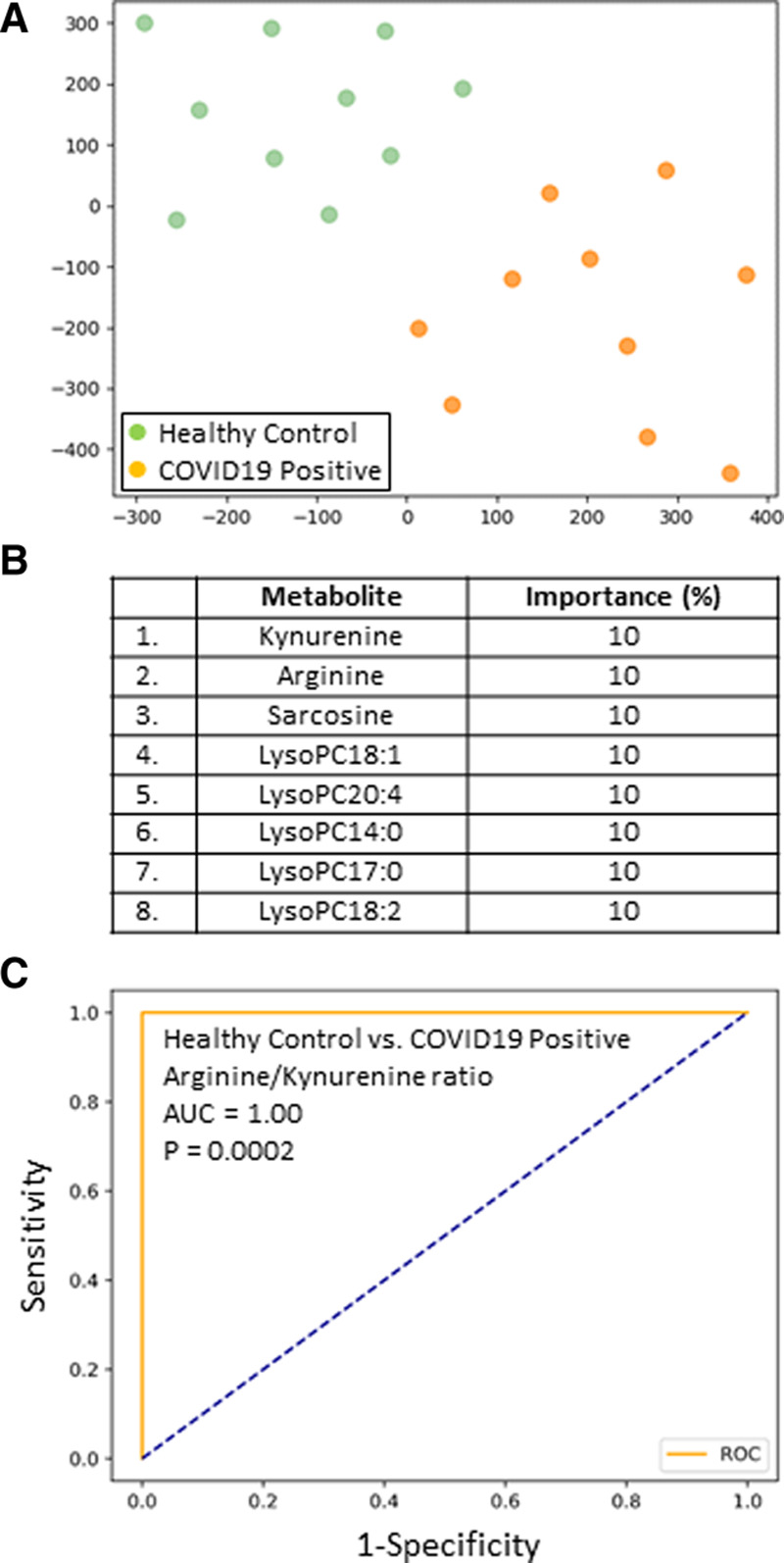
A, Subjects plotted in two dimensions following dimensionality reduction in their respective metabolites by stochastic neighbor embedding. Green dots represent healthy control subjects, whereas orange dots represent age- and sex-matched coronavirus disease 2019 positive (COVID19+) ICU patients (ICU day 1 plasma). The dimensionality reduction shows that based on the plasma metabolites, the two cohorts are distinct and easily separable. The axes are dimensionless. B, Feature classification, demonstrating the top eight plasma metabolites that classify COVID19+ status versus healthy control subjects with their % association. C, Receiver operating characteristic analysis of healthy control subjects versus COVID19+ patients, using an arginine/kynurenine ratio, demonstrates an area-under-the-curve (AUC) of 1.00 (p = 0.0002). The cutoff value is 15.6. The diagonal broken blue line represents chance (AUC 0.50).
A comparison of COVID19+ and COVID19– patient cohorts revealed distinct metabolomes. Feature classification again identified kynurenine as one of the leading metabolites underlying the differences between the COVID19+ and COVID19– cohorts (Fig. 2A). We then determined that an arginine/kynurenine ratio again showed an excellent discriminative ability to determine COVID19 status on ICU day 1 (cutoff ≤ 11.6) via ROC analyses (AUC = 0.98; p = 0.005; Fig. 2B). Figure 2C shows an arginine/kynurenine ratio time plot for the COVID19+ and COVID19– patients over 10 ICU days. The cohorts’ ratios were significantly different on ICU days 1 and 3 (p = 0.005).
Figure 2.
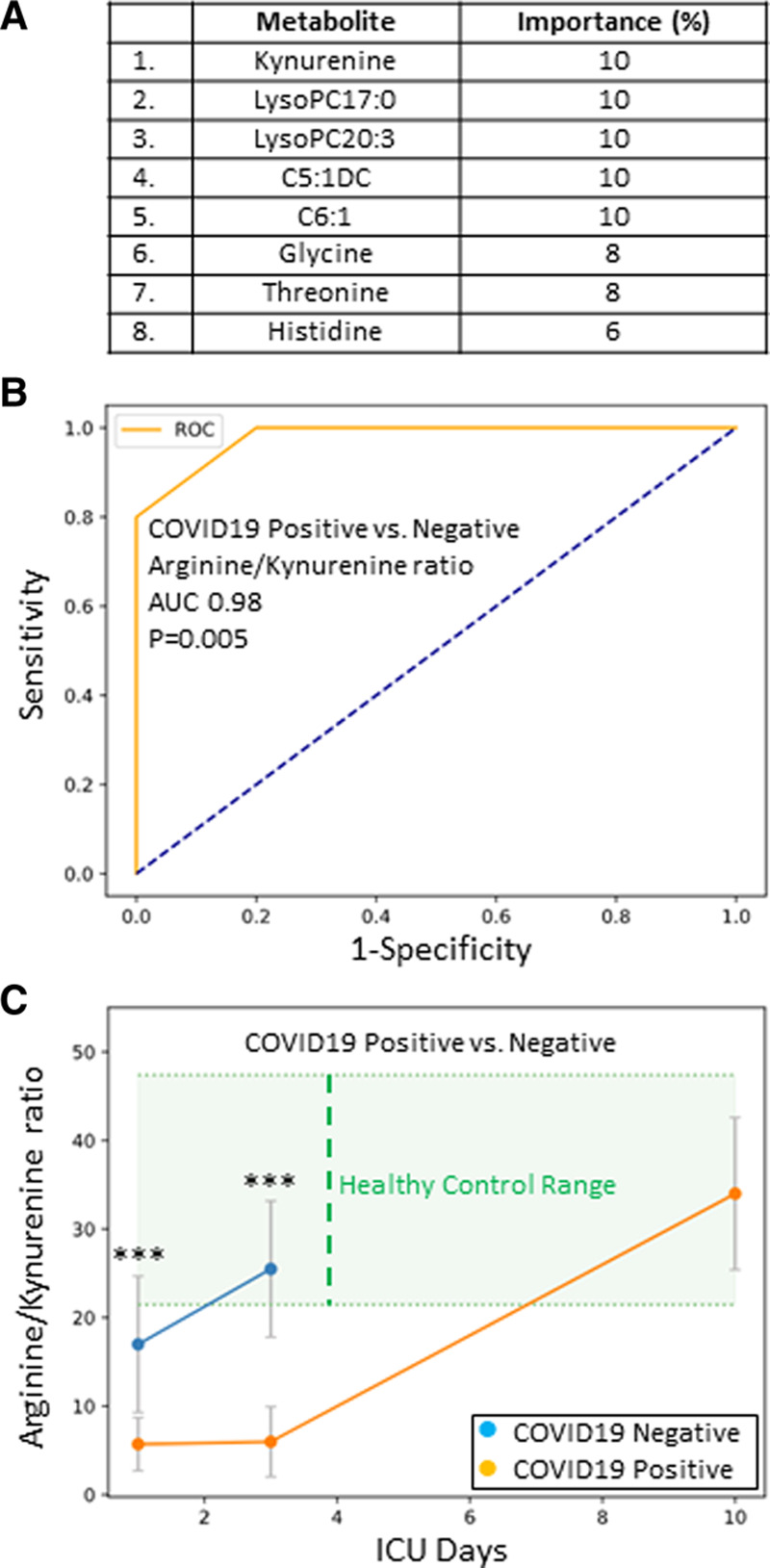
A, Feature classification demonstrating the top eight plasma metabolites that classify coronavirus disease 2019 positive (COVID19+) status versus healthy control subjects with their % association. B, Receiver operating characteristic analysis of COVID19+ versus coronavirus disease 2019 negative (COVID19–) ICU patients, using the arginine/kynurenine ratio, demonstrates an area-under-the-curve (AUC) of 0.98 (p = 0.005). The diagonal broken blue line represents chance (AUC = 0.50). C, A time plot, demonstrating the Arginine/Kynurenine ratio for both COVID19+ (orange dots) and COVID19– (blue dots) patients over 10 ICU days. The two cohorts are significantly different on ICU days 1 and 3 (***p = 0.005). Healthy control range values are represented by green shading.
Figure 3A shows a t-SNE plot for COVID19+ patients that either survived or died, and demonstrates that the outcomes were distinct and separable. To optimize outcome prediction in COVID19+ patients, the number of metabolites was narrowed using feature selection (Fig. 3B). Creatinine was the leading metabolite and could predict death with 100% accuracy on both ICU days 1 (cutoff > 126 μmol/L) and 3 (cutoff > 174 μmol/L). To improve the variation in patient creatinine values, we then tested the ability of a creatinine/arginine ratio to predict death; the corresponding time plot is shown in Figure 3C. Death could be predicted with 100% accuracy on both ICU days 1 (cutoff ≥ 3.4) and 3 (cutoff ≥ 3.7), as the creatinine/arginine ratios were significantly different between the COVID19 patients that lived or died at both time points (p = 0.01). The creatinine/arginine ratios normalized by ICU day 10, regardless of eventual outcome. There were no deaths during the 10 ICU days.
Figure 3.
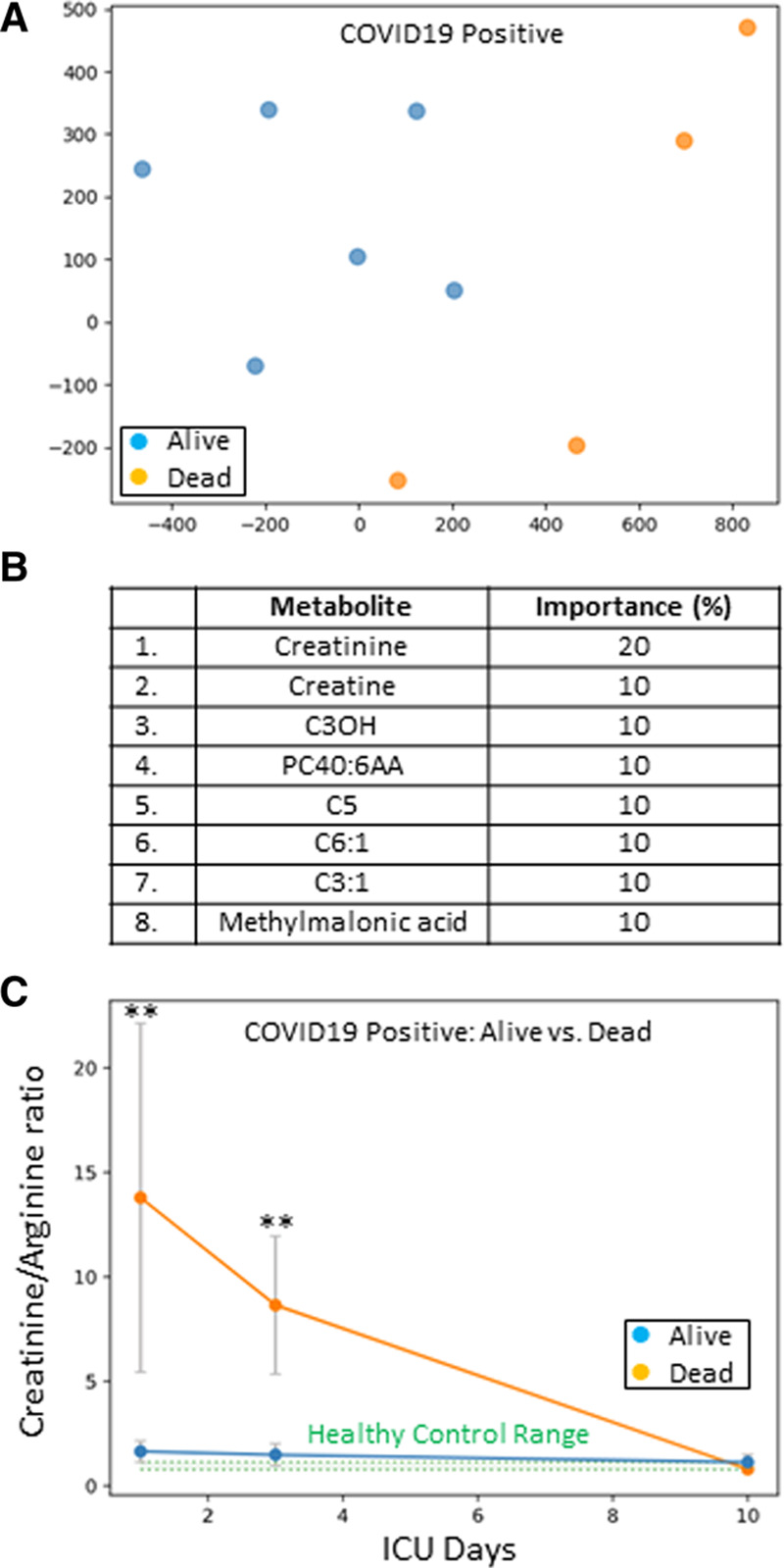
A, Coronavirus disease 2019 positive (COVID19+) ICU patients plotted in two dimensions following dimensionality reduction of their respective metabolites by stochastic neighbor embedding. Blue dots represent COVID19+ ICU patients that survived their ICU stay, whereas orange dots represent COVID19+ ICU patients that died (ICU day 1 plasma). The dimensionality reduction shows that based on the plasma metabolites, the two cohorts are distinct and easily separable. The axes are dimensionless. B, Feature classification, demonstrating the top eight plasma metabolites that classify COVID19+ ICU patient outcome as alive or dead with their % association. Plasma creatinine was the leading outcome predictor metabolite. C, A time plot, demonstrating the creatinine/arginine ratio for COVID19+ ICU patients over 10 ICU days that either survived (blue dots) or died (orange dots). The two cohorts are significantly different on ICU days 1 and 3 (**p = 0.01). Healthy control range values are represented by green shading.
DISCUSSION
In this study, we measured 162 metabolites in plasma obtained from ICU patients, both COVID19+ and COVID19–, as well as age- and sex-matched healthy control subjects. Given the number of metabolites measured, we analyzed the data with the state-of-the-art machine learning. Our exploratory data indicate the presence of a unique COVID19 plasma metabolome dominated by changes in kynurenine, arginine, sarcosine, and LysoPCs. Additionally, we identify that either creatinine alone or a creatinine/arginine ratio predicted ICU mortality with 100% accuracy. Despite the exploratory nature of our study, the data generated suggest that these three metabolites (kynurenine, arginine, and creatinine) could be considered for further investigation as potential diagnostic and prognostic biomarkers for COVID19 and that they may be useful for patient stratification in clinical interventional trials.
Our COVID19+ ICU patients were similar to those reported in earlier cohorts (3–6) with respect to demographics, comorbidities, and clinical presentation. In contrast to COVID19– ICU patients, our COVID19+ ICU patients had a higher frequency of bilateral pneumonia. Previous work by our study group in these same patients has determined a unique inflammatory profile characterized by elevated tumor necrosis factor and serine proteases (9), and a thrombotic profile associated with endothelial activation and glycocalyx degradation (10). By employing targeted proteomics, we also identified six novel protein immune biomarkers that accurately predict COVID19 associated death (12). Taken together with the data from this study, COVID19 represents a severe illness with a unique pathophysiological signature, as well as a high mortality rate. Indeed, in our cohort of COVID19 patients, ICU death was 40% with standardized ICU care.
Our study has identified a unique metabolome in COVID19+ ICU patients that is hypothesis-generating for future diagnostic/prognostic studies. Not only have we provided a rank order listing of metabolites important for COVID19 status, we also identified metabolites that accurately determined COVID19 ICU outcome. The former represents diverse metabolites that influence immune function and survival, whereas the latter represents compromised renal function early in ICU care. Importantly, the metabolites required for COVID19 diagnosis (arginine/kynurenine ratio) and outcome (either creatinine alone or creatinine/arginine ratio) can be easily measured using only MS or immunoassay, making their use as COVID19 biomarkers affordable and easily available. Point-of-care analyses for these metabolites could be rapidly developed, such as a lateral flow immunochromatographic assay. Furthermore, our study raises the possibility that dietary supplementation of tryptophan, arginine, sarcosine, and LysoPCs as adjunctive therapies may aid COVID19 outcome.
COVID19 status relied heavily on increased plasma kynurenine. The essential amino acid tryptophan is metabolized to elevate the energy-producing cofactor nicotinamide adenosine dinucleotide, with kynurenine as the first stable intermediate to be formed (30). Increased degradation of tryptophan, with a consequential increase in kynurenine, occurs during an immune response and is driven by the release of interferon-gamma from the activated T-cells. COVID19 caused intense T-cell activation (31, 32) with an approximate 11-fold increase in plasma interferon-gamma in critically ill COVID19 patients when compared with healthy control subjects (9).
Although plasma kynurenine effectively discriminated COVID19+ patients from healthy control subjects, determination of COVID19 status in ICU patients required further specificity that was optimally provided by an arginine/kynurenine ratio. Arginine, an amino acid precursor for nitric oxide, was significantly depressed in COVID19+ patients. Arginine depletion is likely secondary to the intense requirement for nitric oxide signaling and antiviral activity (33), as well as consumption by the enzyme arginase 1 that represents a macrophage immunoregulatory mechanism (34). As arginine is essential for tissue repair (35), its depletion could potentially delay and/or compromise ICU recovery.
Sarcosine, an amino acid that helped discriminate COVID19+ patients from healthy control subjects, was also significantly depressed. Although not superior to the arginine/kynurenine ratio for diagnosing COVID19 status, sarcosine sequestration may have a critical role in COVID19 pathology. Sarcosine enhances the activity of antigen presenting cells (36) and activates autophagy (37), or the body’s removal of damaged cells and their immunostimulatory debris. As a protective catabolic process during COVID19, autophagy is critical to the antiviral response by direct elimination of virus, the presentation of viral antigens, and the inhibition of excessive inflammation (38). Sarcosine levels decrease with age (37), and the elderly are most susceptible to COVID19 morbidity and mortality.
Depressed plasma LysoPCs also helped discriminate COVID19+ patients from healthy control subjects. The partial hydrolysis of phosphatidylcholines by phospholipase A2 produces LysoPCs, which are subsequently implicated in endothelial activation (39) and phagocytosis of cellular debris (40). Decreased plasma LysoPCs has been observed in sepsis (41), where LysoPCs may aid pathogen elimination, and therapeutic replacement has been suggested to improve sepsis outcome (42).
Acute renal dysfunction is strongly associated with high mortality in ICU patients (43). Plasma creatinine, a marker of renal dysfunction, was an excellent discriminator for COVID19 patients that either lived or died. In our COVID19+ cohort, two patients had chronic kidney disease and two patients required renal replacement therapy. The angiotensin-converting enzyme 2 receptor that is essential for SARS-CoV-2 uptake is highly expressed on tubule epithelial cells (44). Acute kidney injury is reported to occur in up to 37% of COVID19 patients (45) and is secondary to acute tubular injury from direct viral infection (46).
Our data suggest that COVID19 diagnosis (arginine/kynurenine ratio) and outcome (creatinine alone or creatinine/arginine ratio) can be determined with point-of-care measurements of kynurenine, arginine, and creatinine, and that this rapid and affordable biomarker approach may be complimentary to the more expensive and time-consuming diagnostic tools currently employed (e.g., polymerase chain reaction and antigen immunoassay). Furthermore, our study raises the possibility that dietary supplementation of tryptophan (kynurenine precursor), arginine, sarcosine, and LysoPCs may aid COVID19 outcome as adjunctive therapies.
Despite the novelty of the metabolite biomarkers discovered, our study has several limitations. First, we only studied critically ill patients and we cannot determine the full metabolome changes associated with ICU admissions. Second, although our COVD19 study population was limited, we still identified strong associations between the individual metabolites and outcomes and we fulfilled an urgent need for exploratory data to focus future hypothesis-driven studies on larger cohorts. Third, we report only mortality as our primary clinical outcome. Future studies with larger sample sizes can explore whether reported changes in specific metabolites correlate with additional clinical outcomes such as functional status in survivors. Finally, our analyses employed a cross-validation methodology in which the classifier was trained multiple times, each time on a different subset of the data, with the remainder of the data withheld for use only in testing. The reported accuracy is the mean accuracy of all such trials. This is a standard, accepted, technique in the machine learning literature, but should be validated on a larger testing set that is used only once. Overfitting was minimized by using a very small number of trees with a limited depth (29), and the results verified by training a simple linear vector machine and by identifying concordance between the results.
CONCLUSIONS
In summary, we report a unique metabolome in COVID19+ ICU patients, with identification of three metabolites that appear to be accurate diagnostic/prognostic biomarkers for future studies. Given the rapid spread of COVID19 and the critical need for rapid and affordable diagnostics, our data may be invaluable for future testing. In addition, our exploratory data may be useful for guiding resource mobilization and/or goals of care discussion, but only after validation in larger COVID19+ cohorts. Furthermore, patient stratification is critically important for future COVID19 interventional trials.
ACKNOWLEDGMENTS
We thank the entire Lawson COVID19 Study Team for their assistance (Dr. Robert Arntfield, Dr. Ian Ball, Mr. Gordon Barkwell, Ms. Tracey Bentall, Dr. Karen Bosma, Ms. Saoirse Cameron, Ms. Eileen Campbell, Mr. David Carter, Dr. Carolina Gillio-Meina, Dr. Robert Hegele, Ms. Natalya Odoardi, Dr. Ram Singh, Ms. Shannon Seney, Dr. Kelly Summers, and Ms. Sue Tereschyn). We are grateful for the enthusiastic support of the frontline Critical Care Nursing Staff at London Health Sciences Centre.
Footnotes
We acknowledge funding from Western University (Research), the Department of Medicine and Department of Pediatrics at Western University, the Lawson Health Research Institute (https://www.lawsonresearch.ca/), the London Health Sciences Foundation (https://lhsf.ca/), and the AMOSO Innovation Fund.
The authors disclosed a patent pending (Metabolomics Profile of Covid19 Patients; #63065966).
Contributor Information
Collaborators: Robert Arntfield, Ian Ball, Gordon Barkwell, Tracey Bentall, Karen Bosma, Saoirse Cameron, Eileen Campbell, David Carter, Carolina Gillio-Meina, Robert Hegele, Natalya Odoardi, Ram Singh, Kelly Summers, and Sue Tereschyn
REFERENCES
- 1.Johns Hopkins University & Medicine; Coronavirus Resource Center: COVID-19 Map. Available at: https://coronavirus.jhu.edu/map.html
- 2.World Health Organization: WHO Director-General's Opening Remarks at the Media Briefing on COVID-19 - 3 March 2020. Available at: https://www.who.int/dg/speeches/detail/who-director-general-s-opening-remarks-at-the-media-briefing-on-covid-19---3-march-2020
- 3.Bhatraju PK, Ghassemieh BJ, Nichols M, et al. : Covid-19 in critically ill patients in the Seattle region - case series. N Engl J Med 2020; 382:2012–2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhou F, Yu T, Du R, et al. : Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: A retrospective cohort study. Lancet 2020; 395:1054–1062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu C, Chen X, Cai Y, et al. : Risk factors associated with acute respiratory distress syndrome and death in patients with coronavirus disease 2019 pneumonia in Wuhan, China. JAMA Intern Med 2020; 180:934–943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grasselli G, Zangrillo A, Zanella A, et al. : Baseline characteristics and outcomes of 1591 patients infected with SARS-CoV-2 admitted to ICUs of the Lombardy region, Italy. JAMA 2020; 323:1574–1581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tisoncik JR, Korth MJ, Simmons CP, et al. : Into the eye of the cytokine storm. Microbiol Mol Biol Rev 2012; 76:16–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mehta P, McAuley DF, Brown M, et al. ; HLH Across Speciality Collaboration, UK: COVID-19: Consider cytokine storm syndromes and immunosuppression. Lancet 2020; 395:1033–1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fraser DD, Cepinskas G, Slessarev M, et al. : Inflammation profiling of critically ill coronavirus disease 2019 patients. Crit Care Explor 2020; 2:e0144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fraser DD, Patterson EK, Slessarev M, et al. : Endothelial injury and glycocalyx degradation in critically ill coronavirus disease 2019 patients: Implications for microvascular platelet aggregation. Crit Care Explor 2020; 2:e0194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Auld SC, Caridi-Scheible M, Blum JM, et al. : ICU and ventilator mortality among critically ill adults with coronavirus disease 2019. Crit Care Med 2020; 48:e799–e804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fraser DD, Cepinskas G, Patterson EK, et al. : Novel outcome biomarkers identified with targeted proteomic analyses of plasma from critically ill Coronavirus disease 2019 patients. Crit Care Explor 2020; 2:e0189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bujak R, Struck-Lewicka W, Markuszewski MJ, et al. : Metabolomics for laboratory diagnostics. J Pharm Biomed Anal 2015; 113:108–120 [DOI] [PubMed] [Google Scholar]
- 14.Daley M, Dekaban G, Bartha R, et al. : Metabolomics profiling of concussion in adolescent male hockey players: A novel diagnostic method. Metabolomics 2016; 12:185 [Google Scholar]
- 15.Society of Critical Care Medicine: Waiver of Informed Consent in Emergency Situations. Available at: https://www.sccm.org/Communications/Critical-Connections/Archives/2018/Waiver-of-Informed-Consent-in-Emergency-Situations
- 16.Centers for Disease Control and Prevention: Overview of Testing for SARS-CoV-2 (COVID-19). Available at: https://www.cdc.gov/coronavirus/2019-nCoV/hcp/clinical-criteria.html
- 17.Centers for Disease Control and Prevention: CDC 2019 Novel Coronavirus (nCoV) Real-Time RT-PCR Diagnostic Panel - Instructions for Use. Available at: https://www.fda.gov/media/134922/download
- 18.Priestap F, Kao R, Martin CM: External validation of a prognostic model for intensive care unit mortality: A retrospective study using the Ontario Critical Care Information System. Can J Anaesth 2020; 67:981–991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Singer M, Deutschman CS, Seymour CW, et al. : The third international consensus definitions for sepsis and septic shock (Sepsis-3). JAMA 2016; 315:801–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brisson AR, Matsui D, Rieder MJ, et al. : Translational research in pediatrics: Tissue sampling and biobanking. Pediatrics 2012; 129:153–162 [DOI] [PubMed] [Google Scholar]
- 21.Gillio-Meina C, Cepinskas G, Cecchini EL, et al. : Translational research in pediatrics II: Blood collection, processing, shipping, and storage. Pediatrics 2013; 131:754–766 [DOI] [PubMed] [Google Scholar]
- 22.Program NHRP: POLICY: Guidelines for limits of blood drawn for research purposes in the clinical center. M95-9 (rev) 2009 [Google Scholar]
- 23.Foroutan A, Fitzsimmons C, Mandal R, et al. : The bovine metabolome. Metabolites 2020; 10:233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Foroutan A, Guo AC, Vazquez-Fresno R, et al. : Chemical composition of commercial cow’s milk. J Agric Food Chem 2019; 67:4897–4914 [DOI] [PubMed] [Google Scholar]
- 25.Psychogios N, Hau DD, Peng J, et al. : The human serum metabolome. PLoS One 2011; 6:e16957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saude EJ, Slupsky CM, Sykes BD: Optimization of NMR analysis of biological fluids for quantitative accuracy. Metabolomics 2006; 2:113–123 [Google Scholar]
- 27.Ravanbakhsh S, Liu P, Bjorndahl TC, et al. : Accurate, fully-automated NMR spectral profiling for metabolomics. PLoS One 2015; 10:e0124219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van der Maaten L, Hinton G: Visualizing data using t-SNE. J Mach Learn Res 2008; 9:2579–2605 [Google Scholar]
- 29.Tang C, Garreau D, von Luxburg U: When do random forests fail? In: Proceedings of the 32nd International Conference on Neural Information Processing Systems. 2018:2987–2997 [Google Scholar]
- 30.Chen Y, Guillemin GJ: Kynurenine pathway metabolites in humans: Disease and healthy States. Int J Tryptophan Res 2009; 2:1–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.De Biasi S, Meschiari M, Gibellini L, et al. : Marked T cell activation, senescence, exhaustion and skewing towards TH17 in patients with COVID-19 pneumonia. Nat Commun 2020; 11:3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen Z, John Wherry E: T cell responses in patients with COVID-19. Nat Rev Immunol 2020; 20:529–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Akerström S, Mousavi-Jazi M, Klingström J, et al. : Nitric oxide inhibits the replication cycle of severe acute respiratory syndrome coronavirus. J Virol 2005; 79:1966–1969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mondanelli G, Iacono A, Allegrucci M, et al. : Immunoregulatory interplay between arginine and tryptophan metabolism in health and disease. Front Immunol 2019; 10:1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alexander JW, Supp DM: Role of arginine and omega-3 fatty acids in wound healing and infection. Adv Wound Care (New Rochelle) 2014; 3:682–690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dastmalchi F, Karachi A, Yang C, et al. : Sarcosine promotes trafficking of dendritic cells and improves efficacy of anti-tumor dendritic cell vaccines via CXC chemokine family signaling. J Immunother Cancer 2019; 7:321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Walters RO, Arias E, Diaz A, et al. : Sarcosine is uniquely modulated by aging and dietary restriction in rodents and humans. Cell Rep 2018; 25:663–676.e6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Carmona-Gutierrez D, Bauer MA, Zimmermann A, et al. : Digesting the crisis: Autophagy and coronaviruses. Microb Cell 2020; 7:119–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li X, Fang P, Li Y, et al. : Mitochondrial reactive oxygen species mediate lysophosphatidylcholine-induced endothelial cell activation. Arterioscler Thromb Vasc Biol 2016; 36:1090–1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lauber K, Bohn E, Kröber SM, et al. : Apoptotic cells induce migration of phagocytes via caspase-3-mediated release of a lipid attraction signal. Cell 2003; 113:717–730 [DOI] [PubMed] [Google Scholar]
- 41.Mecatti GC, Fernandes Messias MC, Sant’Anna Paiola RM, et al. : Lipidomic profiling of plasma and erythrocytes from septic patients reveals potential biomarker candidates. Biomark Insights 2018; 13:1177271918765137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cunningham TJ, Yao L, Lucena A: Product inhibition of secreted phospholipase A2 may explain lysophosphatidylcholines’ unexpected therapeutic properties. J Inflamm (Lond) 2008; 5:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Clermont G, Acker CG, Angus DC, et al. : Renal failure in the ICU: Comparison of the impact of acute renal failure and end-stage renal disease on ICU outcomes. Kidney Int 2002; 62:986–996 [DOI] [PubMed] [Google Scholar]
- 44.Wysocki J, Lores E, Ye M, et al. : Kidney and lung ACE2 expression after an ACE inhibitor or an Ang II receptor blocker: Implications for COVID-19. J Am Soc Nephrol 2020; 31:1941–1943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hirsch JS, Ng JH, Ross DW, et al. : Acute kidney injury in patients hospitalized with COVID-19. Kidney Int 2020; 98:209–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Farkash EA, Wilson AM, Jentzen JM: Ultrastructural evidence for direct renal infection with SARS-CoV-2. J Am Soc Nephrol 2020; 31:1683–1687 [DOI] [PMC free article] [PubMed] [Google Scholar]


